Abstract
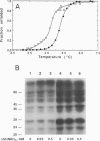
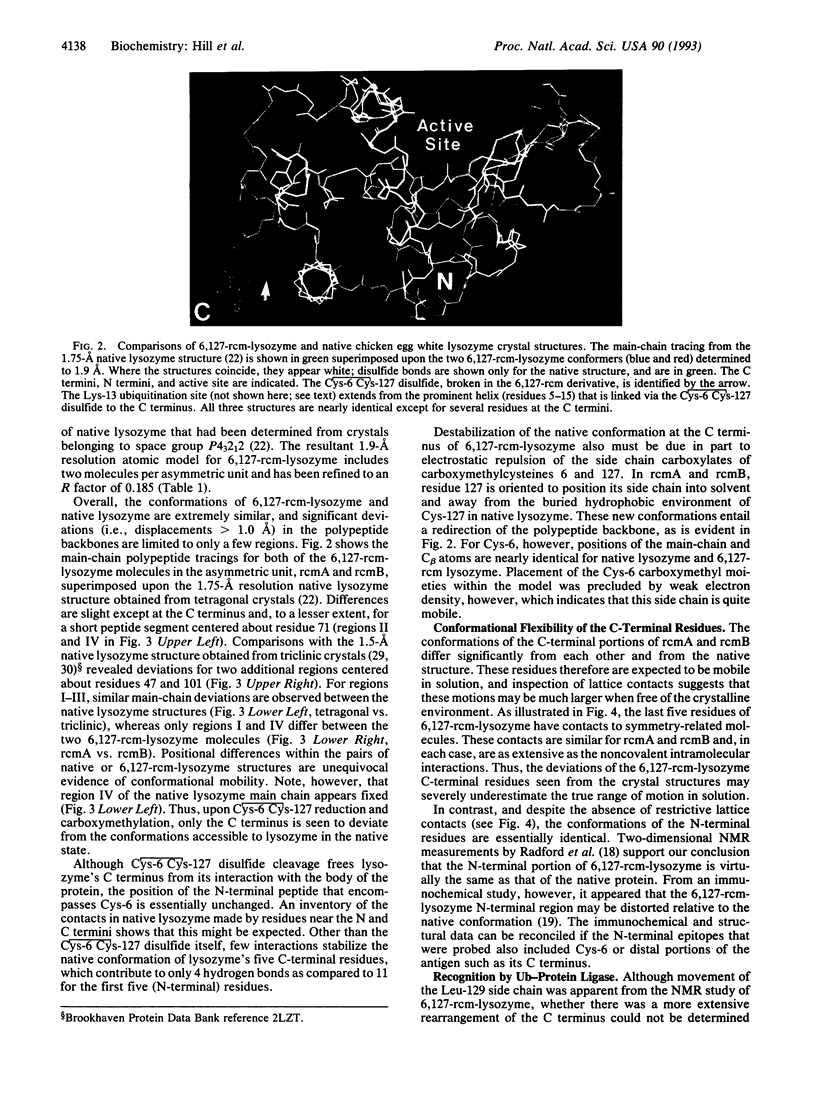
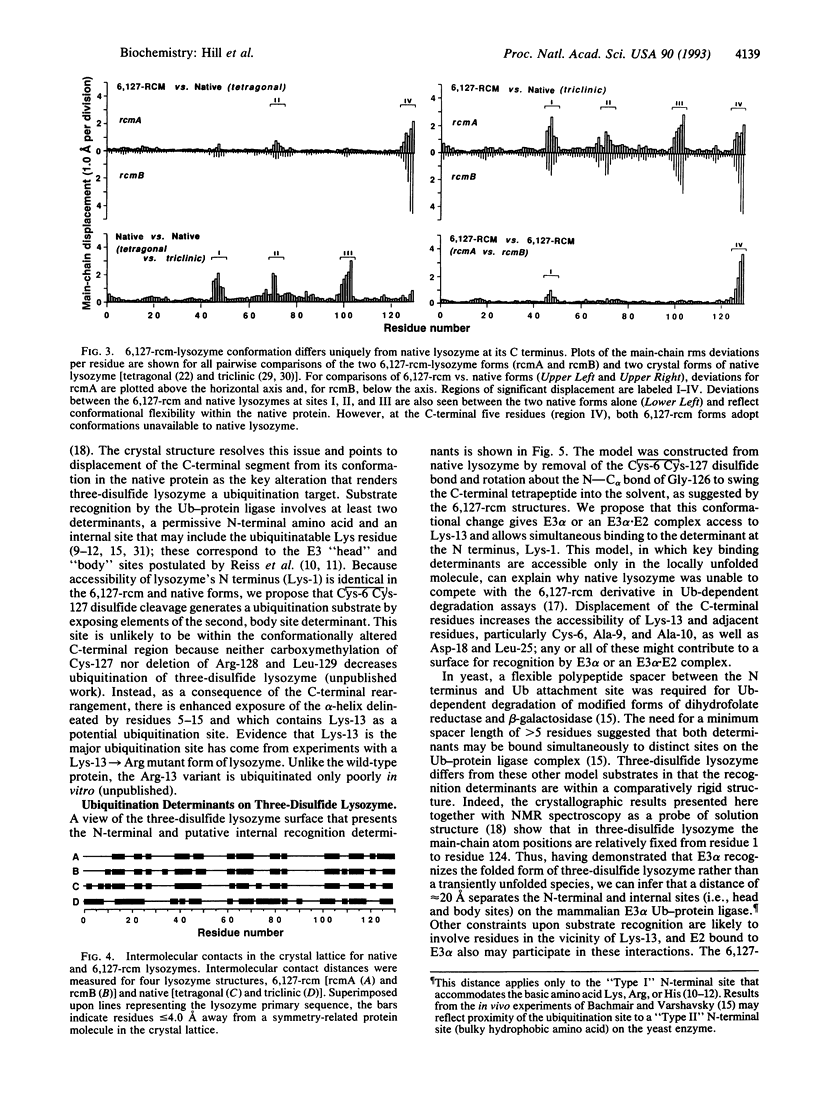
Covalent attachment of ubiquitin marks substrates for proteolysis, but features that identify ubiquitination targets such as chicken egg white lysozyme are poorly understood. Recognition of lysozyme first requires reduction of Cys-6 Cys-127, one of its four native disulfide bonds, and Cys-6,Cys-127-carboxymethylated (6,127-rcm) lysozyme can mimic this three-disulfide intermediate. The 6,127-rcm form of lysozyme is known to retain a substantially native-like conformation in solution, and we demonstrate that it is this folded structure that is recognized for ubiquitination. Because native lysozyme is not a substrate, differences between the native and three-disulfide structures must include features responsible for selective ubiquitination. The 1.9-A resolution crystal structure of 6,127-rcm-lysozyme, reported here, affords a view of this ubiquitin-dependent degradation substrate. Two conformers of 6,127-rcm-lysozyme were obtained in the crystal. These differ uniquely from crystal forms of native lysozyme by displacement of the C-terminal residues. The structures suggest that localized unfolding at the C terminus of three-disulfide lysozyme allows the complex of E3 alpha (ubiquitin-protein ligase) and E2 (ubiquitin-carrier protein) to bind to a surface that includes Lys-1 and the putative ubiquitination site Lys-13. From this we infer that the N-terminal and internal substrate recognition sites on the E3 alpha.E2 complex are separated by approximately 20 A.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmair A., Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989 Mar 24;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- Banerjee S. K., Rupley J. A. Temperature and pH dependence of the binding of oligosaccharides to lysozyme. J Biol Chem. 1973 Mar 25;248(6):2117–2124. [PubMed] [Google Scholar]
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chen Z., Pickart C. M. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990 Dec 15;265(35):21835–21842. [PubMed] [Google Scholar]
- Denton M. E., Scheraga H. A. Spectroscopic, immunochemical, and thermodynamic properties of carboxymethyl(Cys6, Cys127)-hen egg white lysozyme. J Protein Chem. 1991 Apr;10(2):213–232. doi: 10.1007/BF01024786. [DOI] [PubMed] [Google Scholar]
- Dunten R. L., Cohen R. E., Gregori L., Chau V. Specific disulfide cleavage is required for ubiquitin conjugation and degradation of lysozyme. J Biol Chem. 1991 Feb 15;266(5):3260–3267. [PubMed] [Google Scholar]
- Dunten R. L., Cohen R. E. Recognition of modified forms of ribonuclease A by the ubiquitin system. J Biol Chem. 1989 Oct 5;264(28):16739–16747. [PubMed] [Google Scholar]
- Finley D., Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., St John A. C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989 Oct 5;264(28):16700–16712. [PubMed] [Google Scholar]
- Heller H., Hershko A. A ubiquitin-protein ligase specific for type III protein substrates. J Biol Chem. 1990 Apr 25;265(12):6532–6535. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hershko A., Heller H., Eytan E., Reiss Y. The protein substrate binding site of the ubiquitin-protein ligase system. J Biol Chem. 1986 Sep 15;261(26):11992–11999. [PubMed] [Google Scholar]
- Hodsdon J. M., Brown G. M., Sieker L. C., Jensen L. H. Refinement of triclinic lysozyme: I. Fourier and least-squares methods. Acta Crystallogr B. 1990 Feb 1;46(Pt 1):54–62. doi: 10.1107/s0108768189009183. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Olson T. S., Dice J. F. Regulation of protein degradation rates in eukaryotes. Curr Opin Cell Biol. 1989 Dec;1(6):1194–1200. doi: 10.1016/s0955-0674(89)80071-7. [DOI] [PubMed] [Google Scholar]
- Pace C. N., McGrath T. Substrate stabilization of lysozyme to thermal and guanidine hydrochloride denaturation. J Biol Chem. 1980 May 10;255(9):3862–3865. [PubMed] [Google Scholar]
- Pickart C. M., Vella A. T. Levels of active ubiquitin carrier proteins decline during erythroid maturation. J Biol Chem. 1988 Aug 25;263(24):12028–12035. [PubMed] [Google Scholar]
- Radford S. E., Woolfson D. N., Martin S. R., Lowe G., Dobson C. M. A three-disulphide derivative of hen lysozyme. Structure, dynamics and stability. Biochem J. 1991 Jan 1;273(Pt 1):211–217. doi: 10.1042/bj2730211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanadham M., Sieker L. C., Jensen L. H. Refinement of triclinic lysozyme: II. The method of stereochemically restrained least squares. Acta Crystallogr B. 1990 Feb 1;46(Pt 1):63–69. doi: 10.1107/s0108768189009195. [DOI] [PubMed] [Google Scholar]
- Reiss Y., Hershko A. Affinity purification of ubiquitin-protein ligase on immobilized protein substrates. Evidence for the existence of separate NH2-terminal binding sites on a single enzyme. J Biol Chem. 1990 Mar 5;265(7):3685–3690. [PubMed] [Google Scholar]
- Reiss Y., Kaim D., Hershko A. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988 Feb 25;263(6):2693–2698. [PubMed] [Google Scholar]
- Rivett A. J. Eukaryotic protein degradation. Curr Opin Cell Biol. 1990 Dec;2(6):1143–1149. doi: 10.1016/0955-0674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- Stadtman E. R. Covalent modification reactions are marking steps in protein turnover. Biochemistry. 1990 Jul 10;29(27):6323–6331. doi: 10.1021/bi00479a001. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., James M. N. Lysozyme revisited: crystallographic evidence for distortion of an N-acetylmuramic acid residue bound in site D. J Mol Biol. 1991 Jul 20;220(2):401–424. doi: 10.1016/0022-2836(91)90021-w. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992 May 29;69(5):725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]