Abstract
Prostate cancer (PCa) contains phenotypically and functionally distinct cells, and this cellular heterogeneity poses clinical challenges as the distinct cell types likely respond differently to various therapies. Clonal evolution, driven by genetic instability, and intra-clonal cancer cell diversification, driven by cancer stem cell (CSCs), together, create tumor cell heterogeneity. In this review, we first discuss prostate cancer stem cells (PCSCs) and heterogeneity of androgen receptor (AR) expression in primary, metastatic and treatment-failed PCa. Based on literature reports and our own studies, we hypothesize that whereas PCSCs in primary and untreated tumors and models are mainly AR−, PCSCs in CRPCs could be either AR+ or AR−/lo. We illustrate the potential mechanisms whereby AR+ and AR− PCSCs may employ to propagate PCa at the population level, mediate therapy resistance, and metastasize. As a result, targeting AR alone may not be able to achieve long-lasting therapeutic efficacy. Elucidating the roles of AR and PCSCs should provide important clues to designing novel personalized combinatorial therapeutic protocols targeting both AR+ and AR− PCa cells.
Keywords: androgen receptor, prostate cancer, cancer stem cells, prostate cancer stem cells, castration-resistant prostate cancer
I. Cancer stem cells and tumor heterogeneity
Tumors contain genetically heterogeneous cellular clones, which constantly evolve during disease progression and clinical treatment. Clonal evolution, driven by genetic instability of cancer cells, generates cellular heterogeneity and promotes tumor progression. For instance, genome-wide DNA sequencing of three individual prostate tumors revealed the existence of three or more clones within each cancer (Cooper et al., 2015). Even morphologically normal regions could possess as many as ten genetic mutations (Cooper et al., 2015). In untreated primary prostate cancer (PCa), genetic alterations such as TMPRSS2-ERG fusion and PTEN deletion within tumor clones could activate critical signaling pathways such as ERG and PI3K, thus driving clonal evolution (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). In a longitudinal tracking of a CRPC (castration-resistant PCa) patient with 9 prostate tumor foci at the initial presentation, it was found that during the 17 years of tumor progression, only the tumor clones with PTEN, P53 and SPOP mutations gained additional genetic alterations and gave rise to lethal metastatic tumors. Surprisingly, the lethal clone (defined by the presence of the same PTEN, P53, and SPOP mutations) in this patient was found to arise from a morphologically low-grade (Gleason 3) tumor focus rather from the predominant Gleason 4 tumor foci (Haffner et al., 2013). Whole-genome exome sequencing in 50 lethal, and heavily pre-treated metastatic CRPCs also confirmed the monoclonal origin of lethal CRPC (Grasso et al., 2012). These examples highlight the importance of genetically-driven clonal evolution in driving PCa progression.
On the other hand, there is also strong evidence that tumor cells within a genetically identical clone possess different tumorigenic ability and, in most cases, are organized in a hierarchical manner (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Sitting at the apex of this tumorigenic hierarchy is the small subset of stem-like cancer cells, or cancer stem cells (CSCs) that possess high self-renewal and differentiation ability. In other words, CSCs sustain an established tumor clone through unlimited self-renewal and maintain intraclonal heterogeneity through generating both tumorigenic and less or non- tumorigenic cancer cells. Similar to normal hematopoietic stem cells (HSCs), which are among the best-understood adult stem cells, the best-characterized CSCs are CSCs in leukemia or leukemic stem cells (LSCs; Kreso and Dick, 2014). Like HSCs, LSCs are undifferentiated lacking the expression of lineage differentiation markers. Subsequent studies have led to the identification of CSCs in multiple human solid tumors and a common phenotypic feature of these CSCs seems to be the lack of differentiation markers and regulators (e.g., Dubrovska, et al. 2010; Rybak et al., 2015).
In a strict sense, CSCs in human tumors are defined as a population of cancer cells, when prospectively purified out from patient tumors, xenografts, and even long-term cultures, can regenerate and also indefinitely propagate human tumors in immune-deficient mice. In reality, the CSC properties of a candidate population of human tumor cells are best assessed by performing limiting dilution tumor-regeneration assays combined with serial tumor transplantations and cell biological (e.g., clonal in 2D; clonogenic in 3D; sphere formation; single-cell division and differentiation; etc) as well as molecular (e.g., RNA-Seq and ChIP-Seq) characterizations (reviewed in Rycaj and Tang, 2015). The tumor cell population that can initiate or regenerate tumors at low cell doses is considered to be tumor-initiating or tumor-regenerating cells while the tumor cell population that can long-term propagate human xenograft tumors is called tumor-propagating cells (Rycaj and Tang, 2015). Unfortunately, many of the reported CSC populations do not fully satisfy this strict definition. For example, some studies only utilized cell lines to perform in vitro assays without tumor experiments whereas some others only performed tumor experiments without further carrying out serial transplantations. Such shortcomings have created a lot of confusions in the field and led many to even disbelieve the presence of CSCs. Recent lineage tracing studies in genetically driven mouse model tumors (i.e., glioblastoma, and intestinal and skin tumors) have provided definitive evidence for CSCs (Rycaj and Tang, 2015).
II. Prostate cancer stem cells (PCSCs)
The CSC model helps explain the generation of tumor cell heterogeneity from the viewpoint of stem cell maturation and differentiation. PCa is well known to be a highly heterogeneous malignancy with each tumor harboring many tumor clones (Cooper et al., 2015; Haffner et al., 2013). Therefore, it's not surprising that many prostate cancer stem cell (PCSC) populations have been reported (reviewed in Chen et al., 2013 and Rybak et al., 2015). PCSCs are defined, more or less, using a spectrum of in vitro and in vivo assays used to define other CSCs (see above). In vitro, PCSCs preferentially express stem cell and cancer stem cell-associated molecules and self-renewal genes (e.g., Bmi-1, Stat3, Nanog, Sox2, Oct4) and possess high clonal and clonogenic capacities, and in vivo, PCSCs possess higher tumor-initiating and serial tumor-propagating activities than non-PCSCs in immunodeficient mice (Chen et al., 2013; Kroon et al., 2013; Rybak et al., 2015). Three papers, published in 2005, simultaneously provided the earliest proof-of-principle evidence for PCSCs: 1) the Side Population (SP) in the LAPC9 human xenografts was enriched in tumor-initiating cells (Patrawala et al., 2005); 2) ABCG2, a surface pump protein normally involved in cellular detoxification, mediated efflux of androgen in putative PCSCs (Huss et al., 2005); and 3) the CD44+α2β1+CD133+ PCa cells from patient prostate tumors possessed high clonogenic survivability in methylcellulose (Collins et al., 2005).
Our lab, since 2012, has been employing and developing a variety of experimental strategies to elucidate the cellulose basis and molecular regulation of PCa cell heterogeneity, and to link PCa cell heterogeneity to therapy resistance and tumor relapse. In virtually all of our PCSC studies, we have performed tumor-regeneration and, in many cases, serial tumor transplantation assays. Using the SP analysis, we provided the very first piece of evidence that the SP in certain PCa xenograft models is enriched in tumor-regenerating and tumor-propagating cells and thus satisfies the strict definition of CSCs (Patrawala et al., 2005). Using cell surface markers, our systematic studies have provided convincing evidence that the CD44 high-expressing (i.e., CD44+) PCa cell population in most, though not all, PCa models we have studied is significantly enriched in PCSCs with enhanced tumor-regenerating, tumor-propagating, and metastatic capacities (Liu et al., 2011; Liu et al., 2015; Patrawala et al., 2006; Patrawala et al., 2007). Using holoclone assays, we have shown that the PCa cell holoclones, like stem cell-enriched primary keratinocyte holoclones, possess long-term tumor-propagating CSC properties (Li et al., 2008a). Using lentiviral-mediated lineage tracing, we have recently demonstrated that phenotypically undifferentiated PCa cell population that lacks the expression of PSA (i.e., PSA−/lo) harbors self-renewing long-term tumor-propagating PCSCs, which express stem cell gene expression and epigenetic profiles, can undergo authentic asymmetric cell division, and are intrinsically refractory to castration treatments (Liu, et al., 2015; Qin et al., 2012).
Similar to the heterogeneity of CSC populations in other tumor systems (Tang, 2012), the PCSC pool is heterogeneous containing CSC subsets with distinct tumor-regenerating and tumor-propagating capabilities (Liu et al., 2015), potentially explaining many different PCSC populations reported by others (e.g., Collins et al., 2005; Domingo-Domeneck et al., 2012; Dubrovska et al., 2009; Miki et al., 2007; Rajasekhar et al., 2011). Also similar to the undifferentiated nature of LSCs and other CSCs (Tang, 2012), a common phenotypic trait of the reported PCSC populations is the lack of expression of differentiation regulators and markers such as AR (see below), PSA (Qin et al., 2012), and MHC molecules (Domingo-Domeneck et al., 2012).
One of the major unresolved questions related to PCSCs is whether any subpopulation of PCa cells acutely purified from primary patient tumors or CRPCs truly possesses hardcore CSC properties such as regenerating tumors at the single-cell level and enabling serial tumor transplantations. Although patient tumor or early PDX (patient-derived xenograft) derived cells have been demonstrated, in many experimental settings, to possess at least certain CSC properties (especially in vitro), this question has dodged a direct answer mainly due to our current technical difficulty in reconstituting human PCa development in immunodeficient mice (Chen et al., 2013).
III. AR heterogeneity in PCa
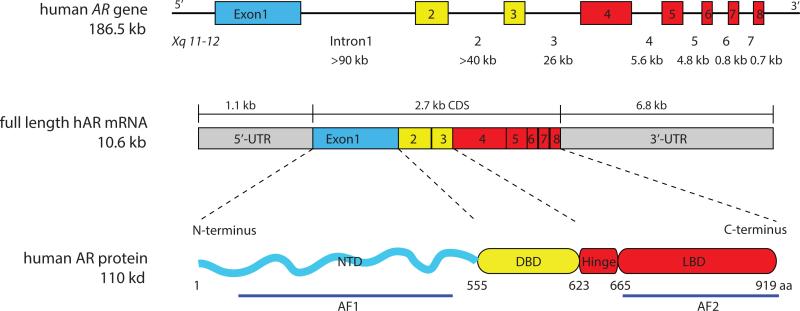
AR is a master regulator of normal prostate differentiation and development. Human AR gene, located on chromosome Xq11-12, encodes a protein with four functional domains: the NH2-terminal domain (NTD), the DNA binding domain (DBD), the hinge domain and the ligand-binding domain (LBD) (Figure 1). The prostate is one of the main organs that express AR, and the AR protein is expressed in the luminal cell layer of the prostatic glands. AR signaling critically regulates development, differentiation, and maintenance of the prostate as documented in both human and animal studies. Somatic mutations of the AR gene lead to malfunction of AR and androgen insensitivity syndrome in human, in which 46 XY individuals present female phenotype and the prostate is absent (Quigley, et al. 1995). The AR NTD knockout male mice all have small immature testes and lack secondary reproductive organs (Kerkhofs, et al. 2009).
Figure 1. Genomic organization of the AR gene and overall domain structure of the AR protein.
The AR gene is mapped to the long arm of X chromosome and spans about 186.5 kb. It contains eight exons interrupted by introns of various lengths (indicated below). The mRNA of AR gene is of 10.6 kb with exon 1 coding for the NTD, exons 2 and 3 the DBD, exons 4-8 the hinge and LBD. The full length AR protein contains 919 amino acids, consisting of a very flexible NTD and constant DBD, hinge domain and LBD. The constitutively active AF1 domain is located in the NTD and the LBD consists of the AF2 domain.
Simanainen et al (2007) established an AR exon 3 knockout mouse model and observed under-developed prostate in the male mice with delayed structural and functional differentiation of the prostate epithelium. There was also increased proliferation in the AR deficient epithelium (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). In another prostate-specific AR knockout mouse model, Wu et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) also reported increased proliferation and less differentiation of the epithelium. These genetic studies suggest that AR promotes prostate differentiation and suppresses epithelium proliferation in the mature prostate; in this way, AR signaling maintains the homeostasis and relative dormancy of mature prostate epithelium. Consistent with this pro-differentiation role of AR, prostate epithelial-specific AR knockout promoted TRAMP tumor development, providing genetic evidence for a tumor-suppressive function of AR (Niu et al., 2008).
Somewhat paradoxically, however, AR expression is frequently overexpressed in PCa and, in fact, AR is thought to be required for prostate tumorigenesis and hence, targeting AR and AR signaling has long been a therapeutic strategy. Androgen-deprivation therapy (ADT) aims to block androgen synthesis (e.g., Abiraterone) or AR functions (e.g., bicalutamide, Enzalutamide). Nevertheless, AR expression has been observed to be heterogeneous in primary, and in particular, in treatment-failed patient tumors. Ruizeveld de Winter et al (1990) examined AR by immunohistochemistry (IHC) staining in 26 primary PCa and found that 7 cases presented a considerable heterogeneity in AR expression and the proportion of AR-expressing cells was decreased in the more aggressive tumors. Similar AR IHC staining by Masai et al (1990) showed that AR expression correlated inversely with grade. Also, Chodak et al (1992) analyzed AR expression in 57 untreated PCa and observed that AR content was significantly higher in differentiated tumors compared to that of poorly differentiated tumors. Our own studies revealed AR− PCa cells to be present in all 9 primary PCa samples we examined representing ~5 - 30% of the total (Liu et al., 2015). Overall, these and many other studies suggest that, although AR− cells may not be dominant in treatment naïve tumors, all primary prostate tumors nevertheless harbor both AR+ and AR− cells or clones (Figure 2, bottom; Liu et al., 2015).
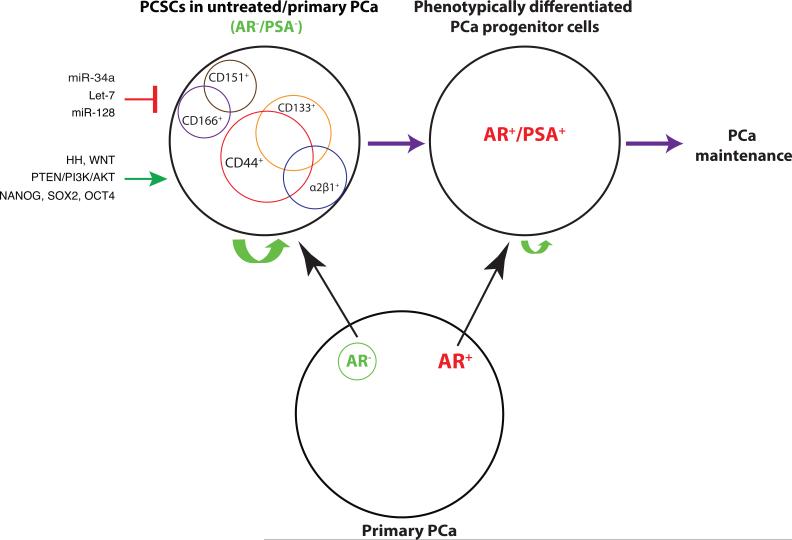
Figure 2. PCSCs in untreated/primary PCa.
Primary PCa contains AR+ PCa cells as the majority with the AR− PCa cells being the minority (below). Depicted on top (left) are several representative PCSC populations reported in primary PCa and untreated prostate tumor models, which are mostly AR− and PSA− but have the capacity to differentiate into more mature, AR+/PSA+ PCa cells (right). The PSA−/lo PCSC population has unlimited self-renewal potential (indicated by a large green arrow) whereas differentiated AR+/PSA+ PCa progenitors cells have more limited self-renewal activity (indicated by a small green arrow) (Liu et al., 2015; Qin et al., 2012). The PCSCs can be positively regulated through HH (Hedgehog), WNT and PTEN signaling pathways, as well as by transcription factors such as NANOG/SOX2/OCT4. On the other hand, several miRNAs including miR-34a, let-7, and miR-128 have been reported to negatively regulate PCSCs.
AR heterogeneity in hormone-refractory PCa has been observed since early 1990's. van der Kwast et al (1991) examined AR expression in CRPC and found that in 13/17 tumors, over 80% of the tumor cells were AR+. However, 3 tumors showed a considerable heterogeneity in AR expression and in 1 sample nearly all tumor cells appeared AR−/lo. Sadi et al (1991) observed similar AR heterogeneity in needle biopsy specimens of 17 patients with Stage D PCa. Ruizeveld de Winter et al (1994) examined AR expression in locally progressive CRPC and found that less differentiated PCa cells tended towards diminished AR expression. Computer qualification of nuclear AR levels in PCa patient samples showed that the AR concentration per cell was significantly more heterogeneous in poor responders (Sadi and Barrack 1993). Our own IHC staining of AR on a tissue microarray of CRPC samples revealed highly heterogeneous AR expression patterns across individuals: there were AR+ as well as AR− CRPC cores, and, within one single CRPC, there were regions that were AR+, AR−, or a mixture of both populations (Liu et al., 2015).
AR expression varies in metastases as well. Shah et al (2004) investigated AR expression by IHC in metastatic lesions of 30 CRPC patients who underwent warm autopsy and observed wide variations in AR expression between tumor samples. Specifically, 31% (83 of 265) of metastatic samples had <50% AR+ cells and 41.5% (100/265) metastases had <10% AR+ cells. Five patient metastases had <1% AR+ cells (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Similarly, Davis et al (2006) reported that both AR+ cells and AR staining intensity decreased in metastatic CRPC cells compared with benign tissues or untreated PCa. Of note, two commonly used PCa cell lines, Du145 and PC3, which were derived from brain and bone metastasis, respectively, and possess have high tumorigenic and metastatic capacities, lack AR expression. ARCaP cells, derived the ascites fluid of a disseminated CRPC, express little AR (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Bone metastases MDA PCa 118a/118b also completely lack AR (and PSA) expression (e.g., Dubrovska, et al. 2010; Rybak et al., 2015).
Similar AR heterogeneity has also been observed in prostatic-specific transgenic mouse models. In ARR2Pb driven c-Myc (i.e., Hi-Myc) model (e.g., Dubrovska, et al. 2010; Rybak et al., 2015), the residual tumors five months post-castration expressed low and heterogeneous levels of cytoplasmic AR compared to the intact mice. These castration-resistant Hi-Myc tumor cells were also quiescent as shown by negative Ki67 staining (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). In prostate-specific Pten-deleted mouse prostate, although most tumor cells expressed AR after 10 weeks’ castration, the expression level was weaker and more diffuse compared to the hormonally intact prostate (e.g., Dubrovska, et al. 2010; Rybak et al., 2015).
AR heterogeneity in CRPCs has a genetic basis. A recent sequencing study of 150 metastatic PCa and CRPCs suggests that genetic alterations of AR (mutations, amplifications) become enriched (~63% patients) in CRPCs compared to those in untreated tumors (Robinson et al., 2015). In addition to mutations in AR itself, alterations of members in the AR signaling pathway were also observed in metastatic CRPCs, including FOXA1 and NCOR1/2, among others. Similarly, by comparing 50 lethal CRPCs and 11 primary cancers, Grasso et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) identified mutations in FOXA1 and MLL2 in CRPCs that likely change the AR signaling in treatment-failed tumors.
IV. PCSCs in primary and untreated PCa: AR negativity and signaling mechanisms
The preceding discussions highlight the presence of AR− PCa cells in untreated PCa (Liu et al., 2015). This is an important point as the AR− PCa cells are expected to not respond well to AR-targeting therapies. This point would be consistent with reports that androgen-independent PCa cells pre-exist in primary tumors, which may become selected during ADT (Finoes RR et al., 2013; Issacs & Coffey, 1981; Liu et al., 2015). Interestingly, in many reported PCSC populations in untreated PCa models or primary tumors, AR expression is often low or undetectable (Figure 2). For example, the CD44+α2β1+CD133+ cells purified from seven human tumor samples (Collins et al., 2005), the ABCG2+ putative PCSCs (Huss et al., 2005), and the CD44+ cells in several PCa xenografts (Patrawala et al., 2006) were all AR−. In fact, the AR−CD44+ PCSCs were shown to be able to differentiate, at the clonal level, into AR+CD44− cells (Patrawala et al., 2006). Gu et al (2007) also showed that the HPET cells (human prostate epithelial cells immortalized by overexpressing hTERT) expressed stem cell molecules such as CD44 and Nanog, could regenerate three prostate epithelial cell types, and were AR negative. Miki et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) showed mutually exclusive expression patterns of CD133 and AR by IHC staining in 16 clinical specimens. Rajasekhar et al (2011) reported both AR and PSA negativity in the TRA-1-60+CD151+and CD166+ PCSC population, which possessed high tumorigenic ability and could generate differentiated AR+ and PSA+ tumors in vivo. The docetaxel-resistant PCSCs that lacked the expression of MHC molecules were also negative for AR and PSA (Domingo-Domeneck et al., 2012). Likewise, the PSA−/lo PCSC population was enriched in AR− PCa cells (Liu et al., 2015; Qin et al., 2012). These and many other studies (reviewed in Liu et al., 2015) suggest that PCSCs in primary and untreated tumors seem to be generally AR−; in other words, AR− (and PSA−) cells are highly enriched in primary and/or untreated PCSC populations (Figure 2). Vice Versa, loss of AR expression has been shown to promote PCSC generation through SATA3 signaling (Schroeder et al., 2014). It remains to be seen whether the AR+ and AR− PCa cells in untreated/primary PCa possess distinct self-renewal, tumor-propagating properties and drug sensitivities as these two populations of PCa cells have not been prospectively separated, purified out, and compared for their biological properties.
PCSCs in untreated PCa remain AR− presumably because these cells are simply less differentiated. Alternatively, molecules such as ABCG2 are preferentially expressed in PCSCs (Huss et al., 2005), which mediates efflux of androgens leading to the degradation of ligand-less AR in PCSCs. At least some of the PCSCs, e.g., SP, CD44+, ABCG2+, and PSA−/lo, have been shown by us to be able to self-renew based on serial tumor-transplantation assays and asymmetric cell divisions using clonal and time-lapse analyses (Qin et al., 2012; Patrawala et al., 2005; Patrawala et al., 2006; Liu et al., 2015). A fraction of PSA−/lo PCa cells can undergo authentic asymmetric cell division regenerating a PSA−/lo daughter cell as well as a differentiated PSA+ cell, which subsequently undergoes rapid proliferation (Liu et al., 2015; Qin et al., 2012). Self-renewal is a shared property for both normal stem cells and CSCs, and, not surprisingly, many molecules and pathways that regulate self-renewal in normal stem cells have been reported to operate in PCSCs (Figure 2). For example, we have shown that NANOG is preferentially expressed in several PCSC populations and its expression is important for CSC properties as its knockdown severely impairs tumor regeneration (Jeter et al., 2009). In contrast, inducible expression of NANOG alone is sufficient to reprogram bulk cancer cells into stem-like cancer cells with enhanced tumor-regenerating and tumor-propagating activities (Jeter et al., 2011). Our results suggest that certain pluripotency molecules may also be functionally important for PCSC self-renewal and other properties. In support, several other studies have similarly implicated OCT4 and SOX2 in conferring on PCSC activities (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Interestingly, reciprocal relationships between AR and NANOG/OCT4/SOX2 have been noted in these studies.
Hedgehog (HH) and WNT signaling often act together and play important roles in regulating self-renewal. The importance of WNT/β-catenin signaling is illustrated by the observations that treatment of LNCaP and C4-2 cells with WNT-3a increased their sphere formation rate and size, with increased nuclear β-catenin accumulation (Bisson and Prowse 2009). Although AR antagonist bicalutamide reduced the sphere size, the sphere formation rate did not change, thus suggesting a role of WNT signaling in PCSC self-renewal independently from AR (Bisson and Prowse 2009). Bmi-1 acts down-stream of HH, and has been shown to be necessary for HH induced self-renewal and several populations of normal stem cells as well as CSCs (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Lukacs et al (2010) investigated the effects of Bmi-1 loss in the presence of over-activated Wnt signaling on murine prostate stem cells (PSCs) and demonstrated that Bmi-1 expression was required for Wnt pathway to modulate self-renewal in the PSCs. In addition, several other signaling molecules and pathways may also be involved in regulating PCSCs. For example, PTEN/PI3K/AKT pathway has been reported to be essential for PCSC proliferation independent of AR status (e.g., Dubrovska, et al. 2010; Rybak et al., 2015).
E-twenty-six (Ets)-related gene (ERG), which is essential to maintain adult HSC self-renewal during stress-induced hematopoiesis (e.g., Dubrovska, et al. 2010; Rybak et al., 2015), is deregulated in most PCa through the most common genetic event TMPRESS2-ERG fusion (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). TMPRSS2-ERG expression is associated with a relative increase in clonogenic PCa cells (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Interestingly, although the expression of TMPRESS2-ERG fusion gene is expected to occur in AR+ PCa cells due to the TMPRESS2 regulation by AR, recent evidence suggests that the TMPRESS2-ERG fusion protein may also be expressed in the AR− PCSCs. Polson and colleagues (2013) demonstrated that in CD133+α2β1+ primary tumor cells with stem cell properties, TMPRSS2-ERG and AR expression was not necessarily concordant. While most of the marker-positive cells were AR negative, they expressed ERG at both RNA and protein levels, which may help maintain the PCSC properties such as self-renewal in the marker positive cells (Polson et al., 2013).
Taken together, the above discussions indicate that many well-known signaling molecules and pathways can regulate and confer the CSC properties in AR− PCSCs (Chen et al., 2013; Rybak et al., 2015). These molecules and pathways represent obvious therapeutic targets, and therapeutics targeting these PCSC-specific signaling nodes could, in principle, be utilized in conjunction with the ADT regimens.
V. PCSCs in CRPC might be AR+ or AR−
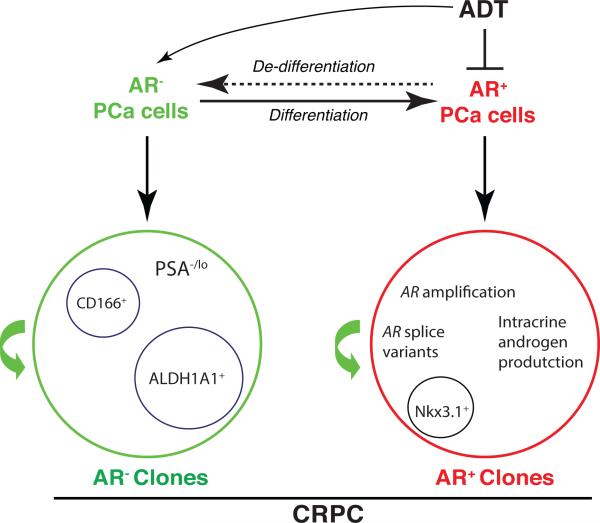
It is well appreciated that AR heterogeneity becomes more pronounced in CRPCs than in the primary tumors (Liu et al., 2015) and activation of alternative AR signaling in PCa cells may promote PCa cell proliferation under androgen-deprived environment (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). What is the cell-of-origin of CRPCs? AR+ or AR− PCa cells? As early as 1981, Isaacs and Coffey, working on the Dunning R3327H rat prostatic adenocarcinoma model, proposed that castration selected for androgen-insensitive cells that pre-existed in the untreated tumors. Craft et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015), working on the LAPC9 xenograft model, also provided histological evidence for outgrowth of the androgen-independent clones in the later stages of CRPC development. Fiñones et al (2013) demonstrated androgen-independent PCa cells in untreated early-stage prostate adenocarcinomas. These androgen-independent and androgen-insensitive PCa cells may not necessarily be AR-, because PCa cells that overexpress AR and splice variants that lack the LBD may also be insensitive or refractory to androgen ablation. Our recent work provided direct evidence for AR− PCa cells in primary patient tumors (Liu et al., 2015). As many PCSCs have been shown to be AR− and to be resistant to castration and other therapeutics (Chen et al., 2013; Liu et al., 2015; Qin et al., 2012; Rybak et al., 2015), it is reasonable to postulate that the AR− PCa cells that pre-exist in untreated tumors could be favored as ‘initiators’ or the cells-of-origin of CRPCs (Figure 3). These AR− PCa cells could be expanded upon ADT-induced elimination of AR+ cells as well as due to the de-differentiation from AR+ PCa cells (Figure 3), much like therapy- or microenvironment-induced de-differentiation of non-CSCs in other tumor systems (Kreso and Dick, 2014; Tang, 2012). As a result, the AR− PCa cells in CRPCs may function as the CSCs for the AR− CRPC clones (Figure 3). The best example is the PSA−/lo PCSC population, which has been evinced to possess significant tumor-regenerating and tumor-propagating activities in fully castrated male mice (Qin et al., 2012). Germann et al (2012) showed that PCa cells expressing stem cell markers such as ALDH1A1 and NANOG became enriched in the BM18 castration model and the castration-resistant stem-like PCa cells had a luminal progenitor phenotype but were negative for AR. Jiao et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) identified a CD166+ cell population in both human and mouse CRPCs, which was enriched in basal stem/progenitor cells that were CK5+/p63+/CK8+/AR−/TROP2hi and displayed enhanced sphere formation and tissue regeneration abilities. Also, studies on NANOG (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) and SOX2 (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) show that PCa cells expressing these molecules are castration resistant and express relatively low levels of AR. These observations raise the possibility that the AR− PCSCs may gain growth advantages in androgen-deficient environment, leading to distinct AR− clones in CRPC (Figure 3).
Figure 3. Hypothetical PCSCs in CRPC.
ADT selectively targets AR+ PCa cells and has been shown to enrich AR− PCa cells, which may result from preferential elimination by ADT of AR+ cells as well as de-differentiation of AR+ PCa cells to AR− cells (top). Clinical CRPCs contain distinct AR+ and AR− clones, both of which might contain their own CSCs. In AR+ clones, PCSCs could have AR amplification or ligand-independent AR signaling pathways to support the self-renewal in androgen deprived environment. Several potential CSC subpopulations in AR− and AR+ PCa cell clones are indicated.
On the other hand, most CRPCs clearly have AR+ cells and clones (Liu et al., 2015). Although these AR+ cells in CRPCs can potentially be derived from the differentiation of AR− PCa cells (Figure 2), it is very likely that at least some AR+ PCa cells can survive androgen deprivation and function as the cells-of-origin as well as CSCs for CRPCs (Figure 3). This is not very difficult to understand because the AR+ PCa cells in most untreated primary tumors constitute the bulk cell population (Figure 2). It is conceivable that due to their abundance, some of these AR+ PCa cells, under the selective pressure from androgen deprivation, may selectively gain genetic alterations such as the AR gene amplification and TMPRSS2-ERG fusion, resulting in expansion of AR+ clones (Figure 3, right). In the resultant AR+ PCa cell clones, AR may likely be still functioning to regulate both conventional as well as new AR target genes (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Regulation of conventional AR targets can be achieved through intratumoral androgen synthesis. Alternatively, AR signaling in the AR+ CRPC clones may be executed through ligand-independent AR splice variants and/or AR crosstalks with activated receptors such as EGFR. In fact, there is evidence that certain AR+ cell population is refractory to castration and can function as the cell-of-origin for PCa in mouse models. Wang et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) showed that castration-resistant Nkx3.1-expressing cells (CARNs) that expressed luminal markers including AR represented a rare population of androgen-resistant cells in the murine prostate that could function as the cells-of-origin for PCa caused by Pten deletion.
Interestingly, expressing wild-type AR at physiological levels in AR− PC3 cells induced growth inhibition (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) whereas knocking down AR in AR-expressing metastatic PCa cells like LNCaP and its derivative C4-2 resulted in growth inhibition, apoptotic cell death and compromised tumor development (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). The contrasting roles of AR in AR− vs. AR+ PCa cell lines imply differential involvement of AR in AR+ and AR− PCSCs in CRPCs. Regardless, the phenotype of PCSCs in CRPCs may well be context-dependent and both AR+ and AR− clones, which possess their own intra-clonal CSCs, likely co-exist in hormone-refractory tumors (Figure 3). Clarifying the precise functions of AR+ vs. AR− PCSCs in CRPC awaits the development of critical experimental tools that can allow the prospective separation of AR+ and AR− CRPC cells.
VI. AR and PCSCs in PCa metastasis
Metastasis is common in CRPC patients. The acquisition of invasive properties through epithelial-mesenchymal transition (EMT), a normal development process, is crucial for the evolution of metastatic population (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). There is accumulating evidence supporting that ADT may induce an EMT in PCa cells (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) and EMT is well-known to promote CSC traits. Studies of Tanaka et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) and Jennbacken et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) showed that N-cadherin was upregulated in castration-resistant LNCaP, LAPC4, and LAPC9 xenograft models. Sun et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) interrogated EMT marker expression in mouse and human CRPC samples and observed overall higher levels of mesenchymal markers in CRPC compared to non-castrated samples. They proposed a negative feedback loop model between ZEB1 and AR to explain the ADT-induced EMT. To some extent, AR signaling may be involved in the EMT switching in PCa cells. The study on AR and ZEB2 suggests that AR may function differently between AR+ and AR− cell lines (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Specifically, ZEB2 expression positively correlate with AR expression in LNCaP cells, but the opposite is true in PC3 and DU145 cell. In addition, the AR splice variants AR3 and ARv567es were shown to promote EMT in PCa cells (e.g., Dubrovska, et al. 2010; Rybak et al., 2015).
Not only do CSCs play an important role in tumor initiation and treatment resistance, they also seem to be involved in distant metastases. Tanaka et al (e.g., Dubrovska, et al. 2010; Rybak et al., 2015) have shown that the castration-resistant, N-cadherin positive PCa cells are enriched in stem cell markers including CD44 and NANOG. Vice versa, Lin−CD44+CD133+Sca-1+CD117+ mouse PSCs express higher levels of mesenchymal markers N-cadherin and vimentin compared to the non stem cells (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). On the other hand, EMT may also suppress the stemness in PCa cells (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). This is not entirely surprising because mesenchymal-epithelial transition (MET) is equally important and required for metastatic colonization. Research on the role of MET in PCa metastasis is very limited.
VII. Clinical Implications and Perspectives
Studies about the potential prognostic role of AR in PCa are controversial, and most evidence suggests that AR is not prognostic in PCa (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Minner et al (2011) examined the AR expression in more than 2800 treatment-naïve PCa patients’ samples and observed no significant correlation between AR expression level and the risk of biochemical recurrence. Studies by Fleischmann et al (2011) in 382 lymph node metastases reported that AR is not prognostic in nodal positive PCa although higher AR does correlate with larger size of metastases. Despite significant improvements in the efficiency of the ADT to block AR signaling, up to now, there is also no clear correlation between androgen signaling ablation and patient prognosis. Study by Ford et al (2003) in 24 CRPC patients showed that 33% of patients have AR amplification and the patients with AR gene amplification had recurrence five months earlier than those without amplification; however, no statistically significant survival disadvantage was observed in the AR amplified patients. More recently, Lu-Yao et al (2014) performed a median 110-months follow-up study of a cohort consisted of 66,717 PCa patients who underwent primary ADT or conservative management and found that primary ADT was not associated with improved long-term overall or disease-specific patient survival. Further, the AR heterogeneity in PCa indicates that targeting AR signaling alone may be of a limited role in preventing disease recurrence in long term.
PCSCs may represent the driving force of tumor progression and metastases. A number of studies have shown that the expression of stem-cell markers has prognostic significance in PCa, as well as other cancer types (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Studies on PSA−/lo PCSCs suggest that intratumoral PSA expression is inversely correlated with the tumor Gleason score and patient survival (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). Multiple studies have shown that the AR− tumor cells are enriched in PCSC populations, implicating a pivotal role of PCSCs in ADT resistance. Hence, targeting PCSCs specifically in an adjuvant setting might be helpful in preventing CRPC. Preclinical studies in PCSCs targeting have provided promising results. For instance, we have demonstrated that microRNA-34a (miR-34a) potently inhibits the PCa progression and metastasis via directly targeting at CD44 (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). We have also reported several other microRNAs including let7b and miR-128 in suppressing PCSC self-renewal and tumor progression (e.g., Dubrovska, et al. 2010; Rybak et al., 2015). At the same time, direct inhibition of WNT, PTEN/PI3K/AKT and others cell-signaling pathways has shown tumor suppressive effect via lowering PCSCs population (e.g., Dubrovska, et al. 2010; Rybak et al., 2015).
Understanding and elucidating the roles of and the interrelationship between AR heterogeneity and PCSCs could offer fresh insight on designing novel therapeutics to target lethal CRPC and metastasis. Recent evidence suggests that in untreated tumors, PCSCs seem to be largely AR− whereas in CRPCs, PCSCs may be either AR+ or AR−. In other words, both AR+ and AR− PCa cell clones co-exist in most CRPCs (Figure 3). In principle, PCSCs, whether AR+ or AR−, are endowed with the fundamental trait of stemness, which is regulated by unique cohorts of genes, epigenetic landscape, and environmental factors (Kreso and Dick 2014). It is high time for us to develop novel therapeutics that target the stemness of PCSCs, which, when used in conjunction with ADT, should help prevent tumor recurrence.
Acknowledgement
We thank other members of the Tang lab for helpful discussions. We apologize to the colleagues whose work was not cited due to space constraint.
Funding
Work in the authors’ lab was supported, in part, by grants from NIH (NCI R01-CA155693), DOD (W81XWH-13-1-0352 and W81XWH-14-1-0575), CPRIT (RP120380), and MDACC Center for Cancer Epigenetics (D.G.T).
Footnotes
Declaration of Interest
The authors declare no conflicts of interest.
Author Contributions
QD and DGT conceptualized the paper; QD wrote the draft; DGT finalized the manuscript.
Reference
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson I, Prowse DM. WNT signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell characteristics. Cell Res. 2009;19:683–697. doi: 10.1038/cr.2009.43. [DOI] [PubMed] [Google Scholar]
- Casey OM, Fang L, Hynes PG, Abou-Kheir WG, Martin PL, Tillman HS, Petrovics G, Awwad HO, Ward Y, Lake R, et al. TMPRSS2-driven ERG expression in vivo increases self-renewal and maintains expression in a castration resistant subpopulation. PLoS One. 2012:7e41668. doi: 10.1371/journal.pone.0041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celia-Terrassa T, Meca-Cortes O, Mateo F, de Paz AM, Rubio N, Arnal-Estape A, Ell BJ, Bermudo R, Diaz A, Guerra-Rebollo M, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest. 2012:1221849–1868. doi: 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rycaj K, Liu X, Tang DG. New insights into prostate cancer stem cells. Cell Cycle. 2013;12:579–586. doi: 10.4161/cc.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Snoek R, Ghaidi F, Cox ME, Rennie PS. Short hairpin RNA knockdown of the androgen receptor attenuates ligand-independent activation and delays tumor progression. Cancer Res. 2006;66:10613–10620. doi: 10.1158/0008-5472.CAN-06-0028. [DOI] [PubMed] [Google Scholar]
- Chodak GW, Kranc DM, Puy LA, Takeda H, Johnson K, Chang C. Nuclear localization of androgen receptor in heterogeneous samples of normal, hyperplastic and neoplastic human prostate. J Urol. 1992;147:798–803. doi: 10.1016/s0022-5347(17)37389-5. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft N, Chhor C, Tran C, Belldegrun A, DeKernion J, Witte ON, Said J, Reiter RE, Sawyers CL. Evidence for clonal outgrowth of androgen-independent prostate cancer cells from androgen-dependent tumors through a two-step process. Cancer Res. 1999;59:5030–5036. [PubMed] [Google Scholar]
- Davis JN, Wojno KJ, Daignault S, Hofer MD, Kuefer R, Rubin MA, Day ML. Elevated E2F1 inhibits transcription of the androgen receptor in metastatic hormone-resistant prostate cancer. Cancer Res. 2006;66:11897–11906. doi: 10.1158/0008-5472.CAN-06-2497. [DOI] [PubMed] [Google Scholar]
- Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, Bonal DM, Charytonowicz E, Gladoun N, de la Iglesia-Vicente J, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22:373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira SM, Garcia-Echeverria C, Schultz PG, Reddy VA. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci USA. 2009;106:268–273. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovska A, Elliott J, Salamone RJ, Kim S, Aimone LJ, Walker JR, Watson J, Sauveur-Michel M, Garcia-Echeverria C, Cho CY, et al. Combination therapy targeting both tumor-initiating and differentiated cell populations in prostate carcinoma. Clin Cancer Res. 2010:165692–5702. doi: 10.1158/1078-0432.CCR-10-1601. [DOI] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Fiñones RR, Yeargin J, Lee M, Kaur AP, Cheng C, Sun P, Wu C, Nguyen C, Wang-Rodriguez J, Meyer AN, et al. Early human prostate adenocarcinomas harbor androgen-independent cancer cells. PLoS One. 2013;8:e74438. doi: 10.1371/journal.pone.0074438. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Rocha C, Schobinger S, Seiler R, Wiese B, Thalmann GN. Androgen receptors are differentially expressed in Gleason patterns of prostate cancer and down-regulated in matched lymph node metastases. Prostate. 2011;71:453–460. doi: 10.1002/pros.21259. [DOI] [PubMed] [Google Scholar]
- Ford OH, 3rd, Gregory CW, Kim D, Smitherman AB, Mohler JL. Androgen receptor gene amplification and protein expression in recurrent prostate cancer. J Urol. 2003;170:1817–1821. doi: 10.1097/01.ju.0000091873.09677.f4. [DOI] [PubMed] [Google Scholar]
- Germann M, Wetterwald A, Guzman-Ramirez N, van der Pluijm G, Culig Z, Cecchini MG, Williams ED, Thalmann GN. Stem-like cells with luminal progenitor phenotype survive castration in human prostate cancer. Stem Cells. 2012;30:1076–1086. doi: 10.1002/stem.1087. [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Yuan J, Wills M, Kasper S. Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer Res. 2007;67:4807–4815. doi: 10.1158/0008-5472.CAN-06-4608. [DOI] [PubMed] [Google Scholar]
- Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, Adejola N, Gurel M, Hicks J, Meeker AK, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, De Marzo AM, Yegnasubramanian S, Epstein JI, Carter HB. Diagnostic challenges of clonal heterogeneity in prostate cancer. J Clin Oncol. 2015;33:e38–40. doi: 10.1200/JCO.2013.50.3540. [DOI] [PubMed] [Google Scholar]
- Huss WJ, Gray DR, Greenberg NM, Mohler JL, Smith GJ. Breast cancer resistance protein-mediated efflux of androgen in putative benign and malignant prostate stem cells. Cancer Res. 2005;65:6640–6650. doi: 10.1158/0008-5472.CAN-04-2548. [DOI] [PubMed] [Google Scholar]
- Isaacs JT, Coffey DS. Adaptation versus selection as the mechanism responsible for the relapse of prostatic cancer to androgen ablation therapy as studied in the Dunning R-3327-H adenocarcinoma. Cancer Res. 1981;41:5070–5075. [PubMed] [Google Scholar]
- Jacob S, Nayak S, Fernandes G, Barai RS, Menon S, Chaudhari UK, Kholkute SD, Sachdeva G. Androgen receptor as a regulator of ZEB2 expression and its implications in epithelial-to-mesenchymal transition in prostate cancer. Endocr Relat Cancer. 2014;21:473–486. doi: 10.1530/ERC-13-0514. [DOI] [PubMed] [Google Scholar]
- Jennbacken K, Tesan T, Wang W, Gustavsson H, Damber JE, Welen K. N-cadherin increases after androgen deprivation and is associated with metastasis in prostate cancer. Endocr Relat Cancer. 2010;17:469–479. doi: 10.1677/ERC-10-0015. [DOI] [PubMed] [Google Scholar]
- Jeter CR, Badeaux M, Choy G, Chandra D, Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ, Tang DG. Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells. 2009;27:993–1005. doi: 10.1002/stem.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ, Tang DG. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–3845. doi: 10.1038/onc.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, Chen D, Li Y, Guo C, Zhang B, et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One. 2012;7:e42564. doi: 10.1371/journal.pone.0042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, Jeter C, Chen X, Vlassov AV, Tang DG. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014;74:4183–4195. doi: 10.1158/0008-5472.CAN-14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008;26:2813–2820. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhofs S, Denayer S, Haelens A, Claessens F. Androgen receptor knockout and knock-in mouse models. J Mol Endocrinol. 2009;42:11–17. doi: 10.1677/JME-08-0122. [DOI] [PubMed] [Google Scholar]
- Kregel S, Kiriluk KJ, Rosen AM, Cai Y, Reyes EE, Otto KB, Tom W, Paner GP, Szmulewitz RZ, Vander Griend DJ. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS One. 2013;8:e53701. doi: 10.1371/journal.pone.0053701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Kroon P, Berry PA, Stower MJ, Rodrigues G, Mann VM, Simms M, Bhasin D, Chettiar S, Li C, Li PK, et al. JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res. 2013;73:5288–5298. doi: 10.1158/0008-5472.CAN-13-0874. [DOI] [PubMed] [Google Scholar]
- van der Kwast TH, Schalken J, Ruizeveld de Winter JA, van Vroonhoven CC, Mulder E, Boersma W, Trapman J. Androgen receptors in endocrine-therapy-resistant human prostate cancer. Int J Cancer. 1991;48:189–193. doi: 10.1002/ijc.2910480206. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Li HW, Chen X, Calhoun-Davis T, Claypool K, Tang DG. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res. 2008a;68:1820–1825. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, Sikes C, Multani AS, Efstathiou E, Lopez A, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J Clin Invest. 2008b;118:2697–2710. doi: 10.1172/JCI33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, Stass SA, Jiang F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2010;90:234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn DE, Yang X, Sun F, Xie Y, Chen H, Jiang R, Chen H, Chumsri S, Burger AM, Qiu Y. A role for OCT4 in tumor initiation of drug-resistant prostate cancer cells. Genes Cancer. 2010;1:908–916. doi: 10.1177/1947601910388271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov IV, Antony L, Dalrymple SL, Becker R, Cheng L, Isaacs JT. PC3, but not DU145, human prostate cancer cells retain the coregulators required for tumor suppressor ability of androgen receptor. Prostate. 2006;66:1329–1338. doi: 10.1002/pros.20483. [DOI] [PubMed] [Google Scholar]
- Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71:5950–5954. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012;72:3393–3404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen X, Chen X, Rycaj K, Chao HP, Deng Q, Jeter C, Liu C, Honorio S, Li H, et al. Systematic dissection of phenotypic, functional, and tumorigenic heterogeneity of human prostate cancer cells. Oncotarget. 2015 Jun 24; doi: 10.18632/oncotarget.4260. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- Lukacs RU, Memarzadeh S, Wu H, Witte ON. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2010;7:682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu-Yao GL, Albertsen PC, Moore DF, Shih W, Lin Y, DiPaola RS, Yao SL. Fifteen-year survival outcomes following primary androgen-deprivation therapy for localized prostate cancer. JAMA Intern Med. 2014;174:1460–1467. doi: 10.1001/jamainternmed.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai M, Sumiya H, Akimoto S, Yatani R, Chang CS, Liao SS, Shimazaki J. Immunohistochemical study of androgen receptor in benign hyperplastic and cancerous human prostates. Prostate. 1990;17:293–300. doi: 10.1002/pros.2990170405. [DOI] [PubMed] [Google Scholar]
- Miki J, Furusato B, Li H, Gu Y, Takahashi H, Egawa S, Sesterhenn IA, McLeod DG, Srivastava S, Rhim JS. Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens. Cancer Res. 2007;67:3153–3161. doi: 10.1158/0008-5472.CAN-06-4429. [DOI] [PubMed] [Google Scholar]
- Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, Simon R, Tennstedt P, Muller J, Scholz L, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, Shah RB, Gaston S, Tomlins SA, Wei JT, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AP, Loughran SJ, Metcalf D, Hyland CD, de Graaf CA, Hu Y, Smyth GK, Hilton DJ, Kile BT, Alexander WS. Erg is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. Blood. 2011;118:2454–2461. doi: 10.1182/blood-2011-03-344739. [DOI] [PubMed] [Google Scholar]
- Niu Y, Altuwaijri S, Yeh S, Lai KP, Yu S, Chuang KH, Huang SP, Lardy H, Chang C. Targeting the stromal androgen receptor in primary prostate tumors at earlier stages. Proc Natl Acad Sci USA. 2008;105:12188–12193. doi: 10.1073/pnas.0804701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003:423302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2-cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006:251696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796–6805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- Polson ES, Lewis JL, Celik H, Mann VM, Stower MJ, Simms MS, Rodrigues G, Collins AT, Maitland NJ. Monoallelic expression of TMPRSS2/ERG in prostate cancer stem cells. Nat Commun. 2013;4:1623. doi: 10.1038/ncomms2627. [DOI] [PubMed] [Google Scholar]
- Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, Calhoun-Davis T, Li H, Palapattu GS, Pang S, et al. The PSA−/lo prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat Commun. 2011;2:162. doi: 10.1038/ncomms1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruizeveld de Winter JA, Trapman J, Brinkmann AO, Boersma WJ, Mulder E, Schroeder FH, Claassen E, van der Kwast TH. Androgen receptor heterogeneity in human prostatic carcinomas visualized by immunohistochemistry. J Pathol. 1990;160:329–332. doi: 10.1002/path.1711600409. [DOI] [PubMed] [Google Scholar]
- Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, Verleun-Mooijman MC, Trapman J, Brinkmann AO, Santerse AB, Schroder FH, van der Kwast TH. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am J Pathol. 1994:144735–746. [PMC free article] [PubMed] [Google Scholar]
- Rybak AP, Bristow RG, Kapoor A. Prostate cancer stem cells: deciphering the origins and pathways involved in prostate tumorigenesis and aggression. Oncotarget. 2015;6:1900–1919. doi: 10.18632/oncotarget.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycaj K, Tang DG. Cell-of-origin of cancer versus cancer stem cells: Assays and interpretations. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-15-0798. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadi MV, Barrack ER. Image analysis of androgen receptor immunostaining in metastatic prostate cancer. Heterogeneity as a predictor of response to hormonal therapy. Cancer. 1993:712574–2580. doi: 10.1002/1097-0142(19930415)71:8<2574::aid-cncr2820710823>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Sadi MV, Walsh PC, Barrack ER. Immunohistochemical study of androgen receptors in metastatic prostate cancer. Comparison of receptor content and response to hormonal therapy. Cancer. 1991;67:3057–3064. doi: 10.1002/1097-0142(19910615)67:12<3057::aid-cncr2820671221>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Herrmann A, Cherryholmes G, Kowolik C, Buettner R, Pal S, Yu H, Müller-Newen G, Jove R. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer Res. 2014:741227–1237. doi: 10.1158/0008-5472.CAN-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- Simanainen U, Allan CM, Lim P, McPherson S, Jimenez M, Zajac JD, Davey RA, Handelsman DJ. Disruption of prostate epithelial androgen receptor impedes prostate lobe-specific growth and function. Endocrinol. 2007;148:2264–2272. doi: 10.1210/en.2006-1223. [DOI] [PubMed] [Google Scholar]
- Snoek R, Cheng H, Margiotti K, Wafa LA, Wong CA, Wong EC, Fazli L, Nelson CC, Gleave ME, Rennie PS. In vivo knockdown of the androgen receptor results in growth inhibition and regression of well-established, castration-resistant prostate tumors. Clin Cancer Res. 2009;15:39–47. doi: 10.1158/1078-0432.CCR-08-1726. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, Chen D, Seo K, Modrusan Z, Gao WQ, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- Sun F, Chen HG, Li W, Yang X, Wang X, Jiang R, Guo Z, Chen H, Huang J, Borowsky AD, et al. Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J Biol Chem. 2014;289:1529–1539. doi: 10.1074/jbc.M113.492140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburrino L, Salvianti F, Marchiani S, Pinzani P, Nesi G, Serni S, Forti G, Baldi E. Androgen receptor (AR) expression in prostate cancer and progression of the tumor: Lessons from cell lines, animal models and human specimens. Steroids. 2012;77:996–1001. doi: 10.1016/j.steroids.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kono E, Tran CP, Miyazaki H, Yamashiro J, Shimomura T, Fazli L, Wada R, Huang J, Vessella RL, et al. Monoclonal antibody targeting of N-cadherin inhibits prostate cancer growth, metastasis and castration resistance. Nat Med. 2010;16:1414–1420. doi: 10.1038/nm.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009a;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009b;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CT, Altuwaijri S, Ricke WA, Huang SP, Yeh S, Zhang C, Niu Y, Tsai MY, Chang C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc Natl Acad Sci USA. 2007;104:12679–12684. doi: 10.1073/pnas.0704940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Gore C, Yang L, Fazli L, Gleave M, Pong RC, Xiao G, Zhang L, Yun EJ, Tseng SF, et al. Slug, a unique androgen-regulated transcription factor, coordinates androgen receptor to facilitate castration resistance in prostate cancer. Mol Endocrinol. 2012;26:1496–1507. doi: 10.1210/me.2011-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Rycaj K, Liu ZM, Tang DG. Cancer stem cells: constantly evolving and functionally heterogeneous therapeutic targets. Cancer Res. 2014;74:2922–2927. doi: 10.1158/0008-5472.CAN-14-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhau HY, Chang SM, Chen BQ, Wang Y, Zhang H, Kao C, Sang QA, Pathak SJ, Chung LW. Androgen-repressed phenotype in human prostate cancer. Proc Natl Acad Sci USA. 1996:9315152–15157. doi: 10.1073/pnas.93.26.15152. [DOI] [PMC free article] [PubMed] [Google Scholar]