Background: PTS2 proteins Gpd1p and Pnc1p are imported into peroxisomes in a PTS2 receptor-dependent manner.
Results: PTS2co-receptor Pex21p is required for peroxisomal piggyback import of Gpd1p and Pnc1p.
Conclusion: PTS2 co-receptors Pex18p and Pex21p enable targeting of distinct cargo proteins under variable stress conditions.
Significance: The life span-regulating Pnc1p is transported into peroxisomes via a novel import route.
Keywords: organelle, peroxisome, protein targeting, protein translocation, protein-protein interaction, Gpd1p, PTS2, Pex21p, Pex7p, Pnc1p
Abstract
Proteins designated for peroxisomal protein import harbor one of two common peroxisomal targeting signals (PTS). In the yeast Saccharomyces cerevisiae, the oleate-induced PTS2-dependent import of the thiolase Fox3p into peroxisomes is conducted by the soluble import receptor Pex7p in cooperation with the auxiliary Pex18p, one of two supposedly redundant PTS2 co-receptors. Here, we report on a novel function for the co-receptor Pex21p, which cannot be fulfilled by Pex18p. The data establish Pex21p as a general co-receptor in PTS2-dependent protein import, whereas Pex18p is especially important for oleate-induced import of PTS2 proteins. The glycerol-producing PTS2 protein glycerol-3-phosphate dehydrogenase Gpd1p shows a tripartite localization in peroxisomes, in the cytosol, and in the nucleus under osmotic stress conditions. We show the following: (i) Pex21p is required for peroxisomal import of Gpd1p as well as a key enzyme of the NAD+ salvage pathway, Pnc1p; (ii) Pnc1p, a nicotinamidase without functional PTS2, is co-imported into peroxisomes by piggyback transport via Gpd1p. Moreover, the specific transport of these two enzymes into peroxisomes suggests a novel regulatory role for peroxisomes under various stress conditions.
Introduction
A hallmark of peroxisomal protein import are the cycling import receptors, which mediate the translocation of natively folded, even oligomerized proteins across the organellar membrane (1–3). Another hallmark of peroxisomes is the involvement of cycling import receptors that shuttle between the peroxisomal membrane and the cytosol (4, 5). Peroxisomal matrix enzymes are post-translationally recognized via peroxisomal targeting sequences of type 1 (PTS1)2 at their extreme C terminus or PTS2-type signals located adjacent to their N terminus. The classical PTS1 consists of the tripeptide SKL-COOH or sequence variants thereof and is bound by the import receptor Pex5p in the cytosol (6–8). The receptor cycle of Pex5p has been determined in detail, and besides cargo recognition, it encompasses at least three additional steps as follows: 1) docking of the receptor-cargo complex at peroxisomal membrane proteins Pex14p or Pex13p (9, 10); 2) formation of a transient pore enabling translocation of the matrix enzyme (11); and 3) release of the receptor from the membrane into the cytosol for another round of import. This export is initiated by receptor ubiquitination followed by receptor dislocation from the membrane to the cytosol in an ATP-consuming process, involving the AAA-ATPases Pex1p and Pex6p (12–14). Besides this typical transport of PTS1 proteins, Pex5p can transport peroxisomal proteins by two alternative pathways. First, cargo proteins without PTS1 may directly bind to the receptor at a site distinct from the PTS1 binding region. One example for such a nonperoxisomal targeting signal protein is the acyl-CoA oxidase Fox1p (15). Second, proteins lacking a functional PTS may oligomerize with another PTS1-containing protein, which is used as a vehicle for peroxisomal import. One of the best studied examples for such piggyback transport is enoyl-CoA isomerase, which is imported together with the PTS1 enzyme Dci1p (16).
The PTS2 consists of a nonapeptide with the consensus sequence (R/K)(L/I/V)X5(H/Q)(L/A/F) (17), which is located near the N terminus, and is recognized by the import receptor Pex7p in the cytosol. In distinction from the PTS1 pathway, the import of PTS2 proteins requires additional auxiliary co-factors. The cargo-bound PTS2 receptor is stabilized and targeted to the peroxisomal membrane by such PTS2 co-receptors (18). In Saccharomyces cerevisiae, the co-receptor function is fulfilled by one of the two redundant peroxins Pex18p or Pex21p (19). Although Pex18p has been identified as a primary binding partner of the Pex7p-Fox3p complex under oleate-inducing conditions, no specific function has yet been assigned to the alternative co-receptor Pex21p. In other organisms only one PTS2 co-receptor exists. In other yeasts Yarrowia lipolytica, Pichia pastoris, and Hansenula polymorpha and in the filamentous fungus Neurospora crassa, Pex20p serves as the corresponding co-receptor of Pex7p (20–24), although in mammals and plants PEX5L fulfills the function of PTS2 co-receptors (25, 26).
In S. cerevisiae, only two proteins are known to contain a functional PTS2. These are the β-oxidation enzyme 3-ketoacyl-CoA thiolase (Fox3p) and the glycerol-3-phosphate dehydrogenase (Gpd1p). For both, the PTS2 sequence is essential and sufficient to target these proteins in a Pex7p-dependent way into peroxisomes (19, 27). Another enzyme, the nicotinamidase Pnc1p, also enters peroxisomes in a Pex7p-dependent manner despite the lack of a PTS (28). For both Gpd1p and Pnc1p, it is not known what function they perform in peroxisomes. Gpd1p performs its known function in the cytosol by producing the osmolyte glycerol from the glycolytic metabolite dihydroxyacetone phosphate under hyperosmolar conditions (29). Pnc1p resides in the cytosol and in the nucleus where other compounds of the NAD+ salvage pathway are enriched (28, 30). The catalytic function of Pnc1p is the conversion of nicotinamide into nicotinic acid, which is important for the redox balance of the cell but also has regulatory function on transcription of genes. This has at least two functional consequences. First, the enzyme initiates NAD+ regeneration (“NAD+ salvage pathway”), and second, it removes nicotinamide, which is a noncompetitive inhibitor of the histone deacetylase Sir2p. The latter effect correlates with the observed life span extension after caloric restriction in S. cerevisiae (28).
Here, we investigated the Pex7p-dependent import pathway of Gpd1p and Pnc1p. We discovered that the constitutively expressed Pex21p, but not the oleate-inducible Pex18p, is required as a co-receptor for PTS2-dependent import of Gpd1p. Our data suggest that the peroxisomal import machinery for PTS2 proteins can adapt to changing environmental conditions by alternative use of co-receptors. Remarkably, Gpd1p forms a dimeric complex with Pnc1p before they are transported together into peroxisomes. This is the first example for a physiologically relevant piggyback transport of the PTS2 pathway.
Experimental Procedures
Strains and Media
S. cerevisiae strains used in this study are listed in Table 1. All strains were grown at 30 °C. YPD medium contained 2% glucose, 2% peptone, and 1% yeast extract. YNBGO medium contained 0.1% glucose, 0.1% oleate, 0.05% Tween, 0.17% yeast nitrogen base without amino acids, 0.1% yeast extract, and 5% ammonium sulfate, adjusted to pH 6.0. Gene deletions and genomic integrations of TEV-protein A as C-terminal fusions were carried out as described previously (31, 32).
TABLE 1.
S. cerevisiae strains used in this study
| S. cerevisiae strain | Description | Source or Ref. |
|---|---|---|
| UTL-7A (wild-type) | MATα, leu2-3, 112ura3-52 trp1 | 58 |
| UTL-7A Δpex7 | pex7::loxP | 4 |
| UTL-7A Δpex18 | pex18::loxP | 43 |
| UTL-7A Δpex21 | pex21::loxP | 35 |
| UTL-7A Δpex18/Δpex21 | pex18::loxP, pex21::loxP | 43 |
| UTL-7A Δgpd1 | GPD1::kanMX6 | This study |
| UTL-7A Pex18-TEV-Protein A | PEX18-TEVcs-Protein A-kanMX6 | 43 |
| UTL-7A Pex21-TEV-Protein A | PEX21-TEVcs-Protein A-kanMX6 | 43 |
| UTL-7A Fox3-TEV-Protein A | FOX3-TEVcs-Protein A-kanMX6 | 43 |
| UTL-7A Gpd1-TEV-Protein A | GPD1-TEVcs-Protein A-kanMX6 | This study |
| BY4742 Δpnc1 | MATα; ura3Δ0; leu2Δ0; his3Δ1; lys2Δ0; PNC1::kanMX4 | Euroscarf, Frankfurt, Germany |
Plasmid Construction
All expression plasmids used in this study code for yeast proteins and are listed in Table 2. The corresponding coding DNA regions were amplified by PCR using genomic DNA of a wild-type strain as a template. Oligonucleotides used for PCR are listed in Table 3. Plasmids pDE1, pDE3, and GPD1_ΔPTS2_pUG35 were derived from pUG35 containing a C-terminal GFP fusion. GPD1_ΔPTS2_pUG35 lacking base pairs 19–45 was generated by overlapping PCR with genomic DNA as template. Plasmids pTSC13 and pDE9 are derived from pUG36 by insertion of LEU2 amplified from pUG-LEU and by replacing GFP with mCherry. Constructs were under the control of a methionine promoter (MET25) and a CYC1 termination region. For expression of plasmid-encoded Pnc1p-GFP and mCherry-Pnc1p, media were supplemented with 0.3 mm methionine to reduce expression levels.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or Ref. | Oligonucleotides |
|---|---|---|---|
| pDE01 | GPD1-GFP | This study | RE3510/RE3567 |
| pDE03 | PNC1-GFP | This study | RE4149/RE4150 |
| pDE09 | mCherry-PNC1 | This study | RE5052/RE4858 |
| pUG34DsRed.SKL | DsRed-SKL | 59 | |
| GPD1_ΔPTS2_pUG35 | GPD1ΔPTS2-GFP | This study | RE3004/RE3005/RE3006 |
| pTSC13-mCherry-Ant1p | mCherry-ANT1 | This study | RE 4659/RE4660 |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide |
|---|
| RE3004, CGCGGATCCATGTCTGCTGCTGCTGAT |
| RE3005, GAACTTCTCTTTCTACCAGCATTATCAGCAGCAGCAGACATGGATCCGCG |
| RE3006, GGTAGAAAGAGAAGTTCCTCTTCTG |
| RE3510, GACTGGATCCATGTCTGCTGCTGCTGATAG |
| RE3511, GATCCTCGAGCTAATCTTCATGTAGATCTAATTC |
| RE3567, GATCGATATCATCTTCATGTAGATCTAATTC |
| RE4149, GACTGGATCCATGAAGACTTTAATTGTTGTTG |
| RE4150, GATCGAATTCTTTATCCACGACATTGATGTTG |
| RE4659, GATCGGATCCATGGTGAGCAAGGG |
| RE4660, GATCCCCGGGTCGCGACTTGTACAGCTCGTCC |
| RE4858, TTATTTATCCACGACATTGATG |
| RE5052, AAGACTTTAATTGTTGTTGATATG |
Subcellular Fractionation
Cells were grown in YNBGO medium with or without 1 m NaCl. Postnuclear supernatants were prepared as described previously (33). Postnuclear supernatants derived from wild-type, Δpex18, Δpex21, and Δgpd1 strains were loaded on linear OptiPrepTM/sucrose (15.5 to 36% (w/v) iodixanol containing 18% (w/v) sucrose) density gradients. Isopycnic density gradient centrifugations were performed according to Cramer et al. (33).
Fluorescence Microscopy
Wide field fluorescence microscopy was performed with a Zeiss Axioskop50 fluorescence microscope (Zeiss). Images were taken with a Princeton Instruments 1300Y digital camera. GFP signal was visualized with a 450–490-nm bandpass excitation filter, a 510-nm dichromatic mirror, and a 515–565-nm bandpass emission filter. DsRed fluorescence was visualized with a 546/12-nm bandpass excitation filter, a 560-nm dichromatic mirror, and a 575–640-nm bandpass emission filter.
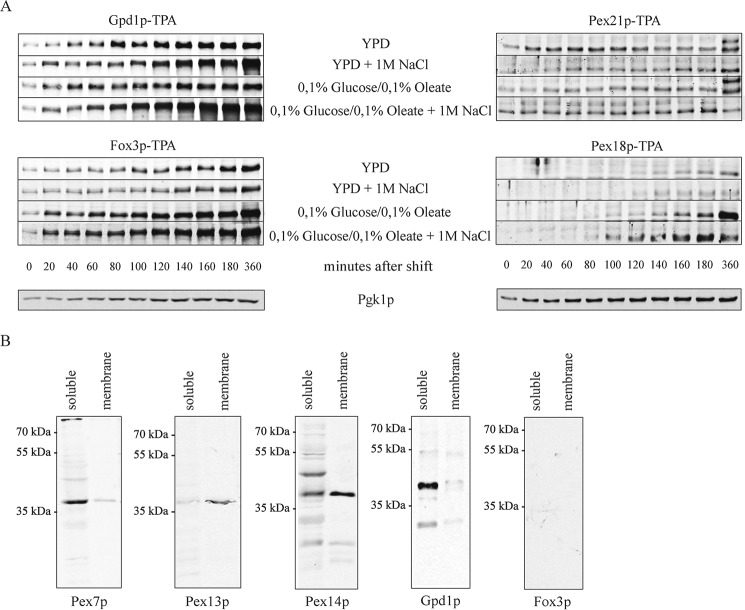
Antibodies and Immunoblotting
For generation of antibodies against Gpd1p, the GPD1 open reading frame was amplified by PCR using genomic DNA of S. cerevisiae strain UTL-7A as template (primers RE3510 and RE3511). The PCR product was digested with BamHI and XhoI and cloned into pGEX4T-2 (GE Healthcare, Munich, Germany), resulting in plasmid pGEX4T-2-GST-GPD1. The plasmid was transformed into Escherichia coli BL21-DE3 Star, resulting in isopropyl β-d-thiogalactopyranoside-inducible expression of GST-tagged Gpd1p. The tagged Gpd1p was purified by affinity chromatography using glutathione-Sepharose 4B (GE Healthcare) according to the manufacturer's protocol. Recombinant proteins were eluted by thrombin cleavage and further purified by size-exclusion chromatography using a Superdex200 column (GE Healthcare). Polyclonal antibodies were raised against the recombinant protein (Pineda, Berlin, Germany).
Immunoblot analysis was performed according to Harlow and Lane (34) with polyclonal rabbit antibodies raised against Pex7p (35), Pex13p (36), Pex14p (9), Fox3p (37), Pcs60p (38), Gpd1p, Porin, and Kar2p (39), Fbp1p (40), protein A (Sigma, Munich, Germany), and GFP-GST (41). Primary antibodies were detected with an IRDye 800CW goat anti-rabbit IgG secondary antibody (LI-COR Bioscience, Bad Homburg, Germany). Semi-quantitative analyses of immunoblot signals were obtained using the “Infrared Imaging System Application Software Version 3.0” (LI-COR Bioscience).
Affinity Purification and Characterization of Protein Complexes
Wild-type cells containing a genomically tagged Pex21p-TEVcs or Gpd1p-TEVcs (tobacco etch virus protease cleavage site)-protein A were grown in YPD supplemented with 1 m NaCl for 16 h. After cell disruption, the cell debris was sedimented (Eppendorf 5810R, 1.500 × g, 10 min), and the supernatant was again centrifuged at 100,000 × g (Sorvall T-647.5) for 1 h at 4 °C. Lysis buffer (20 mm HEPES, 100 mm KOAc, and 5 mm MgOAc, pH 7.5) containing 1% digitonin (Calbiochem) was added to the pellet. The solubilization of membrane proteins was followed by centrifugation at 100,000 × g (Sorvall T-647.5) for 1 h at 4 °C to remove insolubilized membrane proteins. Soluble and/or membrane-bound Gpd1p-TEVcs or Pex21p-TEVcs-Protein A complexes were purified as described earlier (42, 43). The eluate of soluble Gpd1p complexes was subjected to size-exclusion chromatography using a Superose 6 PC3.2/30 column (GE Healthcare).
Sample preparation for protein identification by mass spectrometry was carried out as described in Schrötter et al. (44). LC-MS/MS analysis and data processing were performed as described by Padden et al. (45).
Results
PTS2 Co-receptors Pex18p and Pex21p Are Differently Expressed under Various Stress Conditions
In previous studies, PTS2-dependent import was studied under peroxisome-proliferating growth conditions using oleic acid as the major carbon source (46). The major PTS2-containing protein expressed in oleate-grown cells is the fatty acid oxidation enzyme thiolase, named Fox3p. Although Pex7p is constitutively expressed, Fox3p as well as the co-receptor Pex18p are activated by an inducible “oleate-response element” (ORE) in the promoter region (47–49). Consequently, Pex18p is the preferred PTS2 co-receptor for the import of Fox3p into peroxisomes (19). Nevertheless, Pex21p can rescue the import of Fox3p in a Pex18p deletion strain. Only if both co-receptors were deleted was the import of Fox3p fully compromised.
To characterize alternative peroxisomal import pathways, we analyzed the PTS2-containing Gpd1p, a key enzyme for glycerol production that is essential for growth under hyperosmotic stress conditions (27). To compare the expression of Pex18p, Pex21p, Gpd1p, and Fox3p under different conditions, all four proteins were genomically tagged with protein A. As the tagged proteins fully complemented the mutant phenotype of the corresponding mutants, the tagging did not interfere with the function of the proteins (data not shown). The experimental design allowed the expression of the genes under the control of their own promoters and detection of the proteins with same antibodies. Samples of equal volumes were taken at different time points and analyzed by immunoblotting (Fig. 1A). The steady-state levels of the constitutively expressed control protein Pgk1p (3-phosphoglycerate kinase) correlated well with the cell density profile (data not shown). In contrast, depending on the applied stress conditions, both Gpd1p and Fox3p displayed significant time-dependent alterations. Gpd1p was significantly induced under hyperosmotic conditions (1 m NaCl), and Fox3p was induced in the presence of oleic acid in the medium (Fig. 1A).
FIGURE 1.
Pex21p and Pex18p control distinct cargo-specific PTS2 import pathways. A, time course of induction of Gpd1p-TPA, Pex21p-TPA, Fox3p-TPA, and Pex18p-TPA in different media. Yeast cells expressing genomically tagged protein A (TPA)-fusion proteins under control of the endogenous promoters were grown to the log phase and then shifted to YPD medium supplemented with 2% glucose or oleate-containing medium, each with or without 1 m NaCl as indicated. At each time point, samples of equal volumes were analyzed by immunoblotting with antibodies against protein A. Analysis of the glycolytic enzyme 3-phosphoglycerate kinase Pgk1p of YPD grown cells served as loading control. B, composition of Pex21p-TEV-protein A complexes. Yeast cells expressing Pex21p-TEV-protein A from their genome were grown for 16 h in YPD supplemented with 1 m NaCl. Pex21p complexes were affinity-purified from soluble and membrane fractions after 100,000 × g centrifugation of the cell lysate. Membranes were solubilized with digitonin before affinity purification of membrane complexes. Pex21p-associated proteins were eluted by TEV protease cleavage. SDS samples were subjected to immunoblot analyses using antibodies against Pex7p, Pex13p, Pex14p, Fox3p, and Gpd1p.
In agreement with the proposed role of the PTS2 co-receptor Pex18p in peroxisomal import of Fox3p, the co-receptor was induced in oleate-containing medium (Fig. 1A). In fact, in the absence of oleate, the cellular amount of Pex18p was below the detection level, but a significant increase in the steady-state level was seen when oleic acid was added to the medium. In this respect, the profile correlates well with the profile of Fox3p, the induction of which seem to precede the induction of the co-receptor. In contrast to Pex18p, the co-receptor Pex21p was not induced by oleic acid. Drastic changes of the steady-state concentration of Pex21p were observed neither in oleic acid medium nor under salt stress conditions. Noteworthy, within the first 2 h after shifting the cells into fresh YPD medium, the steady-state level of Pex21p seems to be lower under high salt conditions. However, again in contrast to Pex18p, a significant amount of Pex21p was already clearly detected under all conditions, even in the absence of oleic acid or salt stress. This result suggests that Pex21p is constitutively expressed. As a conclusion, Pex21p is supposed to be responsible for import of PTS2 proteins under conditions when Pex18p is not significantly induced, as in the absence of oleic acid or under salt stress conditions.
To pursue this idea, we analyzed the composition of the cytosolic Pex21p complexes under salt stress conditions for the presence of PTS2 proteins. To this end, cells expressing Pex21p-TPA were grown for 16 h on YPD medium supplemented with 1 m NaCl. Cytosolic and membrane-associated Pex21p complexes were isolated by affinity chromatography and analyzed by immunoblotting (Fig. 1B). Pex7p was detected in the cytosolic complexes, although the docking peroxins Pex14p and Pex13p were highly abundant only in the membrane-associated complex. Gpd1p was clearly identified as a constituent of the cytosolic Pex21p complex (Fig. 1B), although Fox3p, which also was expressed under these growth conditions (Fig. 1A), was not part of the complex (Fig. 1B). Interestingly, Fox3p forms a complex with Pex18p and Pex7p, which as judged by a proteomic analysis does not contain Gpd1p (43).
PTS2 Co-receptor Pex21p Is Essential for the Import of Gpd1p into Peroxisomes
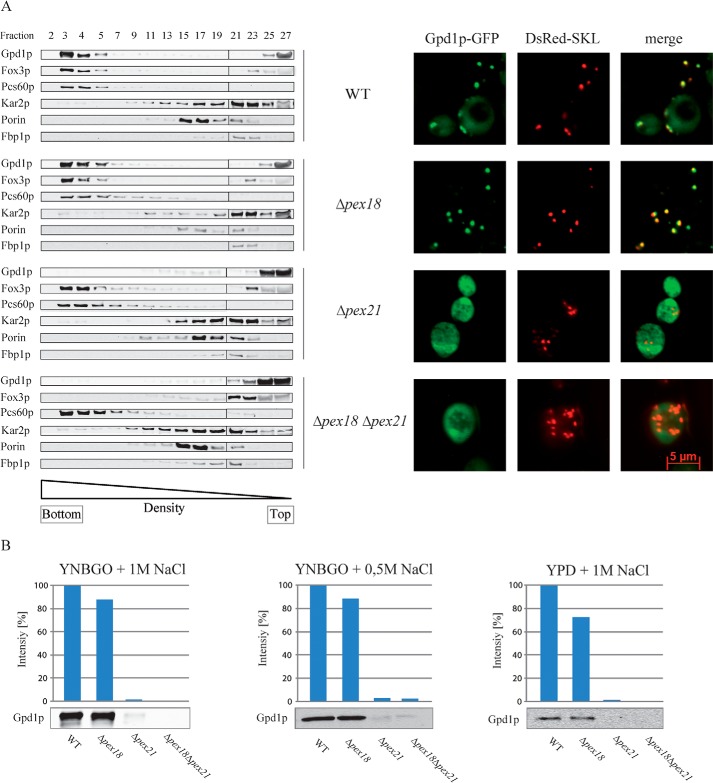
To further analyze the dependence of peroxisomal Gpd1p import on the PTS2 co-receptors, plasmid-encoded Gpd1p-GFP was expressed under hyperosmotic growth conditions in the wild-type strain, single deletion mutants Δpex18 and Δpex21, and double deletion strain Δpex18Δpex21. Postnuclear supernatants were fractionated by isopycnic density gradient centrifugation and analyzed by immunoblotting (Fig. 2A, left panel). As indicated by the presence of the peroxisomal PTS1 protein Pcs60p, peroxisomes were enriched in fractions 3 and 4 at a density of about 1.21 g/cm3. In accordance with previous results (Purdue et al. (19)), the data show that only the deletion of both co-receptors led to complete mislocalization of Fox3p from the peroxisomal fraction to the cytosol (fractions 25–27), although both the Δpex18 and Δpex21 cells exhibited partial import defects. However, the result was different for Gpd1p, which co-fractionated with peroxisomes in wild-type and Δpex18 cells, but it was completely mislocalized to cytosolic fractions in Δpex21 and Δpex18Δpex21 cells. This observation suggests that Pex21p is required for import of Gpd1p. Although Pex18p is present in Δpex21 cells as indicated by the peroxisomal localization of Fox3p, it cannot support peroxisomal import of Gpd1p. This Gpd1p mislocalization in the absence of Pex21p was also found in Pex21p-deficient yeast cells grown under different conditions (Fig. 2B).
FIGURE 2.
Pex21p is required for Gpd1p import into peroxisomes. A, left panel, wild-type (WT) and the indicated mutant strains Δpex18, Δpex21, and Δpex18Δpex21 were grown in oleate medium containing 1 m NaCl for 16 h. Postnuclear supernatants were prepared, and organelles were separated by isopycnic density gradient centrifugation. Fractions were collected from the bottom (fraction 1) of the gradient. Equal quantities of the indicated gradient fractions were analyzed by immunoblotting using antibodies against Gpd1p, Fox3p, Pcs60p (peroxisomal PTS1 matrix protein), Kar2p (endoplasmic reticulum), Porin (mitochondria outer membrane), and Fbp1p (cytosol). A, right panel, plasmid-encoded Gpd1p-GFP and the PTS1 fusion protein DsRed-SKL were expressed in wild-type cells and indicated peroxisomal mutant strains Δpex18, Δpex21, and Δpex18Δpex21. Cells were grown in oleate medium containing 1 m NaCl and analyzed for the intracellular localization of the tagged proteins by fluorescence microscopy. B, comparison of peroxisome-associated Gpd1p in indicated strains under various growth conditions. Semi-quantitative analyses by densitometry were performed from pooled peroxisome-enriched density gradient fractions prepared as described in A. Intensities of wild-type signals were set to 100%.
To validate this result by an independent method, plasmid-encoded Gpd1p-GFP was expressed in the same strains, and its subcellular localization was analyzed by fluorescence microscopy (Fig. 2A, right panel). Microscopic inspection revealed a peroxisomal localization of Gpd1p-GFP as indicated by the congruent punctate pattern obtained with DsRed-SKL in wild-type and Δpex18 cells. In the absence of Pex21p, as in case of Δpex21 and Δpex18Δpex21 cells, GFP-tagged Gpd1p exhibited diffuse cytosolic and nuclear staining without co-localization with the peroxisomal marker protein DsRed-SKL. These results corroborate the biochemical data and demonstrate that Gpd1p is imported into peroxisomes in Pex21p-dependent manner.
Gpd1p Forms a Heterodimeric Complex with Pnc1p
The peroxisome-related function of Gpd1p is still unknown. To identify Gpd1p interaction partners, Gpd1p-TEV-protein A complexes were affinity-purified from soluble fractions and analyzed by SDS-PAGE (Fig. 3, left panel). In comparison with a strain without TEV-protein A (negative control), one additional band was detected in the TEV eluate. This band was identified by mass spectrometry as the nicotinamidase Pnc1p. The Pnc1p is part of the NAD+ salvage pathway and is required for life span extension during caloric restriction (28). The TEV eluate was also subjected to size-exclusion chromatography, which revealed a molecular mass for the Gpd1p-Pnc1p complex of about 60 kDa (Fig. 3, right panel). Taking the molecular masses of monomeric polypeptides (Gpd1p, 42.8 kDa, and Pnc1p, 25 kDa) into account, both proteins form a heterodimeric complex.
FIGURE 3.
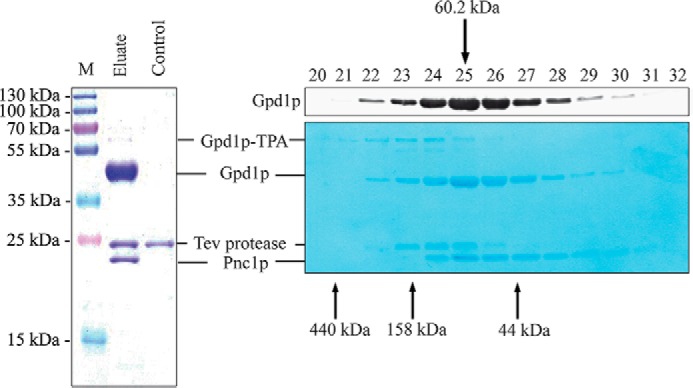
Pnc1p associates with Gpd1p. Gpd1p was affinity-purified from soluble fractions of a strain expressing Gpd1p-TPA under control of its own promoter. Cells were grown on high salt-containing medium and disrupted by glass bead breakage. Soluble fractions were applied to affinity chromatography at an IgG matrix, and Gpd1p-associated proteins were eluted by TEV protease cleavage (left panel). Wild-type cells without genomically integrated TEV-protein A served as a negative control. Gpd1p-associated proteins were separated by SDS-PAGE, and Coomassie-stained polypeptides were identified by mass spectrometry. M, molecular mass marker. Gpd1p complexes were further fractionated by calibrated size-exclusion chromatography (right panels). Fractions were collected, subjected to immunoblot analysis, and either stained with Amido Black dye (lower right panel) or immunodetected using antibodies against Gpd1p (upper right panel). Molecular masses of calibration proteins and calculated mass of the Gpd1p-Pnc1p complex are indicated.
Piggyback Import of Pnc1p into Peroxisomes via Gpd1p-Pex7p-Pex21p Complex
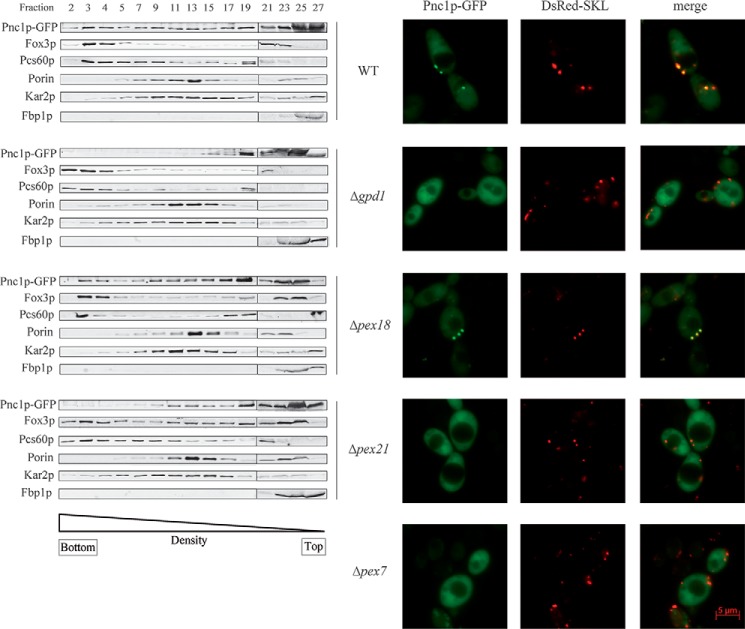
To investigate the Pnc1p import route into peroxisomes, plasmid-encoded Pnc1p-GFP was expressed in the wild-type strain, and subcellular localization of the protein was analyzed by subcellular fractionation studies. To this end, postnuclear supernatants of oleic acid-grown cells were analyzed by isopycnic density gradient centrifugation (Fig. 4, left panel). Most of Pnc1p-GFP was found in cytosolic fractions (fractions 25–27). However, a significant portion was present in fractions 2 and 3, co-fractionating with the peroxisomal matrix proteins Fox3p and Pcs60p. This result indicated that a portion of Pnc1p-GFP was localized in peroxisomes of wild-type cells. This observation was corroborated by fluorescence microscopy (Fig. 4, right panel). In agreement with the results obtained by subcellular fractionation, significant amounts of Pnc1p-GFP were detected in the cytosol and in the nucleus as diffuse fluorescent background labeling all over these cells. However, the study also revealed a congruent punctate fluorescence pattern of Pnc1p-GFP with the peroxisomal marker DsRed-SKL, indicative of a peroxisomal localization of Pnc1p-GFP. The mislocalization of Pnc1p-GFP in Δpex7 cells demonstrated that peroxisomal import of the protein depends on a functional PTS2 import pathway (Fig. 4, right panel).
FIGURE 4.
Peroxisomal Pnc1p import depends on Pex21p and Gpd1p. Cells of wild-type strain as well as Δpex18, Δpex21, and Δgpd1 mutants were grown in oleate medium. The postnuclear supernatant was prepared and further analyzed by density gradient centrifugation (left panel). Fractions were collected from the bottom (fraction 1). Equal volumes of indicated gradient fractions were analyzed by immunoblotting using antibodies against GFP, Fox3p, Pcs60p, Kar2p, Porin, and Fbp1p as indicated. Fluorescence microscopy of plasmid-encoded Pnc1p-GFP and the peroxisomal marker protein DsRed-SKL in cells from wild-type and peroxisomal mutant strains Δpex7, Δpex18, Δpex21, and Δgpd1 (right panel). The data revealed that the peroxisomal localization of Pnc1p depends on the presence of Gpd1p, Pex7p, and Pex21p.
To identify critical constituents of the Pnc1p import pathway into peroxisomes, plasmid-encoded Pnc1p-GFP was transfected in deletion strains Δgpd1, Δpex18, and Δpex21. Postnuclear supernatants derived from cells grown on oleate were analyzed by isopycnic density gradient centrifugation (Fig. 4, left panel). The data show that Pnc1p-GFP was still localized in peroxisome-enriched gradient fractions 2 and 3 in Δpex18 cells, indicated by the co-fractionation with peroxisomal matrix proteins Fox3p and Pcs60p. Interestingly, Pnc1p was not more localized in the peroxisomal fractions in Δpex21 cells. These results were corroborated by fluorescence microscopy analyses, demonstrating that Pnc1p-GFP displayed a congruent fluorescence pattern with the peroxisomal marker in Δpex18 cells but was mislocalized to the cytosol in Δpex21 cells Fig. 4, right panel). A weak cytosolic labeling was also visible in Δpex18 cells, but the intensity resembles background fluorescence seen in wild-type cells and is much less pronounced when compared with cells defective in Pnc1p import. In summary, the import characteristics for Pnc1p, in particular the dependence of peroxisomal import on Pex21p but not on Pex18p, were the same as for its binding partner Gpd1p (Fig. 2).
Subcellular fractionation studies (Fig. 4, left panel) as well as fluorescence microscopy analysis (Fig. 4, right panel) revealed that Pnc1p was mislocalized to the cytosol in cells lacking Gpd1p. Thus, peroxisomal targeting of Pnc1p depends on the presence of Gpd1p. To test for the dependence of Gpd1p targeting in the presence of Pnc1p, plasmid-encoded Gpd1p-GFP and peroxisomal marker mCherry-Ant1p were analyzed in the deletion strain Δpnc1 by fluorescence microscopy (Fig. 5). As shown by the congruent fluorescence pattern, Gpd1p-GFP still co-localized with the peroxisomal marker in Pnc1p-deficient mutant. Thus, peroxisomal targeting of Gpd1p does not require the presence of Pnc1p. Finally, we tested whether the peroxisomal targeting of Pnc1p depends on the presence of the PTS2 of Gpd1p. To this end, we expressed mCherry-Pnc1p or the peroxisomal membrane marker mCherry-Ant1p together with Gpd1p-GFP or Gpd1pΔPTS2-GFP in Δgpd1 mutant cells (Fig. 5B). As expected, Gpd1p-GFP co-localized with mCherry-Ant1p, indicating its peroxisomal localization. Gpd1-GFP also co-localized with mCherry-Pnc1p, as the Gpd1p can target the Pnc1p to peroxisomes. When Gpd1pΔPTS2-GFP was analyzed in Δgpd1 mutant cells, the protein mislocalized to the cytosol. In these cells, the mCherry-Pnc1p is also mislocalized to the cytosol (Fig. 5B). Accordingly, the PTS2 of Gpd1p is required to direct both proteins to peroxisomes. Taken together, the data demonstrate that Pnc1p is imported into peroxisomes in a PTS2- and Pex21p-dependent manner by piggyback transport using Gpd1p as transport vehicle.
FIGURE 5.
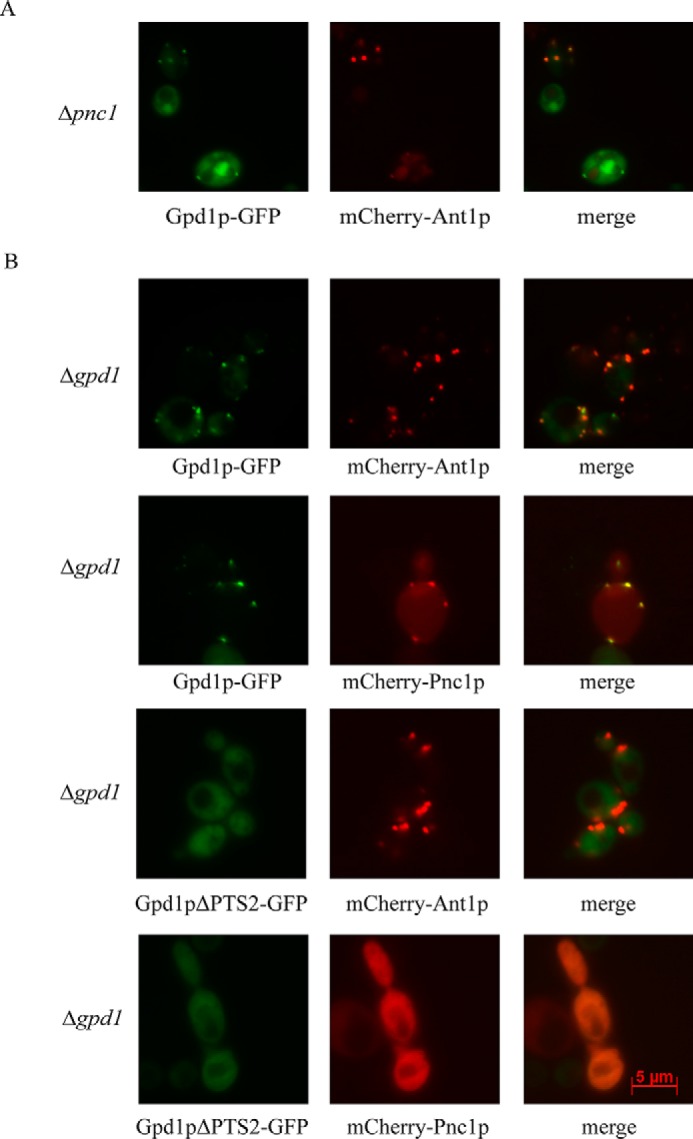
Peroxisomal import of Gpd1p does not depend on Pnc1p but import of both proteins is dependent on PTS2 motif of Gpd1p. A, fluorescence microscopy localization of plasmid-encoded Gpd1p-GFP in a Pnc1p-deficient strain. Gpd1p-GFP shows co-localization with the peroxisomal marker mCherry-Ant1p. B, fluorescence microscopy of plasmid-encoded Gpd1p-GFP, Gpd1pΔPTS2-GFP, mCherry-Pnc1p, and the peroxisomal marker protein mCherry-Ant1p in Δgpd1 mutant cells. The data revealed that the peroxisomal localization of Gpd1p and Pnc1p depends on the presence of the PTS2 motif of Gpd1p.
Discussion
We show that two stress-adaptable import pathways for PTS2 proteins exist in bakers' yeast. The cargo-specific pathways, which depend on the PTS2 receptor Pex7p, involve distinct PTS2 co-receptors. Pex21p is critical for the import of the PTS2 protein Gpd1p and the PTS-free Pnc1p but can also mediate the import of another PTS2 protein Fox3p. In contrast, Pex18p acts as a highly selective PTS2 co-receptor for the import of the oleate-inducible Fox3p. The newly disclosed function for the co-receptor Pex21p cannot be fulfilled by Pex18p. The data establish Pex21p as a more general co-receptor in the PTS2-dependent protein import, although Pex18p is especially important for oleate-induced import of PTS2-proteins.
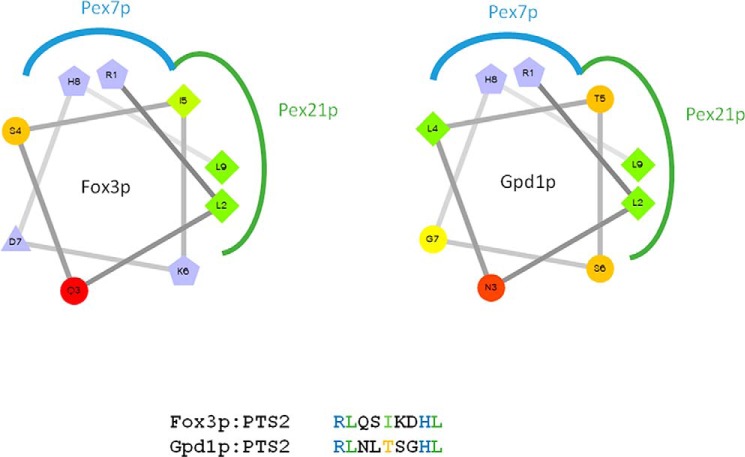
The question arises as to how PTS2 co-receptors selectively discriminate cargo proteins. Recent structural analysis revealed that the PTS2 co-receptors physically interact with the conserved PTS2 nonapeptide. The crystal structure analysis revealed that the co-receptor Pex21p interacts with the hydrophobic backside of the amphipathic PTS2 helix of Fox3p when presented by Pex7p (Fig. 6) (50). Although the well conserved positively charged Pex7p-binding residues (Arg and His) of the PTS2 sequences of Gpd1p and Fox3p are identical, the hydrophobic surfaces of the co-receptor binding areas are different in at least one critical position (Fig. 6). The significance of the hydrophobic residue at position 5 of the PTS2 peptide (RLXXX5XXHL) for efficiency of import was demonstrated previously for plants (51). Fox3p, which is bound by both co-receptors, contains a hydrophobic side chain at this position, whereas Gpd1p, the binding partner of Pex21p, carries the hydrophilic threonine. With respect to cargo selectivity, our results suggest that Pex21p is less selective and can accommodate hydrophobic as well as hydrophilic residues at this position. Post-translational modifications could also contribute to specificity of co-receptor binding of cargo proteins. To this end, it has been shown that phosphorylation of two serine residues close to the PTS2 signal of Gpd1p is important for efficient peroxisomal import (27).
FIGURE 6.
Interface regions of Pex21p, Pex7p, and the amphipathic PTS2 helices of Fox3p and Gpd1p. Helical wheel projections of the PTS2 nonapeptide sequences of Fox3p and Gpd1p were generated by wheel pI v1.4 by D. Armstrong and R. Zidovetzki. Hydrophilic residues are shown as circles, hydrophobic residues as diamonds, and positively charged amino acids as pentagons. Blue color indicates interfaces between Pex7p and the conserved residues arginine (R1) and histidine (H8) of the PTS2 sequence, and the green color shows the interacting regions of PTS2 with the co-receptor.
Peroxisomal localization of Pnc1p has been reported previously (28). However, the Pex7p dependence of import could until now not be reconciled with the obvious lack of a PTS2 motif within the Pnc1p sequence. Here, we show that this is explained by piggyback transport of Pnc1p together with Gpd1p. Piggyback import of heteromeric protein complexes has so far only been observed for PTS1 proteins (16). However, the PTS2 import machinery is also able to accommodate homo-oligomers by usage of a single targeting signal, which was impressively shown for S. cerevisiae thiolase (52) and nonauthentic cargo proteins (51). Interestingly, Gpd1p lacking a functional PTS2 is imported into peroxisomes, probably by hitchhiking onto endogenous Gpd1p (27).
The enzymes Gpd1p and Pnc1p perform known functions in the cytosol and nucleus (Fig. 7A). However, Gpd1p and Pnc1p are also targeted to peroxisomes (28, 53). So far, no peroxisome-specific function has been described for both enzymes, and we speculate that the peroxisomal import of both enzymes serves a stress-induced regulatory function. Under hypertonic growth, a more than 2-fold decrease of mRNA formation is observed (54), and under these conditions, the steady-state level of Pex21p decreases within the first 2 h (Fig. 1). The decrease in Pex21p will slow down the peroxisomal import of Gpd1p (Fig. 7B), which under hypertonic conditions is highly induced. In the cytosol, Gpd1p converts the glycolytic metabolite dihydroxyacetone phosphate into the protective osmolyte glycerol (Fig. 7A). Hyperosmotic stress also elevates expression levels of Pnc1p (27), which initiates NAD+ regeneration from nicotinamide. Thus, both reactions raise the intracellular NAD+ level under hyperosmotic stress conditions. One important recipient of NAD+ under these conditions is the histone deacetylase Sir2p, which prevents osmostress-induced apoptosis-like cell death (55). Moreover, Pnc1p is responsible for the catalytic removal of the nicotinamide, a product of the deacetylating reaction and noncompetitive inhibitor of Sir2p (Fig. 7A). Therefore, the enzymatic activities of Gpd1p in the cytosol and Pnc1p in the nucleus contribute to an increase of sirtuin activity and thereby protect the cell from stress-induced death (56).
FIGURE 7.
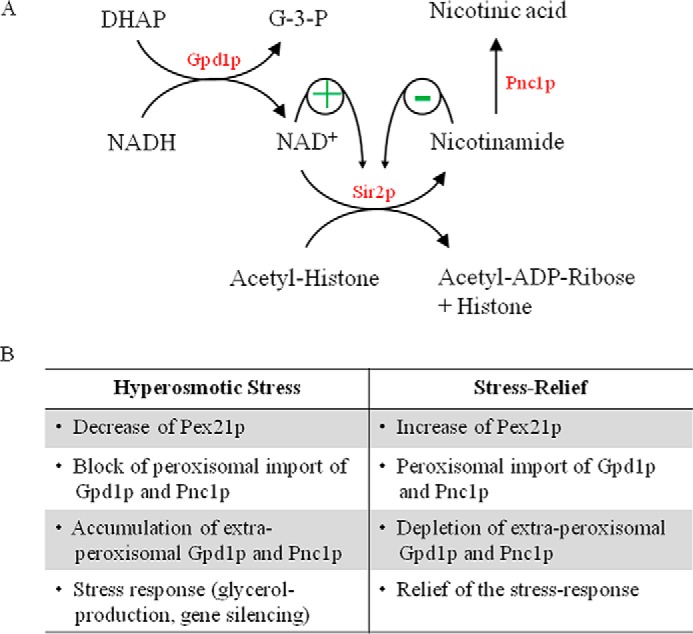
Scheme of metabolic and regulatory relationship between Gpd1p and Pnc1p and regulatory roles of peroxisomes as stress-relief valves. A, Gpd1p converts dihydroxyacetone phosphate and NADH into glycerol 3-phosphate and NAD+. The protein deacetylase Sir2p utilizes NAD+ as co-substrate. Hydrolysis of NAD+ by Sir2p results in the formation of nicotinamide and acetyl-ADP-ribose. Deacetylated histones trigger transcriptional gene silencing and lead to an elongated replicative life span. The nicotinamidase Pnc1p initiates the NAD+ salvage pathway by converting nicotinamide into nicotinic acid. Although high levels of NAD+ activate Sir2p, the breakdown product nicotinamide acts as a potent noncompetitive inhibitor of Sir2p. B, effects of hyperosmotic stress and stress relieve on the steady-state level of Pex21p, peroxisomal localization of Pnc1p and Gpd1p and modulation of the stress response.
To relieve the stress response, Gpd1p and Pnc1p have to be depleted from the cytosol and nucleus. We speculate that this is achieved by an increase in the efficiency of peroxisomal import. Under these conditions, Pex21p is present, and Gpd1p and Pnc1p are sequestered into peroxisomes, spatially separated from their enzymatic substrates (Fig. 7B). In this scenario, Pex21p-dependent import of protein into peroxisomes serves as a controllable pathway to deplete the cytosol from undesired proteins under various stress conditions. Along this line, it was shown that peroxisomal localization of Gpd1p protects cells from damage when several stress-inducing factors, in this case oleate and hyperosmolarity, were combined (27). It also seems noteworthy that gene expression of Pex21p is up-regulated under DNA-damaging conditions (57).
Based on our findings, we propose a novel stress-protective function for Pex21p-mediated protein import. It should be noted that protein translocation into peroxisomes could be an efficient and fast regulatory process because it does not require ATP-consuming unfolding and could remove more than one protein at once. Peroxisomes could serve as a stress-relief valve, which could rapidly down-regulate the steady-state level of enzymes prior to their depletion by proteasomal degradation or transcriptional regulation.
Author Contributions
D. E. performed and analyzed the experiments and wrote the paper. L. C. Z. and J. T. performed and analyzed the experiments. W. S. and R. E. designed the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Thomas Schröter for providing plasmid pTSC13-mCherry-Ant1p.
This work was supported by Deutsche Forschungsgemeinschaft Grant FOR1905 (to R. E.). This work was also supported by a grant from the Marie Curie Initial Training Networks (316723, PerFuMe) (to L.D.C.). The authors declare that they have no conflicts of interest with the contents of this article.
- PTS
- peroxisomal targeting signal
- TEV
- tobacco etch virus
- TPA
- TEVcs-protein A.
References
- 1. Kim P. K., Hettema E. H. (2015) Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427, 1176–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu X., Ma C., Subramani S. (2012) Recent advances in peroxisomal matrix protein import. Curr. Opin. Cell Biol. 24, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schliebs W., Girzalsky W., Erdmann R. (2010) Peroxisomal protein import and ERAD: variations on a common theme. Nat. Rev. Mol. Cell Biol. 11, 885–890 [DOI] [PubMed] [Google Scholar]
- 4. Marzioch M., Erdmann R., Veenhuis M., Kunau W.-H. (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13, 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dodt G., Gould S. J. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: Evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braverman N., Dodt G., Gould S. J., Valle D. (1998) An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7, 1195–1205 [DOI] [PubMed] [Google Scholar]
- 7. Brocard C., Hartig A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 8. Van der Leij I., Franse M. M., Elgersma Y., Distel B., Tabak H. F. (1993) PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 90, 11782–11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., Kunau W.-H. (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83–92 [DOI] [PubMed] [Google Scholar]
- 10. Bottger G., Barnett P., Klein A. T., Kragt A., Tabak H. F., Distel B. (2000) Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell 11, 3963–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meinecke M., Cizmowski C., Schliebs W., Krüger V., Beck S., Wagner R., Erdmann R. (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 12, 273–277 [DOI] [PubMed] [Google Scholar]
- 12. Okumoto K., Noda H., Fujiki Y. (2014) Distinct modes of ubiquitination of peroxisome-targeting signal type 1 (PTS1) receptor Pex5p regulate PTS1 protein import. J. Biol. Chem. 289, 14089–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., Erdmann R. (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 [DOI] [PubMed] [Google Scholar]
- 14. Platta H. W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., Erdmann R. (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schäfer A., Kerssen D., Veenhuis M., Kunau W. H., Schliebs W. (2004) Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol. Cell. Biol. 24, 8895–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang X., Purdue P. E., Lazarow P. B. (2001) Eci1p uses a PTS1 to enter peroxisomes: either its own or that of a partner, Dci1p. Eur. J. Cell Biol. 80, 126–138 [DOI] [PubMed] [Google Scholar]
- 17. Lazarow P. B. (2006) The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763, 1599–1604 [DOI] [PubMed] [Google Scholar]
- 18. Schliebs W., Kunau W. H. (2006) PTS2 co-receptors: diverse proteins with common features. Biochim. Biophys. Acta 1763, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 19. Purdue P. E., Yang X., Lazarow P. B. (1998) Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J. Cell Biol. 143, 1859–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Einwächter H., Sowinski S., Kunau W. H., Schliebs W. (2001) Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfill a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2, 1035–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Léon S., Zhang L., McDonald W. H., Yates J. 3rd, Cregg J. M., Subramani S. (2006) Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 172, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otzen M., Wang D., Lunenborg M. G., van der Klei I. J. (2005) Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2). J. Cell Sci. 118, 3409–3418 [DOI] [PubMed] [Google Scholar]
- 23. Sichting M., Schell-Steven A., Prokisch H., Erdmann R., Rottensteiner H. (2003) Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol. Biol. Cell 14, 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith J. J., Rachubinski R. A. (2001) A role for the peroxin Pex8p in Pex20p-dependent thiolase import into peroxisomes of the yeast Yarrowia lipolytica. J. Biol. Chem. 276, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 25. Matsumura T., Otera H., Fujiki Y. (2000) Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel Pex5-impaired Chinese hamster ovary cell mutant. J. Biol. Chem. 275, 21715–21721 [DOI] [PubMed] [Google Scholar]
- 26. Lee J. R., Jang H. H., Park J. H., Jung J. H., Lee S. S., Park S. K., Chi Y. H., Moon J. C., Lee Y. M., Kim S. Y., Kim J. Y., Yun D. J., Cho M. J., Lee K. O., Lee S. Y. (2006) Cloning of two splice variants of the rice PTS1 receptor, OsPex5pL,and OsPex5pS, and their functional characterization using pex5-deficient yeast and Arabidopsis. Plant J. 47, 457–466 [DOI] [PubMed] [Google Scholar]
- 27. Jung S., Marelli M., Rachubinski R. A., Goodlett D. R., Aitchison J. D. (2010) Dynamic changes in the subcellular distribution of Gpd1p in response to cell stress. J. Biol. Chem. 285, 6739–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003) Nicotinamide and PNC1 govern life span extension by calorie restriction in Saccharomyces cerevisiae. Nature 423, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albertyn J., Hohmann S., Thevelein J. M., Prior B. A. (1994) GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., Lin S. S., Manchester J. K., Gordon J. I., Sinclair D. A. (2002) Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277, 18881–18890 [DOI] [PubMed] [Google Scholar]
- 31. Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972 [DOI] [PubMed] [Google Scholar]
- 33. Cramer J., Effelsberg D., Girzalsky W., Erdmann R. (2015) Subcellular Fractionation: A Laboratory Manual, Cold Spring Harbor Laboratory Press, pp. 97–114, Cold Spring Harbor, NY [Google Scholar]
- 34. Harlow E., Lane D. (1988) Antibodies–A Laboratory Manual, pp. 469–530, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Stein K., Schell-Steven A., Erdmann R., Rottensteiner H. (2002) Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 22, 6056–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Girzalsky W., Rehling P., Stein K., Kipper J., Blank L., Kunau W.-H., Erdmann R. (1999) Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J. Cell Biol. 144, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Erdmann R., Kunau W.-H. (1994) Purification and immunolocalization of the peroxisomal 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. Yeast 10, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 38. Blobel F., Erdmann R. (1996) Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur. J. Biochem. 240, 468–476 [DOI] [PubMed] [Google Scholar]
- 39. Rose M. D., Misra L. M., Vogel J. P. (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 Gene. Cell 57, 1211–1221 [DOI] [PubMed] [Google Scholar]
- 40. Bigl M., Eschrich K. (1994) Overexpression of catalytically active yeast (Saccharomyces cerevisiae) fructose-1,6-bisphosphatase in Escherichia coli. Biol. Chem. Hoppe-Seyler 375, 153–160 [DOI] [PubMed] [Google Scholar]
- 41. Birschmann I., Stroobants A. K., van den Berg M., Schäfer A., Rosenkranz K., Kunau W. H., Tabak H. F. (2003) Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol. Biol. Cell 14, 2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Agne B., Meindl N. M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., Kunau W. H. (2003) Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635–646 [DOI] [PubMed] [Google Scholar]
- 43. Grunau S., Schliebs W., Linnepe R., Neufeld C., Cizmowski C., Reinartz B., Meyer H. E., Warscheid B., Girzalsky W., Erdmann R. (2009) Peroxisomal targeting of PTS2 pre-import complexes in the yeast Saccharomyces cerevisiae. Traffic 10, 451–460 [DOI] [PubMed] [Google Scholar]
- 44. Schrötter A., Pfeiffer K., El Magraoui F., Platta H. W., Erdmann R., Meyer H. E., Egensperger R., Marcus K., Müller T. (2012) The amyloid precursor protein (APP) family members are key players in S-adenosylmethionine formation by MAT2A and modify BACE1 and PSEN1 gene expression-relevance for Alzheimer's disease. Mol. Cell. Proteomics 11, 1274–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Padden J., Megger D. A., Bracht T., Reis H., Ahrens M., Kohl M., Eisenacher M., Schlaak J. F., Canbay A. E., Weber F., Hoffmann A. C., Kuhlmann K., Meyer H. E., Baba H. A., Sitek B. (2014) Identification of novel biomarker candidates for the immunohistochemical diagnosis of cholangiocellular carcinoma. Mol. Cell. Proteomics 13, 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veenhuis M., Mateblowski M., Kunau W.-H., Harder W. (1987) Proliferation of microbodies in Saccharomyces cerevisiae. Yeast 3, 77–84 [DOI] [PubMed] [Google Scholar]
- 47. Einerhand A. W., Kos W. T., Distel B., Tabak H. F. (1993) Characterization of a transcriptional control element involved in proliferation of peroxisomes in yeast in response to oleate. Eur. J. Biochem. 214, 323–331 [DOI] [PubMed] [Google Scholar]
- 48. Einerhand A. W., Voorn-Brouwer T. M., Erdmann R., Kunau W.-H., Tabak H. F. (1991) Regulation of transcription of the gene coding for peroxisomal 3-oxoacyl CoA thiolase of Saccharomyces cerevisiae. Eur. J. Biochem. 200, 113–122 [DOI] [PubMed] [Google Scholar]
- 49. Karpichev I. V., Small G. M. (1998) Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pan D., Nakatsu T., Kato H. (2013) Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p. Nat. Struct. Mol. Biol. 20, 987–993 [DOI] [PubMed] [Google Scholar]
- 51. Flynn C. R., Mullen R. T., Trelease R. N. (1998) Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. Plant J. 16, 709–720 [DOI] [PubMed] [Google Scholar]
- 52. Glover J. R., Andrews D. W., Rachubinski R. A. (1994) Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. U.S.A. 91, 10541–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valadi A., Granath K., Gustafsson L., Adler L. (2004) Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J. Biol. Chem. 279, 39677–39685 [DOI] [PubMed] [Google Scholar]
- 54. O'Rourke S. M., Herskowitz I. (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15, 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vendrell A., Martínez-Pastor M., González-Novo A., Pascual-Ahuir A., Sinclair D. A., Proft M., Posas F. (2011) Sir2 histone deacetylase prevents programmed cell death caused by sustained activation of the Hog1 stress-activated protein kinase. EMBO Rep. 12, 1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurdistani S. K., Grunstein M. (2003) Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4, 276–284 [DOI] [PubMed] [Google Scholar]
- 57. Jelinsky S. A., Samson L. D. (1999) Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. U.S.A. 96, 1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Erdmann R., Veenhuis M., Mertens D., Kunau W.-H. (1989) Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 86, 5419–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuravi K., Nagotu S., Krikken A. M., Sjollema K., Deckers M., Erdmann R., Veenhuis M., van der Klei I. J. (2006) Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119, 3994–4001 [DOI] [PubMed] [Google Scholar]