Background: The Dictyostelium proteome is predicted to encode a vast amount of homopolymeric amino acid tracts, including long polyglutamine tracts.
Results: Proteins with long polyglutamine tracts are soluble in Dictyostelium.
Conclusion: Polyglutamine proteins do not form aggregates in Dictyostelium under normal growth conditions.
Significance: Dictyostelium possess properties that suppress protein aggregation that may be co-opted to address polyglutamine toxicity in human disease.
Keywords: Dictyostelium, neurodegenerative disease, polyglutamine, protein aggregation, protein folding
Abstract
The expression, misfolding, and aggregation of long repetitive amino acid tracts are a major contributing factor in a number of neurodegenerative diseases, including C9ORF72 amyotrophic lateral sclerosis/frontotemporal dementia, fragile X tremor ataxia syndrome, myotonic dystrophy type 1, spinocerebellar ataxia type 8, and the nine polyglutamine diseases. Protein aggregation is a hallmark of each of these diseases. In model organisms, including yeast, worms, flies, mice, rats, and human cells, expression of proteins with the long repetitive amino acid tracts associated with these diseases recapitulates the protein aggregation that occurs in human disease. Here we show that the model organism Dictyostelium discoideum has evolved to normally encode long polyglutamine tracts and express these proteins in a soluble form. We also show that Dictyostelium has the capacity to suppress aggregation of a polyglutamine-expanded Huntingtin construct that aggregates in other model organisms tested. Together, these data identify Dictyostelium as a novel model organism with the capacity to suppress aggregation of proteins with long polyglutamine tracts.
Introduction
Polyglutamine diseases are a group of nine inherited neurodegenerative diseases caused by the expansion of a CAG trinucleotide sequence, which encodes the amino acid glutamine. The resulting protein contains a polyglutamine tract that confers toxic properties to the proteins containing it. Polyglutamine expansion is thought to confer toxicity in a number of ways: altered folding, impaired degradation, abnormal interaction with other proteins, altered subcellular localization, and self-association/aggregation (1). Aggregation of polyglutamine proteins is an overriding theme of this class of diseases, and polyglutamine-expanded proteins aggregate in model organisms tested, including yeast, plants, worms, flies, mice, rats, non-human primates, and human cells and patient samples (2–8).
Upon sequencing of the Dictyostelium genome, it was found that Dictyostelium encode more simple sequence repeats than any genome currently sequenced (9). Simple sequence repeats are so abundant that they encode 11% of the Dictyostelium genome, a 50-fold enrichment over other organisms (9, 10). The number of tandem repeats of trinucleotides (and hexa-, nona-, etc.) are also extremely high in coding regions, resulting in an extraordinary number of repeated amino acid sequences in proteins (9, 10). In coding regions, simple sequence repeats are so common that they occur every 724 bases, resulting in a predicted 9,582 homopolymeric amino acids tracts longer than 10 amino acids (9, 10). Perhaps most surprising, the two most abundant amino acid repeats are polyasparagine and polyglutamine tracts (10). In fact, Dictyostelium encode over 2,000 polyasparagine and polyglutamine tracts that are longer than 20 residues (10). In polyglutamine disease in humans, the pathogenic range for polyglutamine repeats ranges from 21 to 250 glutamines depending on the disease, suggesting that Dictyostelium encode mechanisms to suppress the toxic effects of polyglutamine and other amino acid homopolymers.
Despite the abundance of long, endogenous polyglutamine stretches known to aggregate in other model organisms, the investigation into protein folding/aggregation in Dictyostelium has been virtually nonexistent.2 Here we show that Dictyostelium express a number of proteins that are recognized by an antibody that preferentially recognizes long polyglutamine tracts (>40 glutamines), and these proteins remain soluble. We further show that a polyglutamine-expanded Huntingtin exon 1 protein remains soluble in Dictyostelium, whereas it readily aggregates in yeast and mammalian cells. Together, these data identify Dictyostelium as an organism with an unusual capacity to resist aggregation of proteins with long polyglutamine tracts.
Experimental Procedures
Expression Constructs and Antibodies
Huntingtin exon 1 with 25 or 103 glutamines was cloned into pTX-GFP using KpnI and XbaI for expression in Dictyostelium. Anti-polyglutamine antibody was from Millipore (MAB1574; used at 1:1,000), and anti-GFP was from Life Technologies (A11122; used at 1:1,000). Peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (used at 1:5,000 dilution). β-Actin (Pierce, PA121167; used at 1:1000), α-porin (Invitrogen, 459500; used at 1:1000), and α-tubulin (Developmental Studies Hybridoma Bank, 12G10; used at 1:1000) were used as loading controls for human, yeast, and Dictyostelium cells, respectively.
Primary Tissues
Wild type C57BL/6 mouse embryos (day 13) were collected, and whole body lysates were prepared in NETN buffer (0.5% Nonidet P-40, 150 mm NaCl, 50 mm Tris, and protease inhibitors (Roche Applied Science)).
Mammalian Cell Culture and Transfection
Human embryonic kidney 293 (HEK293) cells were grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) and 1% streptomycin (Life Technologies). Transfections were performed as recommended by the manufacturer with Lipofectamine 2000 (Life Technologies). Rat lysate was obtained from rat insulinoma (832/13) cells, a gift from John Corbett.
Yeast Cell Culture
pYES2/Q25 and pYES2/Q103 plasmids were transformed using the standard lithium acetate protocol into competent JSY5740 WT cells (ura3–52, leu2Δ1, his3Δ200, trp1Δ63) (12). Yeast transformants were grown in glucose complete synthetic medium −Ura at 30 °C to ∼5 OD. Cells were diluted to 0.3 OD and induced for 3 h at 30 °C. Non-transformed cells were grown in YPAD (yeast extract, peptone, agar, dextrose) media. Spheroplasts were prepared as described previously (13).
Dictyostelium Cell Culture, Development, and Transformation
Dictyostelium discoideum AX2 cells were maintained in shaking cultures at 22 °C in HL5 medium. Cells were maintained at a density no greater than 4 × 106 cells/ml. For development, 6 × 108 cells were washed and grown on filter paper soaked with developmental buffer (5 mm Na2HPO4, 5 mm KH2PO4, 1 mm CaCl2, 2 mm MgCl2) at 22 °C. Cells were harvested by vortex and lysed in NETN buffer (14). Protein concentration was determined using BCA. Twenty-five micrograms of protein were run on SDS-PAGE and subjected to Western blotting. Transformations were performed as described previously (15).
Differential Centrifugation and Filter Trap Assay
1 × 107 cells were washed with PBS and lysed with NETN buffer. Samples were centrifuged at 15,000 rpm for 30 min at 4 °C. Supernatant (soluble fraction) was removed and subjected to BCA protein assay. The remaining pellet (insoluble fraction) was washed and resuspended in Laemmli buffer. Ten micrograms of protein and 20 μl of pellet sample were run on SDS-PAGE for Western blotting. For filter trap assays, 8 × 106 cells were similarly lysed and subjected to BCA protein assay. Forty micrograms of protein were diluted with PBS containing 1% SDS, filtered through a 0.2-μm cellulose acetate membrane filter (Sterlitech) using a DHM-48 filter trap hybridization manifold, and analyzed by Western blotting (16).
Immunofluorescence
HEK293 cells were plated on coverslips coated with poly-d-lysine, transfected, and fixed by incubation in 4% paraformaldehyde for 20 min. Cells were washed and stained with DAPI (Life Technologies). Coverslips were mounted with ProLong Gold Antifade reagent (Life Technologies) and imaged with a Nikon Eclipse 90i confocal microscope with a 20× objective. Z-stack images were obtained at 0.5-m intervals at 512 × 512 pixel resolution and merged using Fiji. Live cell imaging of yeast and Dictyostelium cells was performed on 2% low melt agarose pads in galactose medium or starvation buffer, respectively. Slides were prepared as previously described (17).
Pulse Shape Analysis
Transformed Dictyostelium cells were fixed in 1% paraformaldehyde for 1 h at 4 °C and filtered through a 30-μm filter. Samples were analyzed using an LSRII flow cytometer. For each sample, GFP pulse area, height, and width were collected for 100,000–150,000 events. Data were analyzed using FlowJo (version 7.2.1).
Fluorescence Recovery after Photobleaching
Fluorescence recovery after photobleaching (FRAP)3 analysis was performed as described previously (18).
Chaperone Bioinformatics
HSP70, HSP90, and small heat shock proteins (sHSPs) in Homo sapiens, D. discoideum, and Saccharomyces cerevisiae were found through a combination of literature searching and Entrez Gene/UniProt protein IDs and descriptions. Initially, the UniProt sequences of identified human chaperones (19) were used as queries in BLASTP against the NCBI non-redundant protein sequence database selecting for either Dictyostelium or Saccharomyces. These potential chaperones were then further explored through sequence and conserved domain analysis in BLAST. Additional searching for orthologs and paralogs was conducted utilizing InParanoid and MetaPhOrs. Only genes encoding proteins with an α-crystallin domain were considered to be sHSPs (20).
Results
The normal range for polyglutamine tract length in humans is roughly 4–40 glutamines with polyglutamine tracts exceeding this range typically being associated with pathogenesis (21). In contrast to other organisms used to model polyglutamine diseases, the Dictyostelium genome normally encodes proteins with long polyglutamine tracts that reach well into the pathogenic range in humans. For example, the uncharacterized cyclin domain-containing protein encoded by gene DDB_G0302543 contains an uninterrupted tract of 79 glutamines. To determine whether this and other polyglutamine proteins are expressed in Dictyostelium, we utilized the 1C2 antibody (α-polyQ) that preferentially recognizes long polyglutamine tracts (22). Consistent with expression of proteins with long polyglutamine tracts, Dictyostelium lysate showed multiple α-polyQ-positive bands (Fig. 1, A and B), whereas other organisms commonly used to investigate polyglutamine disease did not (Fig. 1, A and B).
FIGURE 1.
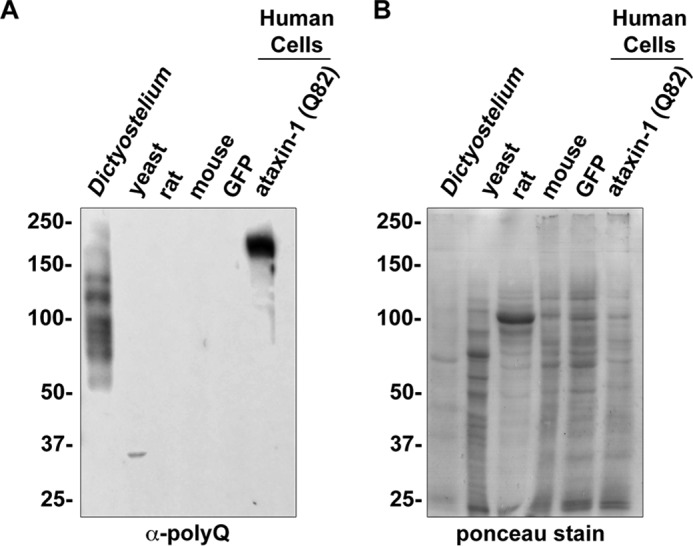
Dictyostelium have multiple α-polyQ-reactive bands. A, Dictyostelium lysate contains multiple 1C2 (α-polyQ)-reactive bands. To obtain equal loading, 40 μg of protein from each organism were separated by SDS-PAGE, transferred to PVDF, and probed with α-polyQ antibody (43). HEK293 cells transfected with either GFP or GFPataxin-1Q82 were used as negative and positive controls. B, Ponceau stain of protein loading in A. Samples from A were analyzed by Ponceau stain prior to Western blotting.
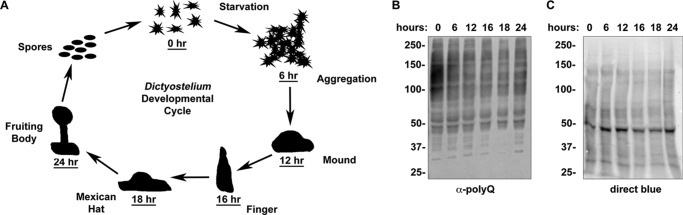
Dictyostelium is termed the social amoeba because of its ability to undergo a developmental process under adverse conditions (Ref. 23 and Fig. 2A). In response to stressors, such as starvation, the unicellular amoeba aggregates and ultimately forms a multicellular fruiting body. Throughout this cycle, the expression of numerous proteins is highly regulated (14). To determine whether proteins with long polyglutamine tracts are expressed in the various developmental stages, we induced development by starvation and collected cells at each developmental time point. The majority of α-polyQ-reactive bands remained present throughout development; however, a small number of bands either appeared or disappeared over the developmental cycle (Fig. 2, A–C). Thus, Dictyostelium expresses proteins with long polyglutamine tracts throughout its developmental cycle.
FIGURE 2.
Dictyostelium express α-polyQ-reactive bands throughout the developmental cycle. A, diagram of the life cycle of Dictyostelium. Dictyostelium exist as a free-living amoebae (0 h) until experiencing stress (e.g. starvation) at which point they aggregate (6 h) and proceed through a developmental process (12, 16, and 18 h) culminating in the formation of a fruiting body (24 h) that can release spores, allowing for transportation to new feeding sites. B, Dictyostelium express polyglutamine proteins throughout their developmental process. Dictyostelium cells were starved (0 h), and cells were collected at the indicated time points. Time points correlate with the different developmental stages outlined in A. C, Direct Blue staining of protein loading in B. Samples from B were analyzed by Direct Blue prior to Western blotting.
Long polyglutamine tracts have been shown to aggregate when expressed in a variety of model organisms, including human cells, rats, mice, flies, worms, and yeast (2–7). Because Dictyostelium encode multiple proteins that have polyglutamine tracts within the pathogenic range, we next wanted to determine whether α-polyQ-reactive bands in Dictyostelium were soluble. To accomplish this, we performed differential centrifugation of Dictyostelium lysate (Fig. 3A). Dictyostelium lysate and human cell lysates overexpressing either GFP or GFPataxin-1Q82 (positive control) were separated to isolate soluble and pellet fractions and analyzed with the α-polyQ antibody. Consistent with the formation of protein aggregates, GFPataxin-1Q82 largely accumulated in the pellet, whereas endogenous α-polyQ-reactive proteins in Dictyostelium remained soluble (Fig. 3A). Similarly, no α-polyQ reactivity was detected in Dictyostelium lysate by filter trap analysis, whereas human cell lysates overexpressing GFPataxin-1Q82 showed intense α-polyQ reactivity (Fig. 3B). Together, these results are consistent with endogenous polyglutamine proteins being soluble in Dictyostelium.
FIGURE 3.
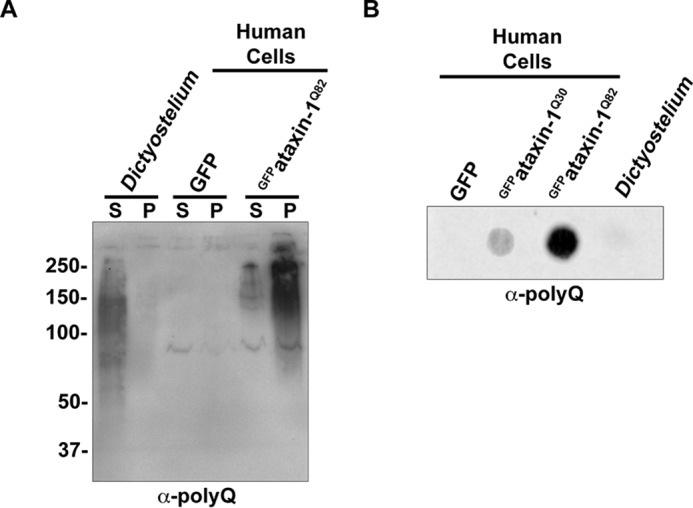
Dictyostelium polyglutamine proteins are soluble. A, differential centrifugation reveals that endogenous α-polyQ-reactive bands are present in the soluble fraction. Dictyostelium lysate and HEK293 cells transfected with either GFP or GFPataxin-1Q82 were separated into supernatant (S) and pellet (P) fractions. Samples were probed with α-polyQ antibody. B, filter trap analysis fails to identify α-polyQ-reactive bands in Dictyostelium. Either Dictyostelium cell lysate or lysate from HEK293 cells transfected with GFP, GFPataxin-1Q30, or GFPataxin-1Q82 was analyzed by filter trap assay.
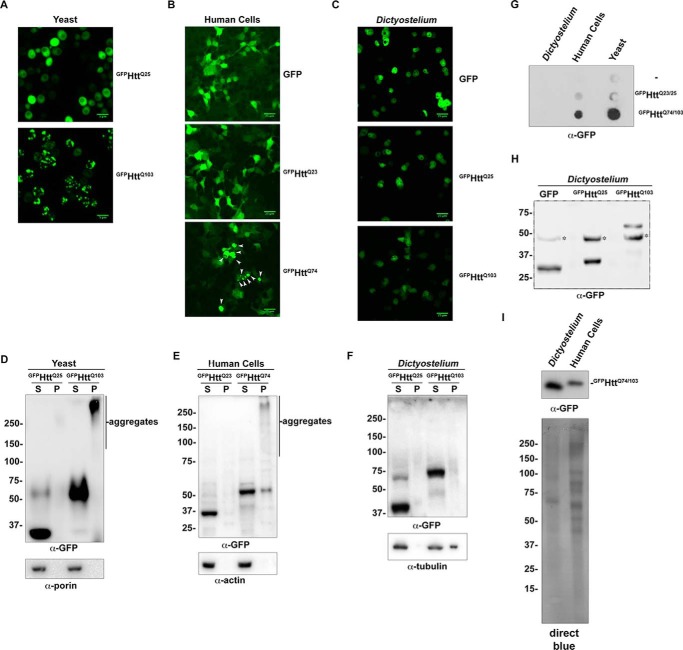
The lack of α-polyQ-reactive bands in the insoluble fractions of the differential centrifugation and filter trap experiments (Fig. 3, A and B) suggests that Dictyostelium is highly resistant to aggregation of polyglutamine proteins. To assess aggregation propensity of a pathogenic human polyglutamine protein, we utilized a Huntingtin exon 1-GFP fusion construct encoding either 25 or 103 glutamines (GFPHttQ25 or GFPHttQ103). In yeast expressing exon 1 of the Huntingtin protein, aggregates form in a polyglutamine length-dependent manner (24, 25). Consistent with previous reports (24, 25), we observed GFP-positive inclusions within 3 h of induction in yeast expressing GFPHttQ103, whereas GFPHttQ25 remained diffuse (Fig. 4A). Similarly, expression of GFPHttQ74 resulted in protein aggregation in human cells, whereas GFP and GFPHttQ23 remained soluble (Fig. 4B and Refs. 26 and 27). Next, we wanted to determine whether GFPHttQ103 would aggregate in Dictyostelium. Constructs encoding GFP, GFPHttQ25, or GFPHttQ103 were expressed in Dictyostelium, and unlike yeast and human cells, we were unable to detect aggregates in Dictyostelium cells expressing GFPHttQ103 by microscopy (Fig. 4C). Biochemical fractionation and filter trap analysis of these lysates confirmed that GFPHttQ103 was present in the insoluble fraction in yeast and human cells, but it remained soluble in Dictyostelium (Fig. 4, D–G). To ensure that GFP was not being proteolytically cleaved from the Htt exon 1 polyglutamine tract, we performed Western blotting analysis of Dictyostelium expressing GFP, GFPHttQ25, and GFPHttQ103. In each sample, GFP or the GFP fusion protein appeared as a single band at the expected molecular weight, indicating that GFP was still fused to Htt exon 1 (Fig. 4H). Importantly, GFPHttQ103 and GFPHttQ74 were expressed to similar levels in Dictyostelium and human cells, confirming that the lack of aggregates in Dictyostelium was not due to insufficient expression of GFPHttQ103 (Fig. 4I).
FIGURE 4.
Dictyostelium are resistant to aggregation of a long polyglutamine tract. A, GFP-expanded Htt exon 1 aggregates in yeast in a polyglutamine length-dependent manner. Yeast were transformed with either GFPHttQ25 or GFPHttQ103 under the control of the GAL1 promoter. Changing the medium from glucose- to galactose-containing medium for 3 h induced expression. Cells were then analyzed by confocal microscopy. B, a GFP-Htt exon 1 construct aggregates in human cells in a polyglutamine length-dependent manner. HEK293 cells were transfected with either GFPHttQ23or GFPHttQ73 for 48 h prior to analysis by confocal microscopy. White arrowheads indicate aggregates. C, polyglutamine expanded Htt exon 1 does not form aggregates in Dictyostelium. Dictyostelium were selected with G418 for 4 days prior to analysis by confocal microscopy. D, biochemical analysis of polyglutamine length-dependent aggregation in yeast. Yeast expressing either GFPHttQ25 or GFPHttQ103 were analyzed by differential centrifugation. E, biochemical analysis of polyglutamine length-dependent aggregation in human cells. HEK293 cells expressing either GFP, GFPHttQ23, or GFPHttQ74 were analyzed by differential centrifugation. F, biochemical analysis of GFPHttQ25 or GFPHttQ103 aggregation in Dictyostelium. GFP, GFPHttQ25, or GFPHttQ103 was transformed into Dictyostelium, and lysates were analyzed by differential centrifugation. G, filter trap analysis of Dictyostelium expressing GFPHttQ103 fails to detect aggregates. Aggregation of normal and expanded polyglutamine Htt exon 1 was compared in yeast, HEK293 cells, and Dictyostelium by filter trap analysis. H, GFP is not cleaved from GFPHttQ25/103 in Dictyostelium. Dictyostelium expressing GFP, GFPHttQ25, or GFPHttQ103 were collected, and 25 μg of lysate were analyzed to ensure that GFP was not being cleaved from the GFPHttQ25/103 proteins. * indicates a cross-reacting band. I, GFPHttQ74/103 is expressed to similar levels in Dictyostelium and human cells. Twenty-five micrograms of cell lysate from Dictyostelium and human cells expressing GFPHttQ103 and GFPHttQ74, respectively, were analyzed by Western blotting. Direct Blue staining was performed to visualize the total protein loaded. Scale bars, 5 μm in A and 25 μm in B and C.
We next wanted to quantitatively assess the presence of polyglutamine aggregates. To accomplish this, we utilized pulse shape analysis on Dictyostelium and human cells expressing the normal and expanded GFPHtt constructs. Although aggregates were readily detected in a polyglutamine length-dependent manner in human cells, no significant population of aggregates was detected in Dictyostelium (Table 1). Together, these data suggest that Dictyostelium are highly resistant to the aggregation of GFPHttQ103.
TABLE 1.
Unbiased, quantitative analysis of GFPHttQ103 aggregation does not detect aggregates in Dictyostelium
HEK293 and Dictyostelium cells expressing either normal or expanded Htt exon 1 constructs were analyzed by pulse shape analysis. n = 3.
| Percentage of cells with aggregates | |
|---|---|
| HEK293-GFPHttQ23 | 0.08 ± 0.04 |
| HEK293-GFPHttQ74 | 10.30 ± 3.76 |
| Dictyostelium-GFPHttQ25 | 0.01 ± 0.01 |
| Dictyostelium-GFPHttQ103 | 0.02 ± 0.01 |
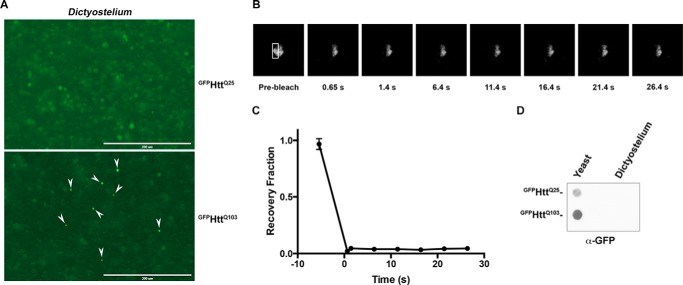
Dictyostelium cultures are typically started anew from stock cultures every 4–6 weeks to prevent the accumulation of mutations (14). We noticed that we would occasionally observe aggregates in older Dictyostelium cultures. To determine whether protein aggregates could form in aged Dictyostelium cells, we transformed freshly thawed cells with either GFPHttQ25 or GFPHttQ103 and cultured them for 5 weeks. We observed a small number of cells that contained large GFP-positive puncta in a polyglutamine length-dependent manner (Fig. 5A). To determine whether these puncta are insoluble, we performed FRAP and observed no recovery (Fig. 5, B and C) up to 10 min after bleaching (data not shown). Five-week-old cultures of Dictyostelium transformed to newly express GFPHttQ103 also had occasional GFP-positive aggregates (data not shown), suggesting that the presence of aggregates depends on the age of the Dictyostelium rather than how long the GFPHttQ103 protein was expressed. The GFPHttQ103 aggregates in aged samples were rare, and the number of aggregates was insufficient to be detected by filter trap assay (Fig. 5D) or differential centrifugation (data not shown). These data indicate that the level of expression of GFPHttQ103 in Dictyostelium is sufficient for protein aggregation to occur and suggest that as mutations accumulate pathways that suppress GFPHttQ103 aggregation may be disrupted.
FIGURE 5.
GFPHttQ103 aggregates can be detected in aged Dictyostelium. A, rare GPF-positive puncta can be detected in aged Dictyostelium cultures. Freshly thawed Dictyostelium cells were transformed with GFPHttQ25 or GFPHttQ103 and maintained for 5 weeks. White arrowheads indicate aggregates. Scale bars, 200 μm. B, GFPHttQ103 puncta are insoluble. A representative image of the FRAP analysis is shown. Boxed area indicates the region of photobleaching. C, FRAP analysis was performed, and samples were allowed to recover for the indicated times. Error bars represent S.E. D, filter trap analysis of Dictyostelium expressing GFPHttQ103 fails to detect aggregates in aged cells. Lysate from aged Dictyostelium expressing GFPHttQ25 or GFPHttQ103 was analyzed by filter trap assay to detect GFPHttQ103. Lysates from yeast expressing either GFPHttQ25 or GFPHttQ103 were used as negative and positive controls.
We next wanted to gain insight into potential differences in the protein quality control network that may be responsible for the unusual resistance of Dictyostelium to polyglutamine aggregation. Molecular chaperones are one class of proteins that have been heavily implicated in preventing polyglutamine aggregation. To determine whether Dictyostelium had any differences in the chaperone profile from yeast and humans, we analyzed the Dictyostelium and yeast genomes for orthologs of human HSP70, HSP90, and sHSPs. Although Dictyostelium had a similar or reduced number of HSP70 and HSP90 homologs compared with human and yeast, they encode for a large number of sHSP proteins (Fig. 6 and Table 2). These data suggest that sHSPs may play a role in suppressing polyglutamine aggregation in Dictyostelium.
FIGURE 6.
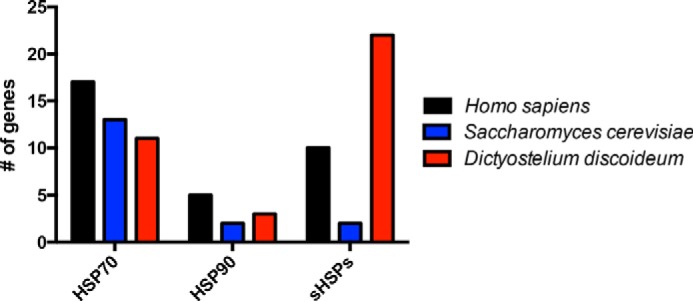
Dictyostelium encode a large number of small heat shock proteins. Bioinformatics analysis of the number of HSP70, HSP90, and sHSP homologs in humans, yeast, and Dictyostelium was performed.
TABLE 2.
sHSP genes in humans, yeast, and Dictyostelium
Gene names for sHSPs are listed. Gene names were collected from UniProt.
| Genes encoding sHSP | ||
|---|---|---|
| Human | Yeast | Dictyostelium |
| HSPB1 | HSP26 | HSPF1 |
| HSPB2 | HSP42 | HSPF2 |
| HSPB3 | HSPG1 | |
| CRYAA | HSPG2 | |
| CRYAB | HSPG3 | |
| HSPB6 | HSPG4 | |
| HSPB7 | HSPG5 | |
| HSPB8 | HSPG6 | |
| HSPB9 | HSPG7 | |
| ODF1 | HSPG8 | |
| HSPG9 | ||
| HSPG10 | ||
| HSPG11 | ||
| HSPG12 | ||
| HSPI | ||
| HSPJ | ||
| HSPK | ||
| HSPL | ||
| HSPM | ||
| DDB_G0280215 | ||
| DDB_G0288861 | ||
| DDB_G0295803 | ||
Discussion
Here we have identified an interesting property of D. discoideum, the ability to resist aggregation of proteins with long polyglutamine tracts. Unlike other commonly used model organisms, Dictyostelium naturally encode proteins that would be expected to aggregate and cause toxicity. Protein aggregation has been studied in a number of model organisms ranging from yeast to human cells. In each of these organisms, introduction of polyglutamine-expanded proteins results in the formation of protein aggregates (Refs. 2–7, 24, and 25; Fig. 4; and Table 1). Alternatively, we have found that endogenous polyglutamine proteins and overexpressed GFPHttQ103 remain soluble in Dictyostelium (Figs. 3 and 4, C, F, and G, and Table 1). How Dictyostelium resist polyglutamine aggregation is largely unknown. Another organism, Plasmodium falciparum has an asparagine-rich proteome that is protected from aggregation by HSP110 during conditions of heat stress (28); however, it is unclear whether other mechanisms that suppress aggregation during normal growth temperatures exist.
Interestingly, unlike P. falciparum, Dictyostelium encode repeats for every single amino acid except tryptophan (9). This raises two questions: 1) what functions do homopolymeric amino acid tracts play in Dictyostelium biology?, and 2) what mechanisms exist to suppress the potentially toxic effects of these repeats? Few studies have been performed to assess the function of these repeats, although they have been found to be enriched in protein kinases, lipid kinases, transcription factors, RNA helicases, and messenger RNA-binding proteins, such as spliceosome components (9). At the amino acid level, glutamine, asparagine, serine, and threonine are the most common repeats, accounting for 80% of the total amino acid repeats in Dictyostelium. This is interesting because all of these amino acids are hydrophilic, and one group of highly hydrophilic proteins, the late embryogenesis abundant proteins, have been shown to protect against polyglutamine aggregation in both human cells and plants (8, 29). In plants, late embryogenesis abundant proteins accumulate to high levels during the final stage of seed maturation, during the time the seed becomes tolerant to desiccation, and during water shortage in vegetative organs (30–33). One unique aspect of Dictyostelium biology is their developmental cycle, which culminates in the formation of spores. Like seeds, Dictyostelium spores are desiccation-resistant, raising a potential role for hydrophilic amino acid repeats protecting against protein aggregation in spores. Consistent with this, glutamine-rich prion-like proteins have been shown to prevent polyglutamine toxicity in yeast (34, 35). Further work to identify the endogenous role of proteins containing amino acid repeats in Dictyostelium is warranted and may provide insight into unique properties of these proteins.
Molecular chaperones are one class of proteins that have been shown to be highly important in countering polyglutamine aggregation. One interesting aspect of the Dictyostelium chaperome is the presence of a large number of sHSPs (Fig. 6 and Table 2). The majority of studies investigating the role of chaperones in regulating polyglutamine aggregation have focused on the larger, ATP-dependent chaperones HSP70 and HSP90, whereas the role of sHSPs is still largely unknown. One sHSP, αB-crystallin, suppresses toxicity of polyglutamine-expanded ataxin-3 but not Htt in Drosophila (36, 37). Biochemical and biophysical studies have revealed insight into the mechanism by which αB-crystallin suppresses ataxin-3 aggregation: instead of binding the polyglutamine tract, αB-crystallin binds the flanking regions to inhibit the initial aggregation step (38). In addition to directly suppressing protein aggregation, sHSPs have also been shown to facilitate autophagic clearance of polyglutamine-expanded Htt via their interaction with Bcl-2-associated athanogene 3 (39). Currently, very little is known about the function of sHSPs in Dictyostelium, and future investigations of sHSP function may provide insight into how Dictyostelium evade polyglutamine aggregation.
We found that a small number of GFPHttQ103 aggregates could be detected in aged Dictyostelium cultures, consistent with sufficient levels of GFPHttQ103 expression for protein aggregation to occur (Fig. 5). One possible explanation is that genetic mutations that accumulate in Dictyostelium cultures with prolonged culturing (14) may disrupt genes that are important for suppressing polyglutamine aggregation. In other organisms, including worms, mice, and humans, levels of key chaperones have been shown to decrease with age (40, 41), and this age-dependent decrease in chaperone levels has been shown to exacerbate polyglutamine toxicity (42). Further work investigating Dictyostelium aging may provide insight into the mechanism utilized to suppress polyglutamine aggregation.
It is thought that suppressing protein aggregation would be therapeutic for numerous neurodegenerative diseases, including the polyglutamine diseases. To our knowledge, Dictyostelium is the first model organism that resists polyglutamine aggregation (Figs. 3 and 4, C, F, and G, and Table 1). This raises the following questions: are Dictyostelium resistant to other types of protein aggregation?, and what mechanisms do Dictyostelium encode to suppress protein aggregation? Moving forward, answering these questions and others may provide insight into the treatment of diseases caused by protein aggregation.
Author Contributions
S. S. and K. M. S. designed the overall study. S. S. and K. M. S. wrote the manuscript. S. S. performed the main work of the study. A. P. and N. G. performed and analyzed the FRAP analysis. K. N. and A. B. participated in select experiments. J. B. S. conducted bioinformatics analysis of chaperones. All authors analyzed the results and reviewed and edited the manuscript.
Acknowledgments
We thank Ray Truant (Addgene plasmid numbers 40261/40262), David Rubinstein (Addgene plasmid numbers 40261/40262), Michael Sherman (Addgene plasmid numbers 1384/1385), and Tom Egelhoff (pTX-GFP) for constructs; Blake Hill and John Corbett for contributing cell lysates; and Sokol Todi and Henry Paulson for helpful comments and critique of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R00 NS073936 from the NINDS (to K. M. S.). This work was also supported by a grant from the Research and Education Program, a component of the Advancing a Healthier Wisconsin endowment at the Medical College of Wisconsin (to K. M. S. and N. G.). The authors declare that they have no conflicts of interest with the contents of this article.
During the preparation of this manuscript, Malinovska et al. (11) also reported that Dictyostelium has an unusual resilience to protein aggregation.
- FRAP
- fluorescence recovery after photobleaching
- sHSP
- small heat shock protein
- Htt
- Huntingtin.
References
- 1. Williams A. J., Paulson H. L. (2008) Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 31, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duennwald M. L. (2013) Yeast as a platform to explore polyglutamine toxicity and aggregation. Methods Mol. Biol. 1017, 153–161 [DOI] [PubMed] [Google Scholar]
- 3. Klement I. A., Skinner P. J., Kaytor M. D., Yi H., Hersch S. M., Clark H. B., Zoghbi H. Y., Orr H. T. (1998) Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95, 41–53 [DOI] [PubMed] [Google Scholar]
- 4. Marsh J. L., Lukacsovich T., Thompson L. M. (2009) Animal models of polyglutamine diseases and therapeutic approaches. J. Biol. Chem. 284, 7431–7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paulson H. L., Perez M. K., Trottier Y., Trojanowski J. Q., Subramony S. H., Das S. S., Vig P., Mandel J. L., Fischbeck K. H., Pittman R. N. (1997) Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19, 333–344 [DOI] [PubMed] [Google Scholar]
- 6. Switonski P. M., Szlachcic W. J., Gabka A., Krzyzosiak W. J., Figiel M. (2012) Mouse models of polyglutamine diseases in therapeutic approaches: review and data table. Part II. Mol. Neurobiol. 46, 430–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu Z., Bonini N. M. (2011) Modeling human trinucleotide repeat diseases in Drosophila. Int. Rev. Neurobiol. 99, 191–212 [DOI] [PubMed] [Google Scholar]
- 8. Liu G., Hu Y., Tunnacliffe A., Zheng Y. (2015) A plant cell model of polyglutamine aggregation: identification and characterisation of macromolecular and small-molecule anti-protein aggregation activity in vivo. J. Biotechnol. 207, 39–46 [DOI] [PubMed] [Google Scholar]
- 9. Eichinger L., Pachebat J. A., Glöckner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Tunggal B., Kummerfeld S., Madera M., Konfortov B. A., Rivero F., Bankier A. T., Lehmann R., Hamlin N., Davies R., Gaudet P., Fey P., Pilcher K., Chen G., Saunders D., Sodergren E., Davis P., Kerhornou A., Nie X., Hall N., Anjard C., Hemphill L., Bason N., Farbrother P., Desany B., Just E., Morio T., Rost R., Churcher C., Cooper J., Haydock S., van Driessche N., Cronin A., Goodhead I., Muzny D., Mourier T., Pain A., Lu M., Harper D., Lindsay R., Hauser H., James K., Quiles M., Madan Babu M., Saito T., Buchrieser C., Wardroper A., Felder M., Thangavelu M., Johnson D., Knights A., Loulseged H., Mungall K., Oliver K., Price C., Quail M. A., Urushihara H., Hernandez J., Rabbinowitsch E., Steffen D., Sanders M., Ma J., Kohara Y., Sharp S., Simmonds M., Spiegler S., Tivey A., Sugano S., White B., Walker D., Woodward J., Winckler T., Tanaka Y., Shaulsky G., Schleicher M., Weinstock G., Rosenthal A., Cox E. C., Chisholm R. L., Gibbs R., Loomis W. F., Platzer M., Kay R. R., Williams J., Dear P. H., Noegel A. A., Barrell B., Kuspa A. (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scala C., Tian X., Mehdiabadi N. J., Smith M. H., Saxer G., Stephens K., Buzombo P., Strassmann J. E., Queller D. C. (2012) Amino acid repeats cause extraordinary coding sequence variation in the social amoeba Dictyostelium discoideum. PLoS One 7, e46150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malinovska L., Palm S., Gibson K., Verbavatz J. M., Alberti S. (2015) Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. Proc. Natl. Acad. Sci. U.S.A. 112, E2620–E2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koirala S., Bui H. T., Schubert H. L., Eckert D. M., Hill C. P., Kay M. S., Shaw J. M. (2010) Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J. Cell Biol. 191, 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zinser E., Daum G. (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11, 493–536 [DOI] [PubMed] [Google Scholar]
- 14. Fey P., Kowal A. S., Gaudet P., Pilcher K. E., Chisholm R. L. (2007) Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2, 1307–1316 [DOI] [PubMed] [Google Scholar]
- 15. Pang K. M., Lynes M. A., Knecht D. A. (1999) Variables controlling the expression level of exogenous genes in Dictyostelium. Plasmid 41, 187–197 [DOI] [PubMed] [Google Scholar]
- 16. Matsumoto G., Stojanovic A., Holmberg C. I., Kim S., Morimoto R. I. (2005) Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J. Cell Biol. 171, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rines D. R., Thomann D., Dorn J. F., Goodwin P., Sorger P. K. (2011) Live cell imaging of yeast. Cold Spring Harb. Protoc. 2011, pdb.top065482 [DOI] [PubMed] [Google Scholar]
- 18. Petersen A., Gerges N. Z. (2015) Neurogranin regulates CaM dynamics at dendritic spines. Sci. Rep. 5, 11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brehme M., Voisine C., Rolland T., Wachi S., Soper J. H., Zhu Y., Orton K., Villella A., Garza D., Vidal M., Ge H., Morimoto R. I. (2014) A chaperone subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 9, 1135–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kappé G., Boelens W. C., de Jong W. W. (2010) Why proteins without an α-crystallin domain should not be included in the human small heat shock protein family HSPB. Cell Stress Chaperones 15, 457–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Orr H. T., Zoghbi H. Y. (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575–621 [DOI] [PubMed] [Google Scholar]
- 22. Trottier Y., Lutz Y., Stevanin G., Imbert G., Devys D., Cancel G., Saudou F., Weber C., David G., Tora L., Agid Y., Brice A., Mandel J.-L. (1995) Polyglutamine expansion as a pathological epitope in Huntington's disease and four dominant cerebellar ataxias. Nature 378, 403–406 [DOI] [PubMed] [Google Scholar]
- 23. Urushihara H. (2008) Developmental biology of the social amoeba: history, current knowledge and prospects. Dev. Growth Differ. 50, Suppl. 1, S277–S281 [DOI] [PubMed] [Google Scholar]
- 24. Krobitsch S., Lindquist S. (2000) Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meriin A. B., Zhang X., He X., Newnam G. P., Chernoff Y. O., Sherman M. Y. (2002) Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 157, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 27. Li S. H., Li X. J. (1998) Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats. Hum. Mol. Genet. 7, 777–782 [DOI] [PubMed] [Google Scholar]
- 28. Muralidharan V., Oksman A., Pal P., Lindquist S., Goldberg D. E. (2012) Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat. Commun. 3, 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y., Chakrabortee S., Li R., Zheng Y., Tunnacliffe A. (2011) Both plant and animal LEA proteins act as kinetic stabilisers of polyglutamine-dependent protein aggregation. FEBS Lett. 585, 630–634 [DOI] [PubMed] [Google Scholar]
- 30. Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dure L. 3rd, Crouch M., Harada J., Ho T. H., Mundy J., Quatrano R., Thomas T., Sung Z. R. (1989) Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol. Biol. 12, 475–486 [DOI] [PubMed] [Google Scholar]
- 32. Garay-Arroyo A., Colmenero-Flores J. M., Garciarrubio A., Covarrubias A. A. (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275, 5668–5674 [DOI] [PubMed] [Google Scholar]
- 33. Hoekstra F. A., Golovina E. A., Tetteroo F. A., Wolkers W. F. (2001) Induction of desiccation tolerance in plant somatic embryos: how exclusive is the protective role of sugars? Cryobiology 43, 140–150 [DOI] [PubMed] [Google Scholar]
- 34. Kayatekin C., Matlack K. E., Hesse W. R., Guan Y., Chakrabortee S., Russ J., Wanker E. E., Shah J. V., Lindquist S. (2014) Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc. Natl. Acad. Sci. U.S.A. 111, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ripaud L., Chumakova V., Antonin M., Hastie A. R., Pinkert S., Körner R., Ruff K. M., Pappu R. V., Hornburg D., Mann M., Hartl F. U., Hipp M. S. (2014) Overexpression of Q-rich prion-like proteins suppresses polyQ cytotoxicity and alters the polyQ interactome. Proc. Natl. Acad. Sci. U.S.A. 111, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bilen J., Bonini N. M. (2007) Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 3, 1950–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carra S., Sivilotti M., Chávez Zobel A. T., Lambert H., Landry J. (2005) HspB8, a small heat shock protein mutated in human neuromuscular disorders, has in vivo chaperone activity in cultured cells. Hum. Mol. Genet. 14, 1659–1669 [DOI] [PubMed] [Google Scholar]
- 38. Robertson A. L., Headey S. J., Saunders H. M., Ecroyd H., Scanlon M. J., Carver J. A., Bottomley S. P. (2010) Small heat-shock proteins interact with a flanking domain to suppress polyglutamine aggregation. Proc. Natl. Acad. Sci. U.S.A. 107, 10424–10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuchs M., Poirier D. J., Seguin S. J., Lambert H., Carra S., Charette S. J., Landry J. (2010) Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem. J. 425, 245–255 [DOI] [PubMed] [Google Scholar]
- 40. Hipp M. S., Park S. H., Hartl F. U. (2014) Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 [DOI] [PubMed] [Google Scholar]
- 41. Morimoto R. I., Cuervo A. M. (2014) Proteostasis and the aging proteome in health and disease. J. Gerontol. A Biol. Sci. Med. Sci. 69, Suppl. 1, S33–S38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang S., Huang S., Gaertig M. A., Li X. J., Li S. (2014) Age-dependent decrease in chaperone activity impairs MANF expression, leading to Purkinje cell degeneration in inducible SCA17 mice. Neuron 81, 349–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eaton S. L., Roche S. L., Llavero Hurtado M., Oldknow K. J., Farquharson C., Gillingwater T. H., Wishart T. M. (2013) Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS One 8, e72457. [DOI] [PMC free article] [PubMed] [Google Scholar]