The transcription factor Zeb2 cooperates with T-bet to control NK cell maturation, viability, and exit from the bone marrow and is essential for rejection of melanoma lung metastasis.
Abstract
Natural killer (NK) cell maturation is a tightly controlled process that endows NK cells with functional competence and the capacity to recognize target cells. Here, we found that the transcription factor (TF) Zeb2 was the most highly induced TF during NK cell maturation. Zeb2 is known to control epithelial to mesenchymal transition, but its role in immune cells is mostly undefined. Targeted deletion of Zeb2 resulted in impaired NK cell maturation, survival, and exit from the bone marrow. NK cell function was preserved, but mice lacking Zeb2 in NK cells were more susceptible to B16 melanoma lung metastases. Reciprocally, ectopic expression of Zeb2 resulted in a higher frequency of mature NK cells in all organs. Moreover, the immature phenotype of Zeb2−/− NK cells closely resembled that of Tbx21−/− NK cells. This was caused by both a dependence of Zeb2 expression on T-bet and a probable cooperation of these factors in gene regulation. Transgenic expression of Zeb2 in Tbx21−/− NK cells partially restored a normal maturation, establishing that timely induction of Zeb2 by T-bet is an essential event during NK cell differentiation. Finally, this novel transcriptional cascade could also operate in human as T-bet and Zeb2 are similarly regulated in mouse and human NK cells.
NK cells are innate lymphocytes with important roles in the defense against intracellular pathogens and in cancer immunosurveillance. They have the capacity to recognize and kill target cells through a limited set of surface receptors and through the release of cytotoxic granules containing perforin and granzymes. NK cell development occurs mainly in the BM. After commitment to the NK cell lineage, NK cells undergo a maturation program (Huntington et al., 2007b). Three maturation intermediates can be defined on the basis of surface expression of CD27 and CD11b: CD11b−CD27+ NK cells (hereafter referred to as CD11b−, the most immature stage), CD11b+CD27+ (double positive [DP]), and CD11b+CD27− (CD27−, the most mature subset), respectively (Kim et al., 2002; Hayakawa and Smyth, 2006). During maturation, NK cells progressively lose their capacity to proliferate, acquire the full set of NK cell receptors as well as cytotoxic arsenal, and modify their trafficking machinery. In particular, they acquire sphingosine-1 phosphate receptor 5 (S1PR5), which allows their egress from the BM and LNs and their circulation through the blood (Walzer et al., 2007; Mayol et al., 2011).
Several transcription factors (TFs) regulate NK cell maturation. The T-box family member Eomesodermin (Eomes) is essential for the early transition from CD11b− to the DP stage (Gordon et al., 2012). Another T-box family member, T-bet, drives terminal NK cell maturation by reducing proliferation (Townsend et al., 2004), up-regulating the expression of S1pr5 mRNA (Jenne et al., 2009), and driving the transition to the CD27− mature stage (Soderquest et al., 2011). Here, in an effort to identify novel TFs involved in NK cell maturation, we screened microarray data for genes up-regulated in mature NK cells and selected Zeb2 (zinc finger E-box–binding protein 2) as a putative regulator of maturation. Zeb2 and Zeb1 are TFs that share a similar protein domain organization and are known as master regulators of epithelial to mesenchymal transition (EMT; Comijn et al., 2001). EMT is a cellular program relevant to embryogenesis whereby epithelial cells are converted into mesenchymal cells (Thiery et al., 2009). Specific inactivation of Zeb2 in embryonic hematopoietic stem cells abrogates early hematopoietic lineage differentiation and affects cellular mobilization (Goossens et al., 2011). However, the in vivo role of Zeb2 in mature cells of the immune system remains unknown. Using conditional KO as well as overexpression mouse models, we show that Zeb2 is essential to promote terminal NK cell maturation and that it functions downstream of T-bet.
RESULTS AND DISCUSSION
Zeb2 is essential for NK cell maturation
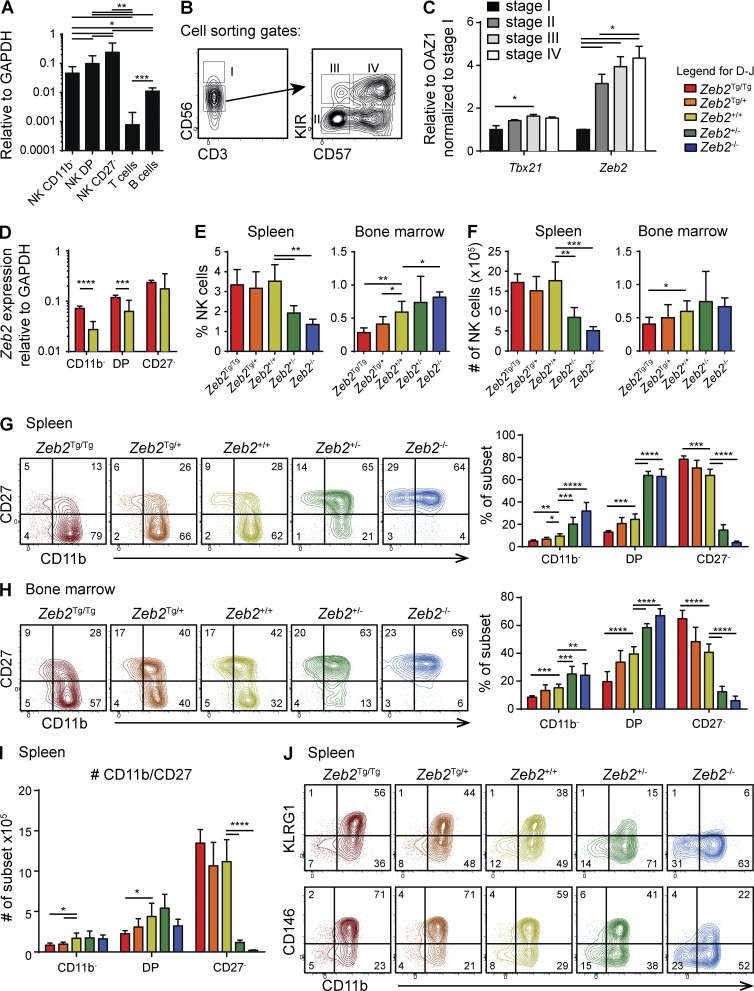
To identify novel genes controlling NK cell maturation, we screened microarray data (Chiossone et al., 2009) for TFs with a higher mRNA expression level in mature CD27− NK cells than in immature CD11b− NK cells. As shown in Fig. S1, the TF that best met this criterion was Zeb2. RT–quantitative PCR (qPCR) analysis of Zeb2 mRNA in NK cell maturation stages further corroborated this point (Fig. 1 A). Zeb2 expression was significantly higher in each of these subsets than in other mature lymphocyte subsets (Fig. 1 A). Next, we measured ZEB2 transcript levels during human NK cell maturation. For this purpose, NK cell maturation intermediates were defined as shown in Fig. 1 B: CD56brightCD3− (stage I), CD56dimCD3−NKG2A+KIR−CD57− (stage II), NKG2A+KIR+CD57− (stage III), and NKG2A−KIR+CD57+ (stage IV; Björkström et al., 2010). Fig. 1 C shows that human ZEB2 mRNA expression was significantly up-regulated in the most mature stages, thus reflecting the mouse NK cell data. ZEB2 expression in human NK cells was linked to that of T-BET, a TF known to regulate late NK cell maturation.
Figure 1.
Zeb2 is required for NK cell terminal maturation. (A) qPCR measurement of murine Zeb2 mRNA expression in sorted spleen WT NK cells of the indicated subsets and in T (CD3+) and B (CD19+) lymphocytes. n > 6 pooled from two independent experiments. (B and C) NK cells were purified from human PBMCs and further sorted by flow cytometry according to the gates shown in B. (C) RNA was then extracted and the level of T-bet, and Zeb2 expression was measured by qPCR. n = 3 sorts (three different donors). (A–C) Bar graphs show the mean ± SD results. (D–J) Flow cytometry analysis on NK cells (CD3−CD122+NKp46+) isolated from the spleen and BM of Ncr1iCre/+ × Zeb2Tg/Tg (Tg/Tg), Ncr1iCre/+ × Zeb2Tg/+ (Tg/+), Ncr1iCre/+ × Zebfl/+ (+/−), Ncr1iCre/+ × Zeb2fl/fl (−/−), and littermate control (+/+) mice. (D) NK cell subsets were FACS sorted from the spleen of the indicated mice, and Zeb2 mRNA levels were determined by qPCR. n ≥ 12 in each group pooled from two independent experiments. (E and F) Bar graphs showing NK cell percentages of viable CD45+ lymphocytes (E) and absolute NK cell numbers (F). (G–J) Analysis of NK cell maturation in the spleen or BM, as assessed by CD11b in combination with CD27 (G–I), KLRG1, or CD146 (J). Numbers on representative contour plots represent the percentage of cells per quadrant. Percentages (G, H, and J) and absolute cell numbers (I) of the indicated subsets among total NK cells are shown. All bar graphs show the mean ± SD results of ≥4 mice in each group representative of at least two independent experiments. For statistics (Student’s t test), Tg mice were compared with littermate controls. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To understand the role of Zeb2 in NK cells, we generated two complementary mouse models. We bred Ncr1iCre/+ mice (Narni-Mancinelli et al., 2011) with Zeb2fl/fl mice (Higashi et al., 2002) or with R26-Zeb2Tg/Tg mice (Tatari et al., 2014). These genetic modifications allow selective inactivation of Zeb2 or transgenic (Tg) Zeb2 expression from the Rosa26 promoter in NK cells, respectively. As the Rosa26 promoter is only moderately active, the total (endogenous + transgene) levels of Zeb2 are significantly up-regulated in CD11b− and DP NK cells (Fig. 1 D) but not in mature CD27− NK cells, which express much higher levels of endogenous Zeb2. We first compared the distribution of NK cells in NK-Zeb2Tg/Tg, NK-Zeb2Tg/+, NK-Zeb2+/+, NK-Zeb2+/−, and NK-Zeb2−/− mice (as defined in Fig. S1 B). The loss of one or two Zeb2 alleles dramatically skewed the normal distribution of NK cells. Zeb2−/− NK cells tended to accumulate in the BM and were significantly decreased in the blood and in peripheral organs such as liver, lung, spleen, LNs, and salivary glands relative to control NK cells (Fig. 1, E and F; and not depicted). Zeb2-overexpressing mice had decreased NK cell numbers in the BM, whereas total splenic NK cell numbers were normal (Fig. 1, E and F). Analysis of CD11b and CD27 expression revealed that NK-Zeb2−/− mice virtually lacked mature CD27− NK cells in all organs, whereas NK-Zeb2+/− mice had a milder but still profound phenotype (Fig. 1, G and H; and not depicted). Reciprocally, expression of one or two copies of the R26-based Zeb2 transgene resulted in increased frequencies of mature CD27− NK cells in the spleen and in the BM, with a similar gene dosage effect (Fig. 1, G and H). The numbers of mature CD27− NK cells tended to increase in the spleen and BM of NK-Zeb2Tg/+ and NK-Zeb2Tg/Tg mice, and the numbers of immature CD11b− and DP NK cells were reduced, showing that Zeb2 Tg expression promoted NK cell maturation (Fig. 1 I and not depicted). To further monitor NK cell maturation in the various mouse models, we also measured the expression of CD146 and KLRG1. KLRG1 and CD146 have previously been shown to associate with terminal NK cell maturation (Huntington et al., 2007a; Despoix et al., 2008). This analysis further confirmed that the frequency of mature NK cells (CD11b+KLRG1+ and CD11b+CD146+) was proportional to the amount of Zeb2 expressed (Fig. 1 J).
Zeb2 is essential for the egress from BM and for the survival of mature NK cells
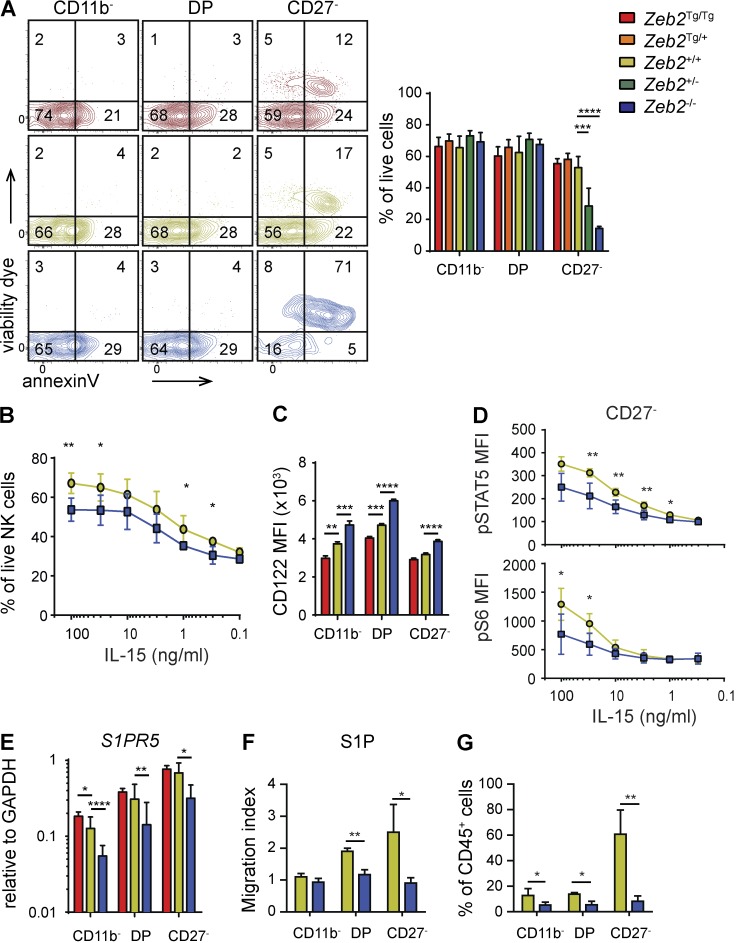
We next investigated whether the absence of CD27− NK cells in the periphery of NK-Zeb2−/− mice was associated with diminished survival, decreased exit from the BM, or both. First, we measured the ex vivo viability of NK cells in the spleen using Annexin V/dead cell staining. We found that mature Zeb2−/− NK cells displayed a decreased viability compared with control and Zeb2Tg/Tg NK cells (Fig. 2 A). Upon overnight culture with IL-15, Zeb2−/− NK cells also displayed decreased viability compared with control NK cells (Fig. 2 B). This was not caused by an impaired expression of IL-15 receptor as CD122 expression was in fact inversely correlated to the amount of Zeb2 expressed, whereas CD132 was normally expressed in the absence of Zeb2 (Fig. 2 C and not depicted). IL-15 signaling was normal in CD11b− and DP NK cells as measured by early STAT5 and S6 phosphorylation (not depicted). However, Zeb2−/− CD27− NK cells responded poorly to IL-15, which could contribute to impair their viability (Fig. 2 D). Thus, Zeb2 is essential for the survival of mature NK cells.
Figure 2.
Zeb2 is indispensable for survival of mature NK cells and for their exit from the BM. (A) Flow cytometry measurement of live/dead staining and Annexin V binding on splenic NK cells. Bar graphs represent the mean percentage viable (live/death−Annexin V−) ± SD results of at least three mice in each group representative for at least two independent experiments. (B) Spleen cells from WT and NK-Zeb2−/− mice were cultured overnight in the presence of IL-15. NK cell viability was then measured as in A. n = 5 representative of three independent experiments. (C) Flow cytometry measurement of CD122 expression levels as mean fluorescent intensity (MFI). The bar graph shows the mean ± SD results of at least four mice in each group representative of at least two independent experiments. (D) Flow cytometry analysis of STAT5 and S6 phosphorylation in gated splenic CD27− NK cells of the indicated mice, after in vitro stimulation with IL-15 for 1 h. Results show the mean ± SD. n = 5 representative of two independent experiments. (E) qPCR measurement of S1pr5 mRNA expression in NK cell subsets sorted from the spleen of mice of the indicated genotype. n > 11 pooled from two independent experiments. (F) Chemotaxis assay of spleen NK cell subsets of the indicated genotype using S1P. n = 4 pooled from two independent experiments. (G) Frequency of sinusoidal NK cells of each subset in the BM of the indicated mice. Mice were injected with anti-CD45 i.v. and subsequently sacrificed 1 min later, leading to CD45 labeling of sinusoidal cells. Bar graphs show the mean ± SD results of four mice in each group representative of at least two independent experiments. For statistics, Tg mice were compared with littermate controls. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001 (Student’s t test).
Second, we measured the mRNA expression of S1pr5, which controls the exit of NK cells from the BM. As shown in Fig. 2 E, Zeb2Tg/Tg NK cells expressed slightly higher levels of S1pr5 than control NK cells, whereas Zeb2−/− NK cells expressed S1pr5 mRNA at a lower level than control NK cells. This correlated with the lower responsiveness of Zeb2−/− NK cells to S1P in in vitro chemotaxis assays (Fig. 2 F) and with the lower frequency of sinusoidal NK cells in the BM of Ncr1iCre/+ × Zeb2fl/fl mice, irrespective of the NK cell subset analyzed (Fig. 2 G). Thus, increased apoptosis and decreased exit from the BM could contribute to the reduction in the number of mature NK cells observed in NK-Zeb2−/− mice.
NK-Zeb2−/− mice are highly susceptible to melanoma outgrowth, but Zeb2−/− NK cells have nearly normal cytotoxicity and cytokine secretion
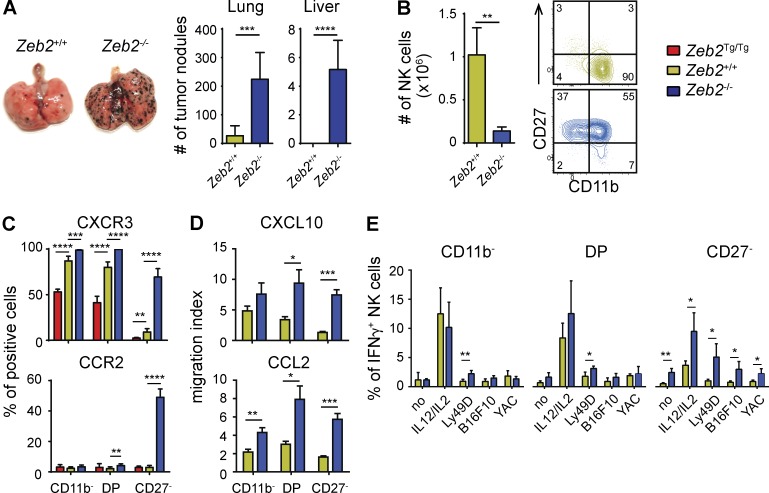
As NK cells have been shown to be important in tumor immune surveillance, we compared the susceptibility of NK-Zeb2−/− and control animals to form B16F10 colony outgrowths after i.v. injection. We observed a marked increase in the number of melanoma nodules in the lungs and livers of NK-Zeb2−/− mice compared with WT littermate controls (Fig. 3 A). We sought to understand the cause of the increased susceptibility of NK-Zeb2−/− mice to melanomas, considering NK cell numbers, trafficking, and cytotoxicity/cytokine secretion as essential factors contributing to NK cell–mediated protection. NK-Zeb2−/− mice have only ∼15% of the normal number of NK cells in the lung, and they lack mature NK cells in this organ (Fig. 3 B). To investigate their trafficking properties, we measured the expression of chemokine receptors. We observed a significant induction of expression of CXCR3, CXCR4, CCR2, and CCR5 in Zeb2−/− NK cells, especially in Zeb2−/− mature CD27− NK cells (Fig. 3 C and not depicted). Correlating with this, Zeb2−/− NK cells migrated better than control NK cells in response to gradients of chemokines that bind to these receptors (Fig. 3 D). Next, we isolated lung mononuclear cells at different time points after B16 injection and followed NK cell degranulation and IFN-γ secretion after in vitro stimulation with different stimuli. The IFN-γ production and degranulation of Zeb2−/− NK cells was normal or even increased at all time points analyzed and for all subsets (Figs. 3 E and not depicted). Trail and FasL expression were also normal on Zeb2−/− NK cells (not depicted). Altogether, our data show no impairment of NK cell effector functions and responsiveness to chemokines in NK-Zeb2−/− mice, suggesting that their higher susceptibility to B16F10 melanoma cells is mostly caused by a low number of NK cells in the periphery.
Figure 3.
NK-Zeb2−/− mice are highly susceptible to B16F10 melanomas. (A) NK-Zeb2−/− and littermate control (NK-Zeb2+/+) mice were i.v. injected with 200,000 B16F10 melanoma cells. Mice were sacrificed 12 d later, and tumor nodules in lung and liver were counted. n = 6 representative for two independent experiments. (B) Absolute lung NK cell numbers of unchallenged NK-Zeb2−/− and littermate control (+/+) mice. FACS plots show a representative CD11b/CD27 profile of gated CD3−CD122+NK1.1+ NK cells as determined by flow cytometry. n ≥ 3 representative for two independent experiments. (C) Flow cytometry measurement of the expression of CXCR3 and CCR2 by gated splenic NK cell (CD3−CD122+NKp46+) subsets. Results show the percentage ± SD of positive cells among NK cells of the indicated subset. n > 5 representative of at least two independent experiments. (D) Chemotaxis assay of spleen NK cells of the indicated genotype using CXCL10 and CCL2. n = 4–8 pooled from two independent experiments. (E) Lung lymphocytes were isolated from day 1 B16-bearing animals and restimulated ex vivo for 4 h in the indicated conditions in the presence of GolgiStop. Their production of IFN-γ was measured using flow cytometry. n = 4 in each group, representative for two independent experiments. All bar graphs represent the mean ± SD results. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Student’s t test).
Tbx21−/− and Zeb2−/− mature NK cells are phenotypically very similar
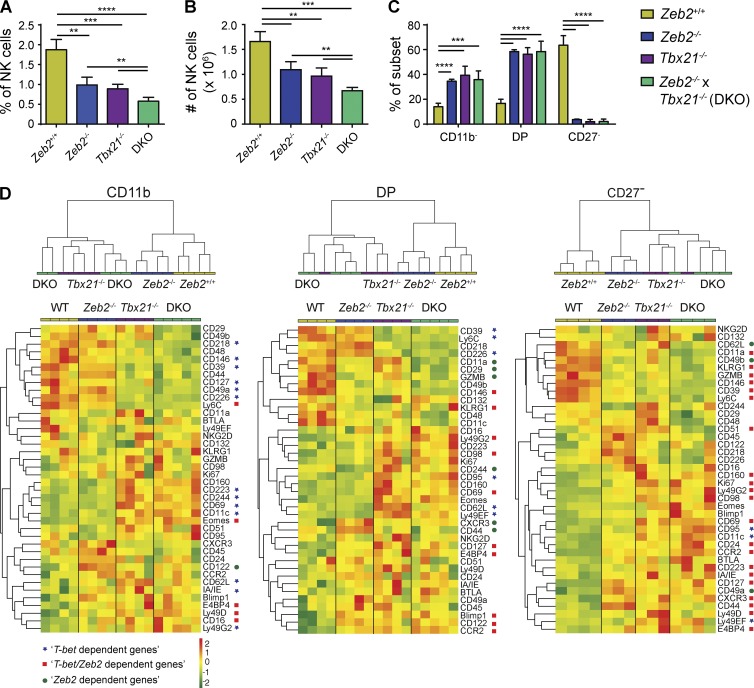
Several traits of Zeb2 mutant NK cells were reminiscent of the phenotype of Tbx21−/− (T-bet KO) NK cells. For example, Tbx21−/− mice lack mature CD27− NK cells (Soderquest et al., 2011), their peripheral NK cells display high rates of apoptosis (Townsend et al., 2004), and they have an impaired capacity to exit BM as a result of decreased expression of S1PR5 (Jenne et al., 2009). These observations prompted us to compare the phenotype of NK cells mutant for either TF in more details. The percentage and number of spleen NK cells as well as the maturation status were very similar in Tbx21−/− and NK-Zeb2−/− mice (Fig. 4, A–C). Combined deficiency of T-bet and Zeb2 (double KO [DKO]) in NK cells did not impair further NK cell maturation but significantly reduced the percentage and number of NK cells compared with deficiency in either TF alone (Fig. 4, A–C). We performed a flow cytometry analysis of 40 cell surface and intracellular proteins and compared subset by subset NK cells of the three genotypes. Fig. 4 D presents the results of the FACS analysis under the form of a global hierarchical clustering of samples and an expression heat map of markers for the three NK cell subsets. For CD11b− and DP NK cells, WT and Zeb2−/− NK cells clustered apart from DKO and Tbx21−/− NK cells. This was the result of a large group of proteins that were either up- or down-regulated in Tbx21−/− and DKO NK cells but not changed in Zeb2−/− NK cells compared with WT NK cells (blue stars; “T-bet–dependent genes”). For CD27− NK cells that express the highest level of Zeb2, Zeb2−/−, DKO, and Tbx21−/− NK cells clustered together, apart from WT NK cells. This effect was the result of a large group of proteins that were coregulated in Zeb2−/−, Tbx21−/−, and DKO NK cells, either up or down-regulated compared with WT NK cells (red squares; “Zeb2/T-bet–dependent genes”).
Figure 4.
T-bet and Zeb2 KO NK cells are near phenocopies. (A–C) Percentage of live lymphocytes (A) and absolute numbers (B) of NK cells in the spleen and their maturation status as assessed by CD11b/CD27 staining, shown as a percentage among NK cells (C). n = 4–5 in each group, pooled from two independent experiments. All bar graphs represent the mean ± SD results. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Flow cytometry analysis of the expression of the indicated genes by gated splenic NK cell subsets isolated from Ncr1iCre/+ × Zeb2fl/fl (Zeb2−/−), Tbx21−/−, NK-Zeb2−/− × Tbx21−/− (DKO), and littermate control (+/+, WT) mice. n = 4–5 in each group. (top) Hierarchical clustering of the indicated samples according to the Euclidean distance calculated on the basis of the MFI of all stainings for CD11b−, DP, and CD27− NK cells. (bottom) MFI of the staining for all markers analyzed, presented as a heat map, after log2 transformation and row-wise centering of the data. Blue stars correspond to genes whose expression is regulated by T-bet and not Zeb2, i.e., significantly different (P < 0.1) between WT and T-bet KO NK cells but not between WT and Zeb2 KO NK cells. Red squares correspond to genes coregulated by T-bet and Zeb2, i.e., genes whose expression is significantly different between WT and T-bet KO and Zeb2 KO NK cells and similar between T-bet KO and Zeb2 KO. Green circles correspond to genes regulated by Zeb2, i.e., genes whose expression is significantly different between WT and Zeb2 KO but not between WT and T-bet KO NK cells.
Thus, several points can be concluded from the analysis in Fig. 4 D: (a) T-bet is already active in immature NK cells, whereas Zeb2 only has limited action in this subset (blue stars); (b) when both TFs are coexpressed at high levels, they have coordinated activities on the expression of many genes (red squares); (c) there is little impact of a combined deficiency compared with lack of T-bet only; (d) several proteins such as CD39, CD146, or CD127 are already regulated by T-bet in immature NK cells and are coregulated by T-bet and Zeb2 in mature NK cells, thus showing that once induced, Zeb2 helps regulate T-bet target genes; and (e) a few genes were also dependent only on Zeb2, especially in DP and CD27− (green circles; “Zeb2-dependent genes”), showing that Zeb2 may have occasional T-bet–independent activity.
T-bet is necessary to induce Zeb2 expression
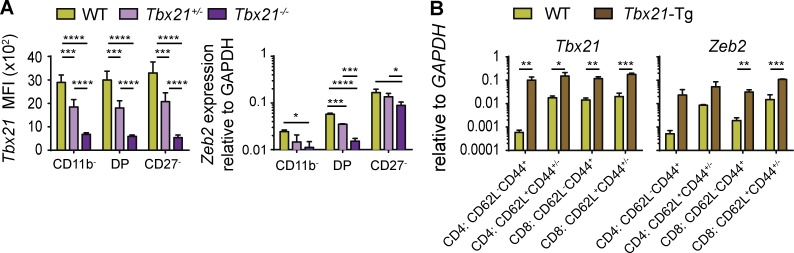
Given the earlier action of T-bet on gene expression in NK cells, we hypothesized that Zeb2 was acting downstream of T-bet. To test this point, we measured Zeb2 mRNA expression in NK cells expressing graded doses of T-bet. NK cell subsets were sorted from Tbx21−/−, Tbx21+/−, and WT mice. The level of T-bet in NK cell subsets from these mice is shown in Fig. 5 A. The steady-state level of Zeb2 transcripts was significantly decreased in Tbx21+/− and even further in Tbx21−/− NK cells compared with controls, showing that T-bet is necessary for optimal Zeb2 expression in NK cells. We also compared the level of T-bet and Zeb2 mRNA in WT versus T-bet Tg mice (T-bet Tg). The latter mice express T-bet under the control of the Cd2 promoter, which is highly active in T cells (Ishizaki et al., 2007). The results in Fig. 5 B show that T-bet expression induced high levels of Zeb2 mRNA in both CD4 and CD8 T cells, irrespective of the subset analyzed (based on CD44/CD62L expression). Thus, T-bet is necessary for optimal expression of Zeb2 in NK cells.
Figure 5.
T-bet is necessary and sufficient to induce Zeb2 expression. (A) Flow cytometry (left) or qPCR analysis (right) of Tbx21 protein or Zeb2 RNA levels in NK cell subsets from spleen suspensions of the indicated mouse strains. n = 3–7 mice in each group representative for two independent experiments. (B) CD4 and CD8 T cells subsets as indicated were FACS sorted from the spleen of CD2–T-bet–Tg and littermate control mice, and their expression of T-bet and Zeb2 were measured by qPCR. n = 3 in each group, two independent experiments. All bar graphs represent the mean ± SD results. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Student’s t test).
Restoration of Zeb2 levels in Tbx21−/− NK cells partially reinstate a wild-type phenotype and antitumor function
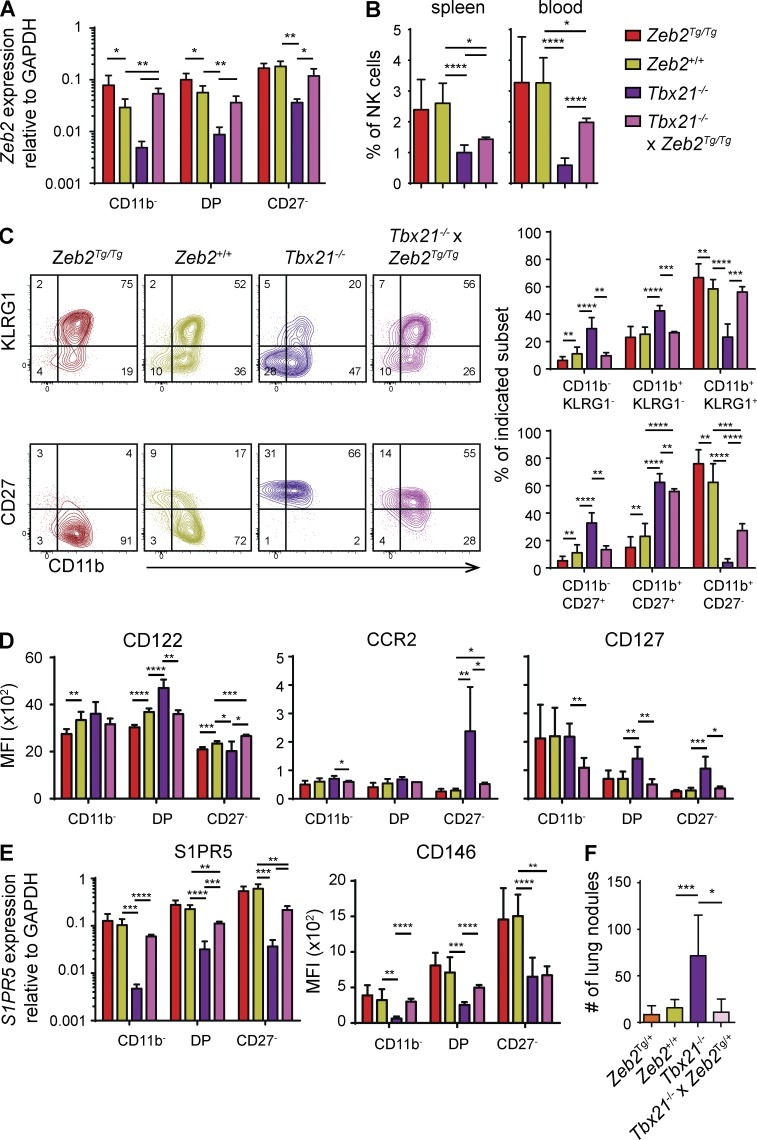
Our results raised the possibility that the lack of Zeb2 induction in Tbx21−/− NK cells contributed in a major way to the observed NK cell phenotype. To test this hypothesis, we restored Zeb2 levels in NK cells in Tbx21−/− mice by crossing R26-Zeb2Tg mice with Ncr1iCre and Tbx21−/− mice. We verified that the Zeb2 transgene restored a physiological level of Zeb2 in the different NK cell subsets (Fig. 6 A) and analyzed the phenotype of NK cells in the different mouse strains generated. Expression of Tg Zeb2 restored a wild-type phenotype when considering the following parameters: maturation as assessed by CD11b/KLRG1 (Fig. 6 C) and expression of CD122, CXCR3, CCR2, and CD127 (Fig. 6 D and not depicted). However, the reexpression of Zeb2 incompletely restored the percentage of peripheral NK cells (Fig. 6 B), NK cell maturation as assessed by CD27/CD11b (Fig. 6 C), and CD146 and S1PR5 expression (Fig. 6 E). The latter observations suggest an essential role of T-bet in repressing CD27 expression and inducing high levels of S1PR5 necessary for NK cell exit from the BM to the blood. To test whether the Zeb2 transgene could restore NK cell function, we also compared the susceptibility of the various mouse strains to form B16F10 nodules. The results shown in Fig. 6 F confirm previous observations that Tbx21−/− mice fail to control B16F10 metastasis (Werneck et al., 2008). However, Tg expression of Zeb2 in NK cells fully restored their capacity to control the tumor.
Figure 6.
Tg Zeb2 expression rescues the maturation defect of Tbx21−/− NK cells. Flow cytometry and qPCR measurement on electronically gated or sorted NK cell (CD3−CD122+NKp46+) subsets isolated from spleens of Ncr1iCre/+ × Zeb2Tg/Tg (NK-Zeb2Tg/Tg), Tbx21−/−, Ncr1iCre/+ × Zeb2Tg/Tg × Tbx21−/− (T-bet−/− × NK-Zeb2TgTg), and littermate controls. n ≥ 3. (A) Zeb2 expression levels as determined by qPCR. (B) NK cell percentages of viable lymphocytes. (C, left) Representative FACS plots showing CD11b expression in combination with CD27 and KLRG1. Numbers represent percentage of cells per quadrant. (C, right) Bar graphs show percentage of the indicated subset among total NK cells. (D) Results show MFI of CD122, CCR2, and CD127. (E) qPCR measurement of S1PR5 on sorted NK cell subsets and MFI of CD146 on gated NK cells. (F) The indicated mice were i.v. injected with B16-F10 melanoma cells. Mice were sacrificed at day 12, and tumor nodules in lung were counted. All bar graphs represent the mean ± SD results. For statistics (Student’s t test), Tg mice were compared with littermate controls, and Tbx21−/− × NK-Zeb2Tg/Tg were compared with Tbx21−/− and littermate controls as indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Altogether, these results indicate that the lack of Zeb2 expression contributes in a major way to the overall phenotype of Tbx21−/− NK cells. However, T-bet also has Zeb2-independent roles in NK cell maturation and homeostasis, further establishing the synergy between these factors during NK cell differentiation.
Concluding remarks
Here, we demonstrate, using an unprecedented panel of mouse models with different Zeb2 allele numbers, that Zeb2 is an essential regulator of the NK cell terminal differentiation program. ZEB2 is also up-regulated during differentiation of human NK cells, suggesting that Zeb2 regulates NK cell differentiation across mammals. Zeb2 not only promotes NK cell terminal differentiation, but also preserves viability of mature NK cells and induces their exit from the BM, all factors contributing to maintain the pool of peripheral mature NK cells, essential for efficient antitumor function in peripheral sites such as the lung.
Our data show that Zeb2 controls NK cell maturation downstream of T-bet. In immature NK cells, T-bet is already active, and Tbx21 deficiency leads to the deregulation of several genes in this subset. When T-bet levels increase, Zeb2 expression is induced in a T-bet–dependent manner, and T-bet and Zeb2 then control the expression of the same genes. This suggests the existence of a feed-forward loop whereby late induction of Zeb2 would help reinforce T-bet action. Such a loop may serve to irreversibly induce terminal NK cell maturation. Another supporting evidence of this model comes from in silico analyses by the ImmGen consortium and the Ontogenet algorithm, which predict T-bet and Zeb2 to coregulate many target genes in NK cells including Klrg1, Mcam (CD146), and S1pr5 (Jojic et al., 2013). In general, feed-forward regulatory loops occur very frequently in transcriptional networks (Gerstein et al., 2012). They are believed to buffer variations in input stimuli and increase robustness of regulatory circuits. The T-bet → Zeb2 loop appears to regulate important aspects of NK cell differentiation such as trafficking and responsiveness to chemokines, proliferation, and survival.
Tg expression of Zeb2 in the absence of Tbx21 partially restored a wild-type NK cell phenotype. This suggests that for many genes and in mature NK cells, Zeb2 and not T-bet is important to control expression of target genes. Yet, the very same genes are dependent on T-bet in immature NK cells, at a stage where Zeb2 is expressed at low and ineffective levels. This suggests that both TFs contribute to transcriptional activation in a cumulative rather than synergistic fashion, a notion arising from the analysis of genome-wide studies (Spivakov, 2014). Other TFs may also cooperate with T-bet and Zeb2 to confer robustness to the regulatory network as Zeb2 is known to recruit several transcriptional cofactors (Conidi et al., 2011). The identification of these transcriptional complexes and of their precise actions will be goals of future studies to better understand the interplay of TF during NK cell maturation.
MATERIALS AND METHODS
Mice
This study was performed in strict accordance with the French and Belgian recommendations for the ethical evaluation of experiments using laboratory animals and the European guidelines 86/609/CEE or the guidelines for animal care at the VIB site Ghent. C57BL/6 mice were purchased from Charles River (L’Arbresle). Ncr1iCre mice were crossed with Zeb2fl/fl mice or R26-Zeb2Tg mice. All strains including Tbx21−/− and CD2–T-bet Tg mice were bred in our animal house. Litters with mice from 8 to 24 wk old were used for analysis.
Blood collection from healthy human subjects
Human peripheral blood was drawn from healthy volunteers, and informed consent was obtained from all participants. PBMCs were then isolated by Ficoll gradient centrifugation, and NK cells were isolated and further sorted as described in Fig. 1 B.
Flow cytometry
Single-cell suspensions of mouse BM, spleen, and lungs were obtained and stained. Human whole blood samples from healthy donors were collected by venous puncture in heparin-containing vials. PBMCs were isolated by Ficoll gradient centrifugation. Intracellular stainings for TFs or granzyme A and granzyme B were performed using Foxp3 kit (eBioscience). Cell viability was measured using Annexin V (BD)/live-dead fixable (eBioscience) staining. Lyse/Fix and PermIII buffers (BD) were used for intracellular staining of phosphorylated proteins. Flow cytometry was performed on a FACSCanto, a FACS LSRII, a FACS Fortessa (all BD), a Navios (Beckman Coulter), or a MACSquant (Miltenyi Biotec). Data were analyzed using FlowJo (Tree Star). Clustering analyses of flow cytometry data and creation of heat maps were performed using the R software. Antibodies were purchased from eBioscience, BD, R&D Systems, Beckman Coulter, or BioLegend. We used mouse antibodies against Blimp1 (5E7), BTLA (8F4), CCR2 (475301), CCR5 (HM-CCR5), CD107a (1D4B), CD11a (M17/4), CD11b (M1/70), CD11c (N418), CD122 (5H4), CD127 (17R34), CD132 (TUGm2), CD146 (ME-9F1), CD16/32 (2.4G2), CD160 (CNX46.3), CD18 (M18/2), CD19 (ebio1D3), CD212 (114), CD218 (P3TUNYA), CD223 (ebioC9B7W), CD226 (10E5), CD24 (M1/69), CD244 (ebio244f4), CD25 (PC61), CD27 (LG.7F9), CD29 (9E67), CD3 (145-2C11), CD39 (24DMS1), CD43 (1B11), CD44 (1M7), CD45 (30F11), CD48 (HM48.1), CD49a (Ha31/8), CD49b (DX5), CD49d (R1.2), CD49e (ebioHma5.1), CD49f (ebioGoH3), CD51 (RMV.7), CD62L (Mel14), CD69 (H1.2F3), CD71 (R17217), CD94 (18D3), CD95 (15A7), CD98 (RL398), CXCR3 (CXCR3-173), CXCR4 (2B11), E4BP4 (52M-E19), Eomes (Dan11mag), GzmA (3G8.5), GzmB (NGZB), IA-IE (M5/114), IFN-γ (XMG1), Ki67 (B56), KLRG1 (2F1), Ly49a (YE1.48.10.6), Ly49D (4E5), Ly49F (HBF719), Ly49G2 (4D11), Ly49H (3D10), Ly6C (AL21), NKG2ACE (20D5), NKG2D (CX5), NKp46 (29A1.4), pS6 (N7.548), pSTAT5 (47/stat5), perforin (ebio-OMAKD), T-bet (ebio4B10), and Trail (N2B2). We used human antibodies against CD3 (UCHT1), CD14 (RMO52), CD19 (HD237), CD56 (HLDA6), CD57 (NC1), NKG2A (Z199), KIR3DL1 (DX9), and KIR2DL1 (HP-MA4). In all panels, a minimum of 50 cells per NK cell subset was analyzed.
Cell sorting and RNA preparation
Lymphocytes were obtained from spleen, and NK cells were enriched using NK cell isolation kit II for mouse (Miltenyi Biotec) or by first incubating for 20 min at 4°C with a cocktail of biotinylated mAbs: rat anti–mouse CD3 (2C11), CD5 (53-7.3), CD19 (ebio1D3), CD24 (M1/69), F4/80 (BM8), TER-119 (ter119), GR1(RB6.8) (eBioscience). After incubation with anti-biotin microbeads (Miltenyi Biotec), NK cells were enriched using magnetic separation. Enriched NK cells were then stained with a life/death marker in combination with anti-CD27, anti-CD11b, anti-CD3, anti-CD19, anti-TCRb, streptavidin, and anti-NK1.1 or anti-NKp46 and anti-CD122 and subsequently sorted into different subsets using a FACSAria Cell Sorter (BD). In some experiments, CD4 and CD8 T cells were sorted using anti-CD3, anti-CD4, anti-CD8, anti-CD44, and anti-CD62L antibodies. Purity of sorted cell populations was >98%, as checked by flow cytometry. Sorted cells were lysed using TRIzol reagent (Invitrogen) or RLT buffer from the RNeasy Micro kit (QIAGEN), and RNA was extracted according to the manufacturer’s instructions.
Quantitative RT-PCR
We used the High-Capacity RNA-to-cDNA kit (Applied Biosystems) or iScript cDNA synthesis kit (Bio-Rad Laboratories) to generate cDNA for RT-PCR. PCR was performed with a Sybr Green–based kit (FastStart Universal SYBR Green Master; Roche) or SensiFast SYBR No-ROX kit (Bioline) on a StepOne plus instrument (Applied Biosystems) or a LightCycler 480 system (Roche). Primers were designed using the Roche software. We used the following primers for mouse qPCR: Zeb2 F, 5′-CCAGAGGAAACAAGGATTTCAG-3′; Zeb2 R, 5′-AGGCCTGACATGTAGTCTTGTG-3′; S1pr5 F, 5′-GCCTGGTGCCTACTGCTACAG-3′; S1pr5 R, 5′-CCTCCGTCGCTGGCTATTTCC-3′; Tbx21 F, 5′-CAACCAGCACCAGACAGAGA-3′; Tbx21 R, 5′-ACAAACATCCTGTAATGGCTTG-3′; Gapdh F, 5′-GCATGGCCTTCCGTGTTC-3′; and Gapdh R, 5′-TGTCATCATACTTGGCAGGTTTCT-3′. The following primers were used for human qPCR: T-bet F, 5′-AGGATTCCGGGAGAACTTTG-3′; Tbet R, 5′-CCCAAGGAATTGACAGTTGG-3′; Zeb2 F, 5′-AGGAGCTGTCTCGCCTTG-3′; Zeb2 R, 5′-GGCAAAAGCATCTGGAGTTC-3′; OAZ1 F, 5′-GGATAAACCCAGCGCCAC-3′; and OAZ1 R, 5′-TACAGCAGTGGAGGGAGACC-3′.
Chemotaxis assays
Spleen cells were suspended in RPMI 1640 supplemented with 4 mg/ml fatty acid–free bovine albumin (Sigma-Aldrich). The same medium was used to prepare S1P (Sigma-Aldrich) at 10−8 M or chemokines at 50 ng/ml (R&D Systems). Cell migration was analyzed in Transwell chambers (Costar) with 5-µm-pore-width polycarbonate filters. Transmigrated cells were stained for CD3, NK1.1, CD27, and CD11b and counted by flow cytometry.
Cell culture and stimulation
Lung or spleen single-cell suspensions were prepared and cultured in the presence of GolgiStop (BD) without or with cytokines (25 ng/ml rmIL-12 and 20 ng/ml rmIL-12 from R&D Systems) or on antibody-coated plates (anti-Ly49D at 10 µg/ml) or with B16F10 or YAC1 cell lines (400,000 cells/well) for 4 h at 37°C. Surface and intracellular stainings were then performed, and IFN-γ production was measured by flow cytometry. For phospho flow analyses, spleen cells were stimulated for 1 h in the presence of graded doses of rmIL-15 (R&D Systems) at 37°C. Spleen cell suspensions were also incubated for 24 h at 37°C with graded doses of rmIL-15 from R&D Systems, and NK cell survival was assessed using Annexin V/live-dead staining.
In vivo labeling of sinusoidal lymphocytes
Mice were injected i.v. with 1 µg anti-CD45 mAb coupled to phycoerythrin (BD). Mice were sacrificed 1 min after antibody injection, and their BM was collected.
Tumor model
B16F10 melanoma cells were resuspended in 1× HBSS and injected into the tail veins of the mice (2 × 105 cells/mouse). The numbers of lung and liver surface nodules were counted under a dissecting microscope 12 d after injection. In some experiments, lungs were collected, and lung mononuclear cells were purified after DNase I/Collagenase A digestion 1 d after B16 injection, divided over 5 wells with different conditions, and NK cell functions were then assayed using a 4-h ex vivo culture as described in "Cell culture and stimulation."
Statistical analyses
Error bars represent the standard deviation. Statistical analyses were performed using two-tailed Student’s t tests or nonparametric tests when appropriate. These tests were run on the Prism software (GraphPad Software). Levels of significance are expressed as p-values (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Online supplemental material
Fig. S1 contains valuable data that further clarify the manuscript and the different mouse lines used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150809/DC1.
Supplementary Material
Acknowledgments
The authors thank the core facilities of the SFR BioSciences Gerland and technicians of the B.N. Lambrecht laboratory and the VIB flow cytometry facility for technical assistance.
The T. Walzer laboratory is supported by the FINOVI foundation, Agence Nationale de la Recherche, European Research council (ERC-Stg 281025), INSERM, CNRS, Université de Lyon, and ENS de Lyon. The G. Berx laboratory is supported by the FWO-V (G.0529.12N), the geconcerteerde onderzoeksacties UGent (GOA-01GB1013W), and the Belgian Federation for the Study Against Cancer. The work is part of the DevRepair (P7/07) IAP-VII network. S. Goossens is a postdoctoral fellow of the Flanders Fund for Scientific Research (FWO). The B.N. Lambrecht laboratory is supported by an ERC grant and B.N. Lambrecht and M.J. van Helden by the Interuniversity Attraction Poles (IUAP-VII/03). The D. Huylebroeck laboratory is supported by the Research Council of KU Leuven (GOA-11/012), FWO-V (G.0954.11N), the Queen Elisabeth Medical Foundation (GSKE 1113), the Interuniversity Attraction Poles (IUAP-VII/07), and Erasmus MC start-up funds. The J.J. Haigh laboratory was supported by Belgium Federation against Cancer, FWO, IUAP, and National Health and Medical Research Council funding bodies.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- DKO
- double KO
- DP
- double positive
- MFI
- mean fluorescent intensity
- qPCR
- quantitative PCR
- TF
- transcription factor
- Tg
- transgenic
References
- Björkström N.K., Riese P., Heuts F., Andersson S., Fauriat C., Ivarsson M.A., Björklund A.T., Flodström-Tullberg M., Michaëlsson J., Rottenberg M.E., et al. 2010. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 116:3853–3864. 10.1182/blood-2010-04-281675 [DOI] [PubMed] [Google Scholar]
- Chiossone L., Chaix J., Fuseri N., Roth C., Vivier E., and Walzer T.. 2009. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 113:5488–5496. 10.1182/blood-2008-10-187179 [DOI] [PubMed] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., and van Roy F.. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 7:1267–1278. 10.1016/S1097-2765(01)00260-X [DOI] [PubMed] [Google Scholar]
- Conidi A., Cazzola S., Beets K., Coddens K., Collart C., Cornelis F., Cox L., Joke D., Dobreva M.P., Dries R., et al. 2011. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFβ/BMP signaling in vivo. Cytokine Growth Factor Rev. 22:287–300. 10.1016/j.cytogfr.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Despoix N., Walzer T., Jouve N., Blot-Chabaud M., Bardin N., Paul P., Lyonnet L., Vivier E., Dignat-George F., and Vély F.. 2008. Mouse CD146/MCAM is a marker of natural killer cell maturation. Eur. J. Immunol. 38:2855–2864. 10.1002/eji.200838469 [DOI] [PubMed] [Google Scholar]
- Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.-K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R., et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature. 489:91–100. 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens S., Janzen V., Bartunkova S., Yokomizo T., Drogat B., Crisan M., Haigh K., Seuntjens E., Umans L., Riedt T., et al. 2011. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 117:5620–5630. 10.1182/blood-2010-08-300236 [DOI] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., and Reiner S.L.. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67. 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y., and Smyth M.J.. 2006. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J. Immunol. 176:1517–1524. 10.4049/jimmunol.176.3.1517 [DOI] [PubMed] [Google Scholar]
- Higashi Y., Maruhashi M., Nelles L., Van de Putte T., Verschueren K., Miyoshi T., Yoshimoto A., Kondoh H., and Huylebroeck D.. 2002. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis. 32:82–84. 10.1002/gene.10048 [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Tabarias H., Fairfax K., Brady J., Hayakawa Y., Degli-Esposti M.A., Smyth M.J., Tarlinton D.M., and Nutt S.L.. 2007a NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J. Immunol. 178:4764–4770. 10.4049/jimmunol.178.8.4764 [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Vosshenrich C.A.J., and Di Santo J.P.. 2007b Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat. Rev. Immunol. 7:703–714. 10.1038/nri2154 [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Yamada A., Yoh K., Nakano T., Shimohata H., Maeda A., Fujioka Y., Morito N., Kawachi Y., Shibuya K., et al. 2007. Th1 and type 1 cytotoxic T cells dominate responses in T-bet overexpression transgenic mice that develop contact dermatitis. J. Immunol. 178:605–612. 10.4049/jimmunol.178.1.605 [DOI] [PubMed] [Google Scholar]
- Jenne C.N., Enders A., Rivera R., Watson S.R., Bankovich A.J., Pereira J.P., Xu Y., Roots C.M., Beilke J.N., Banerjee A., et al. 2009. T-bet–dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J. Exp. Med. 206:2469–2481. 10.1084/jem.20090525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojic V., Shay T., Sylvia K., Zuk O., Sun X., Kang J., Regev A., Koller D., Best A.J., Knell J., et al. Immunological Genome Project Consortium . 2013. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14:633–643. 10.1038/ni.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Iizuka K., Kang H.-S.P., Dokun A., French A.R., Greco S., and Yokoyama W.M.. 2002. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 3:523–528. 10.1038/ni796 [DOI] [PubMed] [Google Scholar]
- Mayol K., Biajoux V., Marvel J., Balabanian K., and Walzer T.. 2011. Sequential desensitization of CXCR4 and S1P5 controls natural killer cell trafficking. Blood. 118:4863–4871. 10.1182/blood-2011-06-362574 [DOI] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y.M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., et al. 2011. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA. 108:18324–18329. 10.1073/pnas.1112064108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderquest K., Powell N., Luci C., van Rooijen N., Hidalgo A., Geissmann F., Walzer T., Lord G.M., and Martín-Fontecha A.. 2011. Monocytes control natural killer cell differentiation to effector phenotypes. Blood. 117:4511–4518. 10.1182/blood-2010-10-312264 [DOI] [PubMed] [Google Scholar]
- Spivakov M. 2014. Spurious transcription factor binding: non-functional or genetically redundant? BioEssays. 36:798–806. 10.1002/bies.201400036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatari M.N., De Craene B., Soen B., Taminau J., Vermassen P., Goossens S., Haigh K., Cazzola S., Lambert J., Huylebroeck D., et al. 2014. ZEB2-transgene expression in the epidermis compromises the integrity of the epidermal barrier through the repression of different tight junction proteins. Cell. Mol. Life Sci. 71:3599–3609. 10.1007/s00018-014-1589-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y.J., and Nieto M.A.. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Townsend M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., and Glimcher L.H.. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 20:477–494. 10.1016/S1074-7613(04)00076-7 [DOI] [PubMed] [Google Scholar]
- Walzer T., Chiossone L., Chaix J., Calver A., Carozzo C., Garrigue-Antar L., Jacques Y., Baratin M., Tomasello E., and Vivier E.. 2007. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat. Immunol. 8:1337–1344. 10.1038/ni1523 [DOI] [PubMed] [Google Scholar]
- Werneck M.B.F., Lugo-Villarino G., Hwang E.S., Cantor H., and Glimcher L.H.. 2008. T-bet plays a key role in NK-mediated control of melanoma metastatic disease. J. Immunol. 180:8004–8010. 10.4049/jimmunol.180.12.8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.