Abstract
Polycomb repressive complex-2 (PRC2) is a histone methyltransferase required for epigenetic silencing during development and cancer. Among chromatin modifying factors shown to be recruited and regulated by long noncoding RNAs (lncRNAs), PRC2 is one of the most studied. Mammalian PRC2 binds thousands of RNAs in vivo, and it is becoming a model system for the recruitment of chromatin modifying factors by RNA. Yet, well-defined PRC2-binding motifs within target RNAs have been elusive. From the protein side, PRC2 RNA-binding subunits contain no known RNA-binding domains, complicating functional studies. Here we provide a critical review of existing models for the recruitment of PRC2 to chromatin by RNAs. This discussion may also serve researchers who are studying the recruitment of other chromatin modifiers by lncRNAs.
Keywords: PRC2, RNA–protein interaction, epigenetic silencing, histone modification, long noncoding RNAs
INTRODUCTION
Polycomb repressive complex-2 (PRC2) is a histone methyltransferase required for epigenetic silencing during development and cancer. It successively adds three methyl groups to lysine 27 of histone H3 (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Müller et al. 2002), providing the H3K27me3 repressive epigenetic mark (for reviews, see Margueron and Reinberg 2011; Di Croce and Helin 2013; Simon and Kingston 2013).
A simple way for PRC2 to locate its binding sites on chromatin would be to recognize specific DNA sequences. Indeed, in Drosophila, Polycomb response elements (PREs) are involved in the recruitment of Polycomb group proteins, possibly with the aid of accessory proteins (for review, see Ringrose and Paro 2007). Although functionally similar DNA sequence elements were identified in vertebrates much later (Sing et al. 2009; Woo et al. 2010; Cuddapah et al. 2012), so far they cannot explain genome-wide association of PRC2 with chromatin. Thus the fundamental question of how mammalian PRC2 identifies its target genes remains open.
Early in the previous decade, PRC2 protein subunits were fully identified to include EZH2, SUZ12, EED, RBBP4, and AEBP2 (Fig. 1), and the complex was shown to deposit the H3K27me3 mark (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Müller et al. 2002). Since then, multiple protein factors have been suggested to be involved in the PRC2 recruitment process (Fig. 2B). In Drosophila, Pleiohomeotic (Pho) DNA-binding protein (Satijn et al. 2001) functions in the recruitment of PRC2 to PREs, and its mammalian homolog YY1 has been suggested to have a role in PRC2 recruitment as well (Caretti et al. 2004). The Drosophila Polycomb group protein Polycomblike (Pcl) interacts with PRC2 (O'Connell et al. 2001; Tie et al. 2003) and is required to generate high levels of H3K27me3 at Polycomb target genes (Nekrasov et al. 2007). The mammalian Pcl homolog PCL2 (MTF2) was shown to associate with PRC2 and to modulate transcription of selected PcG target genes, suggesting that PCL2 is involved in the recruitment of the PRC2 to chromatin (Li et al. 2010, 2011; Walker et al. 2010). PCL1 (PHF1) and PCL3 (PHF19) are two other mammalian homologs of Pcl that were shown to recruit PRC2 to chromatin through interactions with the active chromatin mark H3K36me3 (Ballaré et al. 2012; Brien et al. 2012; Musselman et al. 2012). JARID2, which plays an essential role in embryonic development, interacts with PRC2, regulates its activity, and facilitates its recruitment to chromatin (Peng et al. 2009; Shen et al. 2009; Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010) (for review, see Herz and Shilatifard 2010). PRC2 interacts with DNA methyltransferases DNMT1, DNMT3A, and DNMT3B (Viré et al. 2006; Rush et al. 2009). The DNA methyltransferase 3-like (Dnmt3L) interacts with the PRC2 complex, in competition with the DNA methyltransferases Dnmt3a and Dnmt3b, to maintain low methylation levels at H3K27me3 regions (Neri et al. 2013).
FIGURE 1.
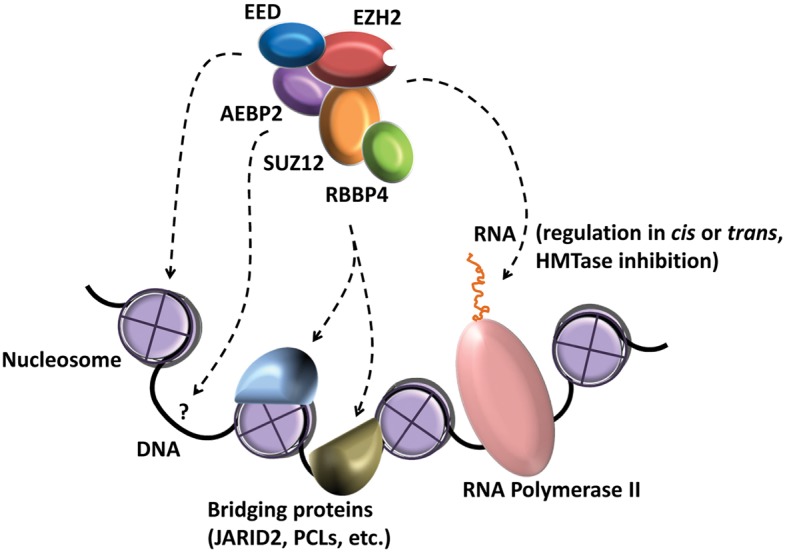
Multiple factors involved in the recruitment of PRC2 to chromatin. PRC2 is a multisubunit histone methyltransferase complex that is regulated and recruited to its target genes through interactions with various factors. These include direct and indirect interactions with nucleosomes, multiple proteins, RNA (which also inhibits the histone methyltransferase activity of PRC2), and possibly DNA. The dashed lines are not meant to indicate which subunit of PRC2 is involved in which potential interaction.
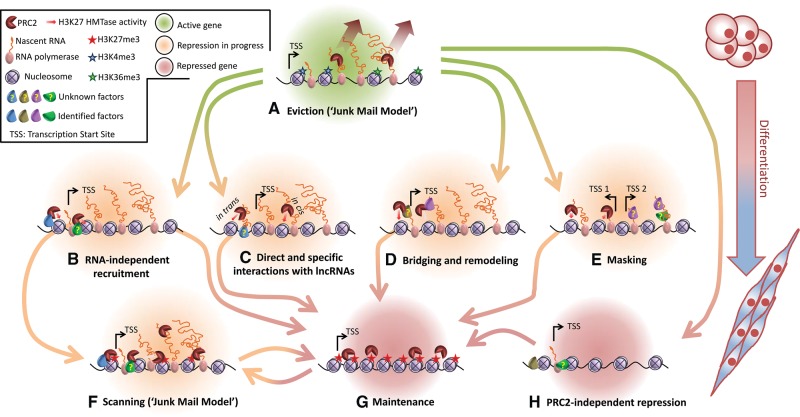
FIGURE 2.
Previously proposed models for the recruitment of PRC2 to chromatin by lncRNA, and RNA in general, compared with RNA-independent recruitment mechanisms. (A) Eviction (“Junk Mail Model”): Promiscuous RNA binding to nascent transcripts leads to eviction of PRC2 from highly active genes (Davidovich et al. 2013; Kaneko et al. 2013) and inhibits its HMTase activity (Cifuentes-Rojas et al. 2014; Herzog et al. 2014; Kaneko et al. 2014b). Simultaneously, H3K4me3 and H3K36me3 active chromatin marks prevent deposition of PRC2 to nucleosomes and inhibit its HMTase activity (Schmitges et al. 2011; Yuan et al. 2011). (B) RNA-independent recruitment: PRC2 is recruited to chromatin through interactions with protein-binding factors, nucleosomes, and/or DNA (for reviews, see Ringrose and Paro 2007; Margueron and Reinberg 2011; Di Croce and Helin 2013; Simon and Kingston 2013; Comet and Helin 2014). (C) Direct and specific interactions with lncRNAs: Site-specific recruitment of PRC2 could occur in cis or in trans (references in text). (D) Bridging and remodeling: Recruitment of PRC2 by RNA can be mediated through protein bridging factors, such as JARID2 (da Rocha et al. 2014; Kaneko et al. 2014a,b), or through RNA structure remodeling, as suggested for ATRX (Sarma et al. 2014). (E) Masking: PRC2 is masked from binding to certain RNA transcripts that are already bound by other factors, thus providing a binding preference (Herzog et al. 2014). (F) Scanning (“Junk Mail Model”): PRC2 interacts with nascent RNA transcripts promiscuously (Davidovich et al. 2013; Kaneko et al. 2013) and scans for repressive epigenetic marks or recruiting factors. Unless deposition to nucleosomes takes place, PRC2 is poised and in check (Kaneko et al. 2014b) while its HMTase activity is inhibited by the RNA. (G) Maintenance: After repression is achieved, PRC2 maintains the repressed epigenetic state through direct binding to nucleosomes carrying H3K27me3 marks (Hansen et al. 2008), which also stimulate its HMTase activity (Margueron et al. 2009). (H) PRC2-independent repression: Transcription shutoff of a Polycomb target gene can take place in a PRC2-independent manner and can lead to subsequent recruitment of PRC2 (Riising et al. 2014). Importantly, most of these models are not mutually exclusive.
Overall, a lack of complete overlap between PRC2 target genes and most of its proposed recruiting factors implied the existence of multiple recruitment mechanisms; possibly by combining relatively weak interactions with multiple factors, the necessary energy to recruit PRC2 could be attained (Margueron and Reinberg 2011). Therefore, it was appealing to assume that other unknown specificity factors were waiting to be discovered, which would allow the mechanism dictating the recruitment of PRC2 to target genes to be fully depicted. This function has been widely proposed to be fulfilled by long noncoding RNAs. In this critical review, we will summarize and analyze evidence previously provided to support the lncRNA recruitment model in the context of recent breakthrough discoveries suggesting alternative models for the recruitment of Polycomb group proteins to chromatin.
The rise of a model: lncRNAs recruit PRC2 for epigenetic repression
Starting in the previous decade, advances in high-resolution tiled microarrays and breakthroughs in next-generation sequencing revealed the overwhelming multitude of noncoding RNAs transcribed from the mammalian genome (for reviews, see Mercer et al. 2009; Ponting et al. 2009; Guttman and Rinn 2012; Rinn and Chang 2012; Kung et al. 2013; Cech and Steitz 2014). Some were long intergenic ncRNAs (lincRNAs), derived from previously unidentified transcription units, whereas others such as bidirectional transcripts from promoters and enhancer RNAs (eRNAs) were new transcripts arising from previously known genetic elements. Some have been inferred to be functional because of sequence conservation across species, and a small but growing number have been shown to be functional by knockout or knockdown approaches but are poorly understood mechanistically. This was the time for the emergence of a new and exciting model to explain how PRC2 identifies its target genes: site-specific recruitment by lncRNAs.
HOTAIR
The lncRNA HOTAIR was shown to be expressed from the HoxC locus in human and mouse cells differentially, at posterior and distal sites (i.e., foot, finger, foreskin, and prostate) (Rinn et al. 2007). RNA interference of HOTAIR, expressed from chromosome 12, led to transcriptional activation of genes at the HOXD locus, spanning >40 kb of chromosome 2. This suggested gene regulation in trans, the first to be demonstrated for any lncRNA. Furthermore, HOTAIR was shown to be required for PRC2 occupancy and for H3K27me3 deposition at target genes within the HOXD locus. Association between PRC2 and HOTAIR, by RNA immunoprecipitation (RIP), was significantly greater than for a few other nonrelevant RNAs that were tested, suggesting binding specificity. Pull-down experiments using biotinylated RNAs showed that purified human PRC2 is associated with HOTAIR RNA, but not with a nonrelevant RNA negative control. Collectively, these results reinforced a model where the lncRNA HOTAIR is expressed from the HoxC locus and recruits PRC2, in trans, to the HoxD locus for epigenetic repression, through direct and specific protein–RNA interactions (Rinn et al. 2007).
How trans-recruitment might occur is an exciting frontier in the field and might involve (for example) triple-helix formation between the lncRNA and the target DNA (Schmitz et al. 2010), base-pairing of the lncRNA to nascent RNA transcribed from the target locus (Lee et al. 2015), or binding of part of the lncRNA to a protein bound to the target locus.
Xist
A short time after HOTAIR, a 1.6-kb ncRNA internal transcript within the Xist gene was discovered in mouse, namely RepA (Zhao et al. 2008). RepA lncRNA, like Xist, contains eight to nine short tandem repeats from the evolutionary conserved A-repeat region of the Xist locus (Nesterova et al. 2001). Importantly, a short synthetic 28-nt RNA with the sequence of a single repeat from within the A-repeat region was shown to bind PRC2 in vitro (Zhao et al. 2008). Qualitative EMSA experiments confirmed binding of this RNA to individually purified EZH2, EZH2-EED heterodimer, and a minimal PRC2 heterotrimeric complex, including EZH2, EED, and SUZ12. The sequence of this RNA construct, which was previously predicted to form a two-hairpin RNA structure (Wutz et al. 2002), was now proposed to be the binding motif of PRC2 within Xist and RepA (Zhao et al. 2008). Point mutations designed to disrupt the secondary structure of the two-hairpin binding motif appeared to prevent PRC2 binding, although the specificity was not quantitated. Therefore, these results supported a model where PRC2 is recruited during the process of X-chromosome inactivation by direct and specific interactions with Xist and RepA RNA. Importantly, this work was the first to suggest a specific RNA-binding motif being responsible for the interactions between PRC2 and a recruiter lncRNA. These results also pointed to EZH2 as an RNA-binding subunit that is sufficient for direct and specific interactions between PRC2 and its RNA-binding partner.
Yet, results obtained within the same work indicated that PRC2 binds the antisense strand of the RNA motif as well. The antisense sequence is a part of Tsix RNA, an Xist RNA antisense transcript that is expressed from the active X chromosome. This observation challenged a simple recruitment model, and therefore a more complex model was suggested: RepA recruits PRC2 for the initiation of X-chromosome inactivation (XCI), while Tsix prevents RepA-PRC2 action in pre-XCI cells by titrating PRC2 away from RepA, by blocking RepA-PRC2 transfer to chromatin, or by preventing PRC2 catalysis (Zhao et al. 2008).
Kcnq1ot1
Another lncRNA that was shown to recruit PRC2 to chromatin around the same time is Kcnq1ot1 (Pandey et al. 2008), a lncRNA with a function in imprinting. Earlier evidence had indicated that Kcnq1ot1 lncRNA was transcribed from the paternal chromosome in mouse and linked to the silencing of multiple protein-coding genes spread over a 1-Mb region within the Kcnq1 domain (Fitzpatrick et al. 2002; Mancini-Dinardo et al. 2006). Moreover, previous works indicated that paternal repression along the Kcnq1 domain is associated with the H3K27me3 repressive mark (Lewis et al. 2004; Umlauf et al. 2004) of PRC2, and the H3K9me3 repressive mark deposited by the G9a HMTase. Then, after the function of PRC2-mediated repression had been recognized for the lncRNA HOTAIR, it was suggested that also Kcnq1ot1 follows a similar mechanism (Pandey et al. 2008). This model was supported by RNA immunoprecipitation of Kcnq1ot1 from nuclear extracts of placental cells, using anti-Ezh2, anti-Suz12, and anti-G9a antibodies. Chromatin RNA immunoprecipitation (ChRIP) confirmed that Kcnq1ot1 is tethered to chromatin, and chromatin oligo-affinity precipitation (ChOP) assays indicated that Kcnq1ot1 specifically interacts with both distant and adjacent genes across hundreds of kilobases within the imprinted locus, suggesting regulation in cis. Collectively, these observations led to a model for how genes, located up to 450 kb from either side of the Kcnq1ot1 promoter, are imprinted in the placenta through recruitment of chromatin modification factors (Pandey et al. 2008).
Braveheart
Braveheart (Bvht) is a lncRNA that is required for cardiovascular lineage commitment in mouse, and it was also shown to associate with PRC2 by RIP experiments (Klattenhoff et al. 2013). Specifically, Bvht was shown to interact with SUZ12 during cardiomyocyte differentiation. A series of Bvht truncations was transcribed in vitro, biotinylated, and used in pull-down experiments to find regions within Bvht that are required for the interaction with PRC2. These observations led to the suggestion that Bvht specifically interacts with PRC2, and they supported a model where Bvht mediates epigenetic regulation of cardiac commitment (Klattenhoff et al. 2013).
Hence, within a few years evidence was accumulated to strongly indicate the association of PRC2 with multiple lncRNAs. In addition to HOTAIR, Xist, RepA, Kcnq1ot1, and Braveheart, as described above, additional PRC2 target transcripts now included the lncRNAs MALAT1 (Guil et al. 2012), both sense and antisense transcripts of H19 (Zhao et al. 2010), ANRIL (Kotake et al. 2011), MEG3 (Zhao et al. 2010; Kaneko et al. 2014a), PINC (Shore et al. 2012), both sense and antisense transcripts of Nespas (Zhao et al. 2010), NEAT1 (Guttman et al. 2011), Air (Zhao et al. 2010), Pint (Marin-Béjar et al. 2013), lncRNA-EBIC (Sun et al. 2014), BLACAT1/linc-UBC1 (which was suggested to act in trans) (He et al. 2013), and COLDAIR from Arabidopsis thaliana (Heo and Sung 2011).
A large PRC2 transcriptome
The rapidly developing field was pushed further forward by a breakthrough discovery, revealing that 20% of the lncRNAs in human cells associated with PRC2 (Khalil et al. 2009). This observation, made using RIP experiments with microarray analysis, indicated for the first time that PRC2 associated with numerous RNA transcripts. Qualitatively, this observation would later be confirmed by multiple independent studies (Kanhere et al. 2010; Zhao et al. 2010; Guil et al. 2012; Kaneko et al. 2013). This work also indicated that PRC2 is preferentially associated with lncRNAs, compared to mRNAs, an observation that was not repeated in later studies that utilized next-generation sequencing-based techniques (Zhao et al. 2010; Guil et al. 2012; Kaneko et al. 2013).
Similar experiments, using high-resolution tiled microarrays, identified a large class of short noncoding RNAs that were transcribed upstream of Polycomb target genes in primary T cells and embryonic stem cells. Many of these RNAs were shown to bind PRC2 (Kanhere et al. 2010). A search for RNA structures related to the previously proposed two-hairpin motif in RepA (Zhao et al. 2008) identified such a motif in 71% of the short ncRNAs transcribed from PRC2-silenced genes compared to 36% of control sequences (Kanhere et al. 2010). In vitro binding experiments, using selected ncRNA transcripts and individually expressed and purified PRC2 subunits EZH2, SUZ12, EED, and RBBP4, confirmed binding to SUZ12. RNA binding was not detected for any other PRC2 subunit, including the previously suggested RNA-binding subunit EZH2 (Zhao et al. 2008). These results led to a model where short ncRNAs transcribed from the vicinity of promoters that are designated for epigenetic repression recruit PRC2 through direct interaction with a two-hairpin motif. PRC2 then introduces the H3K27me3 mark and represses the adjacent promoter. Importantly, the authors designed an experiment to validate the model suggesting that a two-hairpin motif is sufficient to force epigenetic repression in vivo. A set of reporter plasmids was constructed with two-hairpin motifs inserted upstream of a luciferase open reading frame, and control plasmids had the two-hairpin RNA structure disrupted (Kanhere et al. 2010). This was the first attempt, and to our knowledge the only one published, to use point mutations for in vivo validation of the model, suggesting that PRC2 is recruited for epigenetic repression by interactions with a well-defined RNA motif (however, see below).
Around the same time, RNA immunoprecipitation with next-generation sequencing (RIP-seq) was used to reveal the PRC2 transcriptome—namely, the collection of RNA transcripts that associate with PRC2 in mouse embryonic cells (Zhao et al. 2010). Nearly 10,000 transcripts were shown to associate with PRC2. Members of the PRC2 transcriptome included antisense, intergenic, and promoter-associated transcripts, as well as many unannotated RNAs. Many transcripts originated within imprinted regions, oncogenes, tumor suppressor loci, and bivalent domains. Qualitative in vitro binding assays confirmed the binding of selected transcripts to PRC2 and to its purified Ezh2 subunit, including the newly discovered ncRNA target Hes1-as. That RNA included a sequence that was predicted to form a two-hairpin RNA structure, as suggested to recruit PRC2 from other studies (see above). Binding of this RNA by PRC2 in vitro was impaired by the introduction of point mutations designed to disturb the binding motif, but affinities were not quantified. These results led to the suggestion of direct and specific protein–RNA interactions by PRC2, through Ezh2.
The early model for the recruitment of PRC2 by specific interactions with lncRNAs
Collectively, these lines of evidence were sufficiently strong and redundant to promote a general model for the recruitment of PRC2 to chromatin through direct and specific interactions with long noncoding RNAs. According to this model, a lncRNA recruits PRC2 through specific protein–RNA interactions, either directly though PRC2 subunits (Fig. 2C) or by the aid of a bridging protein such as JARID2 (Fig. 2D; da Rocha et al. 2014; Kaneko et al. 2014a,b). Upon recruitment, PRC2 deposits the H3K27me3 mark that leads to repression. Finally, PRC2 utilizes its ability to bind to (Hansen et al. 2008) and be stimulated by (Margueron et al. 2009) previously deposited H3K27me3 marks to spread and maintain the newly formed repressed chromatin domain (Fig. 2G). Despite some isolated warning calls (Brockdorff 2013), this newly established theme exclusively dominated the field.
Promiscuous RNA binding by PRC2
Measurements of the affinity of a protein for its target RNAs and for nonrelevant RNAs are necessary for a quantitative discussion of RNA-binding specificity (Box 1). Admittedly, quantitative binding experiments of PRC2 to RNA are technically challenging, given that PRC2 is a large multiprotein subunit complex, of up to 640 kDa in its dimeric state (Davidovich et al. 2014). Moreover, some of the minimal RNA sequence elements that were previously suggested to be required for PRC2 binding are large, in the range of 80–500 nt (Tsai et al. 2010; Klattenhoff et al. 2013; Wu et al. 2013; Cifuentes-Rojas et al. 2014).
Box 1. Quantification of RNA-binding specificity
Binding specificity is a biophysically meaningful quantitative measure and therefore can be “high” or “low,” rather than “present” or “absent.” Binding specificity can be quantified as the ratio of two dissociation constants (Kds, e.g., in nM) that were independently measured between the protein of interest and two different RNAs (i.e., Kd-fold change = Kd,2/Kd,1), with both experiments performed under the same binding conditions. It can also be quantified as the difference in Gibbs free energies (ΔΔG°, in kJ/mol or kcal/mol), which can be calculated from both dissociation constants as follows: ΔΔG° = RT·ln(Kd,2/Kd,1), where R is the gas constant and T is the temperature (K). RNA binding is promiscuous if a protein binds to many RNAs with small binding specificities, typically Kd-fold change <10 (Davidovich et al. 2014). In cases where RNA not only binds PRC2, but also inhibits its HMTase activity, one can measure Kis for different RNAs and identify the inhibitory mechanism (e.g., competitive, uncompetitive, or noncompetitive inhibition; Lineweaver and Burk 1934).
When the affinity of PRC2 to its target lncRNAs—HOTAIR and the A-repeat from RepA and Xist—was measured in terms of equilibrium dissociation constants, the Kds were found to be in the mid-nanomolar range; surprisingly, affinities within the same range were also measured for nonrelevant bacterial mRNAs (Davidovich et al. 2013, 2015). These observations indicated promiscuous RNA binding by PRC2 in vitro.
Promiscuous binding means binding to many RNAs without the requirement for an obvious protein-binding motif and with affinities that are not enormously different. The observation that a 200-nt mRNA provides multiple binding sites for PRC2 provided additional evidence against the requirement for a specific RNA motif (Davidovich et al. 2014). Note that “promiscuous” is not synonymous with “nonspecific.” For example, if PRC2 were specific for a very short motif such as the dinucleotide G-Pu, then almost all RNAs would have multiple copies of the motif and promiscuous binding would result.
Nonspecific protein–RNA-binding affinity increases with RNA length (Epstein 1979; Kowalczykowski et al. 1986), and PRC2 shows this behavior. Thus, a test RNA can falsely appear to show binding specificity if it is compared to a shorter control RNA. By controlling for RNA length, it was found that both human and mouse PRC2 have a modest three- to eightfold specificity for RepA RNA relative to bacterial mRNAs in vitro (Davidovich et al. 2015), resolving an earlier apparent discrepancy (for review, see Kretz and Meister 2014).
In vitro binding experiments do not address the involvement of other factors that could alter affinity or specificity in vivo. Indeed, the interaction between PRC2 and RNAs has been suggested to be modulated by bridging or regulatory proteins, such as JARID2 (Cifuentes-Rojas et al. 2014; da Rocha et al. 2014; Kaneko et al. 2014a,b), or by post-translational modifications, as shown in the case of Ezh2 T345 phosphorylation (Kaneko et al. 2010). In vivo folding of a target RNA giving a structure that differs from its in vitro conformations could, potentially, increase the affinity of PRC2. This was demonstrated for the RNA helicase ATRX that was suggested to remodel the RNAs RepA and Xist and thus to enhance binding of PRC2 (Sarma et al. 2014). The great abundance of various post-transcriptional modifications throughout the transcriptome (Hodgkinson et al. 2014; Rabani et al. 2014; Alarcon et al. 2015; Sugimoto et al. 2015) could serve as additional determinants to guide PRC2 recruitment, although evidence for this is lacking in the case of PRC2.
Therefore, it was important to assess what is the RNA-binding specificity of PRC2 in vivo. CLIP-seq experiments identified intronic RNA sequences that were associated with EZH2 but failed to identify a binding motif. These findings suggested a role of intronic RNA binding by PRC2 in fine-tuning of gene expression regulation, at the level of transcriptional control (Guil et al. 2012). To overcome potential complications of CLIP, such as low antibody specificity or protein–protein cross-linking at 254 nm, an independent study used photoactivatable ribonucleoside-enhanced CLIP (PAR-CLIP) experiments, using embryonic cell lines expressing epitope-tagged EZH2 (Kaneko et al. 2013). PAR-CLIP experiments use cross-linking at 365 nm (Hafner et al. 2010) (or 312 nm in Kaneko et al. 2013) to eliminate protein–protein cross-linking. Results confirmed promiscuous RNA binding by PRC2 in vivo, with some bias toward the 5′ ends of RNA transcripts (Kaneko et al. 2013). No preference was observed for ncRNAs over mRNAs.
Based on early models, recruitment of PRC2 by lncRNAs would be expected to lead to deposition of the H3K27me3 mark within target loci (Rinn et al. 2007; Zhao et al. 2008, 2010; Kanhere et al. 2010; Tsai et al. 2010). If this were indeed the predominant mechanism in vivo, one would expect to see enrichment for the H3K27me3 mark around promoters of genes transcribing these lncRNAs. Yet, the thousands of RNA transcripts in the PRC2 transcriptome include numerous mRNAs coding for housekeeping proteins and other highly expressed RNAs that are not known to be regulated by Polycomb proteins. Furthermore, the degree of association between PRC2 and RNA, based on RIP-seq, is positively correlated with marks of active chromatin and RNA transcription (Davidovich et al. 2013). Similar results were also obtained when associations of PRC2 with RNA transcripts in vivo were determined by PAR-CLIP (Kaneko et al. 2013). Therefore, RNA transcripts that associate with PRC2 originate mostly from active genes.
Collectively, these results strongly suggest that association of PRC2 with RNA transcripts in vivo does not necessarily lead to repression in cis. Importantly, this strongly implies that PRC2-mediated gene repression by lncRNAs in cis (Pandey et al. 2008; Zhao et al. 2008; Kanhere et al. 2010; Kotake et al. 2011) is unlikely to represent more than a small fraction of PRC2–RNA association events in cells.
Many biological functions require nonspecific or promiscuous protein–nucleic acid binding
The idea that PRC2 binds RNA promiscuously may lead one to wonder whether such protein–RNA interactions could be functional. Although this question has yet to be experimentally established for PRC2, multiple proteins have been previously shown to bind a diverse cohort of nucleic acid partners through functional interactions. Such proteins function in DNA replication and recombination (Jensen et al. 1976), DNA packaging (Agback et al. 1998), RNA folding (Seraphin et al. 1989; Mohr et al. 2002; Huang et al. 2005; Bhaskaran and Russell 2007), and RNA degradation (Chen et al. 2001; Makino et al. 2013). Restriction enzymes can bind noncognate sites with high affinity, often with a Kd that is similar to that of their canonical site (Hinsch et al. 1980; Woodhead and Malcolm 1980). Yet, they are inefficient in cleaving noncognate sites (Lesser et al. 1990) and rather utilize their nonspecific binding activity for scanning DNA (for review, see Pingoud and Jeltsch 2001). Even the MS2 (R17) bacteriophage coat protein, which has been studied for decades as a model system for RNA-binding specificity (Carey et al. 1983a,b; Romaniuk et al. 1987) because of its specific interactions with RNA during translation regulation, has an additional nonspecific binding activity that facilitates its second function of capsid formation (LeCuyer et al. 1995).
What is the function of promiscuous RNA binding by PRC2?
Two mechanisms have so far been suggested for how the intrinsic promiscuous RNA-binding activity of PRC2 can facilitate gene regulation, both of which can coexist. The first is RNA-mediated scanning and eviction, previously referred to as the “junk mail model” (Davidovich et al. 2013) or “poised and in check” (Kaneko et al. 2013, 2014b). The second model suggests selected targeting to any available transcript that is not masked by other factors (Herzog et al. 2014).
RNA-mediated scanning and eviction (the “junk mail model”)
Promiscuous RNA binding by PRC2 may serve as a checkpoint to prevent escape from silencing and therefore function in maintaining the repressed epigenetic state of Polycomb-target genes (Davidovich et al. 2013). In this model, this simple yet robust process of promiscuous RNA binding allows PRC2 to locate active genes using the most direct marker of active transcription—namely, RNA. Promiscuous RNA binding can promote eviction of PRC2 from active genes that should not be repressed (Fig. 2A). Indeed, active chromatin marks H3K4me3 and H3K36me3 were previously shown to inhibit the HMTase activity of PRC2 and to reduce its affinity to nucleosomes (Schmitges et al. 2011; Yuan et al. 2011). Therefore, such an active mark is expected to destabilize deposition of PRC2 to nucleosomes in active genes. In highly active genes, the elongating RNA may serve as decoy to strip PRC2 away from chromatin while the transcription machinery progresses into the gene body (Fig. 2A). Importantly, this model is in excellent agreement with recent multiple independent reports, indicating that RNA inhibits the HMTase activity of PRC2 (Cifuentes-Rojas et al. 2014; Herzog et al. 2014; Kaneko et al. 2014b). As one possibility, RNA may serve as an allosteric effector of HMTase activity.
At the same time, such promiscuous interactions with RNAs may allow scanning for active genes that need to undergo silencing (Fig. 2F). Although the RNA brings PRC2 to the vicinity of the promoter, a given chromatin mark that signals for repression facilitates deposition of PRC2 to chromatin. Chromatin marks that were previously shown to recruit PRC2 are the H2AK119ub mark deposited by PRC1 (Blackledge et al. 2014; Cooper et al. 2014; Kalb et al. 2014) (for review, see Comet and Helin 2014) and the H3K27me3 mark deposited by PRC2 itself (Hansen et al. 2008; Margueron et al. 2009). In summary, upon association with RNA, PRC2 is poised and in check (Kaneko et al. 2014b), but it will not introduce H3K27me3 to chromatin unless it has been deposited to nucleosomes (Fig. 2F).
We termed this the “junk mail model” (Fig. 2A,F) to emphasize the analogy between RNA-mediated scanning by PRC2 and commercial targeting of large populations by advertising mail. In the latter case, commercial organizations use rough and general criteria, such as annual income (Mazzone and Pickett 2011), to target “junk mail” advertisements to the mailboxes of recipients who could potentially, but not necessarily, be their customers. Whether to become functionally involved with the advertiser is the choice of the recipient. In this junk-mail model, PRC2 is the junk mail, chromosomal loci are mailboxes, transcriptional activity is the criterion for delivery, and the response is dictated by the local chromatin context (i.e., active or repressive histone marks and chromatin compaction).
It is important to note that although the junk mail model fits current data, it has not been rigorously tested. Such tests might entail, for example, transcription interference or knock-in of lncRNAs in the vicinity of an mRNA transcription unit, followed by monitoring PRC2-dependent changes in silencing of transcription.
The masking model: preferred binding to exposed transcripts that are not masked by other factors
The promiscuous RNA binding by PRC2 in vitro and in vivo appeared to be at odds with the observations indicating that some transcripts are more efficient than others in binding PRC2 in vivo, as described above. Recently it has been suggested that such an apparent discrepancy could result from the combination of the intrinsic promiscuous RNA-binding activity of PRC2 and competition with other protein factors that bind some RNAs specifically (Fig. 2E; Herzog et al. 2014). Thus, RNA transcripts will have a better chance to compete for PRC2 binding if they are not already bound, or “masked,” by other proteins. Hence, this model (Herzog et al. 2014) provides a simple yet robust explanation for how some binding preferences in vivo can be obtained.
One immediate ramification of this model would be that RNAs that had not evolved to form specific interactions with protein-binding partners would have a better chance to successfully compete for PRC2 binding. Thus, the model indirectly offers a function for poorly conserved RNAs, which include many of the known lncRNAs (Marques and Ponting 2009; Cabili et al. 2011). Even “transcriptional noise” (Elowitz et al. 2002; Swain et al. 2002; Ponjavic et al. 2007; Struhl 2007; McCullagh et al. 2010; Neems and Kosak 2010) can have a role in the recruitment of PRC2!
Alternative explanations for previously obtained results
A general model for Polycomb-mediated repression proposes the following cascade of events: First, PRC2 is recruited to Polycomb target genes and deposits the H3K27me3 mark, and then PRC1 binds to these modified nucleosomes and introduces the repressive H2AK119ub mark (Cao et al. 2002). In parallel, PRC2 binds to its own product (Hansen et al. 2008), the H3K27me3 mark, which also stimulates its histone methyltransferase activity (Margueron et al. 2009). This facilitates spreading and inheritance of Polycomb domains. The missing piece in this puzzle has been the initial recruitment event of PRC2, which has been assumed to take place in a target-specific manner but could not always be explained by PREs. Therefore, it was appealing to assume that certain lncRNAs perform this role. This model was indeed consistent with observations made within most of the works that promoted it. Yet, in most of these studies the model was based on indirect evidence: the association of Polycomb-mediated repression with expression of a given lncRNA, which was shown to bind PRC2.
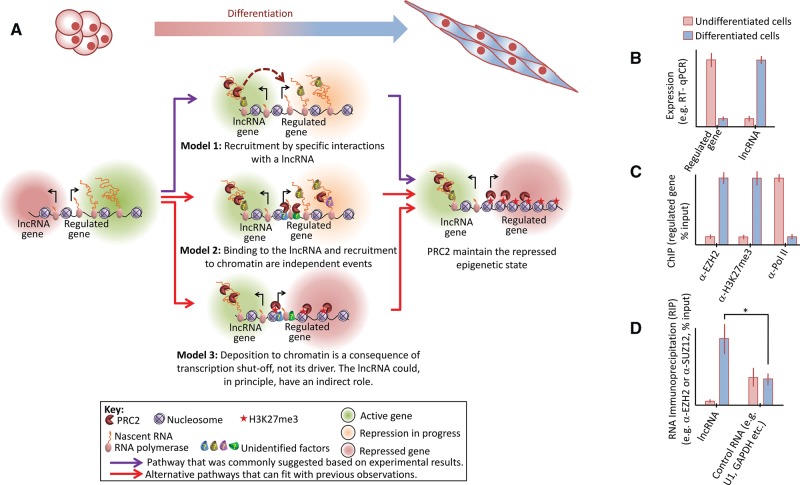
An important piece was added to the Polycomb puzzle more recently, when it was shown that PRC2 could be recruited to chromatin by a PRC1 variant (Blackledge et al. 2014; Cooper et al. 2014; Kalb et al. 2014) (for review, see Comet and Helin 2014). Furthermore, it was previously proposed that large CpG islands depleted of activating transcription factors could recruit PRC2 (Ku et al. 2008). More recently it was also shown that PRC2 may not initiate repression, but it can rather be recruited to nucleosome-free CpG islands of untranscribed genes for the maintenance of the repressed epigenetic state (Riising et al. 2014). These observations and models do not provide a complete mechanistic description for how Polycomb target genes are turned off and whether lncRNAs play an indirect role in this process. Yet, they strongly suggest that some of the indirect observations that were previously used to support early models for lncRNA-mediated recruitment of PRC2 could now have alternative interpretations (Fig. 3).
FIGURE 3.
Alternative explanations for observations that were previously provided to support the recruitment of PRC2 for epigenetic repression through specific interactions with lncRNAs. (A) In undifferentiated cells the differentially regulated gene is active and the lncRNA is repressed or lowly expressed. The cascade of events that could take place through differentiation is illustrated by three models, all of which are compatible with current literature. Model 1 (top, purple arrows): A given lncRNA is differentially expressed and binds PRC2 by specific protein–RNA interactions, either directly or through a bridging factor. This leads to the recruitment of PRC2 to the regulated gene (dashed arrow). Next, PRC2 introduces the H3K27me3 mark. Model 2 (middle, red arrows): The lncRNA is differentially expressed and also binds PRC2, possibly through promiscuous protein–RNA interactions. Yet, the association of PRC2 with the lncRNA does not directly cause its deposition to chromatin. Instead, recruitment of PRC2 to the regulated gene takes place independently by other factors (for reviews, see Ringrose and Paro 2007; Margueron and Reinberg 2011; Di Croce and Helin 2013; Simon and Kingston 2013; Comet and Helin 2014). Model 3 (bottom, red arrows): The differentially regulated gene is repressed in a PRC2-independent manner. The lncRNA could function in this process, but it is not the driving force for the recruitment of PRC2, which only interacts with the RNA through promiscuous interactions. Next, transcription shutoff leads to the recruitment of PRC2 to chromatin (Riising et al. 2014), where it maintains the repressed epigenetic state. In all three models, the end point is identical: In differentiated cells the lncRNA is expressed while the differentially regulated gene is epigenetically repressed, with PRC2 maintaining the repressed epigenetic state. (B–D) Hypothetical in vivo data commonly provided to support the recruitment of PRC2 by specific interactions with lncRNAs (see text for details). (B) Expression analysis supports all three models, but cannot exclude any of them. (C) Chromatin immunoprecipitation (ChIP) for the differentially expressed gene identifies the recruitment of PRC2 to chromatin during differentiation (α-EZH2), deposition of its repressive epigenetic mark (α-H3K27me3), and reduction in RNA polymerase occupancy over the regulated gene (α-Pol II), yet these observations agree with all three models and are therefore not definitive. (D) RNA immunoprecipitation confirms that PRC2 is associated with the lncRNA to a greater extent, compared with a negative control RNA, to a statistically significant degree. Although this observation is consistent with Model 1, it is not definitive and can also be explained by an undetected bridging factor or simply by a higher degree of promiscuous interactions, as previously observed between PRC2 and various nonrelevant RNAs in vivo (Kaneko et al. 2013) and suggested to be the result of competition between PRC2 and other RNA-binding proteins (Herzog et al. 2014).
More specifically, lncRNA-mediated recruitment models have heavily relied on the following logic: PRC2 associates with a given lncRNA, usually shown by RNA immunoprecipitation (Fig. 3D) and, less commonly, validated by qualitative binding assays in vitro. The same lncRNA is also shown to be transcribed from or associated with a PRC2-repressed locus (Fig. 3C). In multiple cases the lncRNA is differentially expressed in parallel to the repression of a regulated gene (Fig. 3B). Therefore, the lncRNA of interest is assumed to recruit PRC2 to chromatin for epigenetic repression by direct and specific interactions (Fig. 3A, Model 1).
These observations are indeed consistent with an RNA recruitment model, yet they would also be expected if lncRNA binding by PRC2 and its deposition to chromatin were independent events (Fig. 3A, Models 2 and 3). Specifically, transcription shutoff is sufficient for the recruitment of PRC2 to many of its target genes (Riising et al. 2014), and PRC2 can be recruited following PRC1 (Blackledge et al. 2014; Cooper et al. 2014; Kalb et al. 2014). PRC2 binds thousands of RNAs, presumably competing with other RNP proteins (Herzog et al. 2014). Collectively, these observations provide an alternative explanation for correlative observations that were previously made to support the recruitment of PRC2 for epigenetic repression by lncRNAs.
For instance, Kcnq1ot1 was suggested to be responsible for the recruitment of PRC2 to imprinted genes (Pandey et al. 2008). Evidence included well-controlled RIP assays, which correlated with the enrichment of H3K27me3 in the Kcnq1 domain of the repressed allele (Lewis et al. 2004; Umlauf et al. 2004). Although it is possible that Kcnq1ot1 recruits PRC2 by direct and specific interactions, one cannot exclude the alternative hypothesis that the recruitment of PRC2 to the Kcnq1 domain is the result of transcription repression, rather than its cause. Under such a scenario, Kcnq1ot1 could have a role in driving this repression process in a PRC2-independent manner, it could recruit PRC2 through a bridging factor, or it might be prone to promiscuous interactions with PRC2 because it is not coated with a large number of other RNA-binding proteins through specific RNA–protein interactions and therefore is not masked from binding. Given these new findings, future mechanistic studies will be required to identify how Kcnq1ot1 and PRC2 function during the repression of the Kcnq1 locus.
Pull-down experiments using biotinylated RNA, in conjugation to immunoblotting, led to the conclusion that PRC2 interacts directly and specifically with Braveheart (Bvht) lncRNA (Klattenhoff et al. 2013). RNA truncations were used to identify regions within Bvht that are required for PRC2 binding. Yet, such truncated RNAs may bind PRC2 with lower affinity simply because they are shorter than the intact RNA or minimally truncated constructs. This possibility is concerning because, as described above, PRC2 binds RNA in a length-dependent manner with lower affinity to shorter RNAs. Of course, it is possible that future studies will reveal well-defined binding motifs for PRC2 within Braveheart RNA, which will be confirmed by point mutations in conjugation with quantitative binding assays in vitro and functional assays in vivo. Until these are discovered, it is impossible to exclude alternative hypotheses such as the presence of a bridging factor, the lack of masking factors, or an indirect cascade of events that leads to gene repression and subsequent recruitment of PRC2.
The idea that lncRNA can be involved in the repression of a Polycomb target gene in parallel to PRC2, rather than actively recruiting PRC2 through direct and specific interactions, was demonstrated for COOLAIR. COOLAIR is an Arabidopsis thaliana lncRNA previously shown to regulate cold-induced Polycomb-mediated gene repression (Swiezewski et al. 2009); however, in this case no physical association with PRC2 and the lncRNA was observed (Heo and Sung 2011) and RIP experiments did not support binding specificity (Csorba et al. 2014). Instead, it was shown that COOLAIR affects transcriptional shutdown independently of Polycomb machinery and H3K27me3 accumulation (Csorba et al. 2014).
It would be unfair to judge early works on PRC2 for not considering these models and observations, as these newly established mechanisms are supported by data that were not available at the time. Yet, in light of this new evidence, future studies will benefit from rigorous mechanistic investigation beyond the mere coincidence of (i) PRC2–lncRNA interactions, (ii) PRC2 deposition to chromatin, and (iii) epigenetic repression.
Identification of binding specificity between a chromatin modifier and lncRNAs: lessons from PRC2
A careful assessment of RNA-binding specificity and its functional consequences in vitro and in vivo is especially important, given recent models for the recruitment of PRC2 in an RNA-independent manner, as described above. Such analysis is also important in light of the growing interest of multiple laboratories in the larger field of lncRNAs and chromatin modifying complexes, which rely on PRC2 as a model system. More than 40 years of RNP research in vitro and in vivo have provided us with great tools to validate functionally relevant RNA–protein interactions, as described below.
Identification of putative target RNAs in vivo
Specific RNA binding requires, by definition, a specific binding motif that distinguishes target RNAs from the rest of the transcriptome (Burd and Dreyfuss 1994; Auweter et al. 2006; Lunde et al. 2007). A binding motif can be a sequence signature, a well-defined structure, or a combination of these. The first step in binding motif discovery is the identification of in vivo RNA-binding partners for a protein. Currently, this process starts with transcriptome-wide CLIP-seq, PAR-CLIP, or, with some limitations, RNA immunoprecipitation.
Reduction of the binding motif within target RNAs to the essential minimum
After the identification of a putative binding motif in vivo, one can reduce it to the minimum RNA sequence or structure that is required for protein binding. Reduction of the binding motif to the essential minimum allows in-depth mechanistic and structural biology analysis. Moreover, reducing the size of the binding motif allows for more accurate quantification of binding affinity and specificity (Box 1), because longer RNAs can lead to increased affinity in the case of nonspecific binding (as discussed above) or decreased affinity if the additional sequences partially occlude the binding motif (Wang et al. 2015). Identification of a minimal and well-defined binding motif can be done in vitro, using a reconstituted system including purified protein and an RNA of interest. Such experiments may include combinations of cross-linking using photoactivatable nucleotides, chemical and enzymatic probing, or systematic RNA truncations and mutagenesis in conjunction with protein–RNA-binding measurements.
Quantification of binding specificity
Quantitation of binding specificity can be done in the form of biophysically meaningful measures, such as Kd-fold change or ΔΔG° (see Box 1). Accordingly, quantification of binding specificity requires quantitative binding assays, where the equilibrium dissociation constant Kd or the rate constants for the dissociation (koff) and association (kon) are measured. Quantitative binding assays have now been applied for PRC2 in multiple studies (Davidovich et al. 2013, 2014, 2015; Wu et al. 2013; Cifuentes-Rojas et al. 2014; Somarowthu et al. 2015) and will be valuable tools to rigorously test putative RNA-binding motifs for PRC2, if these are identified.
Loss of function and compensatory point mutations
A classic test for an RNA motif is to introduce point mutations that disrupt RNA sequence, and in some cases RNA structure, and then measure changes in binding affinity to its protein-binding partner. In the case of high binding specificity, mutation of a critical nucleotide within a binding motif will lead to a large increment in the equilibrium dissociation constant in vitro. In the future, site-specific genome editing techniques could be used to test these mutations in vivo (Doudna and Charpentier 2014; Hsu et al. 2014; Sander and Joung 2014), assaying for changes in PRC2 recruitment and gene silencing.
Identification of evolutionary conservation/covariation
Sequence specificities of RNA-binding proteins often display deep evolutionary conservation, and motifs identified in vitro often correlate well with in vivo RNA-binding data (Ray et al. 2013). In the case of structured RNA motifs, comparative analysis of well-aligned sequences from diverse organisms reveals covariation of the paired bases (e.g., an A → G change on one side of a base-paired stem is compensated by a U → C on the other side). Therefore, a PRC2-binding RNA motif would be expected to demonstrate evolutionary conservation/covariation as well.
What is the evidence for RNA-binding specificity by PRC2?
In principle, divergent models that suggest the recruitment of PRC2 to chromatin by specific (Fig. 2C,D) or promiscuous (Fig. 2A,E,F) interactions with RNAs are not mutually exclusive. Therefore, recent models for transcription regulation through promiscuous RNA binding by PRC2 neither rule out nor replace earlier models. Yet, an important question remains: Was there ever any solid evidence for substantial RNA-binding specificity by PRC2?
As described above, there is no shortness in candidate RNA-binding partners for PRC2, as thousands of them have been proposed. Yet, the more challenging task of motif identification has so far been a limiting step in studying this system.
Considering RNA motifs for PRC2 binding
The two-hairpin motif described above was predicted based on free energy calculations of RNA folding, but it was never confirmed experimentally. Moreover, quantitative binding experiments later showed that the introduction of the predicted two-hairpin element into RNA was insufficient to increase its affinity to PRC2 (Supplemental Fig. S2 of Davidovich et al. 2015). Independently, more complex RNA structures were later experimentally identified in the A-repeat region of Xist and RepA, involving multiple repeats, using NMR (Duszczyk et al. 2011), FRET, and chemical and enzymatic probing (Maenner et al. 2010). The more complex structure of the A-repeat (Maenner et al. 2010) was later suggested to be required for high-affinity interactions with PRC2 (Cifuentes-Rojas et al. 2014). Yet, in a follow-up study we showed that PRC2 binds this RNA with an affinity only three- to eightfold higher than to other nonrelevant RNAs, under different binding conditions and as long as the RNA length is adequately controlled (Davidovich et al. 2015).
A biological role for PRC2-RepA RNA binding is further challenged by observations that PRC2 is dispensable for the initiation of X-chromosome inactivation (Kalantry and Magnuson 2006) and that the A-repeat within Xist RNA is not required for the recruitment of PRC2 to the inactive X chromosome (Plath et al. 2003; Kohlmaier et al. 2004; da Rocha et al. 2014). Furthermore, Xist RNA and PRC2 appear to be spatially separated in vivo (Cerase et al. 2014). Finally, no PRC2 subunits were shown to associate with Xist RNA in vivo, using targeted RNA isolation with mass spectrometry in two independent studies (Chu et al. 2015; McHugh et al. 2015). A third study identified only RBBP7, even after extending the hit list to 200 proteins (Minajigi et al. 2015). Interestingly, RBBP7 and its paralog RBBP4 are associated not only with PRC2 but also with histone deacetylase (HDAC) complexes (Zhang et al. 1997), and they are the only PRC2 subunits that were never proposed to have a function in RNA binding. Future studies will be required in order to determine what is the function of PRC2 in X chromosome inactivation and if RNA binding by PRC2, either specific or not, is a determinant in this process.
The 5′ domain of HOTAIR lncRNA (∼300 nt) was suggested to be responsible for specific interactions with PRC2 using RNA IP experiments (Tsai et al. 2010). A portion of this RNA, containing 89 nt, was later suggested to represent a highly structured minimal binding motif for PRC2 (Wu et al. 2013). Yet, a large part of these 89 nt was later shown to form base pairs with nucleotides external to this region (Somarowthu et al. 2015), therefore complicating their previous assignment as the minimal motif. PRC2 binds the entire 5′ domain of HOTAIR that is required for a stable fold and its partial 300-nt fragment with affinity differences of only twofold (Somarowthu et al. 2015), as could be expected simply given their size difference (see above).
These small PRC2-affinity differences are comparable also to differences in affinities that we previously observed between HOTAIR and nonrelevant RNAs (Davidovich et al. 2013) or RepA and nonrelevant RNAs (Davidovich et al. 2015). As discussed for the A-repeat above, further studies will be required to determine whether such small differences in affinity can lead to biological consequences in vivo. This question of binding specificity is especially interesting in the case of HOTAIR, as this lncRNA acts in trans (Rinn et al. 2007) and targets more than 800 sites genome-wide (Chu et al. 2011), although it is lowly expressed RNA (∼100 copies per cell, much of which is detected in the cytoplasm) (Dodd et al. 2013).
In the case of PRC2, binding motifs could not explain the association of the protein complex with thousands of RNA transcripts, as observed by immunoprecipitation-based approaches (Rinn et al. 2007; Zhao et al. 2008, 2010; Guttman et al. 2011; Heo and Sung 2011; Kotake et al. 2011; Shore et al. 2012; He et al. 2013; Klattenhoff et al. 2013; Sun et al. 2014). In vivo CLIP and PAR-CLIP experiments for EZH2 (Guil et al. 2012; Kaneko et al. 2013) or JARID2 (Kaneko et al. 2014a) did not result in well-defined peaks that could, in principle, lead to the discovery of binding motifs. The only general well-defined binding motif that was ever suggested for PRC2, a two-hairpin structural motif, was initially supported by reporter gene assays in vivo (Kanhere et al. 2010) that were later explained by unintentional promoter mutations in a subset of the plasmids (Supplemental Fig. S3 in Davidovich et al. 2015). The search for RNA motifs that bind PRC2 therefore continues.
Point mutations in conjunction with quantitative binding assays
The introduction of a point mutation into a putative binding motif within an RNA of interest is a valuable tool for rigorous examination of its protein-binding partner, considering both RNA-binding specificity in vitro and functional necessity in vivo. For instance, an AU bulge deletion within the Ku-binding hairpin from the telomerase RNA from yeast significantly reduced its affinity to its binding protein Ku, with Kd increased by >270-fold and ΔΔG >2.9 kcal/mol (Dalby et al. 2013). Bacteriophage MS2 coat protein underwent >1000-fold reduction in affinity (ΔΔG > 3.0 kcal/mol) to its target RNA upon a point mutation in a bulged adenosine within its specific structural binding motif (Romaniuk et al. 1987). The N-terminal RNP domain of U1A protein showed reduction in affinity of >100-fold, or ΔΔG = 2.5 kcal/mol, after a single base modification, adenosine to purine riboside, within its binding site on U1 snRNA (Nolan et al. 1999). These examples demonstrate how a small change in a binding motif leads to a large change in ΔΔG, in the case of high binding specificity. Furthermore, to test the contribution of a base-paired secondary structure element, a deleterious mutation in one partner can be compensated by a complementary mutation in the other (for reviews, see Woese and Pace 1993; Green and Noller 1997).
Admittedly, small apparent binding specificity in vitro is not necessarily a hallmark for nonspecific binding but instead could, in principle, result from a missing protein component (as a post-translational modification or bridging and/or remodeling factor), poor assay conditions, a low complexity motif that might be present also in the control RNA, or a large recognition surface, which is not very sensitive to a given point mutation. Yet, in the case of PRC2, large changes in sequence consistently lead to minor changes in ΔΔG, in the order of 1 kcal/mol or a few-fold in Kd (Davidovich et al. 2013, 2015). This energy difference is on the order of the enthalpy that can be contributed by a single hydrogen bond (Pauling 1960). The only exception found so far is poly(A), which hardly binds to PRC2 (Kaneko et al. 2014b). And although, in a few cases, site-specific mutations were designed to destabilize the predicted two-hairpin motif that was previously suggested to recruit PRC2, no ΔΔG quantification was performed nor compensating mutations used to test for restoration of binding activity.
Evolutionary conservation
The 5′ domain of HOTAIR RNA (Rinn et al. 2007), which was previously suggested to be responsible for the recruitment of PRC2 (Tsai et al. 2010), has poor sequence conservation (Schorderet and Duboule 2011) but high structural conservation as inferred from covariation (Somarowthu et al. 2015). The A-repeat in Xist and RepA RNA is evolutionary conserved (Nesterova et al. 2001; Maenner et al. 2010). Hence, evolutionary conservation of HOTAIR and Xist elements that interact with PRC2 is an appealing hypothesis.
Yet, recruitment of PRC2 takes place even after deletion of the A-repeat within the Xist gene (Plath et al. 2003; Kohlmaier et al. 2004; da Rocha et al. 2014). Deletion of the mouse Hotair gene resulted in homeotic transformation and gene derepression (Li et al. 2013). However, an independent study tested the effect of deletion of a large portion of the HoxC locus in mouse, including the Hotair gene. Surprisingly, little effect was observed, either on the expression pattern or transcription efficiency or on the amount of H3K27me3 coverage, of different HoxD target genes (Schorderet and Duboule 2011). Collectively, in light of these contrary observations, future work will be required to determine the functional significance of the evolutionary conservation observed for HOTAIR and Xist lncRNAs and whether it is related to PRC2 binding.
What elements of PRC2 interact with RNA?
Approximately 60% of the known mRNA-binding proteins contain RRMs (Gerstberger et al. 2014). Other common RNA-binding motifs include the K homology (KH) domain, the DEAD motif, the double-stranded RNA-binding motif (DSRM), and the zinc finger (Gerstberger et al. 2014).
Therefore, it would be tempting to assume that the three zinc fingers in AEBP2 (for review, see Margueron and Reinberg 2011) have a prominent role in RNA binding by PRC2. Yet, AEBP2 is dispensable for RNA binding by PRC2 in the cases of HOTAIR (Tsai et al. 2010) and Xist (Zhao et al. 2008). Moreover, the addition of AEBP2 to PRC2 conferred only a minor increment of approximately twofold in its affinity for RNA (Davidovich et al. 2013). Perhaps the zinc fingers of AEBP2 function mostly in DNA binding (Kim et al. 2009).
SUZ12, which has a single zinc finger, has been demonstrated to have a role in RNA binding (Kanhere et al. 2010; Cifuentes-Rojas et al. 2014). A prominent function in RNA binding was previously assigned to EZH2 (Zhao et al. 2008, 2010; Kaneko et al. 2010; Cifuentes-Rojas et al. 2014). Phosphorylation of Threonine 345 within mouse Ezh2 was shown to increase the affinity of PRC2 to RNA, and mutations in this region reduced its affinity (Kaneko et al. 2010) to a degree that has not yet been biophysically quantified. Intriguingly, EZH2 shares no sequence homology with any known RNA-binding domains in other proteins.
Most of the information regarding PRC2 subunits involved in RNA binding has been based on in vitro binding experiments using individually expressed subunits or complexes where some of the subunits were omitted (Zhao et al. 2008, 2010; Kanhere et al. 2010; Wu et al. 2013). The actual contribution of SUZ12 and EZH2 to the overall RNA-binding activity of the assembled PRC2 complex is not clear. In addition, there are some conflicting observations. For instance, although most works confirmed RNA-binding activity for EZH2 (Zhao et al. 2008, 2010; Kaneko et al. 2010; Wu et al. 2013; Cifuentes-Rojas et al. 2014), this was not always the case (Kanhere et al. 2010). One study detected no interactions between SUZ12 and RNA (Zhao et al. 2010), but two other works found direct RNA-binding activity for this protein (Kanhere et al. 2010; Cifuentes-Rojas et al. 2014). The interpretation of these conflicting observations is further complicated because, to our knowledge, there is no evidence in the current literature supporting the solubility of the individually expressed subunits EZH2 or SUZ12. Thus, one cannot exclude the possibility that at least some of these experiments were performed using soluble aggregates, rather than well-behaved proteins. Indeed, the only work that characterized the solubility of the EZH2-EED heterodimeric complex in vitro, using native gels, found higher-order oligomerization (Wu et al. 2013).
Some proteins, such as ribosomal proteins, evolved unique ways for interacting with their RNA-binding partners (Gerstberger et al. 2014). This could certainly be the case also for PRC2. In any case, the lack of comprehensive knowledge regarding interactions between PRC2 and its RNA-binding partners is limiting mechanistic study at this time.
SUMMARY
Since the discovery that PRC2 interacts with the lncRNA HOTAIR, this field has expanded dramatically and, despite some contradictory evidence and models (for reviews, see Brockdorff 2013; Goff and Rinn 2013; Gendrel and Heard 2014; Kretz and Meister 2014 and above), most of the data accumulated from numerous independent research groups and by different methodologies support a common observation: PRC2 binds RNA. Yet, the biological function of this RNA binding remains unknown. Furthermore, well-defined PRC2-binding motifs within target RNAs have not been verified by the standard criteria of the RNP field. RNA-binding domains within PRC2 protein subunits are still elusive and, assuming that they exist, do not resemble common RNA-binding domains observed in other proteins.
As is always true, the absence of evidence is not evidence of absence. Yet, it is fair to say that the model suggesting recruitment of PRC2 by direct and specific interactions with lncRNAs is still pending validation.
A growing body of evidence indicates that PRC2-mediated repression takes place in orchestration with additional Polycomb group proteins (Blackledge et al. 2014; Cooper et al. 2014), other protein factors (O'Connell et al. 2001; Satijn et al. 2001; Tie et al. 2003; Caretti et al. 2004; Viré et al. 2006; Nekrasov et al. 2007; Peng et al. 2009; Rush et al. 2009; Shen et al. 2009; Landeira et al. 2010; Li et al. 2010, 2011; Pasini et al. 2010; Walker et al. 2010; Ballaré et al. 2012; Brien et al. 2012; Musselman et al. 2012; Neri et al. 2013), surrounding nucleosomes within dense chromatin (Yuan et al. 2012), previously deposited epigenetic marks H3K27me3 (Hansen et al. 2008; Margueron et al. 2009), H2AK119Ub (Kalb et al. 2014) or H3K36me3 (Ballaré et al. 2012; Brien et al. 2012; Musselman et al. 2012), and the transcription state of targeted genes (Riising et al. 2014). RNA can certainly be involved in this process, to an extent that is not fully elucidated mechanistically. Yet, assigning a central role for lncRNAs as a major driving force in the recruitment of PRC2 in a gene-specific manner is premature, as exciting as it would be for those of us immersed in the RNA World.
ACKNOWLEDGMENTS
We thank John Rinn, Olke Uhlenbeck, Dave McKay, and Neil Brockdorff for helpful review of the manuscript. C.D. was supported by the Fulbright Postdoctoral Fellowship and the Machiah Foundation Program. T.R.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article and publication date are online at http://www.rnajournal.org/cgi/doi/10.1261/rna.053918.115.
REFERENCES
- Agback P, Baumann H, Knapp S, Ladenstein R, Härd T. 1998. Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nat Struct Biol 5: 579–584. [DOI] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. 2015. N6-methyladenosine marks primary microRNAs for processing. Nature 519: 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH-T. 2006. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res 34: 4943–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al. 2012. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19: 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran H, Russell R. 2007. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature 449: 1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and Polycomb domain formation. Cell 157: 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. 2012. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19: 1273–1281. [DOI] [PubMed] [Google Scholar]
- Brockdorff N. 2013. Noncoding RNA and Polycomb recruitment. RNA 19: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J, Cameron V, de Haseth PL, Uhlenbeck OC. 1983a. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry 22: 2601–2610. [DOI] [PubMed] [Google Scholar]
- Carey J, Lowary PT, Uhlenbeck OC. 1983b. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry 22: 4723–4730. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. 2014. The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157: 77–94. [DOI] [PubMed] [Google Scholar]
- Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, et al. 2014. Spatial separation of Xist RNA and Polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci 111: 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of xist RNA binding proteins. Cell 161: 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. 2014. Regulatory interactions between RNA and Polycomb repressive complex 2. Mol Cell 55: 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet I, Helin K. 2014. Revolution in the Polycomb hierarchy. Nat Struct Mol Biol 21: 573–575. [DOI] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. 2014. Targeting Polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7: 1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C. 2014. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci 111: 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Roh TY, Cui K, Jose CC, Fuller MT, Zhao K, Chen X. 2012. A novel human Polycomb binding site acts as a functional Polycomb response element in Drosophila. PLoS One 7: e36365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196. [DOI] [PubMed] [Google Scholar]
- Dalby AB, Goodrich KJ, Pfingsten JS, Cech TR. 2013. RNA recognition by the DNA end-binding Ku heterodimer. RNA 19: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. 2014. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell 53: 301–316. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. 2013. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20: 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Goodrich KJ, Gooding AR, Cech TR. 2014. A dimeric state for PRC2. Nucleic Acids Res 42: 9236–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. 2015. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 57: 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20: 1147–1155. [DOI] [PubMed] [Google Scholar]
- Dodd DW, Gagnon KT, Corey DR. 2013. Digital quantitation of potential therapeutic target RNAs. Nucleic Acid Ther 23: 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Duszczyk MM, Wutz A, Rybin V, Sattler M. 2011. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17: 1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186. [DOI] [PubMed] [Google Scholar]
- Epstein IR. 1979. Kinetics of nucleic acid-large ligand interactions: exact Monte Carlo treatment and limiting cases of reversible binding. Biopolymers 18: 2037–2050. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32: 426–431. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. 2014. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol 30: 561–580. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Rinn JL. 2013. Poly-combing the genome for RNA. Nat Struct Mol Biol 20: 1344–1346. [DOI] [PubMed] [Google Scholar]
- Green R, Noller HF. 1997. Ribosomes and translation. Annu Rev Biochem 66: 679–716. [DOI] [PubMed] [Google Scholar]
- Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, Villanueva A, Esteller M. 2012. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol 19: 664–670. [DOI] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300. [DOI] [PubMed] [Google Scholar]
- He W, Cai Q, Sun F, Zhong G, Wang P, Liu H, Luo J, Yu H, Huang J, Lin T. 2013. linc-UBC1 physically associates with Polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta 1832: 1528–1537. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A. 2010. The JARID2-PRC2 duality. Genes Dev 24: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Lempradl A, Trupke J, Okulski H, Altmutter C, Ruge F, Boidol B, Kubicek S, Schmauss G, Aumayr K, et al. 2014. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet 46: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch B, Mayer H, Kula MR. 1980. Binding of non-substrate nucleotides to a restriction endonuclease: a model for the interaction of bam HI with its recognition sequence. Nucleic Acids Res 8: 2547–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, Idaghdour Y, Gbeha E, Grenier JC, Hip-Ki E, Bruat V, Goulet JP, de Malliard T, Awadalla P. 2014. High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science 344: 413–415. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. 2005. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci 102: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Kelly RC, von Hippel PH. 1976. DNA “melting” proteins. II. Effects of bacteriophage T4 gene 32-protein binding on the conformation and stability of nucleic acid structures. J Biol Chem 251: 7215–7228. [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. 2006. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet 2: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Müller CW, Vermeulen M, Müller J. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21: 569–571. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. 2010. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24: 2615–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. 2013. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20: 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. 2014a. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 53: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. 2014b. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28: 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed Polycomb target genes and interact with Polycomb repressive complex-2. Mol Cell 38: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. 2009. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res 37: 2940–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. 2013. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. 2011. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 30: 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Paul LS, Lonberg N, Newport JW, McSwiggen JA, von Hippel PH. 1986. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry 25: 1226–1240. [DOI] [PubMed] [Google Scholar]
- Kretz M, Meister G. 2014. RNA binding of PRC2: promiscuous or well ordered? Mol Cell 55: 157–158. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genetics 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. 2013. Long noncoding RNAs: past, present, and future. Genetics 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12: 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCuyer KA, Behlen LS, Uhlenbeck OC. 1995. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34: 10600–10606. [DOI] [PubMed] [Google Scholar]
- Lee N, Moss WN, Yario TA, Steitz JA. 2015. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 160: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser DR, Kurpiewski MR, Jen-Jacobson L. 1990. The energetic basis of specificity in the Eco RI endonuclease–DNA interaction. Science 250: 776–786. [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Isono K, Yamada D, Endo TA, Endoh M, Shinga J, Mizutani-Koseki Y, Otte AP, Casanova M, Kitamura H, et al. 2011. Mammalian Polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate Polycomb activity both at the Hox gene cluster and at Cdkn2a genes. Mol Cell Biol 31: 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al. 2013. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Reps 5: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineweaver H, Burk D. 1934. The determination of enzyme dissociation constants. J Am Chem Soc 56: 658–666. [Google Scholar]
- Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2010. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol 8: e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino DL, Baumgartner M, Conti E. 2013. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495: 70–75. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20: 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Béjar O, Marchese FP, Athie A, Sánchez Y, González J, Segura V, Huang L, Moreno I, Navarro A, Monzó M, et al. 2013. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 14: R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. 2009. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol 10: R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone J, Pickett J. 2011. The household diary study: mail use & attitudes in FY 2010. U.S. Postal Service, Washington, DC. [Google Scholar]
- McCullagh E, Seshan A, El-Samad H, Madhani HD. 2010. Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nat Cell Biol 12: 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159. [DOI] [PubMed] [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al. 2015. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Stryker JM, Lambowitz AM. 2002. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109: 769–779. [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. [DOI] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff J, et al. 2012. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19: 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neems D, Kosak ST. 2010. Turning down the volume on transcriptional noise. Nat Cell Biol 12: 929–931. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Köcher T, Cohen A, Stunnenberg HG, Wilm M, Müller J. 2007. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J 26: 4078–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg HG, Oliviero S. 2013. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell 155: 121–134. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. 2001. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res 11: 833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan SJ, Shiels JC, Tuite JB, Cecere KL, Baranger AM. 1999. Recognition of an essential adenine at a protein-RNA interface: Comparison of the contributions of hydrogen bonds and a stacking interaction. J Am Chem Soc 121: 8951–8952. [Google Scholar]
- O'Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS. 2001. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem 276: 43065–43073. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246. [DOI] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310. [DOI] [PubMed] [Google Scholar]
- Pauling L. 1960. The nature of the chemical bond and the structure of molecules and crystals: an introduction to modern structural chemistry. Cornell University Press, Ithaca, NY. [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A, Jeltsch A. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res 29: 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641. [DOI] [PubMed] [Google Scholar]
- Rabani M, Raychowdhury R, Jovanovic M, Rooney M, Stumpo DJ, Pauli A, Hacohen N, Schier AF, Blackshear PJ, Friedman N, et al. 2014. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 159: 1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. 2014. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55: 347–360. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk PJ, Lowary P, Wu HN, Stormo G, Uhlenbeck OC. 1987. RNA binding site of R17 coat protein. Biochemistry 26: 1563–1568. [DOI] [PubMed] [Google Scholar]
- Rush M, Appanah R, Lee S, Lam LL, Goyal P, Lorincz MC. 2009. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics 4: 404–414. [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. 2014. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 159: 869–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol 21: 1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. 2011. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Simon M, Boulet A, Faye G. 1989. Mitochondrial splicing requires a protein from a novel helicase family. Nature 337: 84–87. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. 2009. Jumonji modulates Polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AN, Kabotyanski EB, Roarty K, Smith MA, Zhang Y, Creighton CJ, Dinger ME, Rosen JM. 2012. Pregnancy-induced noncoding RNA (PINC) associates with Polycomb repressive complex 2 and regulates mammary epithelial differentiation. PLoS Genet 8: e1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. 2009. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897. [DOI] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. 2015. HOTAIR forms an intricate and modular secondary Structure. Mol Cell 58: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang SB, Jin ZJ, Sun SH, Wang F, Li W. 2014. Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PLoS One 9: e100340. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Swain PS, Elowitz MB, Siggia ED. 2002. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci 99: 12795–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802. [DOI] [PubMed] [Google Scholar]
- Tie F, Prasad-Sinha J, Birve A, Rasmuson-Lestander A, Harte PJ. 2003. A 1-megadalton ESC/E(Z) complex from Drosophila that contains Polycomblike and RPD3. Mol Cell Biol 23: 3352–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874. [DOI] [PubMed] [Google Scholar]
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. 2010. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6: 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwartz JC, Cech TR. 2015. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res 43: 7535–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Pace NR. 1993. Probing RNA structure, function, and history by comparative analysis. In The RNA world (ed. RF G, JF A), pp. 91–117. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. 2010. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell 140: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead JL, Malcolm AD. 1980. Non-specific binding of restriction endonuclease EcoR1 to DNA. Nucleic Acids Res 8: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. 2013. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 52: 9519–9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. [DOI] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. 2012. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337: 971–975. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89: 357–364. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. 2010. Genome-wide identification of Polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]