Abstract
Background:
Advances in early detection and treatment have improved outcomes in patients with colorectal cancer (CRC). However, there remains a need for robust prognostic and predictive biomarkers. We conducted a systematic discovery and validation of microRNA (miRNA) biomarkers in two clinical trial cohorts of CRC patients.
Methods:
We performed an initial ‘discovery' phase using Affymetrix miRNA expression arrays to profile stage III CRC patients with and without tumour recurrence (n=50 per group) at 3-years of follow-up. All patients received adjuvant 5-fluorouracil (5-FU) plus oxaliplatin, that is, FOLFOX, treatment. During ‘validation', we analysed miRNAs using qRT–PCR in an independent cohort of 237 stage II–IV CRC patients treated with 5-FU-based chemotherapy, as well as in normal colonic mucosa from 20 healthy subjects. Association with disease recurrence, disease-free survival (DFS) and overall survival (OS) was examined using Cox proportional hazard models.
Results:
In the discovery cohort, miR-320e expression was significantly elevated in stage III colon cancers from patients with vs without recurrence (95% confidence interval (CI)=1.14–1.42; P<0.0001). These results were then independently validated in stage II and III tumours. Specifically, increased miR-320e expression was associated with poorer DFS (hazard ratio (HR)=1.65; 95% CI=1.27–2.13; P=0.0001) and OS (HR=1.78; 95% CI=1.31–2.41; P=0.0003) in stage III CRC patients.
Conclusions:
In two clinical trial cohorts, a systematic biomarker discovery and validation approach identified miR-320e to be a novel prognostic biomarker that is associated with adverse clinical outcome in stage III CRC patients treated with 5-FU-based adjuvant chemotherapy. These findings have important implications for the personalised management of CRC patients.
Keywords: microRNAs, expression, colorectal cancer, 5-fluorouracil, prognosis, biomarker
Colorectal cancer (CRC) is one of the most common cancers in Western countries and it is the second leading cause of cancer-related deaths among adults (Smith et al, 2010). For patients who undergo potentially curative resection, prognosis is adversely impacted by disease recurrence, which is often associated with distant metastasis (Mina and Sledge, 2011). It is established that CRC patients with stage III disease benefit from adjuvant chemotherapy, whereas such benefit in stage II patients has remained an area of controversy (Bertagnolli et al, 2011, O'Connor et al, 2011). 5-Fluorouracil (5-FU)-based chemotherapy remains the mainstay for treating CRC patients, and its combination with oxaliplatin is considered as the standard of care chemotherapeutic treatment in stage III disease (Kopetz et al, 2008). A major problem in the treatment of CRC is the intrinsic or acquired drug resistance that can ultimately lead to death of the patient (Goldberg et al, 2004). Therefore, prognostic markers, which can facilitate stratification of stage II and III patients that are at high risk for developing tumour recurrence and metastasis, are an imperative need, as availability of such tools may have a significant impact on the clinical management of CRC patients.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that have been implicated in the initiation, progression and metastasis in various human malignancies (Calin and Croce, 2006, Baek et al, 2008). MiRNAs can function as either oncogenes or tumour suppressors, can control the expression of hundreds of downstream gene targets and are resistant to RNAse-mediated degradation making them inherently stable within a variety of clinical specimens—all of these features, which have made them attractive molecular signatures for the development of various disease biomarkers, as well as potential therapeutic targets in cancer (Nicoloso et al, 2009; Spizzo et al, 2009). Interestingly, recent evidence also suggests that expression alterations of specific miRNAs may also be associated with determining chemosensitivity and chemoresistance in human cancer cells (Kagan et al, 1995; Liu et al, 2010; Tomimaru et al, 2010). In this regard, recently, the expression levels of miR-34a, miR-10b, miR-19b and miR-143 were shown to be related to responsiveness to 5-FU-based chemotherapy in CRC cell lines (Borralho et al, 2009; Akao et al, 2011; Kurokawa et al, 2012; Nishida et al, 2012). Such in vitro findings provide a rationale for interrogating the expression of miRNAs in clinical specimens from patients with CRC, which might facilitate development of specific prognostic and predictive biomarkers that can help identify CRC patients who will or will not respond to specific chemotherapies.
The introduction of newer technological platforms including microarrays and next-generation sequencing have revolutionised the approaches aimed towards a better understanding of the molecular events related to tumour progression, distant spread and response to chemotherapies (Boyer et al, 2006; Lujambio et al, 2008). However, till date, only a very small number of studies have attempted genome-wide approaches for the identification of novel prognostic and predictive miRNA biomarkers in clinical specimens from patients with CRC, who have been treated with specific chemotherapies (Schepeler et al, 2008; Schetter et al, 2008). More importantly, to the best of our knowledge, translatability of markers developed is such studies have been hampered due to lack of validation of discovered biomarkers in independent patient cohorts. In view of these shortcomings in the existing literature, we designed the present two phase study: in the first phase, we performed a systematic, genome-wide discovery of miRNAs that may serve as prognostic and predictive biomarkers in patients treated with 5-FU-based chemotherapy. In the second phase, we validated the expression of discovered biomarkers in another large, independent cohort of patients enrolled in a clinical trial where patients were treated with 5-FU-based chemotherapy regimens. Using such an approach, we have identified miR-320e, as a novel prognostic biomarker, which is associated with adverse clinical outcome in stage III CRC patients treated with 5-FU-based adjuvant chemotherapy. These findings have important clinical implications for the personalised management of patients with CRC.
Materials and Methods
Patients
The discovery cohort
A cohort of 100 stage III colon cancer tissues that included 50 patients with tumour recurrence and 50 without recurrence within 3 years of treatment. These patients were enrolled as part of the NCCTG N0147, a phase III trial of FOLFOX alone or combined with cetuximab as adjuvant chemotherapy (Alberts et al, 2012); however, the clinical specimens in this study were limited to the FOLFOX alone study arm. Median patient follow-up on this trial was 4.1 years. Of the 47 patients without tumour recurrence and 50 with recurrence from whom miRNA expression profiling was successfully obtained, 53.2% and 56% were treated with FOLFOX alone, respectively. This study was approved by the institutional review boards of all participating centres, and a written informed consent was obtained from all patients. Data are summarised either as mean with 95% confidence intervals (CIs) on the log2 scale, or these values were exponentiated to generate fold-change with a CI.
The validation cohort
This group included a total of 237 CRC patients (65 stage II, 102 stage III and 70 stage IV), who were enrolled as part of a clinical trial conducted at the Hospital Clinic, Barcelona, Spain. Patients in this clinical trial were enrolled between 1996 and 2008, and the median follow-up was 4.7 years in patients who were alive at last follow-up, and 3.5 years for all patients. All stage II and III patients were treated with 5-FU-based adjuvant chemotherapy following resection of the primary tumour; all stage IV patients were treated with 5-FU and oxaliplatin (Takahashi et al, 2012). Normal colonic mucosa (n=20) from healthy individuals was obtained from the Pathology Department of the Hospital Clinic of Barcelona and included as controls. This study was approved by the Institutional Review Board of Hospital Clinic, Barcelona, Spain, and a written informed consent was obtained from all patients.
RNA extraction
Total RNA including the miRNA fraction was extracted from formalin-fixed paraffin-embedded tissues using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Inc., Austin, TX, USA) according to the manufacturer's protocol.
MiRNA expression microarrays
MiRNA microarray expression profiling analysis included interrogation of ∼2221 miRNAs, using the Affymetrix GeneChip miRNA 2.0 Arrays (Santa Clara, CA, USA). Each sample was labelled using the Genisphere FlashTag Biotin HSR kit (Hatfield, PA, USA). Briefly, 1 mg of total RNA was incubated with ATP and poly-A-polymerase to add a 3′-polyA tail. A ligation reaction was then performed to covalently attach to the miRNA population a multiple-biotin molecule containing a 3′-DNA dendrimer. Labelled samples were subsequently processed according to the manufacturer's instructions. Following hybridisation for 16 h at 48 °C, the arrays were washed and stained in an Affymetrix Fluidics station 450, scanned using an Affymetrix 3000 7G scanner and the expression changes between cases and controls were analysed.
Quantification of miRNA expression by real-time RT–PCR
Expression of miR-320e in the validation cohort was performed utilising the TaqMan reverse transcription–PCR (qRT–PCR) method with the predesigned TaqMan miRNA expression assays (Applied Biosystems Inc., Foster City, CA, USA) using a StepOnePlus Real-Time PCR System (Applied Biosystems). All the experiments were done in duplicate. Results were expressed as 2−ΔCt, and the results were normalised to miR-16. To keep consistent measurements throughout all plates, two independent RNA cell line samples were loaded as internal controls along with each PCR run, and the results from each plate were normalised according to data obtained from these internal controls.
Statistical analysis
The quality of the raw Affymetrix miRNA data was assessed using boxplots and residual MVA plots for each subject pre- and post-normalisation (Eckel et al, 2005). Per-probe and -patient distributions of detection calls were also examined, where signal intensities with detection P<0.06 were considered to be detected. The non-human probes were excluded before normalisation, leaving 2221 human miRNA transcripts for analyses. Data were analysed on the log2 scale, and were normalised using quantile normalisation (Bolstad et al, 2003). Differential expression between tumours with and without recurrence within 3 years was performed via logistic regression by incorporating various variables: both unadjusted and adjusted individually for deficient DNA mismatch repair (dMMR) vs proficient MMR (pMMR) status, presence/absence of a BRAFV600E or KRAS mutation, right vs left side and metastatic regional lymph nodes <4 or >=4. FDR values were calculated, and the significance level was set at FDR q<0.05 (Storey and Tibshirani, 2003). Probes were filtered out of consideration if the standard deviation (s.d.) in cases and controls combined for a given probe was less than the 95th percentile s.d. (0.164) of those probes that were not detected in ≥45% of the samples.
Distributions in the validation cohort are presented as mean (s.d.) or median (range) depending on level of skewness in the data for continuous variables or as counts (percentage) for categorical variables. Nonparametric tests (rank sum, Kruskal Wallace) were used to compare expression between groups. Kaplan–Meier and Cox regression models were used to assess association with time to event outcomes that included overall survival (OS), defined as time from diagnosis to death, and disease-free survival (DFS) defined as time from diagnosis to disease recurrence or death. Owing to the scale of miR320e expression, hazard ratios (HRs) are presented as the change in risk of a person at the 75th percentile relative to a person at the 25th percentile of miR-320e distribution. This can be interpreted as the increase in risk due to a patient moving from the middle of the bottom half of the miR-320e distribution to the middle of the top half of the distribution. Bootstrapping was used to calculate an optimism-corrected c-index, analogous to the area under the receiver operating characteristic curve (Steyerberg et al, 2011). A c-index of 0.5 denotes random predictions, whereas a c-index of 1 denotes perfect predictions.
Results
MiR-320e is frequently upregulated in CRC patients with recurrence to 5-FU-based chemotherapy
The discovery phase
We performed systematic discovery for miRNA biomarkers using Affymetrix microarrays in 100 stage III patients treated with 5-FU-based adjuvant chemotherapy, during the course of NCCTG N0147 trial. High-quality miRNA data were obtained on all but three patients without tumour recurrence at 3 years of follow-up. The median age (range) for these 97 patients was 59 (25–81) years, of which 46 (47.4%) were male. There were 48 (49.5%) primary tumours that were located in the distal colon and 49 (50.5%) tumours that were proximal (Table 1).
Table 1. Characteristics of patients in the discovery and validation cohorts.
| Characteristics | Discovery cohort | Validation cohort |
|---|---|---|
| Age, median (range), years | 59 (25–81) | 65 (32–82) |
| Gender, n (%) | Male, 51 (52.6) | Male, 141 (59.5) |
| Female, 46 (47.4) | Female, 96 (40.5) | |
| TNM, n (%) | II, 0 (0) | II, 65 (27.4) |
| III, 97 (100) | III, 102 (43.0) | |
| IV, 0 (0) | IV, 70 (29.5) | |
| Location, n (%) | Left colon, 48 (49.5) | Left colon, 155 (65.4) |
| Right colon, 49 (50.5) | Right colon, 69 (29.1) | |
| Grade, n (%) | Low (grade 1–2) 68 (70.1) | Well, moderate, 140 (96.5) |
| High (grade 3–4) 29 (29.9) | Poor, 5 (3.5) | |
| Mucinous, n (%) | – | Yes, 25 (16.3) |
| – | No, 128 (83.7) | |
| MMR statusa, n (%) | Deficient 10 (10.3) | Deficient 19 (7.0) |
| Proficient 87 (89.7) | Proficient 249 (91.9) | |
| Follow-up in all patients, median (range) | 3.2 (Range: 0.35–7.1 years) | 4.4 (Range: 0.24–14.4 years) |
Abbreviations: MMR=mismatch repair; TNM=tumour, node, metastasis.
DNA status was determined by MMR protein expression in the discovery cohort and MSI testing in the validation cohort.
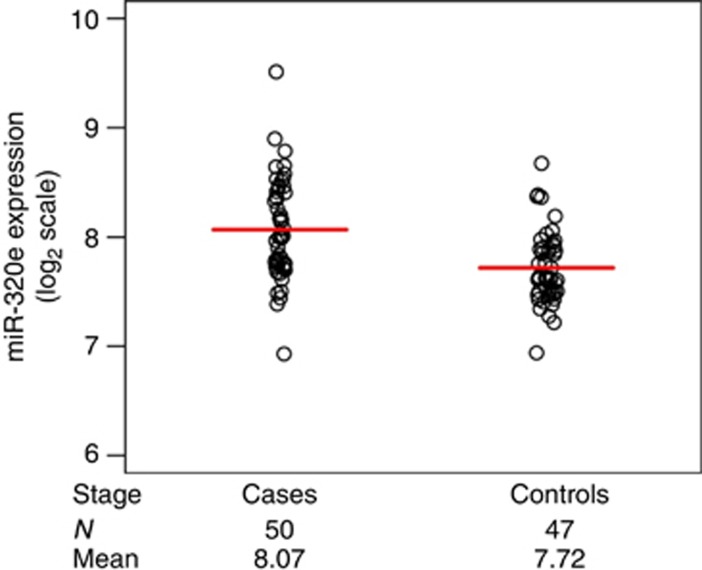
Of the 2221 miRNA probes analysed via microarray analysis, one met statistical significance criteria with unadjusted analyses: miR-320e, with P<0.0001 and a corresponding FDR q-value of 0.0497 (Supplementary Table 1). MiR-320e was upregulated in cases with recurrence vs no recurrence (controls; Figure 1). Specifically, the mean level of miR-320e expression (mean±s.d.) on the log2 scale in patients that recurred was 8.07±0.46 vs 7.72±0.33 compared with patients without recurrence, resulting in a raw fold change of +1.27-fold. After adjusting for sidedness, the mutational status of BRAF and KRAS, and number of metastatic regional lymph nodes (< or ≥4), miR-320e expression was significantly higher in cases vs controls (P=0.0003).
Figure 1.
MiR-320e expression in the discovery cohort. Log2 normalised expression levels for miR-320e in stage III colorectal cancers treated with 5-FU-based chemotherapy. Cases (patients with tumour recurrence) had significantly higher miR-320e expression compared with controls (patients without recurrence). The red line indicates median value for expression.
The validation phase
To further confirm our discovery phase results for miR-320e expression, we perform validation of these data in an independent cohort of 237 patients with stage II–IV CRC. The median (range) age of the patients was 65 (32–82) years, and of these 141 (59.5%) patients were male. Of the 237 CRCs, 69 (29.1%) were located in the proximal colon, 155 (65.4%) were distal and 13 (5.5%) were located in the rectum. In all, 65 (27.43%) of the cases were stage II, 102 (43.04%) were stage III and 70 (29.5%) were stage IV (Table 1).
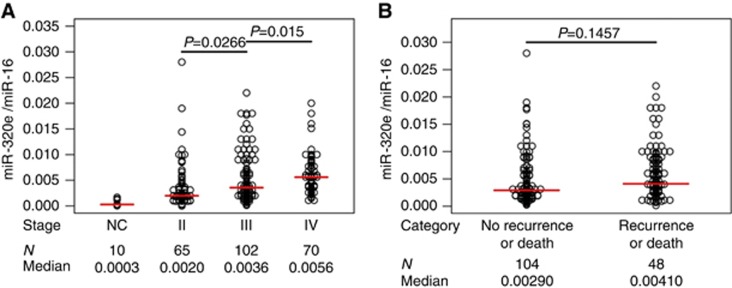
MiR320e results were expressed as 2−ΔCt and normalised with the expression of miR-16. The median level of miR-320e expression in primary CRC tissues was almost 13-fold higher than in normal colonic mucosa (median (range): 0.000286 (0.0000238–0.0017000) vs 0.0037 (0–0.028), P<0.0001)). There was a gradual increasing trend for miR-320e expression with advancing tumour stage (Figure 2A). Specifically, significant miR-320e upregulation was found in CRC patients with lymph node metastases, that is, stage III (0.00357 (0.00013–0.022) vs 0.00200 (0–0.028); P=0.0266) or with distant metastasis, that is, stage IV (0.0056 (0.0011–0.02) vs 0.00200 (0–0.028); P<0.0001) compared with patients without regional or distant metastases, that is, stage II.
Figure 2.
MiR-320e expression in normal colonic mucosa and CRC tissues from the validation cohort. MiR-320e results were expressed as 2−ΔCt and normalised to miR-16. (A) Colonic mucosa vs primary tumours–stage II–IV, miR-320e expression in colonic mucosa from healthy controls (NC) and in stage II, III and IV CRCs; the number of patients (N) and median expression (median) are listed below the graph. (B) Recurrence or death within 3 years vs non-recurrence or death within 3 years–stage II and III, miR-320e expression; the number of patients (N) and median expression (median) are listed below the graph. The red line indicates median value for expression.
Higher-expression miR320e is associated with poor outcome in patients with stage II and III CRC
To determine the potential prognostic significance of miR-320e expression, Cox regression models were utilised with the miRNA expression levels considered as continuous variables. There were 167 stage II and III CRC patients in the validation cohort with sufficient data for these analyses. All of these patients were treated with 5-FU-based adjuvant chemotherapy for 6 months subsequent to tumour resection. Of these patients, 65 out of 167 (38.9%) were stage II and the remainder were stage III. Median follow-up for these patients was 4.4 years (range: 0.24–14.4 years), and was 4.7 years for patients alive at last follow-up (range: 0.69–14.4 years). The 2- and 5-year Kaplan–Meier DFS estimates were 78.32% (72.29, 84.84) and 63.96% (56.58, 72.30), respectively. The 2- and 5-year OS estimates were 91.57% (95% CI=87.44, 95.89) and 78.52% (95% CI=72.15, 85.46), respectively.
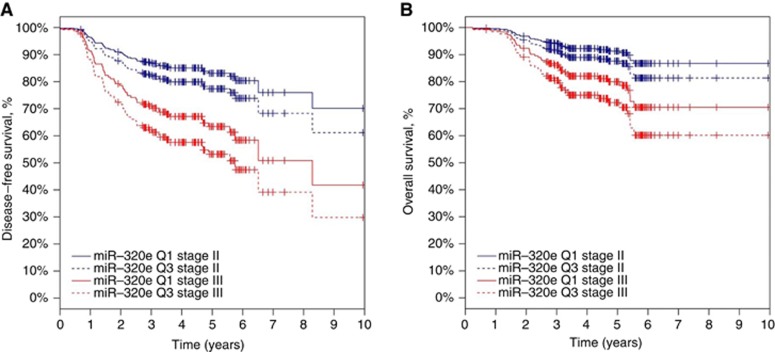
Multivariate models were employed to interrogate the statistical significance of results in these 167 stage II/III patients, and to assess the association of miR-320e expression with DFS and OS, after adjusting for tumour location and TNM stage. After adjustment for TNM stage and tumour location in stage II and III patients, an increase of 0.0052 units of miR-320e expression (that is, an increase from the 25th percentile to the 75th percentile of the miR-320e distribution) was associated with an increased HR for DFS (HR=1.39, 95% CI=1.11–1.74, P=0.0036) and OS (HR=1.46, 95% CI=1.11–1.91, P=0.0059; Table 2, Figure 3). The optimism-corrected c-indices were 0.63 and 0.62 for DFS and OS, respectively.
Table 2. Multivariable analysis of the association between DFS or OS and miR-320e expression in stage II/III CRC patients, adjusting for tumour location and stage.
| Multivariate analysis |
Stage II and III |
||||
|---|---|---|---|---|---|
| Variable | Event/total | HR | 95% CI | P-value | c-index |
|
DFS | |||||
| miR-320e (per 0.0052 unitsa) | 60/167 | 1.39 | 1.11–1.74 | 0.0036b | 0.63 |
| Tumour location (proximal vs distal) | 1.05 | 0.61–1.79 | 0.8606 | ||
| TNM (III vs II) | 2.46 | 1.34–4.50 | 0.0035b | ||
|
OS | |||||
| miR-320e (per 0.0052 unitsa) | 38/167 | 1.46 | 1.11–1.91 | 0.0059b | 0.62 |
| Tumour location (proximal vs distal) | 1.08 | 0.55–2.11 | 0.8251 | ||
| TNM (III vs II) | 2.46 | 1.12–5.37 | 0.0244b | ||
Abbreviations: CI=confidence interval; CRC=colorectal cancer; DFS=disease-free survival; HR=hazard ratio; OS=overall survival; TNM=tumour, node, metastasis.
This is the IQR, representing an increase from the 25th percentile to the 75th percentile of the miR-320e distribution.
P<0.05.
Figure 3.
Predicted DFS and OS functions. Predicted DFS (A) and OS (B) from the Cox regression model containing TNM stage, tumour location and miR-320e expression to visualise the hazard ratios reported in Table 2. Tumour location was set to the left side for calculating the predicted functions. Stage II curves are blue; stage III curves are red. Q1 (solid line) denotes patients with miR-320e expression at the 25th percentile, whereas Q3 (dashed line) denotes patients with miR-320e expression at the 75th percentile. Thus, the curves represent survival functions for patients in the middle of the bottom half of the miR-320e distribution vs those in the middle of the top half of the miR-320e distribution.
When categorising stage II/III the patients into those who had recurred or died within 3 years vs those who were recurrence-free and alive at ≥3 years of follow-up, miR-320e was not significantly different between these groups (0.00410 (0.00013–0.02200) vs 0.00290 (0.00023–0.02800)), P=0.1457; Figure 2B). However, when the cohort was restricted to stage III patients, the level of miR-320e expression was found to be significantly different between patients who had recurred or died within 3 years vs those who were alive and recurrence free after 3 or more years of follow-up (0.00578 (0.00013–0.02200) vs 0.00260 (0.00025–0.01800), P=0.0132).
High miR-320e expression associated with advancing CRC stage
Analysis of the clinicopathological features associated with tumour miR-320e expression levels was also investigated. The variables included in this analysis were age, gender, tumour location, tumour size, tumour grade, mucinous, histology, stage and lymph node metastasis. TNM stage was found to be significantly associated with expression level of miR-320e (P=0.0266). Age was not significantly correlated with miR-320e expression levels (r=0.0626 P=0.4220). Neither tumour location nor lymph node metastasis were significantly associated with the expression level of miR-320e (Table 3). Interestingly, there was a trend towards a positive association between miR-320e and tumour size (P=0.1047) and with lymph node metastasis (P=0.0626); however, these associations were not statistically significant at the 5% level (Table 3).
Table 3. miR-320e expression levels and clinico-pathologic characteristics of stage II and III CRC patients in the validation cohort.
| Characteristics stages II–III | Patient number | MiR-320e expression median (range) | P-value |
|---|---|---|---|
| Age (years) | 67 (32–82)a | ||
|
Genderb | |||
| Male | 96 (57.5%) | 0.00305 (0–0.022) | 0.9638 |
| Female | 71 (42.5%) | 0.00300 (0.00033, 0.028) | |
|
Tumour locationb | |||
| Distal | 113 (67.7%) | 0.00310 (0.00023–0.022) | 0.3618 |
| Proximal | 54 (32.3%) | 0.00225 (0–0.028) | |
|
Tumour sizeb | |||
| <45 mm | 72 (46.8%) | 0.0025 (0.00013–0.028) | 0.1047 |
| >45 mm | 82 (53.2%) | 0.00337 (0–0.019) | |
|
Tumour gradeb | |||
| G1, G2 | 140 (96.6%) | 0.00305 (0.00013–0.028) | 0.1843 |
| G3 | 5 (3.4%) | 0.00090(0.00067–0.009) | |
|
Mucinousb | |||
| Yes | 25 (16.3%) | 0.0033 (0–0.018) | 0.9469 |
| No | 128 (83.7%) | 0.00285 (0.00013–0.028) | |
|
Lymph node metastasisb | |||
| Yes | 95 (61.7%) | 0.0034 (0.00013–0.022) | 0.0626 |
| No | 59 (38.3%) | 0.00200 (0–0.028) | |
|
TNMb | |||
| 2 | 65 (38.9%) | 0.002 (0–0.028) | 0.0266c |
| 3 | 102 (61.1%) | 0.00357 (0.00013–0.022) | |
|
MMR status | |||
| Deficient | 11 | 0.00380 (0.00068–0.018) | 0.5655 |
| Proficient | 155 | 0.00300 (0–0.028) | |
Abbreviations: CRC=colorectal cancer; MMR=mismatch repair; TNM=tumour, node, metastasis.
Age, is represented as median (range).
Evaluated using Wilcoxon Rank Sum test.
P<0.05.
Discussion
This clinical study incorporated both a systematic discovery and an independent validation of a miRNA biomarker, miR-320e, which was found to be associated with CRC recurrence in two clinical trial cohorts of patients treated with 5-FU-based adjuvant chemotherapy. We discovered a number of factors that support miR-320e as a potential and novel prognostic biomarker in CRC patients. First, miR-320e expression was significantly increased in resected primary tumours compared with colon mucosa from healthy controls. We observed a gradual increase in miR-320e expression with advancing TNM stages (II–IV). In this regard, there was a significant increase in miR-320e levels in primary tumours from patients with lymph node (stage III) or distant metastasis (IV), compared with stage II tumours. Second, we were able to corroborate the miR-320e findings in the discovery cohort with those found in the validation cohort. Third, increased miR-320e expression was significantly associated with worse DFS and OS among patients diagnosed with stage II/III CRCs, and in stage III CRCs alone, even after adjusting for lymph node status.
MiR-320e belongs to the miR-320 family of miRNAs that encompass a total of 37 sequences. The best studied family member from this cluster is miR-320a (frequently referred to as miR-320), which was reported to have a tumour-suppressive role in prostate, breast and colon cancer (Schepeler et al, 2008, Bronisz et al, 2011, Hsieh et al, 2013). MiR-320a is encoded within a CpG-rich promoter region of the cell cycle gene POLR3D in the antisense orientation (Kim et al, 2008), and is reportedly regulated by epigenetic methylation silencing (Lee et al, 2009). MiR-320a is located within a chromosomal region at 8p21.3, which is a frequent target for loss of heterozygosity during cancer progression (Kagan et al, 1995). In contrast, miR-320e is an intragenic miRNA located at chromosome 19 q13.32 within intron-3 of the PRKD2 gene, which encodes a kinase D (PKD) serine/threonine protein that appears to have a role in the development and progression of leukemia (Yang et al, 2013) and breast cancer (Onodera et al, 2012). Currently, much less is known about the functional role of miR-320e in human cancer compared with the rest of the miR-320 family, which is not surprising given that our study for the first time has identified its role in CRC through a systematic discovery process. Expression of miR-320e was identified in melanoma and in human embryonic stem cells by next-generation sequencing (Goff et al, 2009, Stark et al, 2010); however, the mechanism responsible for miR-320e deregulation and its functional role remain unknown. To elucidate the mechanisms responsible for miR-320e upregulation, the identification of miRNA gene targets will be crucial to our understanding of the role of miR-320e in CRC. Prominent bioinformatic tools, such as TargetScan, miRBase and DianaLab, have identified five predicted targets for miR-320e, including PP2R2C, IRF6, ONECUT2, CMCL1 and CPEB2 genes–all of which have been shown to display tumour-suppressor activities in different cancer types (Banerjee et al, 2007; Fournier et al, 2010; Botti et al, 2011; Nairismagi et al., 2012; Laudadio et al, 2012; Beukers et al, 2013; Bluemn et al, 2013). Further functional studies are necessary to investigate the mechanistic role of miR-320e in colon cancer progression, metastasis and chemotherapeutic response.
Our findings are in line with some of the previous reports highlighting the potential significance of miRNAs as potential prognostic biomarkers in CRC. The first study in this regard reported that high expression of miR-21 in CRC patients associated with poor survival (Schetter et al, 2008). Since then, although several others overexpressed miRNAs, including miR-10b (Nishida et al, 2012), miR-17-92a cluster (Valladares-Ayerbes et al, 2011), miR-29a (Weissmann-Brenner et al, 2012), miR-31 (Yang et al, 2013) and miR-182 (Liu et al, 2013), as well as few downregulated miRNAs such as miR-143 (Guo et al, 2013) and miR-124 (Jinushi et al, 2014) have been proposed to have prognostic usefulness, currently miR-21 is considered as one of the best miRNA-based prognostic biomarkers in CRC.
Strengths of our study are that tumour specimens used for the discovery cohort are from a randomised clinical trial conducted through a cancer cooperative group in the United States. Thus, they are well annotated and from patients meeting strict eligibility criteria where detailed follow-up data were collected. Importantly, patients received the FOLFOX regimen, which is standard adjuvant therapy for stage III colon cancer patients. In addition, we have replicated our findings in the discovery cohort in an independent European clinical trial cohort. Validation in an independent cohort is considered one of the highest levels of validation (Steyerberg et al, 2011). Together, these strengths mitigate the high risk of false discovery in our discovery cohort resulting from testing over 2221 miRNA probes. Although we present provocative evidence for the clinical relevance of miR-320e in CRC, our study has limitations. There is a need for further verification of the relevance of our findings to CRC patients, as well as studies to understand the functional importance of this miRNA. Because of unavailability of matched serum/blood specimens from patients analysed in this study, we were unable to explore the potential clinical usefulness of miR-320e as a non-invasive biomarker–a concept, which might lead to further enhancement of its diagnostic and prognostic utility in future. Last, our study focused only on the prognostic value of miR-320e, and did not address its potential utility as a predictive biomarker. However, this issue could not be evaluated in our study cohorts that lacked untreated control arms.
In conclusion, our study first reports the clinical and prognostic relevance of miR-320e in patients with CRC. Expression of miR-320e was frequently upregulated, particularly in advanced stage tumours, and was associated with recurrence and survival in CRC in patients treated with 5-FU-based adjuvant chemotherapy. Evidence that miR-320e expression may serve as a novel prognostic biomarker was shown whereby elevated levels were associated with poorer DFS and OS rates. In future studies, bioinformatic and transcriptomic approaches, as well as functional analysis, will be necessary to investigate the mechanistic role of miR-320e in colon cancer.
Acknowledgments
The present work was supported by grants R01 CA72851, CA181572 and CA184792 from the National Cancer Institute, National Institutes of Health to AG and CRB, funds from the Baylor Research Institute and a pilot grant award from the Charles A Sammons Cancer Center to AG. In addition, this work was also supported by a grant from the National Cancer Institute, K05CA-142885 (Senior Scientist Award to F Sinicrope). Lucia Perez-Carbonell was a recipient of a post-doctoral grant from Fundación Alfonso Martín Escudero, Spain. The study was also supported, in part, by grants from the National Cancer Institute to the North Central Cancer Treatment Group (NCCTG; CA-25224), the NCCTG Biospecimen Resource (CA-114740), the Alliance for Clinical Trials in Oncology (CA31946) and the Alliance Statistics and Data Center (CA33601).
Author contribution
LCP, AG, FB and FAS developed the study concept and design, LCP and ALO acquired and analysed the data, FAS, AC and SRA provided the study materials and LP, CRB, FAS, ALO and AG drafted the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License
Supplementary Material
References
- Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T (2011) Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett 300: 197–204. [DOI] [PubMed] [Google Scholar]
- Alberts SR, Sargent DJ, Nair S, Mahoney MR, Mooney M, Thibodeau SN, Smyrk TC, Sinicrope FA, Chan E, Gill S, Kahlenberg MS, Shields AF, Quesenberry JT, Webb TA, Farr GH Jr, Pockaj BA, Grothey A, Goldberg RM (2012) Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: a randomized trial. JAMA 307: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Read CA, Griffiths MH, George PJ, Rabbitts PH (2007) Clonal divergence in lung cancer development is associated with allelic loss on chromosome 4. Genes Chromosomes Cancer 46: 852–860. [DOI] [PubMed] [Google Scholar]
- Bertagnolli MM, Redston M, Compton CC, Niedzwiecki D, Mayer RJ, Goldberg RM, Colacchio TA, Saltz LB, Warren RS (2011) Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer—a study of CALGB 9581 and 89803. J Clin Oncol 29: 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukers W, Hercegovac A, Vermeij M, Kandimalla R, Blok AC, van der Aa MM, Zwarthoff EC, Zuiverloon TC (2013) Hypermethylation of the polycomb group target gene PCDH7 in bladder tumors from patients of all ages. J Urol 190: 311–316. [DOI] [PubMed] [Google Scholar]
- Bluemn EG, Spencer ES, Mecham B, Gordon RR, Coleman I, Lewinshtein D, Mostaghel E, Zhang X, Annis J, Grandori C, Porter C, Nelson PS (2013) PPP2R2C loss promotes castration-resistance and is associated with increased prostate cancer-specific mortality. Mol Cancer Res 11: 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM (2009) MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J 276: 6689–6700. [DOI] [PubMed] [Google Scholar]
- Botti E, Spallone G, Moretti F, Marinari B, Pinetti V, Galanti S, De Meo PD, De Nicola F, Ganci F, Castrignanò T, Pesole G, Chimenti S, Guerrini L, Fanciulli M, Blandino G, Karin M, Costanzo A (2011) Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc Natl Acad Sci USA 108: 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG (2006) Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res 66: 2765–2777. [DOI] [PubMed] [Google Scholar]
- Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ, Martin CK, Li F, Yu L, Fernandez SA, Pécot T, Rosol TJ, Cory S, Hallett M, Park M, Piper MG, Marsh CB, Yee LD, Jimenez RE, Nuovo G, Lawler SE, Chiocca EA, Leone G, Ostrowski MC (2011) Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol 14: 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Therneau TM, Burgoon LD, Boverhof DR, Zacharewski TR (2005) Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics 21: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Fournier A, McLeer-Florin A, Lefebvre C, Duley S, Barki L, Ribeyron J, Alboukadel K, Hamaidia S, Granjon A, Gressin R, Lajmanovich A, Bonnefoix T, Chauvelier S, Debernardi A, Rousseaux S, de Fraipont F, Figeac M, Kerckaert JP, De Vos J, Usson Y, Delaval K, Grichine A, Vourc'h C, Khochbin S, Feil R, Leroux D, Callanan MB (2010) 1q12 chromosome translocations form aberrant heterochromatic foci associated with changes in nuclear architecture and gene expression in B cell lymphoma. EMBO Mol Med 2: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP (2009) Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One 4: e7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts SR (2004) A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol 22: 23–30. [DOI] [PubMed] [Google Scholar]
- Guo H, Chen Y, Hu X, Qian G, Ge S, Zhang J (2013) The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol Cancer 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, Yeh SD, Hong TM, Chen YL (2013) MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 34: 530–538. [DOI] [PubMed] [Google Scholar]
- Jinushi T, Shibayama Y, Kinoshita I, Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H, Iseki K (2014) Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med 3: 1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J, Stein J, Babaian RJ, Joe YS, Pisters LL, Glassman AB, von Eschenbach AC, Troncoso P (1995) Homozygous deletions at 8p22 and 8p21 in prostate cancer implicate these regions as the sites for candidate tumor suppressor genes. Oncogene 11: 2121–2126. [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr., Rossi JJ (2008) MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA 105: 16230–16235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetz S, Freitas D, Calabrich AF, Hoff PM (2008) Adjuvant chemotherapy for stage II colon cancer. Oncology (Williston Park) 22: 260–270, ; discussion 270, 273, 275. [PubMed] [Google Scholar]
- Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M, Rokutan K (2012) Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol 47: 883–895. [DOI] [PubMed] [Google Scholar]
- Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F, Pierreux CE, Odom DT, Peers B, Lemaigre FP (2012) A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 142: 119–129. [DOI] [PubMed] [Google Scholar]
- Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A (2009) Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology 9: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, D'Andrade P, Fulmer-Smentek S, Lorenzi P, Kohn KW, Weinstein JN, Pommier Y, Reinhold WC (2010) mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol Cancer Ther 9: 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Du L, Wen Z, Yang Y, Li J, Wang L, Zhang X, Liu Y, Dong Z, Li W, Zheng G, Wang C (2013) Up-regulation of miR-182 expression in colorectal cancer tissues and its prognostic value. Int J Colorectal Dis 28: 697–703. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M (2008) A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA 105: 13556–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina LA, Sledge GW Jr. (2011) Rethinking the metastatic cascade as a therapeutic target. Nat Rev Clin Oncol 8: 325–332. [DOI] [PubMed] [Google Scholar]
- Nairismagi ML, Vislovukh A, Meng Q, Kratassiouk G, Beldiman C, Petretich M, Groisman R, Füchtbauer EM, Harel-Bellan A, Groisman I (2012) Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene 31: 4960–4966. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA (2009) MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 9: 293–302. [DOI] [PubMed] [Google Scholar]
- Nishida N, Yamashita S, Mimori K, Sudo T, Tanaka F, Shibata K, Yamamoto H, Ishii H, Doki Y, Mori M (2012) MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol 19: 3065–3071. [DOI] [PubMed] [Google Scholar]
- O'Connor ES, Greenblatt DY, LoConte NK, Gangnon RE, Liou JI, Heise CP, Smith MA (2011) Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J Clin Oncol 29: 3381–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Nam JM, Hashimoto A, Norman JC, Shirato H, Hashimoto S, Sabe H (2012) Rab5c promotes AMAP1-PRKD2 complex formation to enhance beta1 integrin recycling in EGF-induced cancer invasion. J Cell Biol 197: 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S, Kauppinen S, Ørntoft TF, Andersen CL (2008) Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 68: 6416–6424. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Cokkinides V, Brooks D, Saslow D, Brawley OW (2010) Cancer screening in the United States, 2010: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer J Clin 60: 99–119. [DOI] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Croce CM, Calin GA (2009) SnapShot: MicroRNAs in cancer. Cell 137: 586–586 e581. [DOI] [PubMed] [Google Scholar]
- Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA, Hayward NK (2010) Characterization of the melanoma miRNAome by deep sequencing. PLoS ONE 5: e9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg EW, Schemper M, Harrell FE (2011) Logistic regression modeling and the number of events per variable: selection bias dominates. J Clin Epidemiol 64: 1464–1465, author reply 1463-1464. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, Boland CR, Goel A (2012) The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One 7: e46684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M (2010) MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer 103: 1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares-Ayerbes M, Blanco M, Haz M, Medina V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Reboredo M, Santamarina I, Figueroa A, Antón-Aparicio LM, Calvo L (2011) Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol 39: 1253–1264. [DOI] [PubMed] [Google Scholar]
- Weissmann-Brenner A, Kushnir M, Lithwick Yanai G et al (2012) Tumor microRNA-29a expression and the risk of recurrence in stage II colon cancer. Int J Oncol 40: 2097–2103. [DOI] [PubMed] [Google Scholar]
- Yang MH, Yu J, Chen N, Wang XY, Liu XY, Wang S, Ding YQ (2013) Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2. PLoS One 8: e85353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Zhang H, Ma L, Peng C, Chen Y, Wang J, Green MR, Li S, Rosmarin AG (2013) GABP transcription factor is required for development of chronic myelogenous leukemia via its control of PRKD2. Proc Natl Acad Sci USA 110: 2312–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.