Abstract
Background:
Renal cell carcinoma is one of the most chemoresistant cancers, and its metastatic form requires administration of targeted therapies based on angiogenesis or mTOR inhibitors. Understanding how these treatments impact the human metabolism is essential to predict the host response and adjust personalised therapies. We present a metabolomic investigation of serum samples from patients with metastatic RCC (mRCC) to identify metabolic signatures associated with targeted therapies.
Methods:
Pre-treatment and serial on-treatment sera were available for 121 patients participating in the French clinical trial TORAVA, in which 171 randomised patients with mRCC received a bevacizumab and temsirolimus combination (experimental arm A) or a standard treatment: either sunitinib (B) or interferon-α+bevacizumab (C). Metabolic profiles were obtained using nuclear magnetic resonance spectroscopy and compared on-treatment or between treatments.
Results:
Multivariate statistical modelling discriminates serum profiles before and after several weeks of treatment for arms A and C. The combination A causes faster changes in patient metabolism than treatment C, detectable after only 2 weeks of treatment. Metabolites related to the discrimination include lipids and carbohydrates, consistently with the known RCC metabolism and side effects of the drugs involved. Comparison of the metabolic profiles for the three arms shows that temsirolimus, an mTOR inhibitor, is responsible for the faster host metabolism modification observed in the experimental arm.
Conclusions:
In mRCC, metabolomics shows a faster host metabolism modification induced by a mTOR inhibitor as compared with standard treatments. These results should be confirmed in larger cohorts and other cancer types.
Keywords: renal cell carcinoma, metabolomics, NMR, metabolic profiles, targeted therapies, mTOR inhibitor
Owing to limited clinical signs, renal cell carcinoma (RCC) is diagnosed at advanced stages and with metastases for ∼15–20% of patients (Rini et al, 2009). The treatment of metastatic RCC (mRCC) usually leads to extremely poor results, with response rates of ∼15–20% and a 5-year survival rate between 5 and 10% (Gupta et al, 2008). mRCC is resistant to a broad range of therapies (Lin et al, 2011), as cytotoxic chemotherapy, radiotherapy, or cytokine therapy. Although their efficacy in delaying tumour growth and progression of the disease is controversial, cytokine therapy drugs have been used as first-line treatment strategy (Gupta et al, 2008; Rini et al, 2009). The development of targeted therapies, especially agents directed to the VEGF pathway, has recently revolutionised the treatment of mRCC, leading to a median overall survival increasing from 13 to 15 to over 25 months. Since 2006, the US Food and Drug Administration and the European Medicines Agency have approved the use of seven targeted therapies for mRCC, five of them being directed to VEGF or its receptors (sunitinib, sorafenib, pazopanib, axitinib, bevacizumab) and two corresponding to inhibitors of the mTOR complex (temsirolimus and everolimus). Numerous clinical trials have been undertaken to understand their effects on renal cell cancer (Patard et al, 2008; Rini et al, 2008; van der Veldt et al, 2010; Rini and Powles, 2013). Several types of targeted therapies acting on different pathways have been proposed (Brekke and Sandlie, 2003; Gerber, 2008; Zhang et al, 2009), and their combination could lead to a synergy promoting better response rate. However, a few clinical trials have demonstrated that the combination of targeted therapies may cause higher toxicity, eventually leading to early termination of phase I trials (Feldman et al, 2009; Patel et al, 2009). The combination of mTOR inhibitors (temsirolimus or everolimus) with bevacizumab in phase I and II has yet shown encouraging results with good responses rates (Zafar et al, 2006; Merchan et al, 2015; Whorf et al, 2008).
In this context, the TORAVA trial was set up to evaluate the effectiveness of combining bevacizumab and temsirolimus as first-line treatment of mRCC, as compared with two other standard therapies based on sunitinib treatment, or a combination of interferon-α and bevacizumab. The results of this trial showed an absence of positive outcome, as the combination of these treatments was responsible for a higher toxicity, and the response as well as the progression-free survival rates were not improved (Négrier et al, 2011). During the TORAVA trial, several translational studies have been set up to highlight predictive markers of the response and clinical outcome (i.e., toxicity).
An increasing number of metabolomic studies in oncology, carried out on diverse types of biological samples (tumour cells, blood serum and so on) aim at highlighting biomarkers to distinguish various cancerous states (Oakman et al, 2011; Jobard et al, 2014), biomarkers of treatment response and toxicity to probe the molecular action of drugs, as well as resistance phenomena (Lodi and Ronen, 2011; Tenori et al, 2012; Weaver et al, 2012; Borgan et al, 2013; Wei et al, 2013).
As concerns RCC, metabolomics has been primarily used for diagnosis (RCC vs healthy subjects) and classification using tumour tissues (Tate et al, 2000), mainly investigating biological fluids such as urine (Perroud et al, 2006; Kind et al, 2007; Kim et al, 2009; Huang et al, 2013), serum (Gao et al, 2008; Zira et al, 2010) or plasma (Lin et al, 2011).
We report here a 1H nuclear magnetic resonance (NMR)-based metabolomic study that investigates the effects of combining two targeted therapies (bevacizumab and temsirolimus) on the serum metabolic profiles of patients as mRCC first-line treatment, in comparison with those receiving the two standard therapies. The objective of this translational study is to explore the action of targeted agents on the metabolism of the host, and specifically the consequence of temsirolimus on the serum metabolic profiles of mRCC patients.
Patients and methods
Patients characteristics
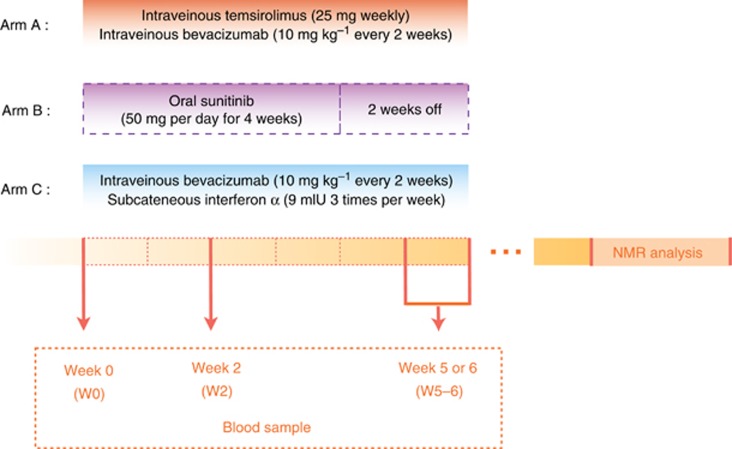
From March 2008 to May 2009, 171 patients with untreated mRCC were enroled in the TORAVA trial. The trial aimed at determining the efficiency and safety of temsirolimus and bevacizumab combination as first-line treatment of mRCC and studying the markers of prognosis. Patients were randomised (ratio 2:1:1) between three different arms: arm A for the combination of bevacizumab (10 mg kg−1 every 2 weeks) and temsirolimus (25 mg weekly); arm B for sunitinib (50 mg per day for 4 weeks followed by 2 weeks off); and arm C for interferon-α (9 mIU three times per week) and bevacizumab (10 mg kg−1 every 2 weeks). The study design is described in details in Figure 1. The arm A was the experimental arm and arm B and C were two standard first-line treatment of mRCC. The study design, patient recruitment, and data-collection methods of the TORAVA trial were described more precisely by Négrier et al (2011). The local ethics committee approved the research protocol. Written informed consent was obtained from each patient before enrolment.
Figure 1.
Study Design of the TORAVA trial. Patients with untreated mRCC were randomised using a 2:1:1 ratio: arm A was administered a combination of bevacizumab and temsirolimus; arm B was treated with sunitinib; arm C received a combination of interferon-α and bevacizumab. Arm A is the experimental arm and the two others arms (B and C) are standard first-line treatments of mRCC. Blood samples were collected at three different times: at baseline (W0), that is, before the first therapy cure; 2 weeks after the start of treatment (W2); and 5–6 weeks after beginning of treatment (W5–6). NMR analyses were performed after completion of the clinical trial.
For each patient, clinicopathological data were recorded including age, gender, BMI, ECOG performance status (PS) (Oken et al, 1982), Memorial Sloan-Kettering Cancer Center (MSKCC) classification (Motzer et al, 2002), evaluation of the response to therapy, tumour type, Fuhrman classification (Fuhrman et al, 1982), and interval between diagnostic and metastasis. Tumour assessment (thoracic, abdominal, and pelvic CT scan, brain MRI or CT scan, and bone scan) was performed at inclusion and every 12 weeks until disease progression. From these data, the best objective response to treatment (complete or partial response) was determined according to the RECIST criteria (Therasse et al, 2000). To characterise the three arms, a descriptive statistical analysis was performed using the analysis of variance for mean and χ2 or Fischer exact test for qualitative data. The significance threshold was set to 0.05 for all tests. Clinico-pathological parameters were compared in order to exclude biases related to patient selection, as summarised in Table 1. No significant differences were observed for all these parameters between the three different arms of the study.
Table 1. Summary of clinicopathological characteristics of TORAVA trial patients.
| Characteristics | Arm A | Arm B | Arm C | P-valuea |
|---|---|---|---|---|
| No. of subjects | 64 (52.9%) | 27 (22.3%) | 30(24.6%) | |
| Age (mean/s.d.) | 59.5±10.7 | 60.5±11.2 | 59.4±8.8 | 0.90 |
| Samples | 0.97 | |||
| W0 | 56 (50%) | 26 (23.2%) | 30 (26.8%) | |
| W2 | 55 (53.9%) | 22 (21.6%) | 25 (24.5%) | |
| W5–6 | 49 (53.8%) | 20 (22%) | 22 (24.2%) | |
| Gender | 0.4 | |||
| Female | 15 (23.4%) | 7 (25.9%) | 11 (36.7%) | |
| Male | 49 (76.6%) | 20 (74.1%) | 19 (63.3%) | |
| Body mass index | 0.34 | |||
| ⩽ 25 | 31 (48.4%) | 15 (55.6%) | 11 (33.3%) | |
| > 25 | 33 (51.6%) | 12 (44.4%) | 19 (63.3%) | |
| ECOG performance status | 0.60 | |||
| 0 or 1 | 60 (93.8%) | 27 (100%) | 29 (96.7%) | |
| 2 | 4 (6.3%) | 0 (0%) | 1 (3.3%) | |
| MSKCC classification | 0.62 | |||
| Unknown | 5 (7.8%) | 2 (7.4%) | 4 (13.3%) | |
| Poor prognosis | 9 (14.1%) | 1 (3.7%) | 4 (13.3%) | |
| Intermediate prognosis | 32 (50%) | 14 (51.9%) | 11 (36.7%) | |
| Favourable prognosis | 18 (28.1%) | 10 (37%) | 11 (36.7%) | |
| Response to treatmentb | 0.94 | |||
| Objective response | 17 (26.6%) | 7 (28.9%) | 9 (30%) | |
| No objective response | 46 (71.9%) | 20 (74.1%) | 20 (66.7%) | |
| No information | 1 (1.6%) | 0 (0%) | 1 (3.3%) | |
| Tumour type | 0.64 | |||
| Conventional renal cell carcinoma (RCC) | 61 (95.3%) | 26 (96.3%) | 30 (100%) | |
| Collecting duct carcinoma | 0 (0%) | 1 (3.7%) | 0 (0%) | |
| Chromophobe RCC | 2 (3.1%) | 0 (0%) | 0 (0%) | |
| Unclassifiable RCC | 1 (1.6%) | 0 (0%) | 0 (0%) | |
| Fuhrman classification | 0.07 | |||
| Grade I and II | 13 (20.3%) | 7 (25.9%) | 12 (40%) | |
| Grade III and IV | 37 (57.8%) | 19 (70.4%) | 15 (50%) | |
| Unknown | 14 (21.9%) | 1 (3.7%) | 3 (10%) | |
| Interval between diagnostic and metastasis | 0.54 | |||
| ⩽12 months | 41 (64.1%) | 16 (59.3%) | 17 (56.7%) | |
| >12 months | 23 (35.9%) | 10 (37%) | 13 (43.3%) | |
| Unknown | 0 (0%) | 1 (3.7%) | 0 (0%) |
Abbreviations: ECOG=Eastern Cooperative Oncology Group; RCC=renal cell carcinoma. The outliers are not included in this table.
P-value calculated using either the χ2 and Fisher exact tests for the proportion or an ANOVA analysis for mean.
Objective response corresponds to at least one objective response at one of the three disease assessment points (week 12, 24, and 48).
Sample collection and preparation
Three hundred and twenty-one serum samples from 121 patients with mRCC were obtained for our metabolomic study. A series of venous blood samples was collected for each patient during the TORAVA trial: one before the first therapy administration (W0), and two on-treatment at two weeks (W2) and 5–6 weeks after the beginning of treatment (W5–6). For the experimental arm, we accessed to 171 samples (W0: 63, W2: 57, W5–6: 51) from 64 patients (52.9% of the total number of patients). For the two other treatments, arm B and arm C, we obtained, respectively, 71 samples (W0: 27, W2: 24, W5–6: 20) from 27 patients (22.3%) and 79 samples (W0: 30, W2: 26, W5–6: 23) from 30 patients (24.6%) (Figure 1). The total number of samples available for any given patient in the trial varied. The reasons for incomplete sample collection were multiple, including inadequate quantity and/or quality of collected serum samples, withdrawal of patients in the trial (e.g., owing to high toxicity of treatment, protocol violation, or request from the patient to stop the trial), or death of patients. Blood samples were recovered from dry tubes (10 ml) and centrifuged after 30 min of sedimentation at 800 g for 10 min. After centrifugation, the supernatant was collected and aliquoted in three cryotubes (1 ml). Cryotubes were stored at −80 °C after collection. For NMR analysis, sera were prepared according to the protocol described by Beckonert et al (2007). Serum samples were thawed at room temperature before use. Then, 200 μl of each was diluted with 400 μl of a 0.9% saline solution (NaCl 0.9% wt/vol, D2O 10% vol/vol) in a microtube, then centrifuged for 5 min at 4 °C at 12 000 g. At last, 550 μl of supernatant was transferred into 5 mm NMR tubes. Samples were kept for <24 h at 4 °C until NMR analysis.
NMR spectroscopy
All NMR spectra were recorded on a Bruker Avance III spectrometer operating at 800.14 MHz for proton, equipped with a 5 mm TXI probe, and automatic sample changer with cooling capacity (4 °C). The temperature was then regulated at 27 °C throughout the NMR experiments. NMR spectral acquisitions for the 321 TORAVA samples were divided randomly into two distinct NMR sessions. One hundred and seventy-five serum samples (W0: 63; W2: 60; W5–6: 52) were analysed in the first batch and 146 for the second batch (W0: 57; W2: 46; W5–6: 43), 2 months apart. For a given patient, samples from the different collection points were randomly distributed between the two NMR sessions. For each session, automatic 3D shimming was performed once on a test serum sample. A test serum sample is a serum sample chosen at random in the cohort with enough volume to prepare an extra tube for NMR calibration purposes. Prior to NMR data acquisition, automatic tuning, and matching, frequency locking on D2O and 1D automatic gradient shimming was performed on each sample. Standard 1H 1D NMR NOESY pulse sequence with water presaturation was applied on each sample to obtain the corresponding metabolic profile. A total of 128 transient free induction decays (FID) were collected for each experiment into 43 588 points over a spectral width of 20 ppm. The acquisition time was set to 1.36 s with a relaxation delay of 2 s. The 90° pulse length was automatically calibrated for each sample at around 10.9 μs. The NOESY mixing time was set to 100 ms. All FIDs were multiplied by an exponential weighting function corresponding to a 0.3 Hz line broadening factor, before Fourier transformation.
All spectra were referenced to the α-glucose anomeric proton signal (δ=5.23 ppm). 1H-NMR spectra were phased and baseline corrected using Topspin 3.1 (Bruker GmbH, Rheinstetten, Germany). After importing all 1D spectra into the AMIX software (Bruker GmbH), spectra were divided into 0.001 ppm-wide buckets to obtain 8500 buckets over the chemical range of 0.5–9 ppm. Residual water signal (4.66–5.05 ppm region) was excluded, and no further normalisation was applied to the spectra. Prior to statistical analyses, 9 out of 321 serum samples in the translational study were excluded because of poor spectral quality or improper collection of the blood in a citrated tube, as detected by NMR.
In addition, 1D 1H-NMR CPMG and 2D NMR experiments (1H–13C HSQC, 1H–1H TOCSY and 1H J-resolved experiments) were recorded on a subset of samples to achieve structural assignment of the metabolite signals. The procedure for metabolite identification exploited knowledge from academic spectral databases such as MMCD (Cui et al, 2008), HMDB (Wishart, 2007), and BMRB (Ulrich et al, 2008) as well as proprietary databases (NMR Suite v. 7.1, Chenomx Inc., Edmonton, Canada; AMIX SpectraBase v. 1.1.2, Bruker GmbH). From analysis of 1D and 2D NMR data, identification of full spin systems allowed unambiguous annotation of 51 metabolites. Corresponding assignments are provided in Supplementary Table 1, and illustrated in Supplementary Figure 1.
Statistics
To build models for sample classification and extract group-specific metabolic signatures, unsupervised and supervised statistical multivariate analyses were conducted using SIMCA-P 13 (Umetrics, Umea, Sweden). Multivariate models were visualised using scores and loadings plots. In a score plot, each point represents a NMR spectrum (i.e., a sample) on the main principal components, whereas the corresponding loadings plot displays the contribution of the NMR variables to the principal components.
An initial principal component analysis (PCA) was carried out to derive the main sources of variance and eventually identify potential outliers from the 1D 1H-NMR data sets (Wold et al, 1987). PCA detected seven serum samples as severe outliers (mainly owing to high concentrations of lipids) that were excluded from further analysis. The final sample set comprised a total of 305 samples.
Orthogonal partial least-squares (O-PLS) analyses were performed to discriminate serum profiles associated with sampling time for each arm by exploiting a supplementary data matrix Y, containing samples class membership (e.g., W0, W2, W5–6 for sampling time) (Trygg and Wold, 2002). The goodness-of-fit parameters R2 and Q2, which relate to the explained and predicted variance, respectively, were used to evaluate the O-PLS model performance. For each O-PLS model, a model validation in MATLAB (The MathWorks Inc., Natick, NA, USA), using homemade O-PLS routines, was carried out by resampling the model 1000 times under the null hypothesis through random permutations of the Y matrix. The decrease in goodness-of-fit R2 and Q2 parameters, when correlation between original model and random models decreased, indicated the good quality of our models. The statistical significance of the calculated model was also assessed by Cross-Validation ANOVA (CV-ANOVA) for each O-PLS model (Eriksson et al, 2008).
In addition, to derive statistically significant associations of individual metabolites, an univariate methodology previously described that couples an automatic binning procedure named statistical recoupling of variables to subsequent ANOVA analysis (Blaise et al, 2009) was used, implemented with MATLAB homemade routines.
Results
Longitudinal discrimination of metabolic profiles in the TORAVA trial
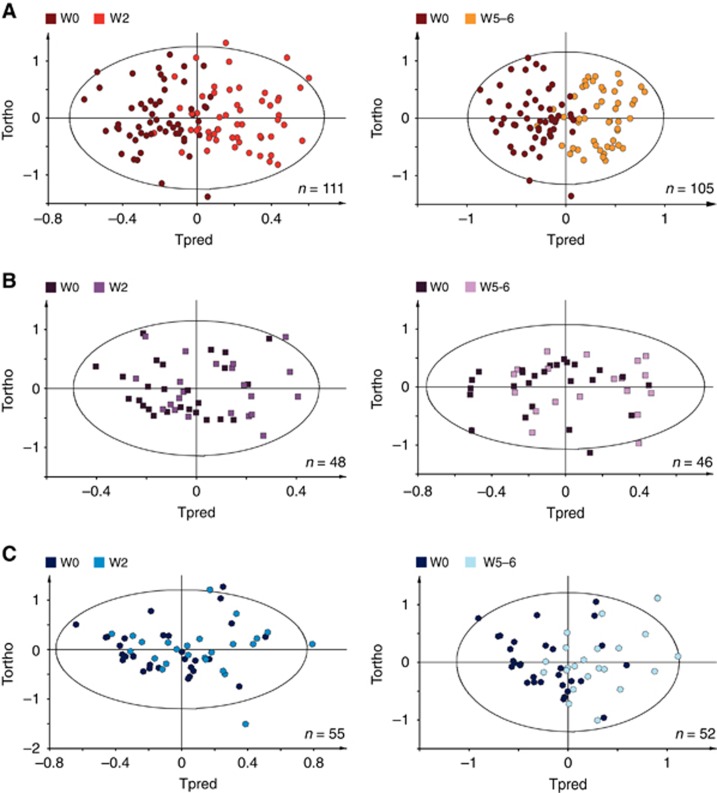
Serum NMR metabolic profiles from mRCC patients were first analysed within each arm of the TORAVA study to probe the specific metabolic response associated with the experimental (A) and reference (B and C) treatments. Supervised multivariate models (O-PLS) were built for the three arms to derive robust statistical models discriminating metabolic serum profiles between W0 and W2 and between W0 and W5–6 (Figure 2).
Figure 2.
Discrimination between pre-treatment and on-treatment serum samples. (A) O-PLS models for arm A, discriminating samples at W0 vs W2 (1+6 components, R2X=0.985, R2Y=0.581, Q2=0.376, CV-ANOVA P-value=1.32 × 10−5) and W0 vs W5–6 (1+6 components, R2X=0.985, R2Y=0.65, Q2=0.462, CV-ANOVA P-value=1.61 × 10−7). (B) O-PLS models for arm B, discriminating samples at W0 vs W2 (1+1 components, R2X=0.816, R2Y=0.147, Q2=−0.188, CV-ANOVA P-value=1) and W0 vs W5–6 (1+1 components, R2X=0.847, R2Y=0.149, Q2=0.058, CV-ANOVA P-value=0.64). (C) O-PLS models for arm C, discriminating samples at W0 vs W2 (1+1 component, R2X=0.919, R2Y=0.124, Q2=0.055, e CV-ANOVA P-value=0.57) and W0 vs W5–6 (1+1 components, R2X=0.935, R2Y=0.319, Q2=0.201, CV-ANOVA P-value=0.029).
For the experimental arm A, a clear discrimination between W0 and W2 (R2X=0.985, R2Y=0.581, Q2=0.376, CV-ANOVA P-value=1.32 × 10−5), and between W0 and W5–6 (R2X=0.985, R2Y=0.65, Q2=0.462, CV-ANOVA P-value=1.61 × 10−7) of the serum metabolic profiles was observed, as illustrated in Figure 2A. Statistical significance for these two models was assessed by high values of goodness-of-fit parameters R2 and Q2, CV-ANOVA P-values<0.05, and model resampling under the null hypothesis (Supplementary Figure 2a & b). Regarding arm B, no significant discrimination was obtained from serum metabolic profiles between W0 and W2, or between W0 and W5–6 (Figure 2B). Finally, for arm C, multivariate modelling of the metabolic profiles between W0 and W5–6 only provided a weak but robust discrimination (R2X=0.935, R2Y=0.319, Q2=0.201, CV-ANOVA P-value=0.029, Figure 2C, Supplementary Figure 2c).
To ensure that the lack of separation between W0 and W2 for arm C was not due to an insufficient number of samples for arm C as compared with arm A that included twice as many patients, a sensitivity analysis was carried out using 1000 O-PLS models calculated from randomly selected subgroups (n=56; 28 samples per class) of metabolic profiles from arm A (Supplementary Figure 3). The distribution of R2 and Q2 obtained for these models shows that even when considering half as many samples as compared with arm C, higher values of R2Y and Q2 were obtained for arm A, confirming an earlier and stronger modification of the host metabolism for the experimental arm.
Metabolic fingerprints associated with targeted therapies for mRCC treatment
Thereafter, individual metabolites significantly associated with the discrimination of pre-treatment and on-treatment samples, respectively, for arm A and C, were identified using univariate analysis of the serum NMR metabolic profiles to understand the metabolic processes involved in the host response to mRCC targeted therapies.
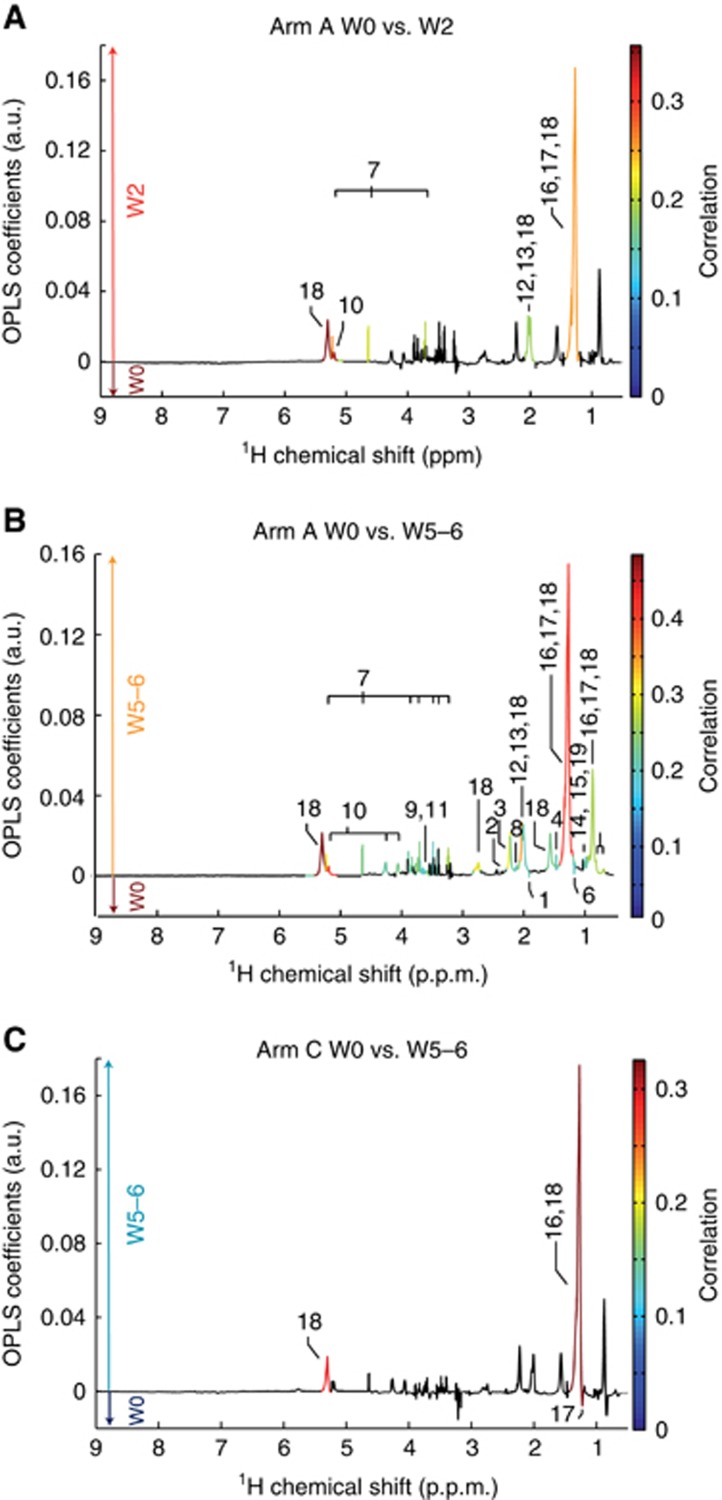
For the experimental arm A, lipids (glycerol backbone of phosphoglycerides and triacylglycerides fatty acids), lipoproteins mainly very low-density lipoproteins and low-density lipoproteins (LDL) as well as glucose and N-acetylglycoproteins (NAC1 & 2) were found in significantly higher concentrations in the serum at W2 as compared with baseline (P<0.05) (Figure 3A). Corresponding fold-changes and P-values are reported in Table 2. After 5–6 weeks of treatment, the same metabolic signature as observed after 2 weeks is obtained for arm A (Figure 3B). However, a larger number of metabolites reach individual statistical significance. Thus, in addition to the metabolites previously identified as statistically significant after 2 weeks of treatment (lipids, lipoproteins (VLDL, LDL), glucose and N-acetylglycoproteins), an increase of the levels of branched chain amino acids (BCAA; isoleucine, leucine, and valine), alanine, glycine, and glutamine, as well as end-products of β-oxidation (acetoacetate, acetone), lipid degradation (glycerol) and cholesterol was observed at W5–6, whereas acetate and ethanol were more concentrated in serum metabolic profiles at baseline. The differences in concentrations of acetoacetate, acetone, glucose, lipids, and lipoproteins remain significant after Benjamini–Hochberg correction for multiple testing. Our results show that several metabolites associated with the combined bevacizumab and temsirolimus treatment are already detected at W2 with a weak discriminative power and subsequently provide a stronger discrimination from baseline metabolic profiles after 5–6 weeks of drug intervention.
Figure 3.
Metabolic fingerprints associated with mRCC targeted therapies. O-PLS loadings plots are represented for arm A: (A) W0 vs W2, and (B) W0 vs W5–6, and for arm C: (C) W0 vs W5–6. Statistically significant individual signals correspond to the coloured spectral regions. Highlighted candidate markers are: (1) acetate, (2) acetoacetate, (3) acetone, (4) alanine, (5) cholesterol (C18, C26, C27), (6) ethanol, (7) glucose, (8) glutamine, (9) glycerol, (10) glycerol backbone of pglys and tags, (11) glycine, (12) N-acetylglycoprotein (NAC1), (13) NAC2, (14) isoleucine, (15) leucine, (16) fatty acids (mainly LDL), (17) fatty acids (mainly VLDL), (18) fatty acids and (19) valine. LDL=low-density lipoprotein; VLDL=very low-density lipoprotein; PGLYs=phosphoglycerides; TAGs=triglycerides.
Table 2. Metabolites identified as significant for arm A and arm C.
|
Arm A |
Arm C |
|||||||
|---|---|---|---|---|---|---|---|---|
|
W0
vs
W2 |
W0
vs
W5–6 |
W0
vs
W5–6 |
||||||
| ID | Name | P-valuea | Fold Change | P-valuea | Fold Change | Variation | P-valuea | Variation |
| 1 | Acetate | 0.7 | / | 0.033 | / | − | 0.6 | − |
| 2 | Acetoacetate | 0.58 | / | 0.0013b | 1.18 | + | 0.69 | + |
| 3 | Acetone | 0.23 | / | 0.0013b | 1.18 | + | 0.23 | + |
| 4 | Alanine | 0.69 | / | 0.023 | / | + | 0.95 | + |
| 5 | Cholesterol | 0.98 | / | 0.006 | / | + | 0.47 | + |
| 6 | Ethanol | 0.73 | / | 0.044 | / | - | 0.9 | - |
| 7 | Glucose | 0.02 | / | 0.022b | 1.12 | + | 0.27 | + |
| 8 | Glutamine | 0.47 | / | 0.039b | 1.11 | + | 0.95 | + |
| 9 | Glycerol | 0.3 | / | 0.029 | / | + | 0.42 | + |
| 10 | Glycerol backbone of PGLYs and TAGs | 0.0003b | 1.22 | 0.018b | 1.15 | + | 0.07 | + |
| 11 | Glycine | 0.36 | / | 0.017 | / | + | 0.60 | + |
| 12 | NAC1 | 0.047 | / | 0.0003 | / | + | 0.25 | + |
| 13 | NAC2 | 0.047 | / | 0.014 | / | + | 0.19 | + |
| 14 | Isoleucine | 0.65 | / | 0.019 | / | + | 0.78 | + |
| 15 | Leucine | 0.11 | / | 0.012 | / | + | 0.88 | + |
| 16 | Fatty acids (mainly LDL) | 0.0075 | / | 0.003b | 1.24 | + | 0.018 | - |
| 17 | Fatty acids (mainly VLDL) | 0.0075 | / | 0.009b | 1.19 | + | 0.018 | + |
| 18 | Fatty acids | 0.0001b | 1.17 | 0.0000002b | 1.29 | + | 0.018 | + |
| 19 | Valine | 0.71 | / | 0.045 | / | + | 0.67 | + |
Abbreviations: LDL=low-density lipoprotein; NAC=N-acetylglycoprotein; PGLYs=Phosphoglycerides; TAGs=Triacylglycerides; VLDL=very low-density lipoprotein. Variation:+corresponds to higher concentration in W5–6 serum metabolic profiles than at baseline;–to lower concentration in W5–6 serum metabolic profiles than at baseline.
P-value without multiple testing correction.
Significant after Benjamini–Hochberg false discovery rate multiple testing correction (q-value <0.05).
The metabolic signature associated with the bevacizumab and interferon-α combination (arm C) at W5–6 is mainly due to an increase of lipids and VLDL lipids but also to the relative decrease of the LDL lipids in serum samples (Figure 3C). Nevertheless, the observed variations for these metabolites do not remain significant after correction for multiple testing (Table 2). Thus, the discriminatory power of individual metabolite is weak and should be further confirmed independently.
Further comparison of the obtained metabolic signatures for arm A and C shows that changes in the metabolic profiles after several weeks of therapy for both arms are quite similar and mainly due to alterations of lipids and carbohydrate metabolism.
To ensure that observed discrimination is due solely to the presence of the treatment, the influence of tumour's characteristics (PS, MSKCC classification, tumour type, Fuhrman classification and interval between diagnostic and metastasis) was analysed for each arm over time. No significant differences were observed for these parameters (Supplementary Table 2), which shows that the impact of tumour characteristics on the serum metabolome of mRCC patients is negligible.
Metabolic changes between the experimental arm and two standard therapies
To compare the metabolic fingerprint of the experimental arm with those of the two standard arms at any given time in the study, further supervised analyses were carried out independently on the serum NMR profiles recorded for the three sampling times (Supplementary Table 3).
As anticipated, no discrimination was observed between the experimental arm and the two other therapies when comparing serum samples collected at baseline (W0), before initiation of the treatments. This lack of separation at inclusion between the three arms emphasises the similarity of the three groups of patients at baseline and confirms the absence of selection biases. After 2 weeks of treatment, a separation trend was observed between arms but is not statistically significant. At W5–6, a weak yet significant discrimination between arm A and B was obtained, validated by a CV-ANOVA P-value of 0.038 and O-PLS model resampling with 1000 iterations (data not shown). Corresponding O-PLS model comparing arms A and C at W5–6 is not validated by the CV-ANOVA P-value and resampling procedure. Similarly, the metabolic profiles of the two standard arms B and C were compared at each sampling time (Supplementary Table 3), and no discrimination was observed, either at baseline or after two and 5–6 weeks of treatment.
Through the univariate analysis used on metabolic serum profiles, statistically significant metabolites for the discrimination of arms A and B after 5–6 weeks of treatment have been identified (Supplementary Figure 4). Lipids, lipoproteins (VLDL and LDL), cholesterol, glucose, the end-products of β-oxidation (acetone, acetoacetate and 3-hydroxybutyrate), N-acetylglycoproteins (NAC1 & 2), BCAA (Valine, Leucine, Isoleucine), alanine and acetate were found in lower concentrations in the serum of patients given sunitinib (arm B) as compared with subjects treated with the experimental arm. None of these differences was yet validated after Benjamini–Hochberg correction for multiple testing. However, the metabolic pattern is very similar to the one established for the longitudinal evolution within arm A (W0 vs W2 or W5–6) in the previous section (Figure 3A and B). This confirms the similarity of the serum metabolic profiles from patients in arm B, at any given time point in the study, with profiles recorded at baseline.
Discussion
During the TORAVA trial, continued treatment over time was limited by the toxicity of the Bevacizumab and Temsirolimus combination that was higher than expected. Thus, the evaluation of this new combination of targeted therapies as first-line treatment for mRCC has failed to demonstrate any benefits for the patients (Négrier et al, 2011).
For several years, a number of studies based on NMR or MS metabolomics approached have focused on the identification of biomarkers of renal cell cancer from biological fluids mainly with urine samples (Perroud et al, 2006; Kind et al, 2007; Kim et al, 2009; Huang et al, 2013) or blood samples (serum/plasma) (Süllentrop et al, 2002; Gao et al, 2008; Zira et al, 2010; Lin et al, 2011). According to these studies, lipoproteins and choline derivatives are major biomarkers correlated with renal cell cancer (Duarte and Gil, 2012). However, no study has focused on the effect of targeted therapy on the metabolic profiles of mRCC patients. The present analysis evaluates the global impact of targeted therapies on the metabolism of these patients.
Our results highlight a significant metabolic signature associated with the experimental combination of bevacizumab and temsirolimus, together with an earlier modification of the metabolism than for patients treated with the two standard therapies. Unlike for the two standard treatment groups, significant metabolites (glucose, N-acetylglycoproteins, lipids, lipoproteins (LDL and VLDL)) were identified in the experimental arm, in which the lipids have only a high discriminatory power, after 2 weeks of therapy. After 5–6 weeks of treatment, a large number of significant metabolites were identified in the experimental arm and some metabolites showed strong discriminatory power. On-treatment samples display higher concentrations of acetoacetate, acetone, alanine, cholesterol, glucose, glutamine, glycerol, glycine, N-acetylglycoproteins, isoleucine, leucine, lipids, LDL and VLDL lipoproteins and valine, and lower concentration of ethanol and acetate, when compared with pre-treatment specimens.
Side effects associated with bevacizumab exposure on the global metabolism have been little described so far in the literature, whereas temsirolimus treatment was already shown to induce hyperglycaemia and hyperlipidemia (hypercholesteremia and hypertriglyceridemia) in RCC patients (Bellmunt et al, 2008). In our study, the available clinical data document at least one of the side effects mentioned above for 59% of the patients in arm A, and the metabolic signature highlighted in this work seems to mainly reflect the effects of mTOR inhibition by the temsirolimus treatment. Indeed, these side effects of temsirolimus correspond to metabolites with the highest discriminatory power in our metabolic signature between samples before and after treatment, that is, lipids, LDL, and VLDL lipoproteins, which allow the transport of cholesterol and endogenous lipids, respectively, end-products of β-oxidation (acetoacetate and acetone), glucose, and glutamine (Table 2). The surplus of lipids due to hyperlipidemia correlates with an excess of ketone bodies (acetoacetate and acetone), which are the end-products of lipid metabolism in the blood (Berg et al, 2002). Likewise, hypercholesterolaemia and hyperlipidemia increase the need of LDL and VLDL for cholesterol and endogenous lipids transport. In addition, the hypertriglyceridemia and hyperglycaemia result in high levels of glucose and glutamine in the serum.
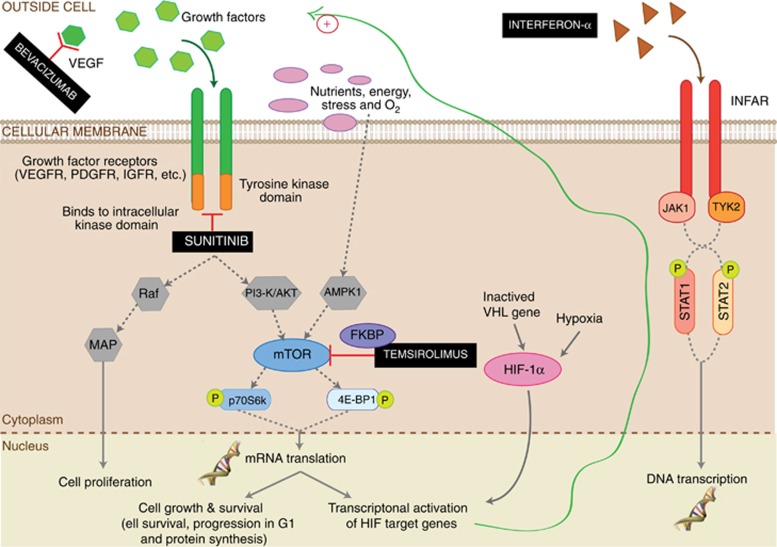
Metabolic abnormalities are to be expected with administration of temsirolimus as the mammalian target of rapamycin (mTOR) regulates a number of metabolic processes including glucose and lipid metabolism (Advani, 2010; Alayev and Holz, 2013). Indeed, mTOR is a central regulator of intracellular pathways involved in tumour cell growth, proliferation, and response to hypoxic stress (Wullschleger et al, 2006; Bellmunt et al, 2008; Alayev and Holz, 2013). As illustrated in Figure 4, Temsirolimus inhibits downstream mTOR signalling by binding to an intracellular protein FKBP-12. The resulting complex arrests the growth of tumour cells and also inhibits angiogenesis (Rini and Atkins, 2009; Advani, 2010; Alayev and Holz, 2013). Meanwhile, bevacizumab, a therapeutic antibody, is designed to directly bind to extracellular VEGF to prevent interaction with VEGF receptor on the surface of endothelial cells, and thereby inhibits VEGF's angiogenic activity, reducing cell growth and metastasis (Hicklin, 2004) (Figure 4).
Figure 4.
Schematic mechanisms of action for drugs involved in the three arms of the TORAVA trial. Bevacizumab, a recombinant humanised monoclonal antibody, binds to human vascular endothelial growth factor (VEGF) and prevents the interaction of VEGF to its receptors on the surface of endothelial cells. Cancer cells tend to overexpress VEGF, which stimulates angiogenesis, facilitating cancer growth and metastasis. Bevacizumab suppresses angiogenesis by inhibition of molecular pathways of MAP and PI3K/AKT. Temsirolimus, an ester analogue of rapamycin, is an inhibitor of mTOR (mammalian target of rapamycin) that binds to intracellular FKBP protein, and the protein–drug complex inhibit the activity of mTOR. mTOR is a crucial regulator of cell growth and proliferation through its ability to integrate signals from nutrients and growth factor stimuli. Inhibition of mTOR activity results in a G1 growth arrest in treated tumour cells. When mTOR is inhibited, its ability to phosphorylate p70S6 kinase (p70S6k) and 4E binding protein-1 (4E-BP1), which are downstream of mTOR in the PI3 kinase/AKT pathway, is blocked. Interferon-α-2a, a sterile protein product for use by injection, binds to type I interferon receptors (IFNAR1 and IFNAR2c), which upon dimerisation activates two JAKs (Janus kinases): JAK1 and TYK2. These transphosphorylate themselves and phosphorylate the receptors. The phosphorylated INFAR receptor then binds to STAT1 and STAT2 (signal transducers and activators of transcription), which dimerise and activate multiple immunomodulatory and antiviral proteins. Sunitinib is an oral, smallmolecule, multi-targeted inhibitor of receptor tyrosine kinases (e.g., platelet-derived growth factor receptors (PDGFR) and vascular endothelial growth factor receptors (VEGFR)), some of which are implicated in tumour growth, pathologic angiogenesis, and metastatic progression of cancer. PI3K: Phosphatidylinositol-4,5-biphosphate-3-kinase; MAP: Mitogen-activating protein kinase 1; AMPK1: AMP-activated kinase-1; Raf: Raf protein kinase; AKT: Anaplastic Lymphoma Kinase; HIF-1α: hypoxia-inducible factor (HIF)-α; VHL gene: von Hippel–Lindau gene.
Similarly, a metabolic signature for the arm C is obtained after several weeks of treatment (5–6 weeks) with interferon-α and bevacizumab combination. It is characterised mainly by a change in lipid concentrations (lipids, LDL, and VLDL lipids) between the two groups. Metabolites identified for this therapy are consistent with side effect that can result from taking interferon-α, mainly hypertriglyceridemia (Sleijfer et al, 2005; Hauschild et al, 2008). Interferon-α, which belongs to a group of cytokines, does not directly kill cancerous cells. Indeed, it boosts the immune system response and reduces growth of cancer cells by regulating the action of several genes that control the secretion of numerous cellular proteins that affect growth (Platanias, 2005) (Figure 4). The broadness of its biological activity often leads to metabolic abnormalities such as alterations of serum lipids and lipoproteins. Thus, it may contribute to hypertriglyceridemia by the increase in lipogenesis and VLDL secretion in the liver (Shinohara et al, 1997).
Based on comparison of the metabolic profiles for the three arms of the TORAVA trial, a significant discrimination is only observed between the experimental arm and arm B (sunitinib) after several weeks of treatment. Interestingly, these two therapies are very different regarding the type of molecules used and their action mechanisms, as illustrated in Figure 4, which can justify this discrimination. As bevacizumab, sunitinib, a small molecule that inhibits the tyrosine kinases of various receptors, is an angiogenesis inhibitor and because of its nature is not associated with metabolic side effects (Coppin, 2008). The observed metabolic fingerprint here is mainly due to the presence of temsirolimus in the experimental arm. Meanwhile, we do not observe any significant separation between the experimental arm (bevacizumab and temsirolimus) and arm C (bevacizumab and interferon-α). As discussed above, the longitudinal metabolic signatures of these two treatment combinations were quite similar after several weeks of treatment and characterised mainly by lipids and lipoproteins. These metabolite variations associated with hyperlipidemia are related to the respective presence of interferon-α (arm C) and temsirolimus (arm A). The lack of separation between the two combinations is therefore not due to the common presence of bevacizumab in the A and C treatments, but rather to the respective presence of temsirolimus and interferon-α that coincidentally produce similar side effects. The absence of discrimination between the two standard arms (B and C) reflects a similar evolution of the metabolic profiles for arms B and C over time (Supplementary Table 3), which confirms the absence of a strong effect associated with VEGF inhibitor treatment in arm C.
We note that access to patients treated with a single drug could ensure that the observed side effects are not associated with synergetic effects from treatment combinations. Unfortunately, these subgroups were not included in the TORAVA clinical trial design. Investigation of additional controls, including treated healthy patients, or individual with untreated mRCC could also provide complementary assessment of our findings. Yet, these type of data are unlikely accessible within a clinical setting, and would rather require in vitro evaluation on a model system.
We also note that most of the observed metabolites, which are mainly associated with side effects of the treatments on the host metabolism, have been identified in previous metabolomic studies describing RCC effects on the metabolism (Gao et al, 2008; Zira et al, 2010). Furthermore, abnormalities in mTOR signalling pathway have been implicated in various pathologies including renal cancer (Wullschleger et al, 2006; Advani, 2010). Temsirolimus-related side effects may probably exacerbate existing metabolic abnormalities common in patient with RCC (Bellmunt et al, 2008). Finally, as metabolic fingerprints obtained in this study are mainly associated to metabolic side effects on the host, the potential to exploit them as treatment response predictors for mRCC seems altogether limited. The analysis of peripheral blood provides a snapshot of the patients overall physiological state that integrates the metabolic composition of several tissues and organs. Here, we focused on the complex interaction between the host and the tumour as well as the global impact of the treatments, which notably reflect direct effects of the drugs on the liver, rather than the tumour metabolism directly.
Our work proposes a metabolic signature for the temsirolimus and bevacizumab combination treatment for patient with mRCC in the first-line setting. Our findings highlight the potential of metabolomic approaches to study treatment effects that are, to better understand their mechanisms of action, predict associated metabolic side effects or toxicity, and potentially predict the clinical response. Bevacizumab and temsirolimus combination caused a strong and rapid modification of the patients metabolism as compared with two conventional treatments. The observed metabolic pattern is conceivably the result of metabolic side effects related to the imbibition of the mTOR protein complex (mTORC1). These results have to be confirmed in larger cohorts with other types of cancer to better understand the effect of mTORC1 signalling inhibitors on the human metabolism.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Advani SH (2010) Targeting mTOR pathway: a new concept in cancer therapy. Indian J Med Paediatr Oncol 31: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alayev A, Holz MK (2013) mTOR signaling for biological control and cancer. J Cell Physiol 228: 1658–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TMD, Bundy JG, Holmes E, Lindon JC, Nicholson JK (2007) Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2: 2692–2703. [DOI] [PubMed] [Google Scholar]
- Bellmunt J, Szczylik C, Feingold J, Strahs A, Berkenblit A (2008) Temsirolimus safety profile and management of toxic effects in patients with advanced renal cell carcinoma and poor prognostic features. Ann Oncol 19: 1387–1392. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L (2002) Fatty Acid Metabolism. In Biochemistry, WH Freeman, (ed.) Freeman, W H: New York, pp 617–732. [Google Scholar]
- Blaise BJ, Shintu L, Elena B, Emsley L, Dumas ME, Toulhoat P (2009) Statistical recoupling prior to significance testing in nuclear magnetic resonance based metabonomics. Anal Chem 81: 6242–6251. [DOI] [PubMed] [Google Scholar]
- Borgan E, Lindholm EM, Moestue S, Mælandsmo GM, Lingjærde OC, Gribbestad IS, Børresen-Dale AL, Engebraaten O, Sørlie TT (2013) Subtype-specific response to bevacizumab is reflected in the metabolome and transcriptome of breast cancer xenografts. Mol Oncol 7: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke OH, Sandlie I (2003) Therapeutic antibodies for human diseases at the dawn of the twenty-first century. Nat Rev Drug Discov 2: 52–62. [DOI] [PubMed] [Google Scholar]
- Coppin C (2008) Sunitinib for advanced renal cell cancer. Biologics 2: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL (2008) Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol 26: 162–164. [DOI] [PubMed] [Google Scholar]
- Duarte IF, Gil AM (2012) Metabolic signatures of cancer unveiled by NMR spectroscopy of human biofluids. Prog Nucl Magn Res Sp 62: 51–74. [DOI] [PubMed] [Google Scholar]
- Eriksson L, Trygg J, Wold S (2008) CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemometr 22: 594–600. [Google Scholar]
- Feldman DR, Baum MS, Ginsberg MS, Hassoun H, Flombaum CD, Velasco S, Fischer P, Ronnen E, Ishill N, Patil S, Motzer RJ (2009) Phase I trial of bevacizumab plus escalated doses of sunitinib in patients with metastatic renal cell carcinoma. J Clin Oncol 27: 1432–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman SA, Lasky LC, Limas C (1982) Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol 6: 655. [DOI] [PubMed] [Google Scholar]
- Gao H, Dong B, Liu X, Xuan H, Huang Y, Lin D (2008) Metabonomic profiling of renal cell carcinoma: high-resolution proton nuclear magnetic resonance spectroscopy of human serum with multivariate data analysis. Anal Chim Acta 624: 269–277. [DOI] [PubMed] [Google Scholar]
- Gerber DE (2008) Targeted therapies: a new generation of cancer treatments. Am Fam Physician 77: 311–319. [PubMed] [Google Scholar]
- Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C (2008) Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev 34: 193–205. [DOI] [PubMed] [Google Scholar]
- Hauschild A, Gogas H, Tarhini A, Middleton MR, Testori A, Dréno B, Kirkwood JM (2008) Practical guidelines for the management of interferon-α-2b side effects in patients receiving adjuvant treatment for melanoma. Cancer 112: 982–994. [DOI] [PubMed] [Google Scholar]
- Hicklin DJ (2004) Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol 23: 1011–1027. [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen Y, Hang W, Gao Y, Lin L, Li DY, Xing J (2013) Holistic metabonomic profiling of urine affords potential early diagnosis for bladder and kidney cancers–Springer. Metabolomics 9: 119–129. [Google Scholar]
- Jobard E, Pontoizeau C, Blaise BJ, Bachelot T, Elena-Herrmann B, Tredan O (2014) A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett 343: 33–41. [DOI] [PubMed] [Google Scholar]
- Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH (2009) Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics 8: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Tolstikov V, Fiehn O, Weiss RH (2007) A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem 363: 185–195. [DOI] [PubMed] [Google Scholar]
- Lin L, Huang Z, Gao Y, Yan X, Xing J, Hang W (2011) LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J Proteome Res 10: 1396–1405. [DOI] [PubMed] [Google Scholar]
- Lodi A, Ronen SM (2011) Magnetic resonance spectroscopy detectable metabolomic fingerprint of response to antineoplastic treatment. PLoS One 6: e26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchan JR, Qin R, Pitot H, Picus J, Liu G, Fitch T, Maples WJ, Flynn PJ, Fruth BF, Erlichman C (2015) Safety and activity of temsirolimus and bevacizumab in patients with advanced renal cell carcinoma previously treated with tyrosine kinase inhibitors: a phase 2 consortium study. Cancer Chemother Pharmacol 75: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Bacik J, Murphy BA, Russo P, Mazumdar M (2002) Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 20: 289–296. [DOI] [PubMed] [Google Scholar]
- Négrier S, Gravis G, Perol D, Chevreau C, Delva R, Bay J-O, Blanc E, Ferlay C, Geoffrois L, Rolland F, Legouffe E, Sevin E, Laguerre B, Escudier B (2011) Temsirolimus and bevacizumab, or sunitinib, or interferon alfa and bevacizumab for patients with advanced renal cell carcinoma (TORAVA): a randomised phase 2 trial. Lancet Oncol 12: 673–680. [DOI] [PubMed] [Google Scholar]
- Oakman C, Tenori L, Claudino WM, Cappadona S, Nepi S, Battaglia A, Bernini P, Zafarana E, Saccenti E, Fornier M, Morris PG, Biganzoli L, Luchinat C, Bertini I, Di Leo A (2011) Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol 22: 1295–1301. [DOI] [PubMed] [Google Scholar]
- Oken MM, Creech RH, Tormey DC, Horton J, Davis TE, McFadden ET, Carbone PP (1982) Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol 5: 649–655. [PubMed] [Google Scholar]
- Patard J-J, Pouessel D, Bensalah K, Culine S (2008) Targeted therapy in renal cell carcinoma. World J Urol 26: 135–140. [DOI] [PubMed] [Google Scholar]
- Patel PH, Senico PL, Curiel RE, Motzer RJ (2009) Phase I study combining treatment with temsirolimus and sunitinib malate in patients with advanced renal cell carcinoma. Clin Genitourin Cancer 7: 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud B, Lee J, Valkova N, Dhirapong A, Lin P-Y, Fiehn O, Kültz D, Weiss RH (2006) Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5: 375–386. [DOI] [PubMed] [Google Scholar]
- Rini BI, Atkins MB (2009) Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol 10: 992–1000. [DOI] [PubMed] [Google Scholar]
- Rini BI, Campbell SC, Escudier B (2009) Renal cell carcinoma. Lancet 373: 1119–1132. [DOI] [PubMed] [Google Scholar]
- Rini BI, Halabi S, Rosenberg JE, Stadler WM, Vaena DA, Ou SS, Archer L, Atkins JN, Picus J, Czaykowski P, Dutcher J, Small EJ (2008) Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol 26: 5422–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini BI, Powles T (2013) Biology and treatment of advanced renal cell carcinoma: a global perspective. Semin Oncol 40: 419–420. [DOI] [PubMed] [Google Scholar]
- Shinohara E, Yamashita S, Kihara S, Hirano K, Ishigami M, Arai T, Nozaki S, KamedaTakemura K, Kawata S, Matsuzawa Y (1997) Interferon alpha induces disorder of lipid metabolism by lowering postheparin lipases and cholesteryl ester transfer protein activities in patients with chronic hepatitis C. Hepatology 25: 1502–1506. [DOI] [PubMed] [Google Scholar]
- Sleijfer S, Bannink M, Van Gool AR, Kruit WHJ, Stoter G (2005) Side effects of interferon-a therapy. Pharm World Sci 27: 423–431. [DOI] [PubMed] [Google Scholar]
- Süllentrop F, Moka D, Neubauer S, Haupt G, Engelmann U, Hahn J, Schicha H (2002) 31P NMR spectroscopy of blood plasma: determination and quantification of phospholipid classes in patients with renal cell carcinoma. NMR Biomed 15: 60–68. [DOI] [PubMed] [Google Scholar]
- Tate AR, Foxall PJ, Holmes E, Moka D, Spraul M, Nicholson JK, Lindon JC (2000) Distinction between normal and renal cell carcinoma kidney cortical biopsy samples using pattern recognition of (1)H magic angle spinning (MAS) NMR spectra. NMR Biomed 13: 64–71. [DOI] [PubMed] [Google Scholar]
- Tenori L, Oakman C, Claudino WM, Bernini P, Cappadona S, Nepi S, Biganzoli L, Arbushites MC, Luchinat C, Bertini I, Di Leo A (2012) Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: a pilot study. Mol Oncol 6: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG (2000) New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 92: 205–216. [DOI] [PubMed] [Google Scholar]
- Trygg J, Wold S (2002) Orthogonal projections to latent structures (O-PLS). J Chemometr 16: 119–128. [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL (2008) BioMagResBank. Nucleic Acids Res 36: D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veldt AM, Haanen JB, van den Eertwegh AJ, Boven E (2010) Targeted therapy for renal cell cancer: current perspectives. Discov Med 10: 394–405. [PubMed] [Google Scholar]
- Weaver Z, Difilippantonio S, Carretero J, Martin PL, Meskini El R, Iacovelli AJ, Gumprecht M, Kulaga A, Guerin T, Schlomer J, Baran M, Kozlov S, McCann T, Mena S, Al-Shahrour F, Alexander D, Wong KK, Van Dyke T (2012) Temporal molecular and biological assessment of an erlotinib-resistant lung adenocarcinoma model reveals markers of tumor progression and treatment response. Cancer Res 72: 5921–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Liu L, Zhang J, Bowers J, Gowda GAN, Seeger H, Fehm T, Neubauer HJ, Vogel U, Clare SE, Raftery D (2013) Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol Oncol 7: 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorf RC, Hainsworth JD, Spigel DR (2008) Phase II study of bevacizumab and everolimus (RAD001) in the treatment of advanced renal cell carcinoma (RCC) — Whorf et al. 26 (15 Supplement): 5010 — ASCO Meeting Abstracts. J Clin Oncol.
- Wishart DS (2007) Current Progress in computational metabolomics. Brief Bioinform 8: 279–293. [DOI] [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P (1987) Principal component analysis. Chemometr Intell Lab 2: 37–52. [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 127: 5–19. [DOI] [PubMed] [Google Scholar]
- Zafar Y, Bendell J, Lager J, Yu D (2006) Preliminary results of a phase I study of bevacizumab (BV) in combination with everolimus (E) in patients with advanced solid tumors — Zafar et al. 24 (18 Supplement): 3097 — ASCO Meeting Abstracts. ASCO Meeting.
- Zhang J, Yang PL, Gray NS (2009) Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 9: 28–39. [DOI] [PubMed] [Google Scholar]
- Zira A, Theocharis SE, Mitropoulos D, Migdalis V, Mikros E (2010) 1H NMR metabonomic analysis in renal cell carcinoma: a possible diagnostic tool. J Proteome Res 9: 1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.