Abstract
MicroRNAs were described to target mRNA and regulate the transcription of genes involved in processes de-regulated in tumorigenesis, such as proliferation, differentiation and survival. In particular, the miRNA let-7 has been suggested to regulate the expression of the KRAS gene, a common mutated gene in non-small cell lung cancer (NSCLC), through a let-7 complementary site (LCS) in 3′UTR of KRAS mRNA. We have reported the analysis performed on the role of the polymorphism located in the KRAS-LCS (rs61764370) which is involved in the disruption of the let-7 complementary site in NSCLC patients enrolled within the TAILOR trial, a randomised trial comparing erlotinib versus docetaxel in second line treatment. In our cohort of patients, KRAS-LCS6 polymorphism did not have any impact on both overall survival (OS) and progression free survival (PFS) and was not associated with any patient’s baseline characteristics included in the study. Overall, patients had a better prognosis when treated with docetaxel instead of erlotinib for both OS and PFS. Considering KRAS-LCS6 status, the TG/GG patients had a benefit from docetaxel treatment (HR(docetaxel vs erlotinib) = 0.35, 95% CI 0.15–0.79, p = 0.011) compared with the TT patients (HR(docetaxel vs erlotinib) = 0.72, 95% CI 0.52–1.01, p = 0.056) in terms of PFS.
Lung cancer is the first cause of cancer-related death in Western countries1. This malignancy is strongly associated with environmental factors and smoking2. The prognosis of patients with Non-Small Cell Lung Cancer (NSCLC) is very poor with a percentage of survivors that is lower than 15% for all stages and lower than 5% in metastatic disease3.
KRAS is one of the most frequently mutated genes in NSCLC, in fact its mutations are present in approximately 20% of this type of tumour. KRAS belongs to the ras family and it encodes a small G protein with intrinsic GTPase activity, which is necessary for protein inactivation, and to tune the downstream effectors involved in pathways such as proliferation and differentiation. Mutations in defined aminoacids determine the loss of intrinsic GTPase activity and the deregulation of downstream pathways4. In addition to mutations, KRAS activity can be altered through a lower protein expression promoted by miRNA binding to its messenger RNA. A polymorphic site in the 3′ untranslated region of KRAS, is able to eliminate the ability of miRNA let-7 to bind to the target. The single nucleotide polymorphism (SNP) (rs61764370), named KRAS let-7 complementary site (KRAS-LCS6), was described as the change of the T-allele to a G-allele. This modification was seen to increase the KRAS expression and to activate the downstream pathways. The KRAS-LCS6 variant is not very common and the G-allele frequency is about 7% in the European population5.
The KRAS-LCS6 was associated with higher cancer risk in triple-negative breast cancer6 and reduced survival in oral cancer patients7. On the contrary, the KRAS-LCS6 SNP was associated with a better outcome in early stage colorectal cancer, but this feature was lost in advanced stages of this disease8. In ovarian cancer the KRAS-LCS6 polymorphism was described to have the opposite role and also no function9,10,11. In lung cancer, the moderate smoker population harbouring the G-allele was shown to have an increased cancer risk5 but the presence of infrequent allele did not reduce the survival rate of patients12.
Since KRAS mutation demonstrated only a little impact on survival, as also reported in TAILOR trial results13, and KRAS activity can be regulated by microRNA, patients stratification based only on KRAS status could not be sufficient to evaluate the role of this biomarker.
Given that the prognostic and predictive role of KRAS-LCS6 polymorphism was not yet investigated in lung cancer, we planned an ancillary study to assess the value of KRAS-LCS6 polymorphism on outcomes within the TAILOR trial, a randomised trial comparing erlotinib versus docetaxel in second line NSCLC.
Results
Between October 2007 and March 2012, 222 eligible patients were enrolled in the TAILOR trial. Among 222 randomised patients (110 to docetaxel and 112 to erlotinib), 218 were fully eligible for the main trial14. Of these, 145 (82.4%) had TT genotype in the KRAS-LCS6 locus, 30 (17.1%) harboured a TG variant whereas only one (0.5%) patient had GG polymorphism (hereafter included in the TG patients group). For the remaining 42 patients, we were not able to collect blood samples. The CONSORT diagram is illustrated in the Supplementary Figure S1. The minor allele prevalence was 10%, consistent with available data. The baseline characteristics of the patients included in the present study according to KRAS-LCS6 polymorphism are illustrated in Table 1.
Table 1. Patient characteristics.
| TT |
TG/GG |
P-value | ||||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| Patients | 145 | 82.4 | 31 | 17.6 | ||
| Age | Median(quartile) | 66.0 (58.8–71.4) | 70.0 (60.9–73.3) | 0.119 | ||
| Sex | Male | 97 | 66.9 | 23 | 74.2 | 0.430 |
| Female | 48 | 33.1 | 8 | 25.8 | ||
| ECOG-PS | 0 | 69 | 47.6 | 18 | 58.1 | 0.241 |
| 1 | 66 | 45.5 | 12 | 38.7 | ||
| 2 | 10 | 6.9 | 1 | 3.2 | ||
| Smoking | Never | 32 | 22.1 | 9 | 29.0 | 0.407 |
| Ex smokers/smokers | 113 | 77.9 | 22 | 71.0 | ||
| Stage at diagnosis | I | 14 | 9.7 | 2 | 6.5 | 0.852 |
| IIA | 4 | 2.8 | 3 | 9.7 | ||
| IIB | 5 | 3.5 | 2 | 6.5 | ||
| IIIA | 25 | 17.2 | 4 | 12.9 | ||
| IIIB | 16 | 11.0 | 3 | 9.7 | ||
| IIIB wet | 6 | 4.1 | 0 | 0.0 | ||
| IV | 75 | 51.7 | 17 | 54.8 | ||
| Grading | G1 | 5 | 5.3 | 2 | 10.0 | 0.933 |
| G2 | 36 | 37.9 | 5 | 25.0 | ||
| G3 | 52 | 54.7 | 13 | 65.0 | ||
| Undifferentiated | 2 | 2.1 | 0 | 0.0 | ||
| unknown | 50 | 11 | ||||
| Histotype | Adenocarcinoma | 100 | 69.0 | 23 | 74.2 | 0.640 |
| Squamous | 34 | 23.5 | 7 | 22.6 | ||
| Bronchoalveolar | 2 | 1.4 | 1 | 3.2 | ||
| Large cells | 2 | 1.4 | 0 | 0.0 | ||
| Other | 7 | 4.9 | 0 | 0.0 | ||
| KRAS status | Wild type | 113 | 77.9 | 20 | 66.7 | 0.190 |
| Mutated | 32 | 22.1 | 10 | 33.3 | ||
| Treatment arm | Docetaxel | 70 | 48.3 | 16 | 51.6 | 0.737 |
| Erlotinib | 75 | 51.7 | 15 | 48.4 | ||
For the TT population the median age at diagnosis was 66 years (interquartile range (IQR): 58.8–71.4 years) whereas it was 70 years (IQR: 60.9–73.3 years) for the TG/GG population. The TT group was predominantly stage IV (51.7%), had adenocarcinoma histology (69.0%), poorly differentiated grade (54.7%), a smoking habit (77.9%), ECOG-PS of 0 (47.6%) and a wild-type status of KRAS (77.9%). Similarly, the TG/GG patients were predominantly stage IV (54.8%), with adenocarcinoma histology (74.2%), poorly differentiated grade (65.0%), smoking habit (71.0%), ECOG-PS of 0 (58.1%) and a wild-type status of KRAS (64.5%).
Although the polymorphism variants were not a stratification marker, the patients were well distributed between the two treatments performed in the main trial. In particular, 48.3% and 51.6% of TT and TG/GG patients respectively were treated with docetaxel. On the other hand, 51.7% of TT and 48.4% of TG/GG patients received erlotinib. None of the characteristics considered were associated with the different genotypes present in the polymorphic site.
Survival outcomes
After a median follow-up of 33.0 months (IQR: 21.4–33.4), 170 patients progressed or died and 150 died.
The baseline characteristics associated with overall survival (OS) were: ECOG-PS (HR(2 vs. 1 vs. 0) = 2.14, 95% CI 1.60–2.85, p < 0.0001) and sex (HR(F vs M) = 0.68, 95% CI 0.47–0.97, p = 0.035). All risk estimate covariates are reported in Table 2. Median OS was 6.8 months (IQR 3.3–20.2 months) in the TT group and 7.3 months (IQR 3.7–15.3 months) in the TG/GG group (unadjusted HR(TT vs TG/GG) = 0.97, 95% CI 0.64–1.47, p = 0.875; adjusted HR(TT vs TG/GG) = 0.82, 95% CI 0.54–1.26, p = 0.373). Figure 1a shows the OS curves according to the KRAS-LCS6 polymorphism.
Table 2. Prognostic evaluation of clinical and histopatological characteristics – Overall Survival.
| HR | Lower 95% HR | Upper 95% HR | P-value | |
|---|---|---|---|---|
| Univariate | ||||
| KRAS-LCS6 (TT vs TG/GG) | 0.97 | 0.64 | 1.47 | 0.875 |
| Age at diagnosis | 1.02 | 1.00 | 1.03 | 0.058 |
| Treatment arm (docetaxel vs erlotinib) | 0.73 | 0.53 | 1.01 | 0.060 |
| Sex (F vs M) | 0.68 | 0.47 | 0.97 | 0.035 |
| Smoking (smoking and ex vs not smoking) | 1.23 | 0.83 | 1.81 | 0.297 |
| Tumour grade | 1.19 | 0.86 | 1.64 | 0.292 |
| Tumour stage (IIIBw/IV vs III vs I/II) | 1.19 | 0.95 | 1.49 | 0.126 |
| ECOG-PS (2 vs. 1 vs. 0) | 2.14 | 1.60 | 2.85 | <.0001 |
| Histotype (squamous vs others) | 1.15 | 0.78 | 1.70 | 0.467 |
| KRAS (mut vs wt) | 1.36 | 0.94 | 1.96 | 0.106 |
| Multivariate | ||||
| KRAS-LCS6 (TT vs TG/GG) | 0.82 | 0.54 | 1.26 | 0.373 |
| Treatment arm (docetaxel vs erlotinib) | 0.73 | 0.53 | 1.01 | 0.060 |
| ECOG-PS (2 vs. 1 vs. 0) | 2.17 | 1.62 | 2.91 | <.0001 |
Figure 1.
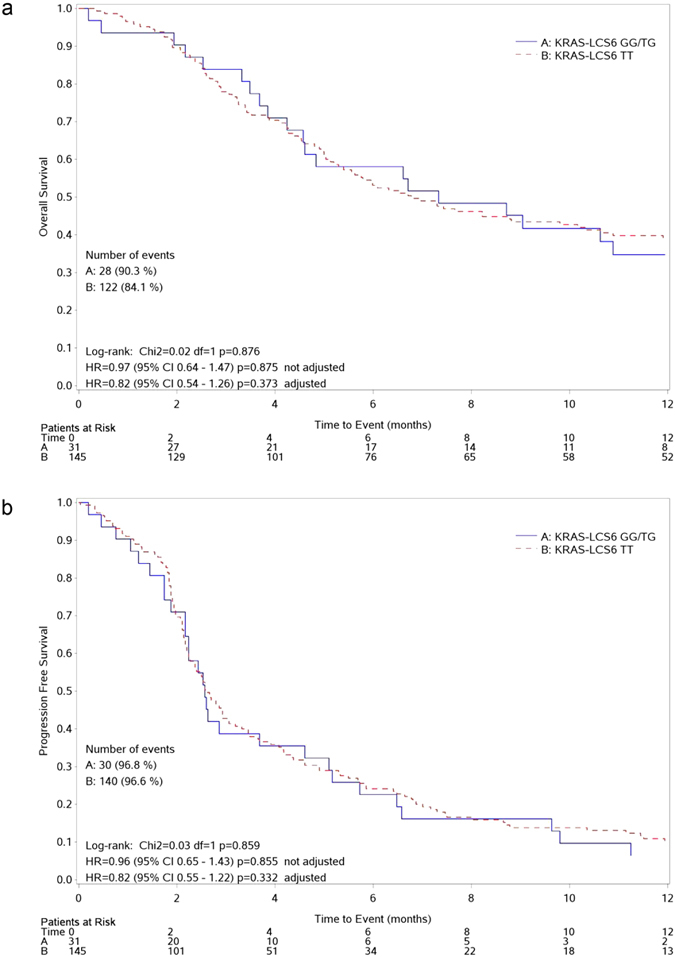
Kaplan-Meier curves for OS (a) and PFS (b) according to KRAS-LCS6 genotype.
ECOG-PS (HR(2 vs. 1 vs. 0) = 1.79, 95% CI 1.37–2.34, p < 0.0001) and treatment arm (HR(docetaxel vs erlotinib) = 0.65, 95% CI 0.48-0.89, p = 0.007) were associated with progression free survival (PFS). All risk estimate covariates are reported in Table 3. Median PFS was the same for both groups: 2.6 months (IQR 1.9–5.9 months) in the TT group and 2.6 months (IQR 1.7–5.7 months) in the TG/GG group (unadjusted HR(TT vs TG/GG) = 0.96, 95% CI 0.65–1.43, p = 0.855; adjusted HR(TT vs TG/GG) = 0.82, 95% CI 0.55–1.22, p = 0.332). Figure 1b shows the PFS curves according to the KRAS-LCS6 polymorphism.
Table 3. Prognostic evaluation of clinical and histopatological characteristics – Progression Free Survival.
| HR | Lower 95% HR | Upper 95% HR | P-value | |
|---|---|---|---|---|
| Univariate | ||||
| KRAS-LCS6 (TT vs TG/GG) | 0.96 | 0.65 | 1.43 | 0.855 |
| Age at diagnosis | 1.01 | 0.99 | 10.2 | 0.267 |
| Treatment arm (docetaxel vs erlotinib) | 0.65 | 0.48 | 0.89 | 0.007 |
| Sex (F vs M) | 0.76 | 0.55 | 1.05 | 0.100 |
| Smoking (smoking and ex vs not smoking) | 1.32 | 0.92 | 1.89 | 0.129 |
| Tumour grade | 1.19 | 0.88 | 1.60 | 0.251 |
| Tumour stage (IIIBw/IV vs III vs I/II) | 1.16 | 0.94 | 1.42 | 0.172 |
| ECOG-PS (2 vs. 1 vs. 0) | 1.79 | 1.37 | 2.34 | <.0001 |
| Histotype (squamous vs others) | 1.22 | 0.85 | 1.74 | 0.278 |
| KRAS (mut vs wt) | 1.04 | 0.73 | 1.48 | 0.822 |
| Multivariate | ||||
| KRAS-LCS6 (TT vs TG/GG) | 0.82 | 0.55 | 1.22 | 0.332 |
| Treatment arm (docetaxel vs erlotinib) | 0.65 | 0.48 | 0.89 | 0.007 |
| ECOG-PS (2 vs. 1 vs. 0) | 1.80 | 1.37 | 2.36 | <.0001 |
Subgroup analyses
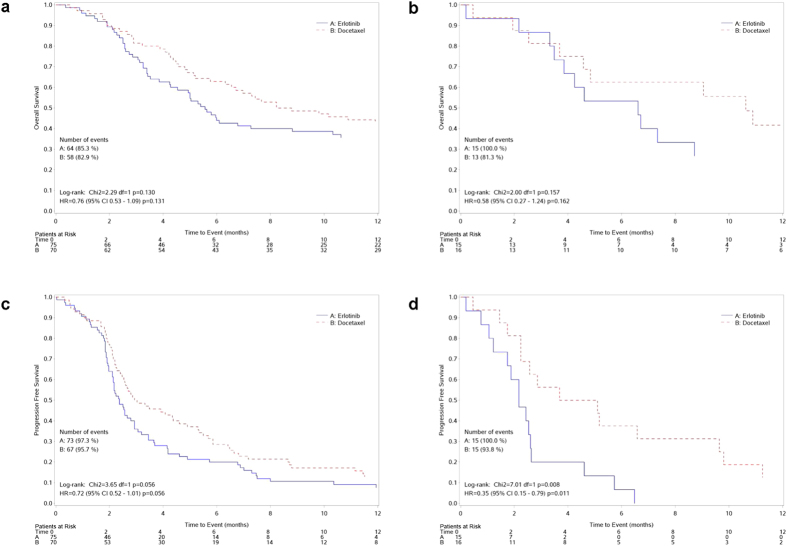
For explorative purposes, we performed a subgroup analysis according to KRAS-LCS6 polymorphism status with the aim of investigating its predictive role on treatment efficacy. In patients with TT polymorphism, although not statistically significant, the risk of death was lower in the docetaxel compared to the erlotinib treated group (HR(docetaxel vs erlotinib) = 0.76, 95% CI 0.53-1.09, p = 0.131). The same was observed for the TG/GG population (HR(docetaxel vs erlotinib) = 0.58, 95% CI 0.27-1.24, p = 0.162). The test of interaction was not statistically significant (p = 0.618). The same was true for patients with TT polymorphism in terms of PFS. The risk of progression was lower in the docetaxel compared to the erlotinib treated group (HR(docetaxel vs erlotinib) = 0.72, 95% CI 0.52-1.01, p = 0.056). On the other hand, we observed a much better PFS in response to docetaxel compared to erlotinib for the TG/GG population (HR(docetaxel vs erlotinib) = 0.35, 95% CI 0.15-0.79, p = 0.011). Again, the test of interaction was not significant (p = 0.133). The curves reporting OS and PFS by treatment in TT and TG/GG patients are reported in Figure 2 while Figure 3 reports the Forest plot for the predictive role of KRAS-LCS6 polymorphism.
Figure 2.
Kaplan-Meier curves reporting OS (upper panels) and PFS (lower panels) in TT (panels (A,C) and TG/GG (panels (B,D) patients according to treatment arm.
Figure 3.
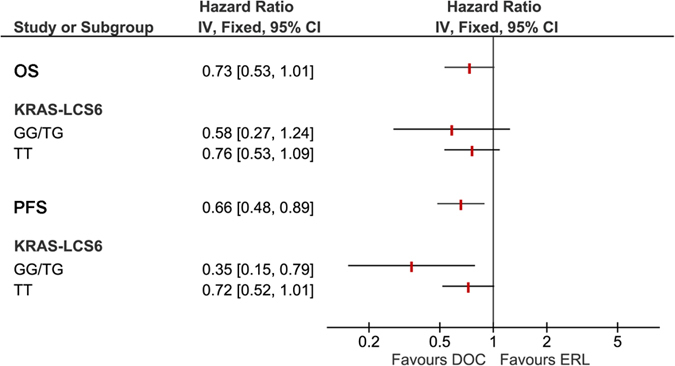
Forest Plots showing the predictive role of KRAS-LCS6 polymorphism.
On the other hand, considering separately the different treatment arm we observed no difference in OS between the two polymorphisms both in docetaxel (HR(TT vs TG/GG) = 1.01, 95% CI 0.55–1.86, p = 0.966) and in erlotinb arm (HR(TT vs TG/GG) = 0.88, 95% CI 0.49–1.58, p = 0.676).
The same was observed in PFS both in docetaxel (HR(TT vs TG/GG) = 1.18, 95% CI 0.67-2.07, p = 0.560) and in erlotinb arm (HR(TT vs TG/GG) = 0.65, 95% CI 0.37-1.15, p = 0.140). The curves reporting OS and PFS by genotypes in the treatment arms are reported in the Supplementary Figure S2.
We performed a second explorative analysis to address the role of KRAS-LCS6 polymorphism in the presence of either wild-type or mutated KRAS. In the presence of a wild-type KRAS both OS and PFS were almost equivalent when the two genotypes were compared (HR(TT vs TG/GG) = 0.93, 95% CI 0.55–1.57, p = 0.792 for OS and HR(TT vs TG/GG) = 0.88, 95% CI 0.54–1.41, p = 0.586 for PFS). When we considered a KRAS mutated background, the TG/GG genotypes seemed to indicate a protective trend in both OS and PFS although not statistically significant (HR(TT vs TG/GG) = 1.29, 95% CI 0.61–2.74, p = 0.501 and HR(TT vs TG/GG) = 1.27, 95% CI 0.60–2.67, p = 0.534 respectively). The test of interaction was not significant for both OS (p = 0.263) and PFS (p = 0.344). The curves reporting OS and PFS by KRAS status and genotypes are reported in Figure 4.
Figure 4.
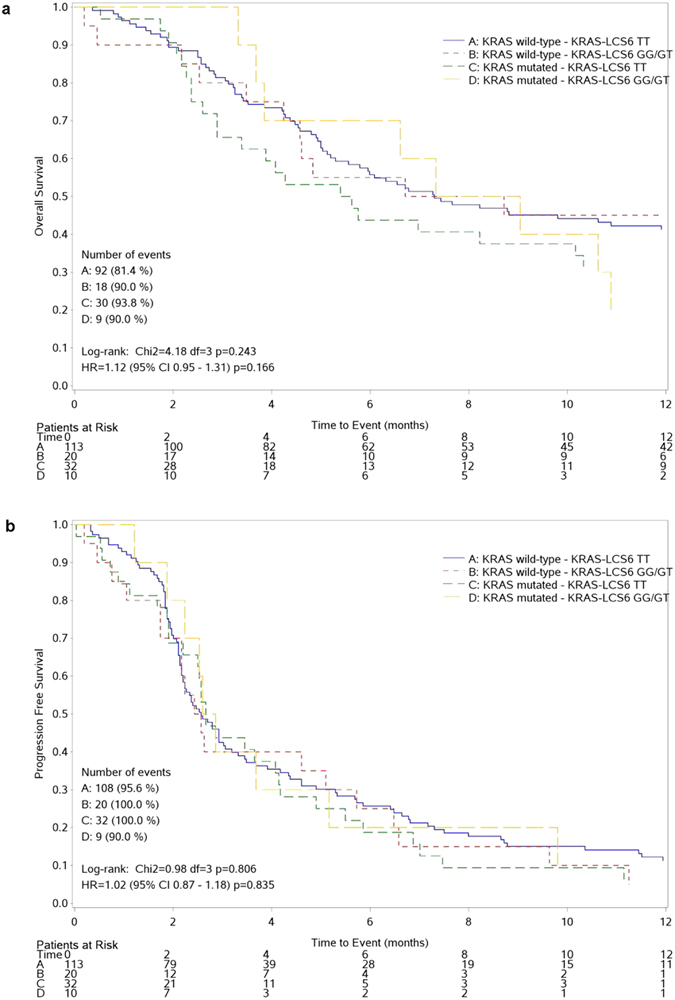
Kaplan-Meier curves reporting OS (upper panels) and PFS (lower panels) by KRAS status and genotypes.
Discussion
In the last two decades, many studies have been published analysing the prognostic and predictive roles of KRAS mutations in sustaining resistance to different types of treatment such as EGFR Tyrosine-Kinase Inhibitors (TKIs) and chemotherapy15,16,17. KRAS mutated patients were indicated to have a worse prognosis and resistance to treatment in different types of cancer but no clear conclusions have been stated for NSCLC18. The data were highly variable since extracted from retrospective studies, which either considered a very small number of patients or evaluated KRAS mutational status only in a subgroup of patients. Another possible reason could be the fact that, in addition to mutations and amplification, KRAS activity can be regulated by microRNA (miRNA), in particular miRNA let-7b5. For these reasons patients stratification based only on KRAS status could not be sufficient to evaluate the role of this biomarker. MicroRNAs let-7 were described as a family of miRNAs able to regulate the expression of some lung cancer oncogenes including KRAS19,20.
In the present work, we analysed the role of the genomic variant present in KRAS-LCS6 within a phase III clinical trial (TAILOR). The TAILOR trial was a non-profit multicentre, open label, randomised trial, conducted in 52 Italian hospitals, comparing erlotinib versus docetaxel in second line NSCLC14. Blood samples were collected with the aim of investigating any association between biomarkers and clinical/histopathological characteristics of the patients and the role of biomarkers possibly involved in the outcomes.
In our study, the genomic variant present in KRAS-LCS6 was not associated with any clinical or histopathological characteristics of the patients included in the study. Furthermore, as already reported by Nelson et al.12, our study confirms that the KRAS mutation prevalence was the same in both the genotype groups. We can support the Nelson hypothesis that occurs despite the up-regulation of KRAS expression, due to the G variant present in the let-7 binding site in the 3’UTR of KRAS, which did not result in any selective pressure for KRAS mutations.
Let-7 was originally identified in Caenorhabditis elegans as a regulator of developmental timing and cellular proliferation21 and, when ectopically expressed in cancer cell lines and xenograft models, miRNA let-7 was able to repress cellular proliferation22,23.
Let-7 expression levels were found to be reduced in NSCLC patients and this decrease has been associated with a worse clinical outcome24.
The same effect of let-7 levels reduction can also be obtained by the lack of miRNA binding site, as happens with the KRAS-LCS6 SNP, but we were not able to confirm the association between polymorphisms and poor prognosis given that, in our study, the TG/GG genotypes did not correlate with any outcome. We have no explanation for the lack of role for this polymorphism in our study. We cannot exclude that the expression levels of miRNA let-7 could be different among patients and nullify the impact of the different genotypes. It is also true that the marked role for KRAS-LCS6 polymorphism has usually been described in the case-control studies assessing cancer risk. In fact, an increased NSCLC risk was described as associated with the polymorphism and this was most evident among people who were light to moderate smokers5.
As reported in the main TAILOR trial and confirmed in this study, patients had a better prognosis in terms of PFS and OS when treated with docetaxel instead of erlotinib.
Because of the lack of the statistical power necessary to demonstrate a predictive effect of KRAS-LCS6, our study can only suggest that KRAS-LCS6 confers a different magnitude of the effect of docetaxel compared to erlotinb on both PFS and OS. In this view, considering KRAS-LCS6 status to stratify patients and perform explorative subgroup analysis, the TG/GG subgroup seemed to benefit more from docetaxel treatment when compared to erlotinib in terms of PFS.
The consideration on statistical power was true also for a second subgroup analysis considering the KRAS status to stratify patients. Patients in KRAS wild-type subgroup with TT genotype had slightly better outcomes whereas in the mutated KRAS population the contrary was observed.
In conclusion, the previous TAILOR results on the superiority of the chemotherapy in the absence of an identified target14 is once more confirmed in all subgroups analysed. Our data suggest that the KRAS-LCS6 polymorphism is not a critical prognostic factor but could identify a subgroup of patients (TG/GG) for which the use of a chemotherapy treatment seems to be extremely important.
Methods
Study design and patients
TAILOR was a non-profit multicentre, open label, randomised trial, funded by the Italian Regulatory Agency AIFA and conducted in 52 Italian hospitals, comparing erlotinib versus docetaxel in second line NSCLC. Details have been published previously13. Within the TAILOR trial we pre-planned a number of ancillary studies including the role of polymorphism on outcomes. Participating hospitals registered all consecutive patients with metastatic, recurrent or inoperable locally advanced NSCLC. Only those with both a EGFR and KRAS centrally determined status were included in the trial. All patients received a first line platinum-based chemotherapy in combination with either vinorelbine, gemcitabine or pemetrexed according to the physician’s decision. Combinations with taxanes and with anti-EGFR agents were not allowed. Patients with EGFR mutations were selectively treated with EGFR Tyrosine-Kinase Inhibitors (TKI) and were excluded from this analysis. All patients had an Eastern Cooperative Oncology Group (ECOG) Performance Status (PS) between 0 and 2 and were at least 18 years of age. Exclusion criteria included any evidence of serious co-morbidities that the investigator judged as a contraindication to the participation in the study, as well as pregnancy and breast-feeding. Research protocol was approved by the Ethics Committee of Ospedale Fatebenefratelli e Oftalmico, Milan (03 October 2007) and all patients who were eligible for participation provided written informed consent with all applicable governing regulations before undergoing any study procedure. All experiments were performed in accordance with the Declaration of Helsinki. The study was registered March 12, 2008 at ClinicalTrials.gov, number NCT00637910.
Samples collection and genotyping
Blood specimens were collected in K2EDTA sample tubes and frozen at −80 °C. DNA was extracted from blood samples using Maxwell 16 DNA Purification Kit (Promega, Milan, Italy). The rs61764370 SNP was genotyped using a TaqMan SNP Genotyping assay (Applied Biosystems, Monza, Milan), based on Real Time PCR technique (ABI 7900, Applied Biosystems). The PCR was carried out in a 384-wells plate with a reaction volume of 5 μL containing genomic DNA (10 ng), 2× TaqMan Genotyping Master Mix (Applied Biosystems), 40× MGB probes and primers. Primers and probe sequences (MGB probes specifically designed for Allelic Discrimination) are property of Applied Biosystems. Thermal cycle conditions were 95 °C for 10 minutes and 40 cycles at 95 °C for 15 seconds and 60 °C for 1 minute. Completed PCR plates were analysed using the Allelic Discrimination Sequence Detection Software (Applied Biosystems).
Statistical methods
Baseline covariate distributions were summarised using descriptive statistics (median and range for continuous variables; absolute and percentage frequencies for categorical variables); Wilcoxon-Mann-Whitney test for continuous covariates and Chi-square test for categorical covariates were used to detect statistical association. Progression Free Survival was defined as the time from the date of randomisation up to the date of first progression or death from any cause, whichever came first. Subjects who had not progressed or died while in the study were censored at the last disease assessment date. Overall survival was defined as the time from the date of randomisation up to the date of death from any cause. Subjects who did not die while in the study were censored at the last follow-up. Survival curves were estimated with the Kaplan-Meier method. Cox proportional hazards models were used for univariate and multivariate (adjusted for ECOG-PS and treatment arm) analysis to estimate the association between KRAS-LCS6 polymorphism and PFS and OS. Results were expressed as Hazard Ratios (HRs) and their 95% confidence intervals (95% CIs). Statistical analyses were carried out using SAS version 9.2 (SAS Institute, Cary, NC).
Additional Information
How to cite this article: Ganzinelli, M. et al. Role of KRAS-LCS6 polymorphism in advanced NSCLC patients treated with erlotinib or docetaxel in second line treatment (TAILOR). Sci. Rep. 5, 16331; doi: 10.1038/srep16331 (2015).
Supplementary Material
Acknowledgments
The study was funded by Agenzia Italiana del Farmaco, AIFA, (MCG) and Italian Association for Cancer Research IG-12915 (MB) and IG-15721 (MCG). EC is recipient of a FIRC fellowship.
Footnotes
Author Contributions M.C.G., M.B. and M.M. conceptualised the study. E.C. collected data. E.R. did the statistical analysis. S.P., F.L., R.L., C.B., M.A.F., O.M., D.F., M.C.L., A.B., G.V., I.P., A.C. and M.G.S. were the main clinical investigators. M.G. and E.C. did the molecular analyses. M.M. and M.B. wrote the paper. All authors reviewed and approved the submitted report on behalf of TAILOR trialists.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Sato M., Shames D. S., Gazdar A. F. & Minna J. D. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2, 327–343 (2007). [DOI] [PubMed] [Google Scholar]
- Herbst R. S., Heymach J. V. & Lippman S. M. Lung Cancer. New England Journal of Medicine 359, 1367–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely G. J., Marks J. & Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 6, 201–205 (2009). [DOI] [PubMed] [Google Scholar]
- Chin L. J. et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 68, 8535–8540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape T. et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol 12, 377–386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B. C. et al. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis 30, 1003–1007 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits K. M. et al. A Let-7 MicroRNA SNP in the KRAS 3′ UTR Is Prognostic in Early-Stage Colorectal Cancer. Clin Cancer Res 17, 7723–7731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiola E. et al. KRas-LCS6 polymorphism does not impact on outcomes in ovarian cancer. Am J Cancer Res 2, 298–308 (2012). [PMC free article] [PubMed] [Google Scholar]
- Pharoah P. D. et al. The role of KRAS rs61764370 in invasive epithelial ovarian cancer: implications for clinical testing. Clin Cancer Res 17, 3742–3750 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner E. S. et al. A KRAS variant is a biomarker of poor outcome, platinum chemotherapy resistance and a potential target for therapy in ovarian cancer. Oncogene 31, 4559–4566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. H. et al. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer 69, 51–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli E. et al. Value of KRAS as prognostic or predictive marker in NSCLC: results from the TAILOR trial. Ann Oncol, (2015). doi: 10.1093/annonc/mdv318. [DOI] [PubMed] [Google Scholar]
- Garassino M. C. et al. Erlotinib versus docetaxel as second-line treatment of patients with advanced non-small-cell lung cancer and wild-type EGFR tumours (TAILOR): a randomised controlled trial. Lancet Oncol 14, 981–988 (2013). [DOI] [PubMed] [Google Scholar]
- Mascaux C. et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 92, 131–139 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardou H. et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. The Lancet Oncology 9, 962–972 (2008). [DOI] [PubMed] [Google Scholar]
- Ying M., Zhu X., Chen K., Sha Z. & Chen L. Should KRAS mutation still be used as a routine predictor of response to EGFR-TKIs in advanced non-small-cell lung cancer? A revaluation based on meta-analysis. J Cancer Res Clin Oncol, (2015). doi: 10.1007/s00432-015-1910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva S. et al. Across the universe of k-ras mutations in non-small-cell-lung cancer. Curr Pharm Des 20, 3933–3943 (2014). [DOI] [PubMed] [Google Scholar]
- Johnson S. M. et al. RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 (2005). [DOI] [PubMed] [Google Scholar]
- Lee Y. S. & Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21, 1025–1030 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000). [DOI] [PubMed] [Google Scholar]
- Johnson C. D. et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67, 7713–7722 (2007). [DOI] [PubMed] [Google Scholar]
- Kumar M. S. et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 105, 3903–3908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64, 3753–3756 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.