Abstract
The actin crosslinking domain (ACD) is an actin-specific toxin produced by several pathogens, including life-threatening spp. of Vibrio cholerae, Vibrio vulnificus, and Aeromonas hydrophila. Actin crosslinking by ACD is thought to lead to slow cytoskeleton failure owing to a gradual sequestration of actin in the form of nonfunctional oligomers. Here we found that ACD converted cytoplasmic actin into highly toxic oligomers that potently “poisoned” the ability of major actin assembly proteins, formins, to sustain actin polymerization. Thus, ACD can target the most abundant cellular protein by employing actin oligomers as secondary toxins to efficiently subvert cellular functions of actin while functioning at very low doses.
Bacterial toxins are the deadliest compounds on the planet. As little as a single molecule of a delivered toxin can compromise vital functions or even kill an affected host cell (1, 2). This is achieved by amplification of a toxin enzymatic activity via signaling cascades (e.g. by cholera, pertussis, and anthrax toxins) or via enzymatic inhibition of vital host complexes present in relatively few copies (e.g. Shiga and diphtheria toxins acting on ribosomes). Such efficiency is crucial because i) the amount of a toxin produced early upon infection is limited by an initially small number of bacterial cells; ii) the host is protected by commensal bacteria; and iii) the host immune system efficiently neutralizes toxins by means of adaptive (antibodies) and innate (e.g. defensins) (3) humoral defense factors.
Owing to its importance in multiple cellular processes, actin is a common target for bacterium- and parasite-produced toxins. Upon delivery to the cytoplasm of host cells via Type I (as part of MARTX toxin) (4) or Type VI (within VgrG1 toxin) (5) secretion systems, the actin crosslinking domain toxin (ACD) catalyzes the covalent crosslinking of K50 in subdomain 2 of one actin monomer with E270 in subdomain 3 of another actin monomer via an amide bond, resulting in the formation of actin oligomers (6, 7). The actin subunits in the oligomers are oriented similar to short-pitch subunits in the filament, except that a major twist of the subdomain-2, required to accommodate such orientation, disrupts the normal inter-subunit interface and precludes polymerization (6).
The currently accepted mechanism of ACD toxicity, via sequestering of bulk amounts of actin as non-functional oligomers, is compromised owing to the high concentration (hundreds of micromolar) of actin in a typical animal cell. Extrapolation of in vitro determined rates of the ACD activity (7) to cellular conditions suggests that a single ACD molecule per cell (i.e. ~ 1 pM) would require over six months to covalently crosslink half of all cytoplasmic actin.
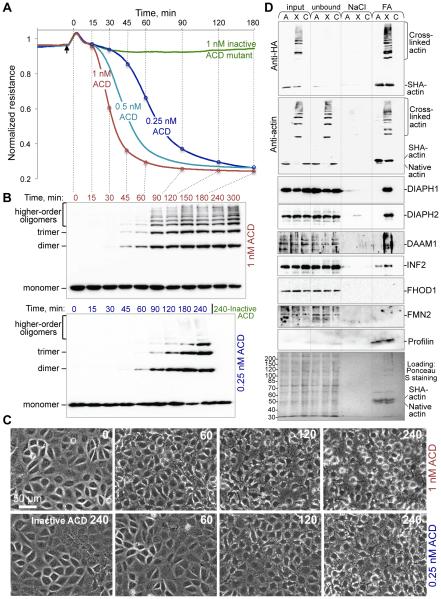
In contrast to these estimations, the integrity of the intestinal cell monolayers was disrupted when only a small fraction of cellular actin (2-6%) was crosslinked by ACD (Fig. 1A-C; fig. S1). To account for such dramatic effects, we hypothesized that the ACD-crosslinked actin oligomers are highly toxic because they can exert an abnormally high affinity to actin-regulatory proteins containing several actin-binding domains. To identify potential high-affinity partners of the actin oligomers, anthrax toxin delivery machinery was used to deliver ACD (8) into HeLa cells transfected with double-tagged (Twin-Strep-tagII and hemagglutinin) actin (SHA-actin; fig. S2) and used for a pull-down assay. Several formins (DIAPH1, DIAPH2, DAAM1, and INF2) preferentially bound to the ACD-crosslinked actin oligomers (Fig. 1D). Treatment of epithelial monolayers with the formin inhibitor SMIFH2 affected the monolayer integrity similar to ACD, whereas the Arp2/3 complex inhibitor CK-666 did not (fig. S3).
Figure 1. Integrity of intestinal monolayers is compromised by low concentration of actin oligomers.
(A-C) Transepithelial electrical resistance (TEER) of IEC-18 monolayers (A) was assessed upon cytoplasmic delivery of LFNACD or a catalytically inactive mutant as a control and correlated with the accumulation of ACD-crosslinked actin species by anti-actin immunoblotting (B) and cell morphology (C). Additional antiactin blots and quantitation of crosslinked actin are presented on fig. S1. (D) SHA-actin pull-down. Lanes A: SHA-actin transfected cells treated with inactive LFNACD (non-crosslinked actin). Lanes X: SHA-actin transfected cells treated with active LFNACD (crosslinked actin). Lanes C: non-transfected untreated cells used as a negative control. “NaCl” and “FA” – fractions eluted from Strep-Tactin beads with 0.5 M NaCl and 50% formamide, respectively. Samples were subjected to immunoblotting and probed with anti-HA, anti-actin, various anti-formin, and anti-profilin antibodies.
Formins are a major family of actin assembly factors involved in numerous actindependent cellular processes. The major functional domains of formins, formin homology domains 1 (FH1) and 2 (FH2), cooperate in nucleation and elongation of actin filaments. A non-covalent FH2/FH2 homodimer nucleates and remains at the polymerizing barbed end to facilitate processive filament elongation while protecting the filament from capping (9). Tandem poly-proline stretches within the FH1 domains bind profilin-actin complexes and accelerate elongation by as much as 10-fold (10-12). FH1 domains of all formins preferentially bound to the oligomers (Fig. 1D) contain 4-14 tandem poly-proline (PP) stretches, which may contribute to strong profilin-mediated interaction with the oligomers.
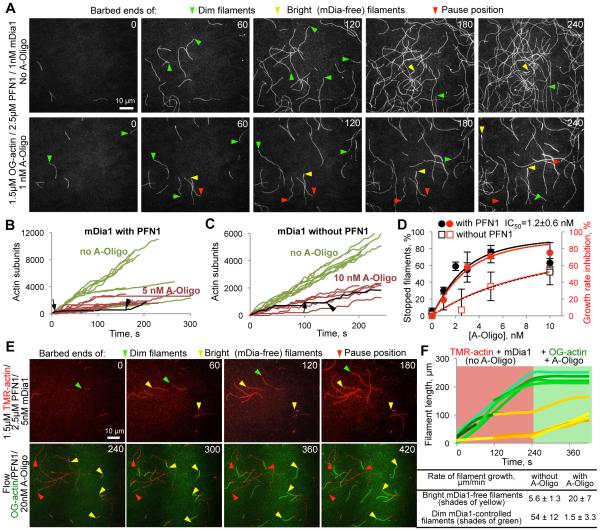
To elucidate the mechanism of formin inhibition, we employed constitutively active FH1-FH2 fragments of mDia1 and mDia2 (mouse orthologues of human DIAPH1 and DIAPH3, respectively) to monitor actin polymerization at the individual filament level by total internal reflection fluorescence microscopy (TIRFM; Fig. 2, 3; fig. S4). In the presence of human profilin-1 (PFN1), the oligomers caused very prominent reversible blocks of elongation of formin-controlled, but not formin-free actin filaments (Fig. 2A-F; fig. S4B,C; Movies S1-5). Formin-controlled filaments were identified by faster growth with a dimmer appearance (Fig. 2A,E) (10), or via direct labeling of formin (Fig. 3A).
Figure 2. Effects of ACD-crosslinked actin oligomers on polymerization of individual filaments controlled by mDia1(14PP).
(A) mDia1(14PP)-mediated polymerization from profilin-actin complexes in the absence (top) and presence (bottom) of actin oligomers (A-Oligo) was monitored by TIRFM. Arrowheads denote actin barbed ends: green – mDia1-controlled (dim and fast); yellow – mDia1-free (bright and slow); red – mDia1-controlled stopped by the oligomers. (B, C) Quantitation of (A): filament elongation plots in the presence (B) or absence (C) of PFN1. Green and red curves describe filament elongation in the absence and presence of oligomers, respectively. Arrows denote the beginning and arrowheads indicate the end of elongation blocks caused by the oligomers on representative curves highlighted in black. (D) IC50 of oligomers determined by TIRFM as percent of stopped filaments (black) or growth rate inhibition (red curves). (E) TMR-labeled actin (red) was polymerized in the presence of mDia1(14PP) and PFN1 without oligomers followed by flow of Oregon Green (OG) actin, oligomers, and PFN1. Arrowheads are as for (A). (F) Quantitation of (E): growth of mDia1-controlled filaments (green traces) and mDia1-free filaments (yellow traces). Better polymerization properties of OG-actin result in faster elongation at the formin-free ends.
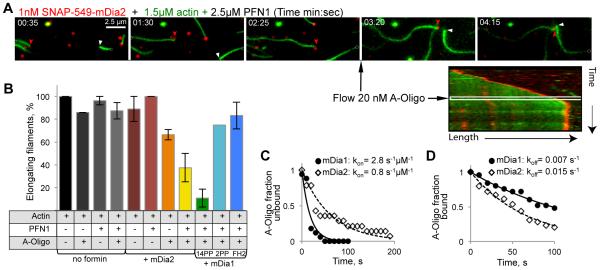
Figure 3. Effects of ACD-crosslinked actin oligomers on polymerization of individual filaments controlled by mDia2 and mDia1 with various FH1 lengths.
(A) OG-actin (green) polymerization in the presence of SNAP-549-mDia2 (red) and PFN1 before and after the addition of oligomers (black arrow) was monitored by TIRFM. Red arrowheads indicate SNAP-549-mDia2 at an actin filament; white arrowheads indicate formin-free filament. Kymograph shows a stalled SNAP-549-mDia2-controlled filament upon addition of oligomers. (B) Effects of oligomers on formin-free filament elongation and elongation controlled by mDia2 and mDia1 formins with various FH1 lengths: 14PP, 2PP, and FH2 (no PP-stretches). (C, D) Oligomer association (kon) (C) and dissociation (koff) (D) rates for mDia1(14PP) and mDia2.
In the presence of PFN1, the fraction of blocked mDia1 formin-associated filaments as well as the inhibition of averaged growth rates depended on the concentration of the added oligomers with an IC50 of 1.2 ± 0.6 (SEM) nM (Fig. 2D), in good agreement with the apparent equilibrium inhibition constant determined kinetically (appKi = koff/kon = 2.5 nM; Fig. 3C,D). After stops (oligomer dissociation), the filaments continued to polymerize with the rates characteristic for formin-controlled filaments (Fig. 2B; fig. S4A). In the absence of PFN1, the inhibition appeared to occur via a similar mechanism, but the overall effect was weaker and the average duration of the blockage events was substantially shorter (Fig. 2C,D). Although a profilin-mediated interaction of the oligomers with the PP stretches of FH1 was not absolutely required, it strongly amplified the efficiency of the inhibition at the elongation stage by contributing to multisite interaction with the oligomers. Thus, mDia1 constructs (fig. S5A) with either removed FH1 domains (FH2 only) or shortened from fourteen (14PP) to two PP-stretches (2PP) showed progressively lower response to inhibition by the oligomers in the presence of PFN1 (Fig. 3B). Similarly, the appKi of oligomers for mDia2 (containing two PP-stretches) was 7.5 fold higher than that for mDia1 and depended on PFN1 (Fig. 3B-D).
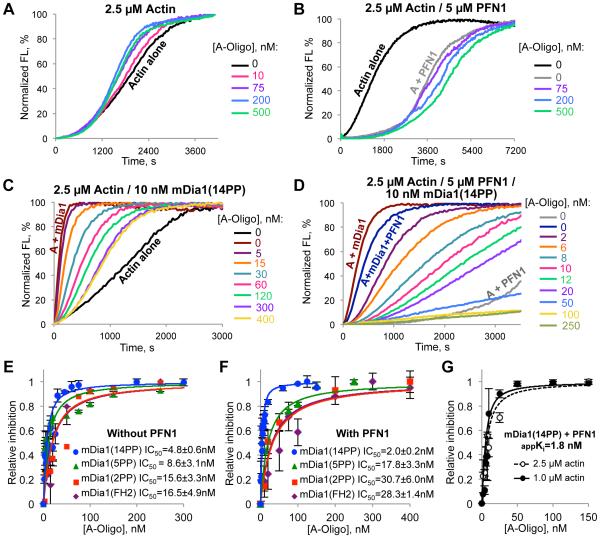
The inhibition of formin-mediated polymerization measured at the individual filament level correlated well with the inhibition observed in bulk pyrene assays (Fig. 4; fig. S5,S6). During spontaneous polymerization in the absence of PFN1, high concentrations (75-500 nM) of the oligomers mildly accelerated the polymerization, while mild inhibition was observed in the presence of profilin (Fig. 4A,B). This is likely because of a low level incorporation of the oligomers into the filaments (6) in the absence, but not in the presence of PFN1 (fig. S5D), leading to filament severing similar to that observed for actin species with impaired inter-subunit surfaces (13).
Figure 4. Actin oligomers inhibit mDia1-controlled actin polymerization in bulk pyrenyl-actin assays.
(A-D) Effects of actin oligomers (A-Oligo) on actin polymerization in the absence (A, B) or presence of mDia1(14PP) (C, D); without (A, C) or with PFN1 (B, D). Normalized FL – pyrene fluorescence expressed in percent of maximum. (E, F) Inhibition of profilin-dependent and independent actin polymerization controlled by various length FH1 mDia1 constructs (14PP, 5PP, 2PP, or FH2 only; see fig. S5A,B and S6) assessed in the absence (E) and presence of PFN1 (F). (G) Apparent Ki for inhibition of mDia1(14PP) by the oligomers in the presence of PFN1 was calculated by measuring IC50 at two different concentrations of actin.
In contrast, the oligomers potently inhibited actin polymerization directed by mDia1 in the presence and, to a lesser extent, absence of PFN1 (Fig. 4C-F; fig. S6). Fitting the inhibition of actin polymerization at 50% of maximum to a binding isotherm equation resulted in an IC50 for the mDia1(14PP) construct equal to 2.0 ± 0.2 (SEM) nM and 4.8 ± 0.6 (SEM) nM in the presence and absence of PFN1 (Fig. 4E,F). The ACD-crosslinked actin dimers purified to homogeneity (fig. S5B) inhibited the mDia1-controlled polymerization less efficiently than the mixture of higher order oligomers (fig. S5F-H), suggesting that the inhibition is amplified via multivalent interactions of the oligomers with mDia1. Accordingly, shortening the FH1 domain progressively decreased the efficiency of inhibition with the IC50 values reaching ~30 and ~16 nM for the mDia1(FH2) constructs in the presence and absence of PFN1, (Fig. 4E,F; fig. S6).
Kinetic modeling (fig. S8) revealed that inhibition of both nucleation and elongation is required to accurately describe the effects of the oligomers on formincontrolled actin polymerization. Using experimentally determined parameter values for inhibition of elongation, good fits to the data (Fig. 4) could be found by assuming that oligomers also inhibit nucleation by binding to free mDia1(14PP) formin with dissociation constants of 0.8 and 5 nM in the presence and absence of PFN1 (fig. S8D,E). Inhibition of nucleation by the oligomers in the absence of PFN1 was also observed experimentally in filament seeding assays (fig. S7) and TIRFM experiments (fig. S4D-G). Similar experiments in the presence of PFN1 were less conclusive owing to the overall lower nucleation ability of formins under these conditions (fig. S7G,H and fig. S4F,G). To improve accuracy, modeling had to account for filament severing owing to incorporation of the oligomers in the absence of PFN1 (Fig. 4A,C; fig. S8C,D).
Bacterial toxins are well known to disorganize the actin cytoskeleton acting via Rho family GTPase controlled signaling pathways (14). Here we found that toxins can not only exploit existing signaling pathways, but also initiate a new toxicity cascade with de novo produced crosslinked actin species as “second messengers”. Owing a unique combination of properties that is neither present in G-nor F-actin (fig. S9A), these new actin species bind with high affinity to formins and adversely affect both nucleation and elongation abilities of these proteins causing their potent inhibition in profilin-dependent and independent manners (fig. S9B). Thus, ACD creates toxic derivatives of actin with a disruptive “gain of function” mode of operation. We propose that the seemingly straightforward original assumption that ACD acts via the accumulation of bulk amounts of non-functional actin is inaccurate, or at least incomplete. The toxin can be highly efficient at very low concentrations by acting on formins and potentially other actin regulatory proteins. This finding calls for the careful re-evaluation of mechanisms employed by other actin-related toxins, both of protein and small-molecule nature.
Supplementary Material
Acknowledgments
We thank M. Vartiainen and T. Viita (University of Helsinki) for donating double-tagged SHA-actin plasmid. This work was supported by OSU startup funds (to DSK), AHA IRG (13IRG14780028 to DSK), NIH (RO1 GM079265 to DRK and RO1 GM098430 to DV). DBH purified proteins, conducted pyrene actin and TIRFM experiments, analyzed data, wrote manuscript; EK conducted cell culture, pull-down and immunoblotting experiments, analyzed data, wrote manuscript; DG purified proteins, conducted pyrene actin assays; CS, JDW and DRK conducted TIRFM and analyzed data; KGB, SRK and NLP conducted TEER; DV performed modeling; DSK coordinated the study, analyzed data, wrote manuscript. The authors declare no conflict of interest. Supplementary Materials contain additional data.
Footnotes
Supplementary Materials:
Materials and Methods
Figures S1-S9
Table S1
References (15-45)
Movies S1-S5
Modeling Program
References
- 1.Tam PJ, Lingwood CA. Membrane cytosolic translocation of verotoxin A1 subunit in target cells. Microbiology. 2007;153:2700–2710. doi: 10.1099/mic.0.2007/006858-0. published online EpubAug (10.1099/mic.0.2007/006858-0) [DOI] [PubMed] [Google Scholar]
- 2.Yamaizumi M, Mekada E, Uchida T, Okada Y. One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 3.Kudryashova E, Quintyn R, Seveau S, Lu W, Wysocki VH, Kudryashov DS. Human defensins facilitate local unfolding of thermodynamically unstable regions of bacterial protein toxins. Immunity. 2014;41:709–721. doi: 10.1016/j.immuni.2014.10.018. published online EpubNovember 20 (10.1016/j.immuni.2014.10.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheahan KL, Cordero CL, Satchell KJ. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc Natl Acad Sci U S A. 2004;101:9798–9803. doi: 10.1073/pnas.0401104101. published online EpubJun 29 (10.1073/pnas.0401104101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. published online EpubSep 25 (10.1073/pnas.0706532104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudryashov DS, Durer ZA, Ytterberg AJ, Sawaya MR, Pashkov I, Prochazkova K, Yeates TO, Loo RR, Loo JA, Satchell KJ, Reisler E. Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin. Proc Natl Acad Sci U S A. 2008;105:18537–18542. doi: 10.1073/pnas.0808082105. published online EpubNov 25 (10.1073/pnas.0808082105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudryashova E, Kalda C, Kudryashov DS. Glutamyl phosphate is an activated intermediate in actin crosslinking by actin crosslinking domain (ACD) toxin. PloS one. 2012;7:e45721. doi: 10.1371/journal.pone.0045721. 10.1371/journal.pone.0045721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. See supplementary materials on Science Online.
- 9.Breitsprecher D, Goode BL. Formins at a glance. Journal of cell science. 2013;126:1–7. doi: 10.1242/jcs.107250. published online EpubJan 1 (10.1242/jcs.107250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. published online EpubJan 27 (10.1016/j.cell.2005.11.038) [DOI] [PubMed] [Google Scholar]
- 11.Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of forminassociated actin filament elongation. Molecular cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. published online EpubFeb 17 (10.1016/j.molcel.2006.01.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. published online EpubOct 29 (10.1016/j.cell.2004.09.039) [DOI] [PubMed] [Google Scholar]
- 13.Kudryashov DS, Phillips M, Reisler E. Formation and destabilization of actin filaments with tetramethylrhodamine-modified actin. Biophys J. 2004;87:1136–1145. doi: 10.1529/biophysj.104.042242. published online EpubAug (10.1529/biophysj.104.042242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemichez E, Aktories K. Hijacking of Rho GTPases during bacterial infection. Experimental cell research. 2013;319:2329–2336. doi: 10.1016/j.yexcr.2013.04.021. published online EpubSep 10 (10.1016/j.yexcr.2013.04.021) [DOI] [PubMed] [Google Scholar]
- 15.Arora N, Leppla SH. Fusions of anthrax toxin lethal factor with shiga toxin and diphtheria toxin enzymatic domains are toxic to mammalian cells. Infection and immunity. 1994;62:4955–4961. doi: 10.1128/iai.62.11.4955-4961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milne JC, Blanke SR, Hanna PC, Collier RJ. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol Microbiol. 1995;15:661–666. doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 17.Cordero CL, Kudryashov DS, Reisler E, Satchell KJ. The Actin cross- linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. The Journal of biological chemistry. 2006;281:32366–32374. doi: 10.1074/jbc.M605275200. published online EpubOct 27 (10.1074/jbc.M605275200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wesche J, Elliott JL, Falnes PO, Olsnes S, Collier RJ. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. published online EpubNov 10 (10.1021/bi981436i) [DOI] [PubMed] [Google Scholar]
- 19.Geissler B, Bonebrake A, Sheahan KL, Walker ME, Satchell KJ. Genetic determination of essential residues of the Vibrio cholerae actin cross-linking domain reveals functional similarity with glutamine synthetases. Mol Microbiol. 2009;73:858–868. doi: 10.1111/j.1365-2958.2009.06810.x. published online EpubSep (10.1111/j.1365-2958.2009.06810.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sliman SM, Eubank TD, Kotha SR, Kuppusamy ML, Sherwani SI, Butler ES, Kuppusamy P, Roy S, Marsh CB, Stern DM, Parinandi NL. Hyperglycemic oxoaldehyde, glyoxal, causes barrier dysfunction, cytoskeletal alterations, and inhibition of angiogenesis in vascular endothelial cells: aminoguanidine protection. Molecular and cellular biochemistry. 2010;333:9–26. doi: 10.1007/s11010-009-0199-x. published online EpubJan (10.1007/s11010-009-0199-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spudich JA, Watt S. The Regulation of Rabbit Skeletal Muscle Contraction. Journal of Biological Chemistry. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 22.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophysical journal. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. published online EpubFeb (10.1529/biophysj.104.047399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudryashova E, Heisler D, Zywiec A, Kudryashov DS. Thermodynamic properties of the effector domains of MARTX toxins suggest their unfolding for translocation across the host membrane. Mol Microbiol. 2014 doi: 10.1111/mmi.12615. published online EpubApr 14 (10.1111/mmi.12615) [DOI] [PubMed] [Google Scholar]
- 24.Kudryashov DS, Cordero CL, Reisler E, Satchell KJ. Characterization of the enzymatic activity of the actin cross-linking domain from the Vibrio cholerae MARTX Vc toxin. The Journal of biological chemistry. 2008;283:445–452. doi: 10.1074/jbc.M703910200. published online EpubJan 4 (10.1074/jbc.M703910200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Molecular biology of the cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chemistry & biology. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. published online EpubNov 25 (10.1016/j.chembiol.2009.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Developmental cell. 2015;32:43–53. doi: 10.1016/j.devcel.2014.10.027. published online EpubJan 12 (10.1016/j.devcel.2014.10.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. published online EpubOct 12 (10.1073/pnas.0405902101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci U S A. 2014;111:4121–4126. doi: 10.1073/pnas.1322093111. published online EpubMar 18 (10.1073/pnas.1322093111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudryashov DS, Grintsevich EE, Rubenstein PA, Reisler E. A nucleotide state-sensing region on actin. The Journal of biological chemistry. 2010;285:25591–25601. doi: 10.1074/jbc.M110.123869. published online EpubAug 13 (10.1074/jbc.M110.123869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 33.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. published online EpubJan 21 (10.1021/bi026520j) [DOI] [PubMed] [Google Scholar]
- 34.Paul AS, Pollard TD. The role of the FH1 domain and profilin in forminmediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. published online EpubJan 8 (10.1016/j.cub.2007.11.062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Courtemanche N, Pollard TD. Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem. 2012;287:7812–7820. doi: 10.1074/jbc.M111.322958. published online EpubMar 2 (10.1074/jbc.M111.322958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero S, Didry D, Larquet E, Boisset N, Pantaloni D, Carlier MF. How ATP hydrolysis controls filament assembly from profilin-actin: implication for formin processivity. J Biol Chem. 2007;282:8435–8445. doi: 10.1074/jbc.M609886200. published online EpubMar 16 (10.1074/jbc.M609886200) [DOI] [PubMed] [Google Scholar]
- 37.Neidt EM, Scott BJ, Kovar DR. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J Biol Chem. 2009;284:673–684. doi: 10.1074/jbc.M804201200. published online EpubJan 2 (10.1074/jbc.M804201200) [DOI] [PubMed] [Google Scholar]
- 38.Yarmola EG, Dranishnikov DA, Bubb MR. Effect of profilin on actin critical concentration: a theoretical analysis. Biophys J. 2008;95:5544–5573. doi: 10.1529/biophysj.108.134569. published online EpubDec 15 (10.1529/biophysj.108.134569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittenmayer N, Jandrig B, Rothkegel M, Schluter K, Arnold W, Haensch W, Scherneck S, Jockusch BM. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell. 2004;15:1600–1608. doi: 10.1091/mbc.E03-12-0873. published online EpubApr (10.1091/mbc.E03-12-0873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- 41.Courtemanche N, Pollard TD. Interaction of profilin with the barbed end of actin filaments. Biochemistry. 2013;52:6456–6466. doi: 10.1021/bi400682n. published online EpubSep 17 (10.1021/bi400682n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gieselmann R, Kwiatkowski DJ, Janmey PA, Witke W. Distinct biochemical characteristics of the two human profilin isoforms. European journal of biochemistry / FEBS. 1995;229:621–628. doi: 10.1111/j.1432-1033.1995.tb20506.x. [DOI] [PubMed] [Google Scholar]
- 43.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. published online EpubAug (10.1016/S0006-3495(01)75731-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmoller KM, Niedermayer T, Zensen C, Wurm C, Bausch AR. Fragmentation is crucial for the steady-state dynamics of actin filaments. Biophys J. 2011;101:803–808. doi: 10.1016/j.bpj.2011.07.009. published online EpubAug 17 (10.1016/j.bpj.2011.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sept D, Xu J, Pollard TD, McCammon JA. Annealing accounts for the length of actin filaments formed by spontaneous polymerization. Biophys J. 1999;77:2911–2919. doi: 10.1016/s0006-3495(99)77124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.