Abstract
Background Plants are hotbeds for parasites such as arthropod herbivores, which acquire nutrients and energy from their hosts in order to grow and reproduce. Hence plants are selected to evolve resistance, which in turn selects for herbivores that can cope with this resistance. To preserve their fitness when attacked by herbivores, plants can employ complex strategies that include reallocation of resources and the production of defensive metabolites and structures. Plant defences can be either prefabricated or be produced only upon attack. Those that are ready-made are referred to as constitutive defences. Some constitutive defences are operational at any time while others require activation. Defences produced only when herbivores are present are referred to as induced defences. These can be established via de novo biosynthesis of defensive substances or via modifications of prefabricated substances and consequently these are active only when needed. Inducibility of defence may serve to save energy and to prevent self-intoxication but also implies that there is a delay in these defences becoming operational. Induced defences can be characterized by alterations in plant morphology and molecular chemistry and are associated with a decrease in herbivore performance. These alterations are set in motion by signals generated by herbivores. Finally, a subset of induced metabolites are released into the air as volatiles and function as a beacon for foraging natural enemies searching for prey, and this is referred to as induced indirect defence.
Scope The objective of this review is to evaluate (1) which strategies plants have evolved to cope with herbivores and (2) which traits herbivores have evolved that enable them to counter these defences. The primary focus is on the induction and suppression of plant defences and the review outlines how the palette of traits that determine induction/suppression of, and resistance/susceptibility of herbivores to, plant defences can give rise to exploitative competition and facilitation within ecological communities “inhabiting” a plant.
Conclusions Herbivores have evolved diverse strategies, which are not mutually exclusive, to decrease the negative effects of plant defences in order to maximize the conversion of plant material into offspring. Numerous adaptations have been found in herbivores, enabling them to dismantle or bypass defensive barriers, to avoid tissues with relatively high levels of defensive chemicals or to metabolize these chemicals once ingested. In addition, some herbivores interfere with the onset or completion of induced plant defences, resulting in the plant’s resistance being partly or fully suppressed. The ability to suppress induced plant defences appears to occur across plant parasites from different kingdoms, including herbivorous arthropods, and there is remarkable diversity in suppression mechanisms. Suppression may strongly affect the structure of the food web, because the ability to suppress the activation of defences of a communal host may facilitate competitors, whereas the ability of a herbivore to cope with activated plant defences will not. Further characterization of the mechanisms and traits that give rise to suppression of plant defences will enable us to determine their role in shaping direct and indirect interactions in food webs and the extent to which these determine the coexistence and persistence of species.
Keywords: Herbivory, plant defence, herbivore, jasmonate, salicylate, induction, suppression, manipulation, detoxification, sequestration, resistance, community interactions, facilitation, adaptation, plant–animal interaction
INTRODUCTION
At a first glimpse, plants seem subject to the caprices of their environment, being constantly confronted with stresses such as drought, heat or ultraviolet radiation while frequently being challenged by pathogens or herbivores. However, these stresses have posed selection pressures on plants that have resulted in numerous adaptations, ranging from stress tolerance and resistance to the ability to manipulate their environment. Many of these adaptive traits have multiple functions. For example, plants are equipped with a waxy cuticle that prevents evaporation of water, but also forms an efficient barrier against phytopathogens (Eigenbrode and Espelie, 1995). Moreover, many species have epidermal leaf hairs (trichomes), some of which carrying a special gland at the tip, and for which a broad diversity of functions have been reported. For example, they form structural barriers that hinder small arthropods in their mobility (Simmons and Gurr, 2005), but also guide light to the leaf surface of the leaf so that it can be used optimally for photosynthesis (Wagner et al., 2004) while filtering out UV-A and -B (Karabourniotis and Bornman, 1999). Moreover, glandular hairs also secrete protective coatings that prevent fungal spores from germinating (Shepherd et al., 2005), and contain glues and toxins that obstruct and intoxicate plant-surface-dwelling arthropods when ruptured (Glas et al., 2012). Preformed structural barriers are referred to as ‘constitutive defences’ and they augment the so-called induced defences, which become operational only in response to an attack.
Defence against herbivores requires measures different from defence against pathogens. In general, phytopathogens are less mobile than herbivores, and migration across or through their host plant is often passive and occurs over relatively short distances. For example, plant-infecting viruses usually need plant proteins for tissue-to-tissue transport, plant-infecting fungi can only relocate by growing longer hyphae and plant-infecting flagellar bacteria can only travel short distances by taxis in fluid media. Hence, the infected host can attack such relatively immobile pathogens on the spot; at the site of infection, plants will often respond by producing structural reinforcements (e.g. cell wall thickening and callose deposition) or toxins (e.g. phytoalexins or alkaloids), or by initiating programmed local cell death (apoptosis) to isolate and possibly kill the pathogen (Dangl and Jones, 2001). Programmed cell death, orchestrated by the hypersensitive response, is highly efficient in preventing pathogens from spreading and is therefore one of the most common anti-parasite defence strategies found in nature. Herbivores, however, are not very susceptible to this isolate-and-kill strategy because they are mobile. Therefore, anti-herbivore defences generally come down to a go-away-or-die strategy or a slow-them-down strategy, and these two strategies share many physiological characteristics. In both instances, plants will mount a sequence of defence programmes that serve to interfere with herbivore growth and development on the one hand and to reallocate resources on the other, in order to delay growth of the herbivores into larger individuals, stages or populations, which consume more plant tissue. For example, well-known anti-herbivore defence proteins include proteinase inhibitors (PIs) and polyphenol oxidases (PPOs), both believed to interfere with digestive processes in the herbivore gut (Zhu-Salzman et al., 2008). Hence, the simultaneous reallocation of resources may not only serve to rescue resources so that the plant can use them later for growth and reproduction, but may also serve to deprive the herbivore of food. This may be an effective strategy, especially when the plant is attacked by small herbivores or relatively immobile (immature) stages. Resource allocation is characterized by reallocation of nutritious carbon-containing (Schwachtje et al., 2006; Babst et al., 2008) and nitrogen-containing (Newingham et al., 2007; Gomez et al., 2012) substances to either the reproductive tissues or the storage organs, such as roots (Anten and Pierik, 2010). Hence, it can give rise to rapid flowering or to a period of dormancy (Stowe et al., 2000) to regrow later, depending on the life-history characteristics of both plant and herbivore. Consequently, herbivores that decide to stay on defended plants select plant tissues where defences are lower (Paschold et al., 2007; Shroff et al., 2008; Stork et al., 2009) and/or increase their feeding intensity to gain sufficient biomass, thus compensating for the decreased efficiency of food conversion (Gomez et al., 2012). Plants, in turn, often initiate systemic responses (Pieterse et al., 2009) to decrease the chance that the herbivore will simply move to undefended tissues (Paschold et al., 2007) and furthermore produce secondary metabolites to constrain compensatory feeding responses (Steppuhn and Baldwin, 2007).
The sequence of defence programmes executed by plants under attack appears not to be fully hard-wired, suggesting a certain degree of herbivore-specific tailoring by the plant, and while the early responses seem usually aimed at rescuing the attacked tissue, they may shift towards senescence and tissue death (reminiscent of the hypersensitive response) after a couple of days (Steinbauer et al., 2014). It is unknown whether, how and when the plant decides to switch from the ‘rescue’ response to the ‘scorched earth’-like tactic and whether this is characterized by a partial (Kahl et al., 2000) or complete induced shut-down of local metabolic activity, e.g. to initiate senescence (Gross et al., 2004). Moreover, it is unclear whether, and if so for how long, tissues that are being sacrificed are supported by photosynthates (Ferrieri et al., 2013) and by defensive products from distal tissues (Nabity et al., 2009). From the herbivore’s point of view, resource depletion at the feeding site may represent a more difficult problem to deal with than toxins, because they cannot develop resistance to an absence of nutrients. However, some herbivores such as gall-makers have evolved abilities to manipulate plant resource flows and turn their feeding site into a sink for resources (Tooker et al., 2008), but also leaf-cutters and trenchers (Dussourd and Denno, 1991) that prevent plants from transporting defence compounds to the feeding site and possibly also from transporting resources away from it.
Whereas the role of resource allocation remains under-studied, induced plant defences and their effects on herbivores have been analysed in great detail. In this review, we evaluate what determines the susceptibility of arthropod herbivores to plant defences and which herbivore traits counteract these defences. We focus on the induction and suppression of plant defences and outline how traits determining the susceptibility of parasites to plant defences can give rise to exploitative competition and facilitation in the ecological communities inhabiting the plant. First, we describe the diversity of herbivore feeding modes and life styles with which plants have to cope, and how plants detect the presence of herbivores through saliva and oral secretions.
HERBIVORE FEEDING AND PLANT DEFENCE STRATEGIES
Herbivore feeding styles
Early in evolution, herbivorous arthropods were mandibulate and chewed on vegetation (Budd and Teldford, 2009). After vascular plants had emerged, many different forms of feeding evolved, such as sap-sucking, leaf-mining, gall-forming and nectar-feeding (Grimaldi and Engel, 2005). Among the arthropods, many differently shaped mouthparts can be found which are suitable for chewing, siphoning, piercing–sucking and sponging, or a combination of these. Chewing insects, such as many lepidopteran larvae, but also beetles and grasshoppers, have two mandibles, surrounded by sensory organs, which they use for crushing or cutting food while mixing it with salivary secretions before swallowing it. This type of feeding removes relatively large quantities of leaf material. Siphoning insects, typically adult lepidopterans, feed by sucking without piercing. Often, these insects have a long coiled proboscis that can be extended to reach food such as nectar. Siphoning itself does not cause much damage, although flowers may be damaged severely by an agile siphoning insect (Kessler et al., 2008). The piercing–sucking arthropods feed by means of stylets, which are needle-shaped mouthparts. Stylets are common among insects and mites, although they probably evolved independently in these groups and possibly even more than once among mites (Manton and Harding, 1964; Caravas and Friedrich, 2010). The insect stylet is a single anatomical structure, often equipped with two separate channels, one for releasing saliva and the other for ingesting food (Miles, 1999). Mite mouthparts are composed of two or more stylets, which can be grouped together in pairs to form a piercing structure with an internal duct, which probably serves to release saliva (Ragusa and Tsolakis, 2000). While stylet-feeding insects such as whiteflies, psyllids and aphids usually feed from vascular tissue (Miles, 1999), herbivorous mites feed on the contents of mesophyll cells (Park and Lee, 2002). Insects such as thrips are usually also considered stylet–mesophyll feeders, but have an asymmetrical mouth cone, i.e. they have a single mandibular stylet, which is used for piercing and rasping plant tissue, after which their acinial stylet is inserted in the wound to take up food (punch-and-suck feeding) (Coll and Guershon, 2002; Kindt et al., 2003). Finally, sponging insects have non-functional mandibula but an extended food channel that ends in a sponge-like labellum. They secrete saliva, which is dabbed onto their (solid) food to liquefy it, after which the labium channels the food to the oesophagus. This mode of feeding is typical of many species of flies (Vijaysegaran et al., 1997). Thus, arthropods have diverse feeding styles and, especially among insects, different life stages can make use of distinct modes of feeding.
Different herbivore life stages feed differently
Different feeding styles accompany diverse life styles of herbivorous arthropods. These can differ considerably across stages of the life cycle, especially in holometabolous arthropods, but also in many hemimetabolous species. For example, adult thrips are winged and feed mostly from plant tissues. They deposit their eggs under the epidermis of the host plant and the wingless larvae that emerge from these eggs move across the plant surface to more protected areas and feed from pollen, nectar and/or leaf tissue until they develop into the nymphal stage (the third larval stage) and move into the soil to pupate (Broadbent et al., 2003). Many other herbivores also spend part of their life cycle inside the host plant and/or the soil. Many lepidopteran and dipteran species have larval stages called leaf miners; they live inside a leaf or in needle tissue, where they feed and find protection. The adults of leaf miners are free-living but deposit their eggs underneath the leaf cuticle, where the larvae usually stay to graze the mesophyll until they develop into pupae. This feeding behaviour causes the formation of distinct blotches or serpentine tunnels on plant leaves (Connor and Taverner, 1997). In addition, leaf-rollers are moth larvae that feed and pupate within the protection of rolled-up leaves; these leaves are either cut and folded or rolled up and wrapped by the larvae in their silk to create a shelter and feeding site (Gaston et al., 1991). Leaf miners and leaf-rollers usually cause minor damage, i.e. mostly aesthetic, but can cause defoliation and fruit malformation when densities are too high (Witzgall et al., 2008). Another group of herbivores are the stem and root borers, which deposit their eggs in stems or roots and whose larvae consume these tissues. They can be very damaging, not only because they often cause stunted growth, but also because they weaken the plant structure. Some species of borers have specialized in specific plant tissues such as fruits and seeds (frugivores) or bark (Mainali, 2014). Whereas leaf rollers physically create their shelter, galling insects induce somatic plant tissues to form domatia (Stone and Shonrogge, 2003). Many species of gall midges (Harris et al., 2003) and gall mites (Van Leeuwen et al., 2010a) manipulate the plant into producing tumorous outgrowths by mitosis to form hollow structures in which they spend a substantial part of their life cycle and which often serve as a feeding site, reminiscent of galls induced by root-knot nematodes (Caillaud et al., 2008) or Agrobacterium (Zhu et al., 2000). Many eriophyids do not induce protective cavities but other types of external malformations, like tufts or hairy outgrowths (erineum), in which they can seek shelter, while others induce deterioration rather than formation of plant leaf hairs (Karioti et al., 2011; Van Houten et al., 2013). Finally, most sap feeders, such as aphids, whiteflies and psyllids, spend their whole life cycle on the plant surface. However, the juvenile stages of these homopterans can be relatively immobile and sometimes cover themselves with honeydew for protection (VanDoorn et al., 2015). Because phloem lacks essential amino acids, many homopteran species possess bacteriomes which harbour symbiotic bacteria that provide these and vitamins in return for nutrients (Schwemmler, 1989). Thus, plants encounter a wide range of different attackers and are therefore in need of defence systems that allow a degree of tailoring.
Plant defence theories
Understanding the mechanisms that plants have evolved to defend themselves and identification of the ecological drivers of this evolution have been major challenges during recent decades. Whereas research focusing on plant physiological aspects of these defences mostly worked from a scenario in which a single plant species is attacked by a single species, it has become increasingly clear that the diversity of ecological interactions within plant-inhabiting communities is an important determinant of the evolution of plant defence strategies. This notion gave rise to several theoretical frameworks revolving around the central dilemma that plant defences require resources that would otherwise be available for growth and reproduction (Mattson, 1980; Stamp, 2003). These frameworks can be classified into two groups: one of these attempts to explain the distribution of plant defences based on defensive function and plant life history, whereas the other group attempts to explain this based on resource availability. The first theoretical framework is the optimal defence theory, which hypothesizes that plants with limited resources will defend different tissues differently, depending on the chance they will be attacked, the fitness value of the tissue and the cost of the defence. It was suggested that such a strategy will impose an ‘evolutionary dilemma’ leading to selection for defences at intermediate levels when plants are frequently attacked by generalists and specialists, because the latter will generally be more resistant to the plant’s defences than the former (Zangerl and Rutledge, 1996). This theory is related to the plant apparency hypothesis (Feeny, 1976), which poses that plants that are attacked by a relatively large diversity of herbivores will need to display a larger diversity of defences than plants that are targeted by few species. It assumes that, compared with short-lived species, long-lived species are more likely to encounter generalists as well as specialists during their lives and hence have been under selection to display a broader range of defences. Third is the carbon:nutrient balance hypothesis, which hypothesizes that plant defences are constrained by nutrient variation in the environment and posits that the C:N ratio will dictate which secondary metabolites are synthesized (Bryant et al., 1983). This hypothesis predicts that changes in available nutrients will change the palette of defences. The fourth is the resource availability hypothesis, which posits that defence strategies are determined by the inherent growth rate of the plant, which is assumed to be constrained by resource availability (Coley et al., 1985). This theory implies that a trade-off between growth rate and defences will restrict species to particular habitats. Finally, the fifth theory is based on the growth–differentiation balance hypothesis and it subsumes elements of the previous hypotheses (Herms and Mattson, 1992). It posits that the ecological costs of the physiological trade-off between growth and secondary metabolism (defence) vary across environments. Since plants must protect their acquired resources, which are needed for growth in order to be able to compete for new resources, natural selection has shaped their secondary metabolism to be flexible and their life histories to vary across environments. Hence, this theory aims to explain patterns of phenotypic and genetic variation in secondary metabolism in response to environmental variation and resource gradients. It needs to be said that it has been difficult to generate testable hypotheses from these defence theories, not only because the magnitudes of the costs and benefits of defences have proved to be very difficult to measure, but also since they often fail both to distinguish among evolutionary, ecological and physiological levels of analysis and to clearly distinguish between genetic and environmental influences.
Plant defence strategies
Plant defences are often divided into three basal strategies: deterrence (antixenosis), resistance (antibiosis) and tolerance (a compensation strategy to reduce the detrimental effects of herbivory). Deterrence traits are usually constitutively expressed and can emanate from colours, odours or textures (such as hairs) that demotivate a herbivore from feeding on the plant, or from the absence of feeding stimuli that otherwise would stimulate the attacker. Resistance traits are those that can injure or kill a herbivore or slow its development and reproduction. Finally, tolerance comes from those traits that do not primarily serve to negatively interact with the herbivore, but to compensate for damage through changes in assimilation rate, compensatory growth, phenological shifts, resource allocation or morphological changes. These three strategies are not mutually exclusive and can overlap mechanistically and functionally. Hence, it will often be difficult to tell these three strategies apart (Stout, 2013) and it is doubtful whether deterrence as a stand-alone defence strategy will be evolutionarily stable since it offers ample opportunities for herbivores to adapt.
Constitutive plant defences
Plants cannot simply accumulate all the defences that have emerged during the course of evolution within a ‘super-genotype’ because defensive structures, compounds or processes such as the inducible defences (Baldwin, 1998) cost energy to form and maintain. Hence, only those defences for which selection pressure has been constant and strong enough have been retained. Moreover, the optimal defence theory predicts that plants with limited resources are selected to arrange the relatively costly and less costly defences across tissues based on the fitness value of these tissues. Moreover, assuming that the induced defences are overall less costly than constitutive defences, this theory predicts that tissues with a higher probability of being attacked will rely more on constitutive defences, whereas tissues with a lower probability of being attacked will depend more on induced defences. Indeed, the reproductive parts of wild parsnip (Pastinaca sativa) had the highest levels of constitutive furanocoumarins with low inducibility and had the highest probability of being attacked, while in the roots, which were less frequently attacked, constitutive levels were relatively low but highly inducible (Zangerl and Rutledge, 1996).
Discriminating experimentally between constitutive and induced defences is often not easy since there can be considerable overlap. For example, the size and density of physical barriers such as spines and plant hairs (Glas et al., 2012), which are commonly considered to function as constitutive defences, can also be increased by induction (Traw and Dawson, 2002). For example, formation of more and longer thorns was induced by giraffes feeding on acacia (Acacia seyal) trees (Milewski et al., 1991). Moreover, not only are leaf hairs mechanical barriers (Pott et al., 2012), but glandular trichomes (Tissier, 2012) are important production sites of a wide variety of constitutive and induced secondary metabolites with defensive functions, including terpenoids, phenylpropenes, flavonoids, methylketones, acyl sugars and defensive proteins, and some of these compounds are inducible. For instance, the emission of terpenoids (Van Schie et al., 2007) and the production of acyl sugars and defensive proteins (Hare and Walling, 2006) can be induced in trichomes by treating plants with methyl jasmonate, a compound that activates the jasmonate (JA) signal transduction cascade, also induced by herbivores. Even mere contact of insects with a plant was found to suffice to induce the expression of PIs in glandular trichomes (Peiffer et al., 2009). The contents of glandular trichomes, such as sticky acyl sugars and polyphenols, can be excreted or released after they are ruptured by insect movement, and cause entrapment of small herbivores, often followed by death (Simmons et al., 2004). Trichomes of tobacco (Nicotiana tabacum) also produce defensive proteins that are secreted to the leaf surface and inhibit germination of oomycete spores (Shepherd et al., 2005). Conversely, some pathogens, like Pseudomonas syringae on tomato, have adapted to use trichomes as their habitat (Schneider and Grogan, 1977), and damaged trichomes may be used as an entry point by these bacteria (Huang, 1986). Thus, trichomes are important components of both the constitutive and the inducible defence system, and trichome secretions function to hinder herbivore feeding and the germination of fungal spores. Other forms of constitutive defence involve the plant’s waxy cuticle; this primarily serves to prevent the evaporation of water (Buschhaus and Jetter, 2012), but wax morphology and chemistry contribute to a plant’s resistance by restraining herbivore foraging behaviour (Eigenbrode and Shelton, 1990). Also, leaf toughness, which includes cell wall lignification, is known to deter herbivore feeding (Choong, 1996) and is positively correlated with resistance to pathogens (Bhuiyan et al., 2009).
Induced plant defence
Induced defences are often subdivided into direct and indirect defences. Direct defence includes the activation or production of antifeedants, such as toxins and inhibitors of digestion, which negatively affect the growth and/or survival of herbivores (Howe and Jander, 2008). The existence of induced defences has been known for more than 100 years (early work is reviewed by Chester, 1933). A well-known example of a herbivore-induced plant defence is increased PI gene expression and enzyme activity. Although most plant PIs have regulatory roles in the plant’s endogenous protein metabolism, the herbivore-induced PIs inhibit proteases in the gut of herbivores, thereby decreasing the plant’s palatability and increasing its resistance (Hartl et al., 2010). Defences may also be induced in the phloem (Will et al., 2013). For instance, feeding on rice by the brown planthopper (Nilaparvata lugens) induces the deposition of callose on the plant’s sieve plates to block further transport of sap through the attacked phloem tissues (Hao et al., 2008). Another mechanism of such sieve tube occlusion is dependent on large protein bodies called forisomes, which can seal off sieve elements in a Ca2+-dependent manner (Furch et al., 2007) to obstruct herbivore feeding.
Indirect defence refers to plant traits that enhance attraction or arrestment of natural enemies of the herbivore, such as predators and parasitoids (Sabelis et al., 2001). Often, this type of defence is inducible. That natural enemies of herbivores use plant odours for locating prey has been suggested several times (reviewed by Vinson, 1976), and Dicke and Sabelis (1988) outlined a framework for the mode of action and the evolution of indirect defence strategies, mediated by so-called infochemicals, which forms the basis for our current view of the phenomenon. Since then, induced indirect defences have been reported for many plant species under laboratory conditions, including Arabidopsis (van Poecke et al., 2001), cotton (De Moraes et al., 1998), tomato (Kant et al., 2004) and maize (Schnee et al., 2006). In 1999, Thaler showed that indirect defences can act in the field while, in 2001, Kessler and Baldwin showed that plant volatiles can establish indirect defences under natural conditions. They supplemented Nicotiana attenuata plants with synthetic volatiles and some of these increased the natural predation of herbivore eggs and repelled adult moths. In a later study with transgenic plants that were silenced for genes involved in volatile production, the same group showed that indirect defences can actually promote a plant’s fitness under natural conditions (Schuman et al., 2012). Moreover, it was found that hyperparasitoids also respond to herbivore-induced plant volatiles; volatiles released by plants infested with parasitized caterpillars attracted more hyperparasitoids than volatiles emitted by plants infested with healthy caterpillars (Poelman et al., 2012). Indirect defence is known to occur below ground as well. A well-known example is the release of the volatile β-caryophyllene by maize roots into the soil when attacked by larvae of the beetle Diabrotica virgifera virgifera; this compound was shown to function as an attractant for entomopathogenic nematodes that attack the beetle larvae (Rasmann et al., 2005). Finally, restoring this function in maize varieties deficient in the release of β-caryophyllene from roots also increased attraction of the nematodes (Degenhardt et al., 2009).
Volatiles are not the only means by which plants can increase the abundance of natural enemies in their vicinity. Natural enemies can be arrested by providing them with food, e.g. extrafloral nectar (Pemberton and Lee, 1996) or food bodies (Fischer et al., 2002). Also, dead insects entrapped on sticky plants were shown to attract predatory insects such that overall herbivore damage decreased and fruit production increased (Krimmel and Pearse, 2013). Finally, an alternative means by which plants establish indirect defence is to provide shelter (domatia) such as cavities or tufts of hair, for small natural enemies, which these can use to moult and/or to protect their eggs (Walter, 1996).
HERBIVORE DIGESTION, SALIVA AND REGURGITATION
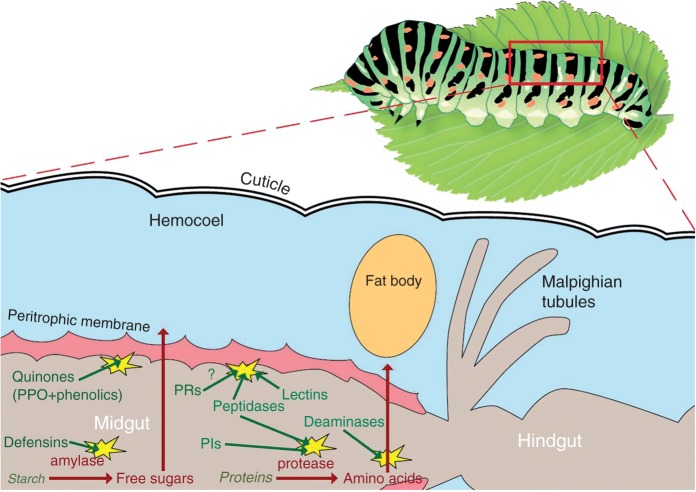
A herbivore’s host-plant range is closely linked to its digestive physiology (Pearse et al., 2013). The midgut and salivary glands are important organs for establishing interaction between a phytophagous insect and its host plant (Shukle et al., 2010). For example, the ability of insects to tolerate ingested tannins, which are among the most abundant secondary plant metabolites, is determined by a variety of biochemical and physiological features of their midgut (Fig. 1). These include the release of surfactants and antioxidants, the maintenance of a high pH and the formation of a protective peritrophic membrane envelope lining the midgut such that it functions as a barrier (Barbehenn and Constabel, 2011). The more a herbivore can maintain flexible and diverse digestion and detoxification pathways, the more host plants may eventually become available to it, as is the case for the spider mite Tetranychus urticae, which has been found to live on well over 1000 plant species (Dermauw et al., 2013a).
Fig. 1.
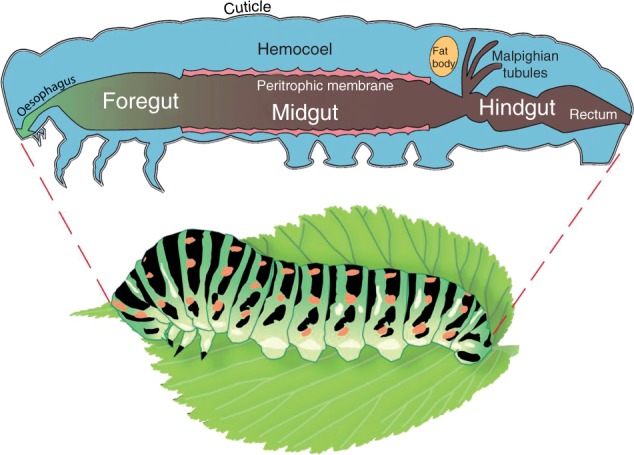
The herbivore digestive system. The insect digestive system has two openings, the mouth and the anus. The digestive tract has three sections: the foregut, the midgut (stomach) and the hindgut. Food is stored in the foregut and mixed with saliva to help digestion; sometimes this mixture is regurgitated, but most digestion takes place in the midgut. Nutrients are absorbed in the midgut and the hindgut. Nitrogen-containing waste from metabolic processes is excreted from the Malpighian tubules and the remaining waste from the anus. The midgut is lined with a semipermeable membrane, called the peritrophic membrane, which is composed of proteins and chitin and allows the passage of liquids while blocking the passage of solid food particles and microorganisms.
Herbivore symbionts to aid digestion
In many cases, herbivores rely on the help of microorganisms to overcome some of the problems that come with feeding on plant tissues. Phloem is a troublesome food source for insects, not only because it lacks essential components, but also because its sugar content and osmotic pressure are high. Many phloem feeders have adapted tolerance to this and display sucrose transglucosidase activity in their gut, which can transform the excess ingested sugar into long-chain oligosaccharides, which in turn can be excreted as honeydew (Douglas, 2006). Especially homopteran insects such as aphids, which live solely on plant sap (i.e. predominantly phloem), have evolved associations with microbes to compensate for the deficiencies in this food source. Phloem is not only rich in sugars but also lacks several amino acids and vitamins. Hence, insects such as aphids have evolved intimate endosymbiotic relationships with bacteria, which supply them with essential amino acids and vitamins and receive other nutrients in return (Engel and Moran, 2013). These bacteria seldomly reside within the gut, but instead are usually found in specialized cells called bacteriocytes (Schwemmler, 1989).
Chewing insects in particular have to deal with the poorly digestible cell wall components that they ingest. The major structural components of the primary plant cell wall are cellulose, hemicellulose and pectin, forming a complex and organized structure, and proper degradation of these components requires a range of enzymes, including cellulases, hemicellulases and pectinases (Cosgrove, 2005; Vilanova et al., 2012). Microbial symbionts present in the digestive tract of insects are known to contribute significantly to the digestion of plant cell wall components. The prevailing view was that herbivorous insects are completely dependent on these symbionts, but it was found that some herbivorous insects also produce their own endogenous plant cell-wall-degrading enzymes (Caldéron-Cortés et al., 2012). These enzymes are usually secreted by the epithelial cells of the insect’s midgut and move forward to the foregut or are secreted by the salivary glands (Terra and Ferreira, 1994). In some cases, these endogenous cell-wall-degrading enzymes have been acquired by insects from microbes by horizontal gene transfer. Xylanases, for example, were transferred from gammaproteobacteria to the mustard leaf beetle Phaedon cochleariae or its ancestor (Pauchet and Heckel, 2013). Other cases of horizontal gene transfer that enable arthropods to feed on plants or plant tissues with an unfavourable nutrient composition show that such transfers may be important drivers of herbivore evolution. Examples of these have been found for the spider mite T. urticae (Grbić et al., 2011; Wybouw et al., 2012) and the coffee berry borer beetle Hypothenemus hampei (Acuña et al., 2012).
The importance of amino acid availability
Herbivorous animals have a lower tissue carbon-to-nitrogen ratio than plants. Hence, they must eat an excess of carbon-rich plant material to acquire sufficient nitrogen, making nitrogen one of the central determinants of herbivore foraging behaviour and population growth (Fagan et al., 2002). Proteins (i.e. amino acids) are the main macronutrients containing nitrogen and are commonly considered to strongly affect the growth rates of arthropod herbivores (Mattson, 1980). The nutritional value of a host plant is not solely based on its protein quantity, as protein quality is hypothesized to be equally important. Protein quality depends on the essential amino acid composition and can be quantified based on which essential amino acids have the lowest abundance relative to the composition required by a herbivore (Barbehenn et al., 2013).
Dietary protein is broken down into peptides and amino acids in the midgut region of the insect digestive tract, a process catalysed by the abundant gut proteases. There are two types of proteases: proteinases, which cleave protein chains at specific peptide bonds, and exopeptidases, which remove amino acids from the C- or N-terminus of a protein (Jongsma and Bolter, 1997). Such free amino acids, but also di- or tripeptides, are subsequently absorbed via transporter proteins in the midgut. Plants, in turn, have evolved mechanisms to disturb the uptake of amino acids by herbivores as an integral part of their defence system (Chen et al., 2005; Yang et al., 2013). For example, essential amino acids in the insect gut can be bound to quinones, highly reactive molecules generated by the ingested plant material. This compromises the plant’s nutritional value, thereby reducing insect performance (Felton, 2005). In addition, the digestion of dietary plant proteins by the insect’s gut proteases may be hampered by ingested plant PIs, which rapidly accumulate in herbivore-damaged plant tissues. In turn, some herbivores have evolved novel proteases that are largely insensitive to plant PIs or have adapted by increasing their overall protease gene expression levels and thereby the total amount of gut protease activity (Jongsma and Bolter, 1997).
Herbivore saliva and regurgitant
Herbivore oral secretions play an important role in plant–herbivore interactions. Oral secretions are a mixture of secretions from the labial and mandibular salivary glands and regurgitant (Vadassery et al., 2012). Regurgitation is the expulsion of material from the herbivore’s oesophagus and this gut reflux is usually composed of partially digested food and gut juices. Some species regurgitate to digest food, e.g. some species have life stages that cannot chew and that repeatedly draw regurgitated liquid in and out of their proboscis, whereas for others regurgitation may have evolved as a defence mechanism against natural enemies (Rhainds et al., 2011). Chewing caterpillars may regurgitate during feeding (Vadassery et al., 2012), but their tendency to do so differs across species of herbivore and host plant. Regurgitant can be collected from caterpillars by gently squeezing them (Peiffer and Felton, 2009) and regurgitant of several herbivore species has been shown to contain components that alter the plant defence response when applied to wounded tissues. Digestive enzymes in regurgitant may emanate from the gut or from the saliva (Afshar et al., 2013; Chen et al., 2013) and some constituents of the saliva of caterpillars (Musser et al., 2002) and aphids (Rodriguez and Bos, 2012), were found to modulate the host’s defence responses.
Elicitors of defences from herbivore saliva and regurgitant
An elicitor of plant defences can be any substance that provokes a specific defence-related response in a host plant after exposure to it. Four groups of elicitors have been distinguished in the literature: plant-derived (endogenous) elicitors, herbivore-derived elicitors, conjugates of these two, and synthetic elicitors.
Plant-derived elicitors are molecules produced or released upon injury or infection and are responsible for induction or amplification of the plant’s defence responses (either local or systemic) against the attacking organism. Such molecules can include cell wall fragments, phytohormones, reactive oxygen species (ROS) and peptides (Albert, 2013; Gozzo and Faoro, 2013). Many elicitors act in concert or in sequence with other elicitors or signalling molecules (Kessler and Baldwin, 2002). Six groups of plant-borne peptide elicitors that play a role in plant–herbivore interactions have been identified: (1) peptides derived from preproteins such as systemin; (2) hydroxyproline-rich systemin (HypSys); (3) a group referred to as ‘plant elicitor peptides' (Peps); (4) cryptic peptides, derived from ‘preproteins' that have their own (unrelated) primary functions, such as inceptin, which is derived from a chloroplastic ATP synthase; (5) SubPep, which is derived from a subtilisin-like protease (Yamaguchi and Huffaker, 2011); and (6) CAPE1 (CAP-derived peptide 1), which is derived from PR-1 (Pathogenesis-related protein 1) (Chen et al., 2014). All these plant peptides were shown to act upstream of a subset of defence responses and to modulate these responses. Receptors for the Peps have been identified (Yamaguchi et al., 2010). Inceptins are disulphide-bridged peptides, originally described from cowpea (Vigna unguiculata), and are derived from the chloroplast ATP synthase γ subunit following digestion by proteolytic enzymes in the gut of the fall armyworm (Spodoptera frugiperda) larva. Hence, inceptin is considered a plant-derived elicitor and it induces and amplifies local and systemic defence responses (Yamaguchi et al., 2011). Inceptin recognition shows that plants have evolved the means to sense insects not only directly via their secretions or movements, but also indirectly by monitoring the emergence of catabolic products indicative of an insect that successfully feeds and digests (Schmelz et al., 2009). Elicitor activity of inceptins seems to be specific to legumes in the genera Phaseolus and Vigna (Yamaguchi et al., 2011). Interestingly, the inceptin-related peptide of the velvet bean caterpillar, Anticarsia gemmatalis, has a C-terminal truncation and does not induce but rather antagonizes defences (Schmelz et al., 2012). This indicates that plants and herbivores may be involved in an arms race reminiscent of plants and pathogens. The elicitor 2-hydroxyoctadecatrienoic acid (2-HOT) is generated during Manduca sexta feeding on N. attenuata by a dioxygenase from the plants using plant linolenic acid as a substrate. However, this conversion occurs mainly locally at the feeding site, possibly because the dioxygenase has a high pH optimum and may increase its activity under the alkaline conditions at the feeding sites or potentially in the insect’s mouth during chewing and regurgitation (Gaquerel et al., 2009). Also, plant hormones, such as the linolenic acid-derived jasmonic acid (JA) and the phenolic salicylic acid (SA), elicit specific defences when applied to plants as pure compounds. Moreover, plants that are attacked by herbivores release distinct volatile blends that can contain methyl jasmonate (the volatile form of JA) and methyl salicylate (the volatile form of SA), which, together with a small subset of additional volatiles, are potent elicitors of defences in systemic uninduced leaves of the same plant or in nearby plants (Kessler and Baldwin, 2002).
It may not come as a surprise that pure (synthetic) plant defence signalling molecules, or substances closely related to them, elicit such defences when applied manually to uninduced plants. However, molecules derived directly from herbivores have also been found to elicit specific defence-related processes. Such elicitors can originate from organs associated with feeding (gut, salivary glands, etc.) and can be present in saliva, regurgitant or other secretions such as honeydew. In lepidopterans, saliva is proposed to be a more important source of elicitors than regurgitant, whereas the opposite may be true for Coleoptera (Kim et al., 2011). Herbivore saliva has been studied in detail (Miles, 1999). Some lepidopteran larvae also possess a ventral eversible gland, whose secretions have been associated with silk strengthening, defences against predators and the production of anti-aggregation pheromones (Zebelo and Maffei, 2012). The secretions may, however, also interact with the plant host because the tip of the everted gland can reach the mouthparts of the larvae, allowing them to mix with the oral secretions. Research by Zebelo and Maffei (2012) suggests that the ventral eversible gland of Spodoptera littoralis might contain elicitors that are able to trigger early plant defences in Arabidopsis. Phloem-feeding herbivores deposit secretions onto the leaf surface when attaching their stylet and coat the stylet trajectory with a protective sheet. Subsequently, they inject saliva in a pierced vascular bundle (Hogenhout and Bos, 2011). Two salivary proteins (Mp10 and Mp42) of the green peach aphid Myzus persicae were found to act as elicitors in Nicotiana benthamiana and to reduce aphid fecundity when expressed in plants. Furthermore, Mp10 overexpression in N. benthamiana resulted in chlorosis and local cell death. The involvement of the plant chaperone protein SGT1 suggest that aphid elicitor recognition is mediated by proteins encoded by R (Resistance) genes: these are sensory proteins that are known to recognize pathogen elicitors. Induction of chlorosis was not observed in tomato, implying that the Mp10 response may be specific for N. benthamiana. Mp10 was also able to suppress the ROS response induced by the well-known bacterial elicitor flagellin 22, but not by the putative insect elicitor chitin (Bos et al., 2010). Hence, mechanisms of recognition and signal transduction of aphid salivary proteins are unclear at this stage (Hogenhout and Bos, 2011).
The interface between the plant and the feeding insect will often contain a mixture of substances of both herbivore and plant origin. Sometimes plants specifically recognize substances that originate from the plant but only after they are processed by the herbivore, reminiscent of inceptin. Some lepidopteran larvae harbour fatty acid amino acid conjugates (FACs) in their digestive system. The fatty acid moiety of these conjugates originates in the plant (Felton and Tumlinson, 2008) and is conjugated to amino acids, such as glutamine, in the insect gut. These conjugates may play a primary role in the regulation of glutamine supply for nitrogen assimilation (Yoshinaga et al., 2010). The oral secretions and regurgitant of caterpillar larvae may also contain FACs, which can act as elicitors of specific defence responses after recognition by the host plant (Bonaventure et al., 2011). The FACs have a broad taxonomic distribution in insects (Felton and Tumlinson, 2008). The first FAC found was named volicitin [N-(17-hydroxylinolenoyl)-l-glutamine]. Together with several related substances, such as N-linolenoyl-l-glutamine and N-linolenoyl-glutamic acid, it constitutes a significant fraction of the elicitor pool of lepidopteran larvae, responsible for the production and release of induced plant volatiles (Alborn et al., 1997). Apart from inducing volatiles, FACs are also known to induce an increase in activity of the salicylic-induced protein kinase and the wound-induced protein kinase in N. attenuata leaves when attacked by caterpillars or treated with oral secretions (Wu et al., 2007). Other elicitors of plant volatiles with a basal fatty acid moiety are caeliferins. These are disulphooxy fatty acids originally isolated from Schistocerca americana regurgitant and were found in grasshoppers of the suborder Caelifera. These compounds elicit defence responses in corn (Alborn et al., 2007) and Arabidopsis (Schmelz et al., 2009). Furthermore, synthetic caeliferin A16:0 was shown to strongly induce ethylene production in Arabidopsis (O’Doherty et al., 2011). However, application of synthetic caeliferin A16:0 to puncture wounds in Arabidopsis did not induce any of the responses observed on treatment with grasshopper oral secretions (Schäfer et al., 2011). Regurgitant of Pieris brassicae caterpillars also contains the enzyme β-glucosidase. This molecule is the first reported herbivore-associated elicitor and triggers the same emission of volatiles in cabbage plants as that induced by feeding caterpillars (Mattiacci et al., 1995). Another well-known enzyme from caterpillar saliva with elicitor properties is glucose oxidase (GOX). In some cases, however, GOX was found to act neutrally or in favour of the herbivore (Tian et al., 2012; Musser et al., 2002).
Together, these elicitors activate defensive responses of the host plant. Hence, they are referred to as herbivore-associated molecular patterns (HAMPs), a term that covers all herbivore-derived signalling compounds that might come into contact with host plants during any stage of their life cycle and elicit defence reactions (Felton and Tumlinson, 2008). The HAMPs are presumably recognized by pattern recognition receptors that evolved to recognize conserved, generally occurring pathogen- and herbivore-derived molecules or motifs, but so far no specific HAMP receptors have been identified (Erb et al., 2012), with the exception of a putative volicitin receptor (Truitt et al., 2004). However, the additional perception of specific individual herbivore-associated elicitors may allow the plant to distinguish the type of attacking herbivore (Poelman et al., 2011).
Non-oral elicitors of defences
HAMPs also include those elicitors that do not directly result from feeding activities. These include the secretions from the ventral eversible gland of S. littoralis, but the fluids secreted by female pea weevils (Bruchus pisorum), which are used to attach the eggs to the plant surface and can also contain substances perceived by the plant. These fluids contain mono- and bis-(3-hydroxypropanoate) esters of long chain α,ω-diols (‘bruchins’) and increase cell division and induce neoplasm formation in several legume hosts. Also, benzyl cyanides from the oviposition fluids of mated female Pieris rapae can elicit transcriptional changes in defence-related genes (Fatouros et al., 2008). The presence of feeding herbivores can also be detected by the plant through components present in the excreted honeydew (VanDoorn et al., 2015). In addition to HAMPs, non-molecular signals can also alert a host plant. For example, herbivore larvae can betray their presence to plants by their crawling, which stimulates the synthesis of 4-aminobutyrate (GABA), while imprints of their footsteps lead to increases in chlorophyll fluorescence or superoxide production. This possibly represents early defence signalling events (Hall et al., 2004). Plant trichomes can also operate as sensors of herbivore movements after being touched (Peiffer et al., 2009).
REGULATION OF PLANT DEFENCES AGAINST HERBIVORES
Detection of herbivores
Plants recognize herbivores by their molecular patterns or their elicitors. It is hypothesized that polyphosphoinositides generated at the plasma membrane play an important role as second messengers, just as they do during pathogenesis (Munnik and Nielsen, 2011). The most rapid measured responses are ion (e.g. Ca2+ and K+) fluxes across the plasma membrane, followed by changes in the plasma membrane potential. Subsequently, a protein kinase cascade can activate the production of ROS such as hydrogen peroxide by activating an NADPH-dependent oxidase. Hydrogen peroxide can have a direct effect on herbivores or enter the cell, thus changing its redox status. The rapid increase in cytosolic Ca2+ can also give rise to increased nitric oxide-mediated processes that precede the upregulation of JA levels (Zebelo and Maffei, 2015). These responses occur not only locally, but also in unattacked neighbouring cells and in distal tissues. Herbivory, the application of oral secretions to wounded leaves and aphid probing have been shown to give rise to membrane depolarization due to an electrochemical gradient between the interior and the exterior of the attacked plants cells. This membrane depolarization can travel with a speed of up to 40 cm s–1 through the entire plant and mutant plants with attenuated wound-induced surface potential changes exhibit a reduced JA response in distal leaves (Mousavi et al., 2013). Moreover, ablation of the ventral eversible gland of S. littoralis reduced this depolarization as well as the Ca2+ and hydrogen peroxide bursts and downstream defence responses (Zebelo and Maffei, 2012). The relationship between Ca2+ levels, ion channel activity and the oxidative burst is correlative but may depend more strongly on K+ than on Ca2+. Additional to these rapid electric signals (Mousavi et al., 2013), slower chemical signals are also transmitted to distal tissues, either via the vascular tissues (Schilmiller and Howe, 2005) or via the air (Sugimoto et al., 2014) (Fig. 2).
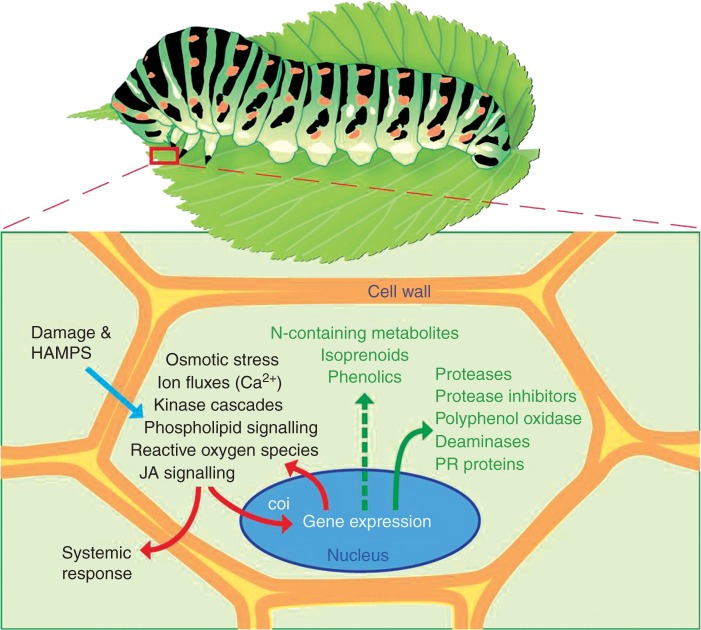
Fig. 2.
The plant defence response. Mechanical damage and exposure to oral secretions from herbivores lead to osmotic stress, cause ion fluxes and elicit signalling cascades leading to rapid accumulation of jasmonates (JAs). Jasmonoyl isoleucine diffuses into the nucleus, where it associates with a protein complex including its receptor encoded by the coi (coronatine insensitive) gene. Binding to this complex initiates degradation of transcriptional suppressor proteins, thereby stimulating defence gene expression. Jasmonate also stimulates increased accumulation of secondary metabolites (phytotoxins). Together, these changes make plant tissues less palatable. Upstream processes are shown in black, the defence substances produced by the plant in green.
Defence-regulating plant hormones
Three phytohormones play a primary role in regulating defence responses (Pieterse et al., 2009): JA (Wasternack and Hause, 2013), SA (Vlot et al., 2009) and the volatile ethylene (Adie et al., 2007). The central roles of JA and SA are substantiated by the fact that biosynthesis mutants are hypersensitive to a wide range of attackers. Several other phytohormones are known to play a secondary role in plant defence by modulating it, including abscisic acid (ABA) (Dinh et al., 2013), auxin (Kazan and Manners, 2009), cytokinin (Choi et al., 2011), gibberellic acid (GA) (Yang et al., 2012) and brassinosteroids (Nakashita et al., 2003) and possibly strigolactones (Torres-Vera et al., 2013). In concert with these hormones, a small set of signalling peptides, such as systemin (Ryan, 2000) and the Peps from Arabidopsis (Huffaker et al., 2006), are also involved in orchestrating plant defences. The peptide systemin of tomato, Solanum lycopersicum (Pearce et al., 1991), functions upstream of JA biosynthesis and may facilitate priming of the plant’s JA response (Kandoth et al., 2007). Moreover, homologues from different plant species were found to have different (non-defence related) functions (Schmidt and Baldwin, 2006). In contrast, some of the Peps appeared to have similar functions across different plant species, because Pep3 from maize (Zea mays) induces accumulation of JA and ethylene as well as their downstream responses, including the emission of volatiles (Huffaker et al., 2013).
Jasmonate as a regulator of plant defences against herbivores
Jasmonic acid regulates the core defences of dicots against herbivorous arthropods (Howe and Jander, 2008) and necrotrophic pathogens (Glazebrook, 2005). The biosynthesis of JA was elucidated by Vick and Zimmermann (1984) and seems quite conserved across species. In tomato, JA biosynthesis was shown to take place in the chloroplast and peroxisomes of the phloem companion cells (Howe, 2004). Briefly, the first step in JA biosynthesis comprises the formation of α-linolenic acid, which is released from the galactolipids of chloroplast membranes by the action of one or more phospholipases, although it is still unclear which roles the different lipase candidates play in α-linolenic acid formation during different plant–herbivore interactions (Wasternack and Hause, 2013). Subsequently, α-linolenic acid is converted via three enzymatic steps into 12-oxophytodienoic acid (OPDA), and dinorOPDA is also formed in Arabidopsis (Stintzi et al., 2001). OPDA is then imported into the peroxisomes, where it is converted by OPDA reductase OPR3, followed by three cycles of β-oxidation into JA. Finally, JA diffuses into the cytosol, after which a range of JA conjugates and derivatives are formed (Yan et al., 2013), among which is jasmonoyl isoleucine (JA-Ile), which is the main bioactive form of JA (Fonseca et al., 2009). Although JA-Ile has a well-established role in regulating defence gene expression, OPDA may also function as such independently (Taki et al., 2005).
Before induction, JA-dependent responses are constitutively blocked due to repressor proteins, called jasmonate ZIM domain (JAZ) proteins, bound to transcription factors that otherwise would promote defence gene expression (Thines et al., 2007), including several MYC (Chini et al., 2007; Fernández-Calvo et al., 2011) and MYB (Qi et al., 2011) transcription factors. The JAZ proteins have two types of functional domain: ZIM domains and Jas domains. The ZIM domains establish homo- or heterodimerization among individual JAZ proteins but also interactions with additional (co)-suppressors, such as TOPLESS and NINJA (Pauwels et al., 2010). The Jas domains establish the interaction with the transcription factors, which prevents these from functioning. Transcriptional (de)repression also regulates the synergistic action of JA and ethylene since JAZ proteins repress not only the transcriptional activity of the ethylene-stabilized transcription factors EIN3 and EIL1 but also interfere with their transcription by promoting histone acetylation. However, induced JA-Ile interrupts the interaction between the JAZ proteins and EIN3/EIL1 to enhance their transcriptional activity (Zhu et al., 2011) by promoting the ubiquitination–degradation of JAZ proteins via a protein complex called the SCFCOI1 complex. Hence, activation of JA-responsive genes largely is obtained by derepression of transcription.
In Arabidopsis, the JA responses downstream of SCFCOI1 are executed via two different branches: one branch that is dependent on MYC transcription factors (referred to as the MYC branch) (Dombrecht et al., 2007) and the other depending on transcription factors like ETHYLENE RESPONSE FACTOR1 (ERF-1) and OCTADECANOID-RESPONSIVE ARABIDOPSIS 59 (ORA59), which is referred to as the ERF/ORA59 branch (Pré et al., 2008; Zhu et al., 2011). These branches are known to antagonize each other: the MYC2 transcription factor suppresses expression of ERF-dependent JA-responsive genes and vice versa (Lorenzo et al., 2004; Dombrecht et al., 2007). The levels of JA in Arabidopsis leaves can start to rise within 30 s after wounding (Glauser et al., 2009). The burst is transient: levels decrease again after a few hours (Reymond et al., 2000; Schittko et al., 2000); however, two consecutive bursts have been observed in S. nigrum (VanDoorn et al., 2011). In N. attenuata, large veins can constrain the spatial spread of JA bursts, and while a second elicitation can suppress a (second) burst, a third elicitation can induce it again (Stork et al., 2009). Subsequently, induction of JA accumulation can also occur in distal leaves (Glauser et al., 2008). Spatiotemporal variability in JA accumulation may be a defensive tactic by itself because it makes it difficult for herbivores to anticipate which tissues are defended poorly and which strongly (Stork et al., 2009). Finally, it has been shown that plants synchronize the JA response with the feeding activities of a generalist herbivore across day–night cycles (Goodspeed et al., 2012), although different plant ecotypes challenged by different kinds of herbivores may exhibit different circadian interactions (Jander, 2012).
Ethylene as a regulator of plant defences against herbivores
Ethylene is a gaseous hormone and is involved in development, senescence and defence against necrotrophic pathogens (Chen et al., 2005). Endogenous ethylene concentrations in plant tissues depend on the activities of two biosynthetic enzymes, 1-aminocyclopropane-1-carboxylic acid synthase (ACS) and 1-aminocyclopropane-1-carboxylic acid oxidase (ACC oxidase), which convert S-adenosyl-Met to ethylene, but also on the rates of outward diffusion and metabolization (Wang et al., 2002). Transcription factors that control ethylene-responsive genes are constitutively repressed by proteins such as JAZ (Zhu et al., 2011) and ethylene perception controls the ethylene response. Arabidopsis contains five ethylene transmembrane receptors, located in different organelles (Kendrick and Chang, 2008). These receptors are active in the absence of ethylene (Hua and Meyerowitz, 1998), and suppress the ethylene response by constitutively stimulating phosphorylation of the ethylene signalling hub EIN2 (ETHYLENE INSENSITIVE2), leading to its degradation (Qiao et al., 2009). Upon binding to ethylene, the receptors become inactive, allowing unhindered accumulation of EIN2 in the cytosol. This initiates degradation of the ethylene transcriptional repressors and thus the activation of ethylene-responsive genes in the nucleus (An et al., 2010).
Salicylate as a regulator of plant defences against herbivores
Salicylate mediates defences against biotrophic pathogens (Glazebrook, 2005) and phloem-feeding herbivores (Kaloshian and Walling, 2005). During pathogen infections, defence responses can spread systemically, so are also expressed in uninfected tissues, and this is referred to as systemic acquired resistance. Several candidate signals have been reported to play a role in systemic acquired resistance, including the SA-derivative methyl salicylate. However, SA is the central local regulator because plants that are unable to accumulate SA are often highly susceptible to pathogen infections (Dempsey and Klessig, 2012). In rice (a monocot), the JA and SA pathways are thought to regulate a common set of defence genes that are effective against both biotrophic and necrotrophic pathogens (De Vleesschauwer et al., 2013). Salicylate is derived from chorismate, the end-product of the shikimate pathway. From there it can be synthesized in plants via at least two distinct biosynthetic routes. The first route delivers SA in two steps and depends on the enzymes isochorismate synthase, which is induced upon pathogen infection (Wildermuth et al., 2001), and isochorismate pyruvate lyase. The second route depends on the phenylpropanoid pathway. This is a pathway responsible for a variety of products, such as flavonoids and lignins, but also for SA, and there may be parallel sub-branches within the branch leading to SA (Boatwright et al., 2013). Which of these pathways or branches determines induced SA levels most strongly may also differ across plant species. Once formed, SA may be modified further by glucosylation, methylation or amino acid conjugation. Most of these derivatives are inactive and may serve to fine-tune local and systemic SA accumulation and function or may provide safe storage. Methyl salicylate is inactive but easier to transport to distal tissues, either actively via the phloem or passively via the air (Dempsey et al., 2011).
A central role is played by the NONEXPRESSOR OF PR GENES (NPR) protein family. It has recently been discovered that NPR3 and NPR4 are SA receptors, whereas NPR1 acts a master regulator of SA-mediated responses (Yan and Dong, 2014). NPR1 proteins are constitutively present in the cytosol of the cell as oligomers (Tada et al., 2008) and their concentration increases upon induction (Spoel et al., 2009). Accumulation of SA causes an increase in the levels of reduced glutathione (the antioxidant form of glutathione), thereby changing the redox status of the cell, i.e. the balance between oxidants and antioxidants (Spoel and Loake, 2011), and this generates NPR1 monomers by the thioredoxin-catalysed reduction of monomeric disulphate bridges. Subsequently, NPR1 monomers migrate into the nucleus (Mou et al., 2003; Tada et al., 2008). Without NPR1, the expression of the SA-responsive genes is repressed by TGA transcription factors. After NRP1 has arrived in the nucleus, a portion of it is phosphorylated. Phosphorylated NPR1 binds to the TGA transcription factors and this complex allows the expression of target genes such PR-1. Unphosphorylated NPR1 may assemble together with different transcription factors and give rise to TGA-independent expression of other target genes. After a round of transcription initiation, the NPR1 protein complexes are degraded via the proteasome and new monomeric NPR1 proteins need to enter the nucleus from the cytosol to keep the response going (Mukhtar et al., 2009).
Hormonal crosstalk in plant defences against herbivores
The distinct defence signalling pathways that are regulated by phytohormones interact directly and indirectly, forming complex networks, and these interactions can be additive, antagonistic or synergistic (Koornneef and Pieterse, 2008). Of all the interactions that occur between hormonal defence signalling pathways, crosstalk between the JA and SA pathways has received most attention, after it was discovered that SA can inhibit the plant’s wound response (Doherty et al., 1988) and indications were found for the opposite (Sano et al., 1994). Under most conditions, crosstalk between SA and JA is antagonistic (Thaler et al., 2012), but when applied to plants in specific ratios synergistic interactions were also observed (Mur et al., 2006), and other plant hormones may modulate this crosstalk (Robert-Seilaniantz et al., 2011). Suppression of the JA response by SA occurs downstream of JA-Ile perception, depending on ORA59 (Van der Does et al., 2013). Other regulators include NPR1 and some of its interacting partners, including the TGA and WRKY transcription factors (Pieterse et al., 2012). Nuclear localization of NPR1 is not required for the suppression of JA signalling, and it has therefore been suggested that the role of NPR1 in mediating JA/SA crosstalk depends on a function executed in the cytosol (Spoel et al., 2003). The adaptive value of the JA/SA (antagonistic) crosstalk is not clear. It has frequently been suggested that it allows plants to fine-tune the balance between different defensive strategies, depending on the type of attacker, or in case the plant is attacked by multiple attackers, depending on the timing and sequence of infestation (Pieterse and Dicke, 2007). However, whether JA/SA antagonism is adaptive remains an open question (Thaler et al., 2012).
MOLECULES USED BY PLANTS TO RESIST HERBIVORES
The collective hormonal responses and their interactions induced by herbivores determine which defences are established in which host plant tissues and to what extent. Induced plant defences upon herbivory are seldom lethal: the fact that herbivores can move away from defended tissues will usually prevent them from ingesting a fatal dose. Hence, plant defences induced by herbivores will cause them to depart or, alternatively, slow down their development and population growth because bigger herbivores, or higher herbivore densities, consume and reproduce more, thus causing more damage. Many of these herbivore-induced plant defences rely on the direct antagonistic action of enzymes that interfere with feeding activities, (gut) digestive processes and gut integrity (Carlini and Grossi-de-Sa, 2002) (Fig 2).
Defence proteins
Protease inhibitors
Most organisms produce a range of enzymes belonging to the protease class (also called ‘peptidases’). Proteases are enzymes that perform proteolysis on target proteins and can thereby regulate enzymatic activities (Rawlings et al., 2014). A subset of these proteases are the endopeptidases, which cleave the chain of amino acids of the target protein, and are referred to as ‘proteinases’. The active site of a protease can be centred on a particular amino acid, e.g. a serine protease has its active site at a serine, and these ‘active site’ amino acids are used for their classification. Proteases themselves are regulated by PIs, which are also commonly occurring enzymes across the tree of life. Most plants upregulate a subset of their PIs, often measured as PI activity, upon herbivory, and some of these are associated with resistance to herbivores (Ryan, 1990; Lison et al., 2006). These typical herbivore-induced plant PIs are believed to act on proteases in the herbivore’s gut and have been suggested to have a dual role: to reduce the efficiency of proteinase activity during the herbivore’s digestion of plant proteins but also to protect co-ingested defensive proteins of the plants against herbivore proteases (Macintosh et al., 1990). Proteases and their inhibitors often form couples: a serine protease can be inhibited by a serine PI, although these functional annotations can be less strict than the name suggests. PIs can have different modes of action, but in general mimic the substrate of the protease and establish a strong bond with the enzyme, thus creating a protease–PI complex and delaying or blocking its proteolytic activity (Bateman and James, 2011). Different plant PI families have been associated with defence against specific families of herbivores (Ryan, 1990). For instance, serine PIs have a primary role in defence (Hartl et al., 2010) and affect the performance of some lepidopteran species (Duan et al., 1996; Yeh et al., 1997). In contrast, cysteine PIs are effective against coleopteran species such as the southern corn rootworm, Diabrotica undecimpunctata howardi (Fabrick et al., 2002), and the spider mite T. urticae (Santamaria et al., 2012).
Peptidases
Several peptidases/proteases are associated with plant defence responses, but it is not always clear what their functions in defence responses are and whether they directly interact with the physiology of attackers or have regulatory roles within the plant’s defence network (Harrison and Bonning, 2010). While subtilisin-like proteinases are associated with anti-pathogen defences and depend on SA signalling (Jorda and Vera, 2000), cysteine proteases (such as papain) are typically induced by herbivory and are associated with disruption of the peritrophic matrix in the insect gut (Pechan et al., 2002; Fescemyer et al., 2013). Some insects have evolved adaptations to protect the peritrophic membrane against these proteases (Li et al., 2009; Zhu-Salzman and Zeng, 2015). Interestingly, overexpression of a cotton cysteine protease in Arabidopsis improved the effect of plant-delivered small RNAs, designed to trigger gene silencing via RNAi in Helicoverpa armigera, possibly by increasing the permeability of the peritrophic matrix (Mao et al., 2013). Finally, herbivory induces strong JA-dependent expression of leucine aminopeptidase (LAP) genes. The LAPs are exopeptidases that hydrolyse the terminal amino acid (leucine, methionine and arginine) residues of proteins (Gu et al., 1999). Herbivore-induced accumulation of LAPs in the chloroplasts depends on the JA pathway and it is assumed that they have a direct role in defence against plant consumers, probably by inactivating herbivore digestive enzymes by proteolysis (Fowler et al., 2009; Lomate et al., 2013). However, they may also have regulatory roles in the plant itself and modulate the expression of defence genes (Scranton et al., 2013).
Amino acid degrading proteins
Digestion of proteins in the intestinal lumen results in free amino acids and peptides, available for absorption by the insect. Transepithelial transport of (essential) amino acids in the alimentary canal (predominantly the midgut) often requires energy. Some plant defensive enzymes degrade such amino acids in the arthropod gut before they can be taken up. Manduca sexta feeding on tomato (S. lycopersicum) was found to induce an arginase and a threonine deaminase, which degrades the amino acids arginine and threonine respectively, which could be found back in the gut of the larvae. Interestingly, the optimal pH for enzymatic activity of both enzymes was found to be similar to the pH found in the midgut of M. sexta. Moreover, threonine deaminase was inactive in tomato itself due to an activation domain that was cleaved off in the herbivore gut, thereby activating the enzyme (Chen et al., 2005; Gonzales-Vigil et al., 2011). This suggests that some plant-defensive enzymes have evolved to be activated only after having reached the herbivore gut.
Polyphenol oxidases
The PPOs have been associated with plant defences: they are among the major gene/enzyme families induced by wounding, herbivory or after application of JA or JA derivatives (Thaler et al., 1996; Constabel and Ryan, 1998; Stout et al., 1998). While PPOs can be found in diverse phylogenic groups (Tran et al., 2012), such as green algae, mosses and gymnosperms, Arabidopsis does not have them. Their mode of action and their site of action during plant–herbivore interactions is still under debate. Polyphenol oxidases catalyse the oxidation of ortho-oriented dihydroxy phenolic compounds, thereby generating quinones, which are highly reactive molecules that can either spontaneously polymerise or damage proteins, amino acids and nucleic acids via an alkylation reaction (Constabel and Barbehenn, 2008). Polyphenol oxidases have been implicated in at least two distinct defence responses. First, polyphenolics on the plant surface hinder crawling herbivores. For instance, Yu et al. (1992) and Kowalski et al. (1993) found a correlation between PPO activity, i.e. phenol polymerization activity, in glandular trichomes of different solanaceous species and the entrapment of small arthropods between such trichomes. Second, PPO-mediated generation of quinones in the herbivore’s gut, i.e. after ingesting and mixing plastidial plant PPOs in combination with vacuolar plant phenolics, damages amino acids and proteins and thus interferes with food digestion (Constabel and Barbehenn, 2008).
Activity of PPOs has been associated with defence against herbivores such as coleopterans (Castañera et al., 1996) and lepidopterans (Felton et al., 1992a). Moreover, PPO overexpression in different plant species was also shown to increase resistance to several herbivores. Overexpression of a potato PPO in tomato increased resistance to the common cutworm Spodoptera litura (Mahanil et al., 2008) and overexpression of a poplar (Populus trichocarpa) PPO in Populus tremula increased resistance to the forest tent caterpillar Malacosoma disstria. Interestingly, this poplar PPO is latent in plant cells, whereas it was fully active when isolated from the caterpillar’s frass, suggesting activation in the insect’s gut (Wang and Constable, 2004). However PPO overexpression does not always have an effect on herbivores (Barbehenn et al., 2007), indicating that PPO effectiveness could depend on specific physiological conditions in the herbivore gut (Felton et al., 1992b) or on the availability of specific PPO substrates. Finally, other oxidases, such as peroxidase and lipoxygenase, may play a similar functional role in plant defences by creating potent electrophiles or interfering with the accumulation of essential nutrients (Zhu-Salzman et al., 2008).
Lectins
Plant defensive lectins are associated with the disruption of several processes involved in the digestion of food and nutrient uptake in herbivores (Michiels et al., 2010). Lectins comprise a diverse family of proteins that bind specifically with mono- and oligosaccharides (Komath et al., 2006). Although the mode of action is not well understood, there are several cases in which herbivorous insects that propagated on artificial diets with lectins or on plants that overexpressed lectin genes showed delayed or impaired development (Gatehouse et al., 1996; Sauvion et al., 2004). Due to their high affinity with oligosaccharides, it is assumed that lectins can interact with glycoproteins in the digestive tract of the herbivore and bind to the insect’s intestinal epithelial cells or its peritrophic membrane and disrupt these tissues (Macedo et al., 2004).
Pathogenesis-related proteins
Pathogenesis-related (PR) proteins include a wide variety of proteins with diverse functions, predominantly associated with resistance to pathogens. These proteins can be classified into 17 families and they are often used as defence marker genes, though not all of them are functionally understood: most of them are classified as a glucanase, chitinase, thaumatin, PI or peroxidase. They are defined as pathogen-induced proteins and for the majority of them evidence is largely lacking that they play a significant role in anti-herbivore defences. For example, PR-2 proteins have β-1,3-endoglucanase activity and are associated with (fungal) cell wall degradation. However, some of these proteins could also be active against herbivores. The families PR-3, 4, 8 and 11 are chitinases (Sels et al., 2008). Insect chitinases, key enzymes for arthropod morphogenesis, have insecticidal activity when delivered via the plant (Kramer and Muthukrishnan, 1997). However, the effects of plant chitinases on insects are less clear. Carnivorous plants secrete chitinases to digest arthropod prey (Paszota et al., 2014) and purified chitinases from mulberry latex were found to have insecticidal activity (Kitajima et al., 2010). In addition, overexpression of a poplar chitinase in tomato inhibited the development of Colorado potato beetles (Lawrence and Novak, 2006). Moreover, larvae of Orgyia antiqua showed a lower growth rate when feeding on transgenic birch (Betula pendula) expressing a sugar beet chitinase (Vihervuori et al., 2013), but transgenic plants in the field were more susceptible to aphids (Vihervuori et al., 2008). Thus, there are indications that plant chitinases are active against insects and have the potential to damage the exoskeleton and chitin-rich peritrophic membrane of arthropods, but these activities could be limited to some species and life stages of herbivores. Other PR proteins could also have insecticidal activities. PR-1 is found in the digestive fluids of pitcher plants (Buch et al., 2014) and PR-2 in the digestive fluid of sundew, although possibly in order to utilize pollen grains, fungal spores or detritus as nutritional source (Michalko et al., 2013). However, there is not much evidence for herbivore-induced foliar PR proteins other than chitinases that affect herbivores directly, although PR proteins are ingested by them and can be found in their frass (Chen et al., 2007).
Small cysteine-rich defence proteins
There are two families of small cysteine-rich proteins that are suggested to play a role in plant defence against herbivores: defensins (e.g. PR-12) and cyclotides. Defensins are small cysteine-rich proteins, commonly synthesized in plants but also by animals. They are proteins of 45–54 amino acids that contain eight conserved cysteine residues and are similar to thionins (Thomma et al., 2002). Most defensins operate by binding to cell membranes, resulting in pore-like membrane defects, causing efflux of essential ions and nutrients. Plant defensins are predominantly active against fungi (Stotz et al., 2009), but some defensins inhibit α-amylase activity and have no effect on fungi (Osborn et al., 2009). α-Amylase is a typical insect gut enzyme and there are indications that particular defensins reduce the digestibility of plant material (Shade et al., 1994; Shiau et al., 2006). Cyclotides are peptides of typically 28–37 amino acids, derived from longer precursor proteins, such as metallocarboxy peptidase inhibitors (Cavallini et al., 2010). They contain six conserved cysteine residues connected by three intermolecular disulphide bonds that form a knotted structure. Their mode of action is poorly understood, but is also associated with disrupting membrane integrity. Several cyclotides were found to exhibit insecticidal activities (Poth et al., 2011; Pinto et al., 2012), although they may also find applications as therapeutic drugs (Smith et al., 2011) (Fig. 3).
Fig. 3.
Mode of action of plant defence proteins in the herbivore gut. Herbivores utilize plant material predominantly to obtain sugar and amino acids. They digest proteins into amino acids via proteases and starch/sucrose into free sugars via amylases/invertases. Plants produce special proteins that are co-ingested and interfere with digestive processes in the gut. Defensins inhibit α-amylase activity. Deaminases degrade amino acids. Proteinase inhibitors and peptidases inhibit the arthropod’s proteases. Peptidases, lectins and possibly some PR proteins damage the peritrophic membrane. Plant polyphenol oxidases in combination with plant phenolics are believed to generate quinones in the arthropod gut. These quinones may damage soluble and membrane proteins and DNA. Digestive processes of the herbivore are shown in red and counter-measures of the plant in green.
Defence metabolites
While defensive plant proteins play a significant role in direct interactions between herbivores and plants, the role of non-protein secondary metabolites is just as big. The term ‘secondary’ is used to contrast them with metabolites that are directly involved in growth, development or reproduction, although it is not always possible to determine the precise physiological role of each metabolite. Across the plant kingdom, there is a staggering diversity of secondary metabolites, and they can be distinct for small phylogenetic groups. Many secondary metabolites have been implicated in plant defences or are stress-related. Despite the rich diversity of secondary metabolites, their biosynthetic origins allow them to be classified into three basal groups: (1) the phenolics; (2) the isoprenoids; and the (3) N-containing compounds (Fig. 2).
Phenolics
Phenolics consist of an aromatic ring with one or more hydroxyl groups. Two pathways are responsible for the majority of plant phenolics: (1) the phenylpropanoid pathway (Cheynier et al., 2013), which converts the aromatic amino acid phenylalanine into phenolics, and (2) the acetate/malonate (polyketide) pathway (Quideau et al., 2011). Single phenolics can be polymerized to form polyphenols and both can be subjected to additional modifications, giving rise to a vast quantity (>9000) of chemically diverse metabolites, which include benzoquinones, phenolic acids (such as SA), coumarins, flavonoids, lignins and tannins (Balasundram et al., 2006). Phenolics have various functions in primary metabolism; for instance, they protect plants from UV radiation (Landry et al., 1995) or form, as lignins, an integral part of the secondary cell wall in vascular plants (Boerjan et al., 2003). Furthermore, flavonoids are crucial for reproduction because they are required for pollen development (Van der Meer et al., 1992) and provide many of the visual and volatile cues used by flowers (Hoballah et al., 2007) and fruits (Jaakola et al., 2002) to attract pollinators and seed dispersers, respectively.
Plants use phenolics to resist attacks from herbivores because of their deterrent (Kessler et al., 2012b) and toxic (Lindroth and Peterson, 1988) nature. Hence, they are often constitutively present at or near the cell surface or stored as inactive compounds away from activating enzymes (e.g. in vacuoles or specialized cells, or bound to the cell wall) (Pourcel et al., 2007). Inactive phenolics can be activated when a herbivore disrupts plant cells, thereby mixing them with activating enzymes such as glycosidases, PPOs and peroxidases to produce toxic (free) phenolics and quinones (Constabel and Barbehenn, 2008). On top of this, herbivory can induce accumulation of phenolic compounds and PPO and peroxidase activity (Stout et al., 1999; Constabel et al., 2000), thereby amplifying the defence response. Upregulation of chemical defences can coincide with the deposition of lignin at the site of infection or attack, creating an extra physical barrier, which is especially effective against small organisms such as nematodes and relatively immobile arthropods (Valette et al., 1998). Finally, some volatile phenolics, such as methyl salicylate, are involved in the indirect defence response (Ament et al., 2004, 2010).
Isoprenoids
Isoprenoids constitute the largest and structurally most diverse class of metabolites with over 55 000 known structures. The universal isoprene [C5] precursors isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) are produced via two pathways in plants: the cytosolic/peroxisomal mevalonate (MVA) and the plastidial 2-methylerythritol 4-phosphate (MEP) pathway (Lange et al., 2000; Sapir-Mir et al., 2008). DMAPP (C5) is a precursor for the synthesis of cytokinins and isoprene. It also serves as the substrate for successive head-to-tail condensations with one or more IPP units to obtain precursors of longer-chain isoprenoids (Akhtar et al., 2013). Isoprenoids are building blocks for several phytohormones and are essential components of membranes (Schaeffer et al., 2001) and the photosynthetic (Fraser et al., 2000) and respiratory (Ducluzeau et al., 2012) machinery. Terpenoids are derived from isoprenoid precursors through the action of terpene synthases and subsequent modifications lead to a large diversity of molecules (Schilmiller et al., 2009), some of which are volatile and emitted in large quantities (Kant et al., 2009). Volatile terpenoids are present in relatively large quantities in most plant-derived scent bouquets (Knudsen et al., 2006). As such, they are cues for pollinators (Wright et al., 2005) and seed dispersers (Hodgkison et al., 2007), but also function in responses against biotic and abiotic stresses (Gershenzon and Dudareva, 2007). Since volatile terpenoids are highly lipophilic, they can penetrate plasma membranes and increase their permeability (Sikkema et al., 1995) to exert direct toxic (and repellent) effects on arthropods (Bleeker et al., 2012; Vaughan et al., 2013). Moreover, terpenoids serve as key compounds in the indirect defence against herbivores as attractants of their natural enemies both above- and below-ground (Kant et al., 2009). Their production and release is regulated in a spatiotemporal manner (Köllner et al., 2004) through defence-associated phytohormones such as JA and SA (Van Poecke and Dicke, 2002). Although considered highly effective (Kessler and Baldwin, 2001), terpenoid-mediated indirect defence can also backfire as it might attract even more herbivores (Carroll et al., 2006) and can prime defence responses in competing neighbouring plants (Kessler et al., 2006) and guide parasitic plants to a new host (Runyon et al., 2006).
Nitrogen-containing compounds
Nitrogen-containing secondary metabolites are thought to have primarily defensive purposes. Alkaloids constitute the biggest subgroup (>12 000 compounds). They bear at least one nitrogen atom in a heterocyclic ring and have been identified in ∼20 % of all plant species (Levin, 1976). Several amino acids as well as purine nucleotides and isoprenoids are precursors of alkaloids (Itkin et al., 2013). Their biosynthesis is often initiated in roots, followed by phloem and usually xylem transport (Courdavault et al., 2014). Alternatively, the final steps of their de novo biosynthesis can take place above ground (Miettinen et al., 2014). Alkaloids are constitutively present in plants, but their production and transport can increase upon herbivory (Baldwin, 1988) and exogenous application of methyl jasmonate (Baldwin, 1996). They can react with DNA, membranes and enzymes, and are therefore potent toxins for many organisms, including arthropods (Wink et al., 1998). Interestingly, the nectar of some plants contains sublethal amounts of alkaloids, which not only protects them from nectar robbers (Kessler et al., 2008), but also improves their reproductive success by manipulating the behaviour of their natural pollinators (Kessler et al., 2012a).
Many plant species accumulate cyanogenic glucosides (α-hydroxynitrile glucosides), which protect them against herbivores because they can release volatile hydrogen cyanide (HCN), which inhibits cellular respiration (Way, 1984). Because plants are vulnerable to high concentrations of HCN as well, cyanogenic glucosides are stored in vacuoles, whereas the HCN-liberating enzymes β-glucosidase and α-hydroxynitrile lyase are localized in plastids (Thayer and Conn, 1981), the apoplast (Frehner and Conn, 1987) or in intracellular protein bodies (Swain et al., 1992). Upon herbivory, the cyanogenic glucosides become exposed to β-glucosidases. Depending on the pH, the resulting α-hydroxynitriles will either dissociate spontaneously into HCN or will be enzymatically converted to it by α-hydroxynitrile lyases (Siritunga et al., 2004).
Like cyanogenic glucosides, glucosinolates are derived from amino acids (Hansen et al., 2001). They are N- and S-containing defensive metabolites mainly found in Brassicaceae (Halkier and Gershenzon, 2006). Glucosinolates can be activated by the enzyme myrosinase (Husebye et al., 2002), from which they are separated by compartmentalization. Herbivory mixes the two (Barth and Jander, 2006), thereby triggering the production and release of various reactive hydrolysis products, mainly isothiocyanates and nitriles (Bones and Rossiter, 1996), which can be directly toxic and repellent to herbivores (Lazzeri et al., 2004) but also attract specialist herbivores (Beran et al., 2011) and serve as attractants of parasitoids (Mumm et al., 2008). Consistently, glucosinolate biosynthesis can be induced by herbivory (Hopkins et al., 2009) and is controlled by JA, SA and ethylene (Schweizer et al., 2013) (Fig. 3).
PLANT DEFENCES IN NON-ATTACKED TISSUES
Induced plant defensive substances accumulate not only locally at the feeding site, but also in undamaged tissues, and these ‘systemic’ responses have distinct spatiotemporal dynamics. There are several cases showing that distinct defence-associated changes can be observed in tissues neighbouring the damaged area within 1 min after wounding or herbivory (Schittko et al., 2000; Glauser et al., 2009), and these changes are typically the signalling events upstream of the actual defence response. A steady increase in the levels of JA and JA-Ile can be observed after a few minutes of damaging, with a peak after ∼30 min (Glauser et al., 2008; Stork and Baldwin, 2009; VanDoorn et al., 2011). The extent to which the JA from this JA burst is synthesized de novo or released from storage is not always clear; it probably differs across different plant species and with the number of elicitations. The JA burst is preceded by a fast signal (Schittko et al., 2000), which most probably is electric, i.e. transmitted via membrane depolarizations, and which also travels to distal leaves (Glauser et al., 2009; Koo et al., 2009; Mousavi et al., 2013). However, molecular signals also travel from leaf to leaf, via either the phloem or the xylem (Malone and Alarcon, 1995; Rhodes et al., 1999). However, the identity of these signals is still elusive. Grafting experiments with tomato plants showed that JA biosynthesis, but not perception, was required for initiating the systemic signal while the downstream defence responses required perception (Schilmiller and Howe, 2005). Peptides such as systemin and Peps, and phytohormones such as ABA, auxin and cytokinins (Soler et al., 2013), are closely associated with local and systemic signalling, but especially JA and usually JA derivatives play a critical role (Wu and Baldwin, 2010).
Temporal and spatial heterogeneity of signalling within and between leaves
The spatiotemporal patterns of systemic responses differ across plant species, depending on differences not only in their size or age but also in plant-specific (vascular) architecture. Because not all leaves of a plant are connected to the same degree, the induction of defensive compounds is higher in undamaged leaves with the most direct vascular connection to the attacked leaf, the so-called orthostichous leaves (Orians et al., 2000), and these are less vulnerable to herbivores than leaves with a less direct connection (Viswanathan and Thaler, 2004). Heterogeneity in the levels of defensive compounds in damaged tissues can stimulate herbivores to move to other areas in an attempt to avoid the induced defences (Shroff et al., 2008). For example, caterpillars of M. sexta might be able to escape from the induced resistance of a plant through ‘induced movement’ (Paschold et al., 2007). Also, the foraging behaviour of the generalist insect Helicoverpa armigera depends on JA: the larvae leave induced areas quickly and move to non-induced distant parts. Interestingly, Plutella xylostella, which is known to be resistant to defensive chemicals in some Brassicaceae plants, did not display this behaviour (Perkins et al., 2013). Finally, heterogeneity of leaf systemic response (Stork et al., 2009) or plant systemic response due to heterogeneity of vascular connections was suggested to be adaptive because it makes it more difficult for herbivores to learn which tissues will be least defended. Rodriguez-Saona and Thaler (2005) used normal tomato plants and JA-deficient (def-1) tomato plants to analyse local and systemic induced JA responses in relation to patterns of caterpillar feeding damage. They observed that the systemic response in leaves with a stronger vascular connection was stronger than in leaves with a weaker connection, but at a similar physical distance from the damaged area. Hence the extent to which a herbivore can avoid induced defences is determined by the strength of the vascular connection of the induced leaf and the newly selected (distal) leaf.
It was shown that electric and vascular signals can act in synergy with airborne signals to optimize the systemically expressed resistance within a plant (Heil and Ton, 2008). Moreover, exposure of plants to relatively high amounts of synthetic volatiles was shown to induce defences. For example, exposure of tomato plants to methyl jasmonate results in the accumulation of PIs in systemic leaves of the plant, in neighbouring tomato plants and even in plants of different species, such as tobacco and alfalfa (Farmer and Ryan, 1990). Similarly, methyl salicylate may act as an airborne signal to activate resistance in uninfected tissues of an infected plant and in neighbouring plants (Shulaev et al., 1997). In contrast, corn plants previously exposed to the natural volatiles emitted by neighbouring plants accumulated more JA when damaged mechanically or induced with caterpillar regurgitant compared with corn plants not pre-exposed to volatiles (Engelberth et al., 2004). It has been shown that plant volatiles elicit responses not only in neighbouring plants but also in their own distal tissues (Heil and Silva Bueno, 2007) and, apart from elicitation, volatiles can also ‘prime’ defences: priming means that the actual defence response is not established yet but is preprepared in such a way that it is displayed faster and/or more strongly upon actual induction by a herbivore later on (Kessler et al., 2006; Heil et al., 2007). Airborne signalling has at least two advantages over vascular signalling. First, priming the induction of plant defences through the air overcomes vascular restrictions resulting from the plant’s orthostichy; second, airborne signalling can reach distal plant parts faster than compounds that are transported through vascular tissues (Heil and Ton, 2008). This is especially relevant for bushy plants in which signals transported via the vascular system have to travel long distances. For example, systemic induced resistance in sagebrush depends on air contact, possibly due to restrictions in vascular connections (Karban et al., 2006). Similarly, undamaged leaves of hybrid poplar (Populus deltoides × nigra) exposed to volatiles from wounded leaves with little or no vascular connection were primed to defend against larvae of the gypsy moth, Lymantria dispar (Frost et al., 2007). Thus, there is ample evidence that plant volatiles can facilitate signalling between leaves with weak vascular connections and facilitate priming in synergy with signals transmitted directly via plant tissues.
HOW HERBIVORES COPE WITH DEFENCES
Plants and herbivores have coevolved for over 400 million years. While plants have evolved signalling networks to regulate induced defences and diversity in their palette of secondary metabolites, herbivores have been under pressure to evade defences (reviewed in Alba et al., 2011). Hence, behavioural adaptions have evolved that allow herbivores to avoid defended plant tissues (Paschold et al., 2007; Shroff et al., 2008; Perkins et al., 2013) as much as possible or to dismantle defensive structures such as trichomes (Cardoso, 2008) and latex channels (Rodrigues et al., 2010). However, herbivores have also evolved a variety of mechanisms to cope with deterrent substances produced by their host plants. Given the enormous economic impact of herbivore resistance to agrochemicals, a large part of our knowledge of adaptations to xenobiotics comes from the field of pesticide resistance (Despres et al., 2007). However, the mechanisms at play are similar to those that enable them to resist defensive phytotoxins, and a functional overlap between adaptation to agrochemicals and to phytotoxins has been suggested (Dermauw et al., 2013a).
Two general mechanisms allow herbivores to cope with the xenobiotics from their environment: mechanisms that decrease exposure (pharmacokinetic responses) and mechanisms that decrease sensitivity (pharmacodynamic responses). Pharmacokinetic responses comprise a variety of adaptations that reduce uptake, increase catabolism and allow sequestration, whereas the pharmacodynamic response types comprise adaptations at the level of interactions between allelochemicals and their target-site(s) (Taylor and Feyereisen, 1996; Kennedy and Tierney, 2013). Together, these mechanisms determine the level of tolerance of herbivores to xenobiotics (Fig. 4).
Fig. 4.
The herbivore resistance response. Plant secondary metabolites can have diverse target sites. Two general mechanisms allow arthropod herbivores to cope with plant secondary compounds: mechanisms that decrease exposure via detoxification, transport and/or sequestration (shown in this figure) but also mechanisms that decrease sensitivity (see main text). Detoxification (indicated as ‘Detox’ in this figure) usually occurs in three phases: in phase I, enzymes such as P450s and CCEs (carboxyl/cholinesterases) modify the metabolite; in phase II GSTs and UGTs conjugate it and in phase III the conjugated metabolites are transported out of the cell, typically by ABC transporter and SLC family proteins. The midgut, fat body and Malpighian tubes are the insect tissues where these detoxification phases occur (Harrop et al., 2014). Metabolites are excreted via faeces and urine. Some metabolites, or modified metabolites, are stored in the cuticle or in other organs and are used by the arthropod for its own protection. Plant substances are indicated in green and arthropod responses in red.
Pharmacokinetic responses
Mechanisms of decreased exposure
The detoxification of xenobiotics usually occurs in three phases. In phase I, the xenobiotic is modified by reactions that incorporate a nucleophilic functional group (a hydroxyl, carboxyl or amine group) and this often results in a more polar/hydrophilic substance. In phase II, the metabolite resulting from phase I is conjugated to endogenous molecules such as glutathione or a sugar molecule, which further increases the compound’s polarity/hydrophilicity. In phase III, the phase II conjugated xenobiotic is transported out of the cell by cellular transporters. In several cases, these transporters can also act as a first line of defence, preventing allelochemicals entering the cell by rapid efflux without the need for modifications (sometimes this is referred to as phase 0).
Enzymes that operate during phase I are often cytochrome P450 monooxygenases (P450s) and carboxyl/choline esterases, whereas enzymes such as glutathione-S-transferases (GSTs) and UDP-glycosyltransferases (UGTs) typically operate during phase II. Finally, transport of phase II metabolites out of the cell is often performed by ATP-binding cassette (ABC) and solute carrier (SLC) family proteins. Adaptations that allow herbivores to enhance their detoxification of particular target xenobiotics often occur via mutations that increase the production of specific detoxification enzymes and transporters as well as by mutations that improve their catalytic or transport properties (Brattsten, 1992; Despres et al., 2007; Kennedy and Tierney, 2013).
Phase I detoxification
A wide range of allelochemicals, including furanocoumarins, terpenoids, glucosinolates, flavonoids and alkaloids can be metabolized by P450s of herbivorous arthropods (Despres et al., 2007; Feyereisen, 2012). The involvement of P450s within the lepidopteran genus Papilio in furanocoumarin resistance is one of the first documented examples relating to plant–arthropod interactions (Berenbaum, 1982). The P450s are by far the best studied enzymes of phase I, and have been associated with resistance to plant allelochemicals and pesticides in a wide range of species (Feyereisen, 2012; Schuler, 2012). Although esterases have been linked to pesticide resistance in a number of cases, their role in plant allelochemical defence remains elusive (Despres et al., 2007; Li et al., 2007). Notably, esterases were found not always to operate via hydrolysis but also to confer resistance to pesticides by sequestration, i.e. by binding to the target substance without modifying it. Remarkably, esterase genes were duplicated in some aphid species, which gave rise to elevated expression, such that 3 % of their total protein content consisted of these enzymes (Devonshire and Sawicki, 1979; Devonshire and Moores, 1982). It is well imaginable that such esterases are involved in resistance or tolerance to phytotoxins as well.
Phase II detoxification
Conjugation of xenobiotics by GSTs has been linked to allochemical tolerance in a number of cases, although most evidence has been obtained from in vitro assays. The expression of GSTs in arthropod herbivores can be induced by a number of allelochemicals, and these enzymes are generally believed to make up an important component of the overall pharmacokinetic response. Some GSTs of the polyphagous insect Spodoptera frugiperda are able to metabolize a variety of thiocyanate conjugates. Moreover, the diversity of glucosinolate-derived thiocyanates that insects such as the larvae of S. frugiperda, Trichoplusia ni and Anticarsia gemmatalis can metabolize correlates with their host plant range (Li et al., 2007), whereas the major glucosinolate-derived thiocyanate was found to be a glutathione-conjugated derivative in several generalist insect herbivore species (Schramm et al., 2012). Recently, a role for GSTs in tolerance to glucosinolates was also suggested for the whitefly Bemisia tabaci and in the mustard-feeding specialist Scaptomyza nigrita (Elbaz et al., 2012; Gloss et al., 2014). Finally, an enzyme called GST16 inactivates the phytohormone OPDA in the gut of Helicoverpa armigera by isomerization to inactive iso-OPDA (Dąbrowska et al., 2011). However, it is unclear whether this OPDA modification has adaptive significance (Shabab et al., 2014).
The second important class of conjugation enzymes is that of the UGTs, which convert lipophilic aglycones into more hydrophilic glycosides by conjugation with UDP-glucose. UGTs have been shown to be involved in resistance of lepidopterans to the alkaloid capsaicin and the detoxification of benzoxazinoids (Ahn et al., 2011; Wouters et al., 2014), and they may be widespread enzymes that allow herbivorous arthropods to adapt to xenobiotics (Ahn et al., 2014). Some insects have evolved traits that in principle allow them to prevent the activation of protoxins, such as glucosinolates, by plant enzymes. For example, aphids prevent glucosinolates from mixing with myrosinases by avoiding rupturing myrosinase-containing plant cells. Moreover, the insect degrades these glucosinolates in the gut and conjugates the breakdown product to ascorbate, glutathione and cysteine. However, artificial feeding assays suggested that these conjugates also have an antifeeding effect in M. persicae. Hence, glucosinolates may also have defensive functions independent of myrosinase, via post-ingestive breakdown processes occurring in the aphid (Kim et al., 2008).
Phase III detoxification
While phases I and II have been characterized in some detail, phase III has been documented in far less detail (Sorensen and Dearing, 2006; Dermauw and Van Leeuwen, 2014). Gaertner et al. (1998) suggested that the efflux of nicotine and other alkaloids by the Malpighian tubules, the main excretory organs of insects, of M. sexta is mediated by an ABC transporter. This finding was based on the observation that verapamil, a known inhibitor of the ABC transporter, blocks nicotine transport in the Malpighian tubules of M. sexta. In addition, Petschenka et al. (2013) found an ABC transporter that probably functions as cardenolide efflux carrier, thereby protecting the nervous tissues of lepidopterans. Other transporter families are also implicated in mediating the efflux of plant allelochemicals. Govind et al. (2010) showed that transporters of the major facilitator superfamily, which include many transporters of the SLC family, were downregulated in M. sexta larvae after feeding on wild tobacco mutants that were unable to produce JA. Interestingly, other members of the same superfamily were upregulated in arthropod herbivores that were transferred from their preferred host to a more challenging, less suitable host (de la Paz Celorio-Mancera et al., 2013), e.g. in the non-insect arthropod T. urticae (Dermauw et al., 2013b). Regardless of the identity of the transporters or transport mechanisms, a number of cases have been described in which the evolution of transport systems has determined herbivore success. For example, the ability to selectively transport plant glycosides has been suggested to stand at the basis of the evolution of life styles and host ranges of leaf beetles (Kuhn et al., 2004; Strauss et al., 2013).
Miscellaneous resistances to xenobiotics
Some insects, especially host-plant specialists, have developed resistance mechanisms that differ from those described above. For example, a flavin-dependent mono-oxygenase in arctiid moths was shown to be involved in detoxifying pyrrolizidine alkaloids (Sehlmeyer et al., 2010; Wang et al., 2012). These enzymes usually have general functions in an insect’s primary metabolism but here evolved, after gene duplication, to catalyse N-oxidation of pyrrolizidine alkaloids (Langel and Ober, 2011). Moreover, specialist crucifer-feeding insects have developed the means to redirect the formation or to overcome the toxic action of glucosinolate breakdown products (Pentzold et al., 2013) by evolving novel enzymes and proteins into general detoxification pathways. Pieris rapae was shown to use a nitrile-specifier protein to divert the degradation of glucosinolates in the toxic isothiocyanates to less toxic nitriles (Wittstock et al., 2004; Stauber et al., 2012). Another specialist, the diamondback moth, Plutella xylostella, desulphates glucosinolates and thereby generates inactive metabolites (Ratzka et al., 2002).
Other examples of adaptations to xenobiotics in a number of lepidopterans include the detoxification of cyanogenic glucosides by converting cyanide into β-cyanoalanine (Zagrobelny and Møller, 2011). It was shown recently that the β-cyanoalanine synthase genes of lepidopterans as well as mites stand at the basis of this adaptation and were originally obtained from bacteria by horizontal transfer into the ancestral genomes (Wybouw et al., 2014). This enzyme might also be crucial for countering the negative effects of glucosinolate in P. rapae because their breakdown also generates cyanide (Stauber et al., 2012; Wybouw et al., 2014). In addition, some lepidopterans have evolved the means to convert cyanide into nitrogen (Engler et al., 2000; Pentzold et al., 2014).
Sequestration
Some herbivores make use of plant allelochemicals for their own defence against predators by storing ingested chemicals in specialized tissues or in the integument. A variety of mechanisms have been described that allow herbivores to exploit these toxic substances without suffering from their latent detrimental effects. Compounds can be sequestered either directly or after biotransformation, such as by oxidation and conjugation. More than 250 insect species have been shown to sequester plant metabolites covering a wide range of chemical classes, such as alkaloids, cyanogenic glucosides, glucosinolates, isoprenoids and cucurbitacins (Nishida, 2002; Opitz and Müller, 2009). In some cases, insects have evolved the ability to synthesize these compounds de novo by convergent evolution of the biosynthetic pathways (Jensen et al., 2011). Also, some insect species have evolved additional means to efficiently utilize sequestered compounds for their own defence. For example, some aphid and flea beetle species have evolved a specific myrosinase that allows them to convert glucosinolates into their toxic breakdown products in a similar way as plants do and to release these as toxic and repellent volatiles into the air (Beran et al., 2014; Rahfeld et al., 2014).
Pharmacodynamic responses: mechanisms of decreased sensitivity
While many adaptations of herbivores to xenobiotics depend on mechanisms that directly or indirectly divert these substances from their target sites, some adaptations are due to reduced target-site sensitivity. Although alterations in the target site of pesticides have been associated with herbivore resistance to pesticides (Ffrench-Constant, 2013; Van Leeuwen et al., 2010b; Feyereisen et al., 2015), few studies have linked such mutations to insect resistance to plant allelochemicals. This lack of documented target-site resistance to phytochemicals is probably due to the fact that many of these have multiple modes of action and our knowledge of these mechanisms is limited (Berenbaum, 1987; Despres et al., 2007). The best documented example of target-site insensitivity to phytotoxins is that of amino acid substitutions in the α subunit of Na+/K+-ATPase from four different insect orders (Lepidoptera, Coleoptera, Heteroptera and Diptera), conferring resistance to the plant toxin ouabain, a cardenolide produced by several members of the Apocynaceae. Remarkably, the same substitution was found in different insect species, all of which specialized in cardenolide-containing plant species, suggesting that this replacement must have evolved independently several times (Agrawal et al., 2012; Dobler et al., 2012; Zhen et al., 2012; Dalla et al., 2013). Finally, tolerance to l-canavanine, a non-proteinogenic amino acid of leguminous plants, can also be considered as a target-site-based resistance mechanism in some insect species. The toxicity of l-canavanine is caused by its incorporation into proteins, replacing l-arginine. Insects such as the bruchid beetle Caryedes brasiliensis have specialized in feeding on the l-canavanine-rich seeds of Dioclea megacarpa and have evolved an arginyl-tRNA synthetase that can discriminate between l-canavanine and l-arginine, thereby effectively avoiding the adverse biochemical effect of l-canavanine (Rosenthal et al., 1976; Leisinger et al., 2013). In addition to target-site insensitivity, mechanisms of decreased exposure to l-canavanine have also been reported for the tobacco budworm Heliothis virescens (Melangeli et al., 1997) (Fig 4).
Plant defence suppression by herbivores
Because many defensive actions of plant are induced or maintained by ongoing physiological processes, some herbivores have evolved the means to interfere with these processes. In this way, these herbivores may manipulate resource flows (Clark and Harvell, 1992) or suppress defences (Musser et al., 2002). The suppression of defences is distinct from defence avoidance, such as ‘stealthy feeding’ or vein cutting, which serve to prevent detection or to prevent physical contact with defensive substances. However, these two processes will not always be easy to separate experimentally (Karban and Agrawal, 2002; Stireman and Cipollini, 2008; Alba et al., 2011). Suppression of defences is characterized by lowering the rate of production of defensive compounds. Hence, suppression can operate upstream or downstream of a defensive pathway and block it altogether or dampen it to intermediate levels, although assessing the latter can be difficult without having a suppression-free control experiment as a benchmark (Alba et al., 2015). Finally, downregulation of plant defences qualifies as suppression when it is paralleled by an increase in (reproductive) performance of the herbivore (Table 1).
Table 1.
Defence suppression by arthropod herbivores
| Arthropod | Common name | Plant | Defence suppressed | Mode of action | Biological readout | Reference |
|---|---|---|---|---|---|---|
| Helicoverpa zea (Insecta) | Corn earworm | Nicotiana tabacum | Nicotine production | Salivary enzyme glucose oxidase | Caterpillar weight gain | Musser et al., 2002, 2005 |
| Helicoverpa zea (Insecta) | Corn earworm | Solanum lycopersicum | JA defence response | Salivary enzymes | Wu et al., 2012 | |
| Spodoptera exigua (Insecta) | Beet armyworm | Medicago truncatula | Genes involved in terpenoid biosynthesis | Saliva-derived factors | Bede et al., 2006 | |
| Spodoptera exigua (Insecta) | Beet armyworm | Arabidopsis thaliana | JA defence | SA–JA antagonism | Weech et al., 2008 | |
| Spodoptera littoralis, Pieris brassicae (Insecta). | African cotton leafworm, cabbage butterfly | Arabidopsis thaliana | Wound-inducible genes | Unknown | Larval weight gain | Consales et al., 2012 |
| Pieris rapae, Spodoptera exigua (Insecta) | Small white; beet armyworm | Arabidopsis thaliana | Green leaf volatiles | Unknown | Feeding choice | Savchenko et al., 2013 |
| Manduca sexta (Insecta) | Tobacco hornworm | Nicotiana attenuata | Nicotine production | Ethylene | Kahl et al., 2000; Voelckel et al., 2001 | |
| Frankliniella occidentalis (Insecta) | Western flower thrips | Arabidopsis thaliana | JA defence | SA–JA antagonism | Host plant preference | Abe et al., 2012 |
| Mayetiola destructor (Insecta) | Hessian fly | Triticum spp. | Protease inhibitors, lectins | Unknown | Liu et al., 2007; Wu et al., 2008 | |
| Bemisia tabaci (Insecta) | Silverleaf whitefly | Arabidopsis thaliana | JA defence response | SA–JA antagonism | Nymphal developmental speed | Zarate et al., 2007 |
| Bemisia tabaci (Insecta) | Silverleaf whitefly | Phaesolus lunatus | (E)-β-ocimene production, JA defence | SA–JA antagonism | Attraction of predatory mites, preference, oviposition | Zhang et al., 2009 |
| Bemisia tabaci (Insecta) | Silverleaf whitefly | Nicotiana tabacum | JA defence | Vectored virus | Population growth | Zhang et al., 2012 |
| Bemisia tabaci (Insecta) | Silverleaf whitefly | Solanum lycopersicum | JA defence | Saliva via endogenous bacterium | Survival, oviposition | Su et al., 2015 |
| Megoura viciae (Insecta) | Aphid | Vicia fabae | Phloem clogging | Ca2+ chelators | Behaviour | Will et al., 2007 |
| Acyrthosiphon pisum (Insecta) | Pea aphid | Vicia faba | Unknown | Salivary effector protein C002 | Survival, behaviour | Mutti et al., 2008 |
| Myzus persicae (Insecta) | Green peach aphid | Nicotiana benthamiana | Oxidative burst | Salivary effector proteins C002, Mp10, Mp42 | Reproductive performance | Bos et al., 2010 |
| Myzus persicae (Insecta) | Green peach aphid | Arabidopsis thaliana | Unknown | Salivary effector proteins C002, PIntO1, PIntO2 | Reproductive performance | Pitino and Hogenhout, 2013 |
| Macrosiphum euphorbiae (Insecta) | Potato aphid | Nicotiana benthamiana, Solanum lycopersicum | Unknown | Salivary effector proteins Me10, Me23 | Reproductive performance | Atamian et al., 2013 |
| Myzus persicae (Insecta) | Green peach aphid | Nicotiana tabacum, Nicotiana benthamiana, Arabidopsis thaliana | Indole glucosinolate, callose, hydrogen peroxide | Salivary effector protein Mp55 | Reproductive performance | Elzinga et al., 2014 |
| Brevicoryne brassicae (Insecta) | Cabbage aphid | Brassica oleracea | JA defence | Interference with JA signal transduction | Caterpillar developmental speed, caterpillar weight gain | Soler et al., 2012 |
| Macrosteles quadrilineatus (Insecta) | Aster leafhopper | Arabidopsis thaliana | JA defence | Vectored phytoplasma | Reproductive performance, oviposition rate | Sugio et al., 2011 |
| Phenacoccus solenopsis (Insecta) | Cotton mealybug | Gossypium hirsutum | Gossypol production, JA defence response | Unknown | Development al speed, adult weight gain, feeding choice | Zhang et al., 2011 |
| Leptinotarsa decemlineata (Insecta) | Colorado potato beetle | Solanum lycopersicum | Proteinase Inhibitors | Unknown | Lawrence et al., 2011 | |
| Leptinotarsa decemlineata (Insecta) | Colorado potato beetle | Solanum lycopersicum | JA defence | Oral bacteria | Larval weight gain | Chung et al., 2013 |
| Tetranychus urticae (Acari) | Two-spotted spider mite | Solanum lycopersicum | JA and SA defence, terpenoids and methyl salicylate | Unknown | Oviposition rate | Kant et al., 2008 |
| Tetranychus evansi (Acari) | Red spider mite | Solanum lycopersicum | JA and SA defence, terpenoids and methyl salicylate | Unknown | Oviposition rate, population growth | Sarmento et al., 2011a, Alba et al., 2015 |
| Aculops lycopersici (Acari) | Tomato russet mite | Solanum lycopersicum | JA defence | Unknown | Population growth | Glas et al., 2014 |
Suppression of plant defences is a well-known phenomenon in plant pathogens such as pathogenic bacteria (Abramovitch et al., 2006), rust fungi (Voegele and Mendgen, 2003), oomycetes (Kamoun, 2006) and viruses (Kasschau and Carrington, 1998). However, nematodes and mites were also found to suppress defences. Several phytophagous nematode species interfere with host plant resistance (Haegeman et al., 2012) and the cyst nematode Meloidogyne incognita was found to suppress SA- and JA-dependent systemic acquired resistance in Arabidopsis thaliana (Hamamouch et al., 2011). In addition, two spider mite species were found to suppress the defences downstream of both JA and SA simultaneously in tomato (Kant et al., 2008; Sarmento et al., 2011a; Alba et al., 2015), whereas an eriophyid mite was found to suppress only the downstream JA defences, independently from hormonal crosstalk (Glas et al., 2014). Other examples of suppression are from insects and can be attributed to hormonal crosstalk in the majority of cases. Hemipteran phloem feeders such as the mealybug Phenacoccus solenopsis (Zhang et al., 2011) and the whitefly B. tabaci were found to suppress JA defences (Zhang et al., 2009), possibly by inducing antagonist SA defences (Zarate et al., 2007; Walling, 2009). In addition, the aphid Megoura viciae inhibits defensive phloem clogging (Will et al., 2007), whereas other aphid species were found to suppress the oxidative burst (Bos et al., 2010). The leafhopper Macrosteles quadrilineatus suppresses JA defences indirectly via an effector derived from a vectored phytoplasma (Sugio et al., 2011). Also, chewing larvae of several lepidopteran species have been found to interfere with induced defences (Bede et al., 2006). On A. thaliana, larvae of Spodoptera exigua inhibit JA-mediated defence responses via the systemic acquired resistance pathway (Weech et al., 2008) whereas Pieris brassicae suppresses defences independently of the JA and SA pathways (Consales et al., 2012). In addition, Helicoverpa zea was found to suppress nicotine accumulation in N. tabacum (Musser et al., 2002, 2005) and to suppress JA- and ethylene-regulated genes via salivary enzymes in tomato (Wu et al., 2012). This is reminiscent of the downregulation of nicotine by M. sexta feeding on N. attenuata, although it may reflect a plant-adaptation rather than a herbivore-adaptive trait (Kahl et al., 2000; Voelckel et al., 2001). Moreover, the Colorado potato beetle, Leptinotarsa decemlineata, suppresses transcription of PI genes in tomato (Lawrence et al., 2011). Finally, larvae of virulent strains of the hessian fly, Mayetiola destructor, secrete substances via their vestigial mouthparts into plant tissues, thereby suppressing the expression of PI and lectin genes (Stuart et al., 2012). Thus, evidence for suppression of plant defensive processes is found across herbivorous insects, plant-eating mites and nematodes.
Whereas induction of plant defences often results from elicitor or HAMP recognition, the suppression of induced or constitutive defences is often attributed to so-called effector molecules. These molecules are especially well known from phytopathogens and had been discovered already in the 1970s (Shiraishi et al., 1978), but the notion that herbivores may secrete molecules with similar properties arose after the discovery that GOX from the saliva of H. zea caterpillars counteracts the production of nicotine in N. tabacum (Musser et al., 2002). Although our knowledge of herbivore effectors is still limited, a staggering diversity of effectors from pathogens (Deslandes and Rivas, 2012; Rovenich et al., 2014) and nematodes (Mitchum et al., 2013; Kazan and Lyons, 2014) has been discovered. Roughly, these effectors comprise the following functional groups (although these are not mutually exclusive).
Metabolites secreted into the host to manipulate particular physiological processes such as hormonal signalling. Some strains of P. syringae produce the JA mimic coronatine, which puts JA defences into overdrive to suppress SA defences (Zhao et al., 2003).
Enzymes that interfere with the host’s ability to control infected tissues encoded by transgenes inserted into the host’s genome by the pathogen. The best known example is Agrobacterium tumefaciens, which injects its transfer DNA (which carries genes that facilitate gall formation) into plant cells, where it integrates into the host’s genome and is expressed by the host (Zhu et al., 2000).
Enzymes secreted by pathogens to perform a metabolic conversion in the host that affects its defences. Fungi such as Septoria lycopersici produce tomatinase, which not only detoxifies the defensive alkaloid tomatine but also generates hydrolysis products that suppress the hypersensitive response (Bouarab et al., 2002).
Secreted proteins that interfere with transcription factors or that act as transcription factors of defence genes of the host. The first is the case for an effector called SAP11, produced by phytoplasmas vectored by leafhoppers. SAP11 destabilizes the host’s CIN-TCP transcription factors, leading to downregulation of the JA response (Sugio et al., 2011). The second is the case for the transcription activator-like (TAL) effectors produced by the Xanthomonas bacterium. TAL effectors are translocated to the host’s nucleus, where they modulate the expression of specific target genes to facilitate the infection (Boch and Bonas, 2010).
Secreted proteins that interfere with host receptors involved in defences. For example, the P. syringae HopF2 effector suppresses plant immunity by targeting the R gene co-receptor BAK1 (Zhou et al., 2014).
Secreted proteins that interfere with defence signalling cascades downstream of receptor recognition. This is the case with AvrB of P. syringae, which phosphorylates the signalling hub RIN4 to block PAMP (Pathogen-Associated Molecular Pattern) triggered immunity (Mackey et al., 2002).
Secreted proteins that manipulate proteasome functioning in defensive processes. This is the case for the E3 ubiquitin ligase AvrPtoB of P. syringae, which initiates degradation of a kinase that is essential for innate immunity (He et al., 2006).
Secreted proteins that perform proteolysis of plant defence proteins. This was found for an extracellular PI of Phytophthora infestans, which targets a tomato PR protein (Tian et al., 2004) and for AvrRpt2 of P. syringae, which acts as protease to eliminate RIN4 (Axtell et al., 2003).
Secreted proteins that interfere with host vesicle trafficking during immune responses. This is the case for HopM1 of P. syringae, which mediates degradation of the vesicle trafficking regulatory protein MIN7 (Nomura et al., 2011).
Secreted proteins that interfere with RNAi, such as the 2b protein of the cucumber mosaic virus. This protein protects the virus against RNAi-mediated degradation and also suppresses SA defences (Ji and Ding, 2001) and JA defences induced by their aphid vectors (Westwood et al., 2014).
Secreted small RNAs that manipulate a host’s RNAi machinery, such as a small RNA transferred by Botrytis cinerea, which silences the protein Argonaute 1 in Arabidopsis and tomato and thereby suppresses host immunity (Weiberg et al., 2013).
Effectors may be powerful weapons whereby pathogens and nematodes interfere with plant defences, but plants have evolved a range of R genes in return, which specifically serve to recognize effectors and bypass suppression (Bergelson et al., 2001). These molecular sensors are used in plant breeding to obtain pathogen-resistant crops but R gene resistance is often broken again shortly after its introduction due to counter-adaptations in pathogens (Bent and Mackey, 2007). Hence, plant breeders have redirected their focus to the effector targets, also referred to as susceptibility genes or S genes (van Schie and Takken, 2014), since effector-resistant breeding targets are promising tools for obtaining more durable resistance (Gawehns et al., 2013).
Effectors involved in plant defence suppression by herbivores
There are indications that plant-defence-suppressing herbivorous arthropods also secrete effectors via their saliva into their host, similar to pathogens and nematodes. The first of such salivary components that was discovered was the enzyme GOX, which is the most abundant molecule in the oral secretions of the caterpillar of H. zea (Musser et al., 2002). This enzyme catalyses the oxidation of glucose to d-gluconic acid and thereby generates hydrogen peroxide. The amount of GOX applied to T. tabacum plants correlates with an increase in SA and a decrease in the accumulation of nicotine. Possibly, GOX suppresses or attenuates JA and ethylene responses by crosstalk with SA (Diezel et al., 2009; Eichenseer et al., 2010). GOX has been found in many more caterpillar species (Eichenseer et al., 2010) and other herbivorous insects, such as aphids and non-herbivores such as honeybees (Iida et al., 2007; Harmel et al., 2008). A comprehensive study by Eichenseer et al. (2010) showed large variation in GOX activity within families and subfamilies of 88 caterpillar species, but these activities depended on the host plant species as well. Moreover, a recent report showed that H. zea GOX elicits the JA pathway in tomato (Tian et al., 2012). Taken together, these studies suggest that some plants, such as tomato, may have evolved a recognition mechanism for GOX, resembling R-gene-mediated recognition of effector proteins in plant–pathogen interactions.
Advances in genomics and proteomics have greatly facilitated the discovery of more effector proteins in insects. After the Acyrthosiphon pisum (peach aphid) salivary glands were sequenced, the first aphid effector was discovered. This protein is a 22-kDa salivary-secreted protein of unknown function called C002 (Mutti et al., 2008). RNAi-mediated knockdown of C002 expression affected A. pisum foraging and feeding behaviour and reduced aphid fitness. Bos et al. (2010) used N. benthamiana to ectopically express C002 from M. persicae (green peach aphid) and showed that aphid fecundity increased on these plants. Transient overexpression of a second aphid protein, Mp10, sufficed to suppress the flagellin-triggered oxidative burst in N. benthamiana, but aphid reproduction was lower on these plants. A subsequent study characterized two additional aphid proteins, Mp1 (PIntO1) and Mp2 (PIntO2), which correlate positively with aphid fecundity on Arabidopsis. Interestingly, the performance of M. persicae did not improve on Arabidopsis plants expressing the A. pisum orthologues of both effectors (Pitino and Hogenhout, 2013). Finally, two putative effectors of Macrosiphum euphorbiae were found, Me10 and Me23, both of which increased aphid fecundity on N. benthamiana, whereas only Me10 increased their fecundity on tomato (Atamian et al., 2013).
Research on gall midges has provided independent evidence for a role of effector proteins in plant–herbivore interactions. Early larval stages of the hessian fly, M. destructor, are plant parasites. When they colonize wheat (Triticum spp.), the larvae induce feeding cells in their host, which provide them with food until they develop into adults (Harris et al., 2006). More than 30 hessian fly resistance genes have been found in wheat, some of which are predicted to encode typical R proteins (Liu et al., 2005). On resistant wheat, hessian fly larvae are unable to induce feeding cells, but instead induce a hypersensitive-like response that prevents them from eating (Harris et al., 2010). One M. destructor gene, vH13, functions as an avirulence factor on wheat carrying the H13 resistance gene. In contrast, larvae from populations that are virulent on H13 wheat did not express vH13, while RNAi-mediated knockdown of vH13 in avirulent larvae made some of them virulent (Aggarwal et al., 2014). These data suggest that vH13 may function as an effector on non-resistant wheat varieties.
Thus, there are indications that herbivores may make use of effectors, just as pathogens do. This notion is strengthened by the existence of anti-herbivore R genes such as Mi-1, Vat and Bph14 (Rossi et al., 1998; Dogimont et al., 2008; Du et al., 2009). The high diversity found among pathogen effectors discourages the use of protein homology as a strategy to identify herbivore effectors (Rep, 2005). Nevertheless, most effector proteins share structural features that can be easily recognized, such as an amino-terminal signal peptide, the absence of transmembrane domains and a small protein size. Furthermore, effectors that operate in the plant apoplastic space are usually rich in cysteine residues (Rooney et al., 2005). Several studies have exploited these common properties to find novel effector-encoding genes from sequenced pathogen genomes or transcriptomes. Comprehensive datasets on herbivore transcriptomes and proteomes (Grbić et al., 2011; Su et al., 2012; DeLay, 2012) will probably give rise to the discovery of new effectors in the near future (Table 1).
HOW COMMUNITY INTERACTIONS SHAPE THE ADAPTIVE PROCESS
Plants are often attacked by a diverse community of enemies, including herbivores and pathogens. Interspecific competition within phytophagous communities can be direct (e.g. interference through fighting) or indirect (e.g. using a resource deprives others of also using it) (Denno et al., 1995). Because induction and suppression of defences manifest themselves in distal tissues (Kant et al., 2008; Alba et al., 2015), it is predicted that herbivores and pathogens will also interact indirectly through the changes in the plant elicited by their feeding (Ohgushi, 2005). These interactions may lead to decreased performance, but also to facilitation: herbivores were sometimes found to benefit from the presence of conspecific or heterospecific attackers (Faeth, 1986; Harrison and Karban, 1986; Karban, 1989).
Indirect interactions in phytophagous communities via induced defences
Responses induced by one particular species can result in resistance to another (Long et al., 2007; Poelman et al., 2008a; Mouttet et al., 2013), whereas the order of herbivore arrival on the plant can influence the performance and number of herbivore species occurring later in the season (Van Zandt and Agrawal, 2004; Viswanathan et al., 2005; Erb et al., 2011). Hence, inducible plant defences can be major determinants of ecological interactions; in particular, defences depending on JA and SA appear to play important roles in determining community composition. For example, JA-deficient wild tobacco plants in the field were colonized by herbivores that normally ignore these plants (Kessler et al., 2004), and studies in which plants were sprayed with synthetic phytohormones to assess the effect of induced defences on their ecology showed that such artificial induction can decrease the abundance of herbivores when JA is applied (Thaler et al., 2001) or of pathogens when SA or SA mimics are applied (Inbar et al., 1998). However, treating tomato plants with JA increased the growth of the pathogen P. syringae, whereas induction of SA responses enhanced the performance of the beet armyworm (S. exigua) (Thaler et al., 1999). Moreover, when JA and SA responses were induced simultaneously the performance of the cabbage looper caterpillar (T. ni) increased, but not that of the thrips Frankliniella occidentalis, the spider mite T. urticae and M. sexta larvae (Thaler et al., 2002). In addition to these field studies, experiments have also been carried out in the laboratory with model organisms to reveal the mechanisms that underlie indirect interactions. Using Arabidopsis mutants impaired in the JA, ethylene or SA-pathway, de Vos et al. (2006) found that feeding by the JA-inducing caterpillar P. rapae did not, contrary to expectation, induce resistance against the necrotrophic fungus Alternaria brassicicola. In contrast, it did reduce disease symptoms caused by P. syringae, but this effect was not (exclusively) dependent on JA, ethylene or SA. It also reduced infectiousness of the biotroph turnip crinkle virus due to an ethylene-primed SA response (de Vos et al., 2006). In addition Thaler et al. (2010) showed that P. syringae induced JA, SA and increased the activity of PIs in tomato plants and thereby reduced the growth of S. exigua caterpillars. Conversely, infection with tobacco mosaic virus induced only an SA response, causing induced susceptibility to S. exigua caterpillars and induced resistance to aphids (M. persicae). Herbivores feeding on different plant organs can also affect each other through the induction of plant responses (Soler et al., 2013). Larvae of P. rapae grew more slowly on plants that were also infested with root-feeding nematodes (Pratylenchus penetrans) and this correlated with higher foliar glucosinolate levels (Van Dam et al., 2005). Similarly, Soler et al. (2005) found that P. brassicae larvae developed more slowly on wild mustard (Brassica nigra) plants that were infested with the cabbage root fly (Delia radicum), and this in turn affected developmental rates of Cotesia glomerata, which is a parasitoid of the herbivore. Systemic signals that have been proposed to be involved in above–belowground interactions include the phytohormones JA, ABA, ethylene, auxin and cytokinin, but for none of these has a clear role been unequivocally established. This suggests that the simultaneous induction of different defences may affect community members differently.
Indirect interactions in phytophagous communities via suppressed defences
Not only induction, but also suppression of defences can affect interactions between herbivores and pathogens. In principle, the benefits of suppression by a single herbivore species can be shared by other species in the community. Within the spider mite species T. urticae, there is intraspecific variation in the ability to induce and suppress defences (Kant et al., 2008). Some spider mites are very sensitive to the JA defences they induce in tomato plants. However, when these mites share their feeding site with other types that can suppress JA defences, their reproductive performance increases dramatically (Alba et al., 2015). In addition, suppressor mites of the species Tetranychus evansi as well as non-suppressor mites of the species T. urticae perform better when they are introduced on leaves already suppressed by T. evansi than on uninduced control leaves (Sarmento et al., 2011a,b). Similarly, whiteflies (B. tabaci) were found to suppress JA defences, and this was shown to improve the performance of spider mites on Lima bean (Zhang et al., 2009) and to disrupt the attraction of natural enemies (Zhang et al., 2009, 2013a). In turn, some parasitoids have adapted to locate hosts using SA-induced volatiles, the emission of which is not suppressed by whiteflies (Zhang et al., 2013b). Soler et al. (2012) showed that the cabbage aphid (Brevicoryne brassicae) inhibited the production of JA in cabbage (Brassica oleracea) plants, and this led to increased growth and development of the large cabbage white butterfly (P. brassicae) on plants that had been co-infested with the two species. Finally, field-grown tomato plants were frequently infested with the two-spotted spider mite T. urticae and the tomato russet mite, Aculops lycopersici; the first of these induces JA and SA defences simultaneously, whereas the second induces only SA and suppresses the JA response. Spider mites had much higher reproductive performance on plants infested with russet mites, an effect that was not due to the russet mite-suppressed JA response but to the antagonistic effect of the doubled SA response induced by both species. However, this same SA response inhibited infection by P. syringae. Hence, the overall effect of selective suppression one type of defence is determined by the presence of competing plant parasites and the distinct palette of defences these induce (Glas et al., 2014).
Indirect interactions among plant defence, herbivores and predators
During recent decades, the study of interactions between plants and other organisms has developed from the investigation of relatively simple interactions between plants and herbivores to that of more complex multitrophic interactions and their importance for the structure of communities of plants and arthropods. In nature, plants are part of complex food webs in which the sum of direct and indirect interactions between organisms, either allies or enemies, determines the selection pressures on plants. Plant defences against herbivores play an important role in these interactions (Hairston et al., 1960, Price et al., 1980, Janssen et al., 1998, Poelman et al., 2012). Plant defences comprise not only traits that interfere with herbivores directly (Walling, 2000), but also traits that operate indirectly by facilitating foraging predators and host-seeking parasitoids. These indirect plant defences often rely on the release of volatile compounds into the atmosphere, which can act as cues for prey-searching natural enemies (Dicke et al., 1990; Turlings et al., 1991).
Plant toxins ingested by herbivores may interfere with their natural enemies. Hence plant defence may actually provide a herbivore with additional protection. While sequestration is adaptive and entails storage of ingested plant toxins in specialized tissues or eggs (Duffey, 1980; Tooker and De Moraes, 2007), in principle any plant substance present anywhere in a herbivore can serve as antipredator protection. For example, tomatine incorporated in the diet of H. zea resulted in prolonged larval development, reduced pupal eclosion and size, while it reduced adult longevity of its parasitoid Hyposoter exiguae (Campbell and Duffey, 1979). Nicotine in the diet of M. sexta decreased survival of its parasitoid Apanteles congregates (Barbosa et al., 1982), while there is evidence that plants stop producing nicotine when attacked by nicotine-tolerant herbivores (Kahl et al., 2000; Voelckel et al., 2001), possibly to facilitate parasitoids. Alternatively, sometimes predators may themselves evolve resistance to plant toxins such as nicotine (Kumar et al., 2014). In addition, glucosinolates were found to affect the performance of the second and the third trophic level (Poelman et al., 2008b; Hopkins et al., 2009; Kos et al., 2012). Furthermore, Kauffman and Kennedy (1989) found that a high concentration of the methyl ketone 2-tridecanone in a particular accession of Lycopersicon hirsutum was toxic to the herbivore H. zea but much more to the herbivore’s parasitoid Campoletis sonorensis. Moreover, the stinkbug Podisus maculiventris reared on M. sexta caterpillars avoided prey that had been fed on tomatine and chlorogenic acid (a phenolic) in an artificial diet (Traugott and Stamp, 1997). This shows that not only growth, development and mortality but also the behaviour of natural enemies can be modulated by plant toxins ingested by their prey. There are also indications that JA-dependent plant metabolites can constrain herbivore predation. Thaler (1999) showed that treating tomato with JA increased parasitism of S. exigua by H. exiguae in the field, but the parasitoids developed more slowly on these caterpillars and gained less weight than control wasps. In addition, Kaplan and Thaler (2010) observed lower predation of the caterpillar of M. sexta by the predaceous stinkbug P. maculiventris on plants overexpressing JA-related induced defences in field experiments. However, manual application of JA forces the constitutive display of defences that are normally induced, and this may result in overestimation of the magnitude of the predator response.
Another layer of complexity is added when communities harbour one or more herbivore species that can suppress plant defences (Alba et al., 2011). Suppression of defences by one species can facilitate another competing species (Sarmento et al., 2011b; Alba et al., 2015). For instance, suppression of defences by specialized strains of the two-spotted spider mite T. urticae (Kant et al., 2008) by the red spider mite T. evansi (Sarmento et al., 2011b) or by the tomato russet mite A. lycopersici (Glas et al., 2014) benefits not only these species themselves but also defence-sensitive competing species inhabiting the same leaf or plant. Hence, suppression of plant defence can indirectly mediate competition between herbivores, forcing the suppressor to adopt strategies to reduce competition with opportunistic herbivores. For instance, the spider mite T. evansi produces more web in the presence of T. urticae, and this behaviour results in elimination of the competitor (Sarmento et al., 2011b). Suppression of plant defences also reduces the emission of induced plant volatiles (Rodriguez-Saona et al., 2003; Kant et al., 2008; Zhang et al., 2009; Schwarzberg et al., 2011; Sarmento et al., 2011a), although this does not always reduce the attraction of predators (Sarmento et al., 2011a). Moreover, suppression of plant defences by spider mites can potentially backfire in the presence of predatory mites, because these prefer the eggs of prey derived from plants with suppressed JA defences to those from plants with activated JA defences (L. M. S. Ataide, University of Amsterdam, Netherlands, unpubl. res.).
Like suppression, induction also affects the performance of competitors. When plants are simultaneously attacked by more than one herbivore species, the palette of plant defences these induce together will determine their mutual interactions and those with their natural enemies. For example, cutting and trenching prevent the plant from transporting photosynthates away from, and defence compounds towards, the tissue where the herbivore is feeding (Dussourd and Denno, 1991; Delaney and Higley, 2006; Oppel et al., 2009). This allows the herbivore to exploit a nutrient-rich part of the plant without having to deal with elevated plant defences. However, if the isolated tissue is large enough, other herbivores can profit from this resource as well, without having to invest in cutting a vein or digging a trench. Similarly, density-dependent feeding efficiency is also observed with gregarious feeding (Prokopy and Roitberg, 2001). For example, gregarious aphids can create sinks in plant tissue, which are preferentially supplied with nutrients by the plant compared with parts where individual aphids feed (Larson and Whitham, 1991). Also, the adult mass of froghoppers, a predictor of fecundity, peaks at intermediate juvenile group size (Wise et al., 2006). In this way, gregarious feeding provides clear benefits because multiple herbivores can feed more efficiently than single herbivores. Interactions with the third trophic level may also shift when multiple herbivore species attack a plant simultaneously. For instance, when cabbage plants are simultaneously attacked by more than one herbivore species, the blend of volatiles released from these plants is different from that released by a singly infested plant. This new blend is less attractive to natural enemies of one of the herbivores and consequently adults of these herbivores preferred to oviposit on cabbage plants previously attacked by the other herbivore species, thus reducing the risk of parasitism of their offspring (Shiojiri et al., 2002).
Finally, induced plant defences can also mediate indirect intra- or interspecific interactions among plants, and this possibly arises from competition between plants for enemy-free space. It has long been known that non-attacked plants close to attacked plants can be warned of imminent attacks through the volatile cues released by the neighbours (Baldwin and Schultz, 1983; Bruin et al., 1992; Karban and Maron, 2002; Baldwin et al., 2006). These volatiles prime the defences of neighbouring plants such that herbivores attacking these plants will trigger the induction of defences more rapidly and usually more strongly than in non-primed plants (Engelberth et al., 2004). Since well-defended plants will deflect herbivores onto their less defended neighbour plants (McNickle and Dybzinski, 2013), the ability of plants to eavesdrop on attacked neighbour plants and be primed to an imminent herbivore attack can be considered an adaptation to increase the plant’s competitive ability.
The examples mentioned above illustrate the importance of incorporating the indirect interactions between plants, herbivores and natural enemies in the study of the evolution of plant defences. Besides the interaction between plants and their herbivores, many other organisms can benefit or suffer, either directly or indirectly, from the defensive products induced by herbivores. The net effects of plant defences on plant fitness thus depend not only on the interaction of the plant with the inducing herbivore, but also on effects on plant fitness through the interaction web associated with the plant. The true fitness effects of plant defences can therefore only be evaluated in the natural environment in which the various interacting species have evolved.
Evolutionary consequences of defence suppression
Defence suppression is perhaps the most striking example of a herbivore strategy that affects the performance of other herbivores living on the same host plant. There are indications that defence suppression is not always restricted to the site of the suppressor, and may act systemically throughout leaflets (Kant et al., 2008; Glas et al., 2014; Alba et al., 2015). For example, the performance of a non-suppressing strain of the spider mite T. urticae increased when feeding from a leaflet co-infested with a suppressor T. urticae strain (Kant et al., 2008) or with the suppressor species T. evansi (Alba et al., 2015), even when spatially separated from one another through a lanolin barrier. In addition, the performance of such non-suppressing T. urticae mites also increased on plants on which suppressor mites had been feeding previously but had been manually removed (Sarmento et al., 2011a). This demonstrates that the beneficial effects of defence suppression for competing herbivores are not restricted to the suppressor’s feeding site or to the moment at which the suppressor is feeding. This raises a key question: why did herbivores evolve the ability to suppress plant defences beyond their feeding site, while the alternative, i.e. the evolution of resistance to plant defences, seems an equally effective but competitively much more attractive trait for herbivores living in communities?
There is no straightforward answer to this question yet, but ecological theory does allow speculation on the adaptive benefits of systemic and long-lasting defence suppression. First of all, herbivores may simply have to work hard to keep the monopoly of their feeding site, and limit the negative effects of interspecific competition as much as possible. For example, T. evansi produces massive amounts of dense webbing, impenetrable for competing species (Sarmento et al., 2011b) and interferes with the reproduction of competitors (Sato et al., 2014), whereas tomato russet mites (A. lycopersici) may avoid competitors by feeding preferentially between the dense leaf-hair forests of tomato (van Houten et al., 2013; Glas et al., 2014). These strategies will be effective against competing herbivore species, but not against intraspecific competition. One potential benefit for herbivores of extending plant defence suppression beyond their own feeding site could be kin selection (Hamilton, 1964): when the individuals that profit from the suppressed plant defence are the suppressor’s relatives, inclusive fitness benefits for the suppressor can outweigh the costs of resource investment. Indeed, when individuals are more related to their neighbours, kin selection can theoretically increase such ‘helping’ behaviour, even when taking the negative fitness effects of stronger competition among kin into account (Mitteldorf and Wilson, 2000). However, a defence-suppressed area on a host plant is a public good (Rankin et al., 2007) when accessible to all conspecifics, and this will allow unrelated individuals to take advantage of the situation as well. These ‘cheaters’ do not have to invest resources in suppressing plant defence but do take the benefit of increased performance. In general, the evolutionary stability of public goods depends on the degree of relatedness at the local site as well as the probability of unrelated individuals dispersing to this site. A potential model system for studying defence-suppressed host plants as public goods is the defence-suppressing spider mite T. evansi. It usually feeds and reproduces in the same area, which increases the chance that individuals are surrounded by relatives. Indeed, high relatedness is frequently observed in T. evansi populations (Boubou et al., 2012).
Apart from intraspecific competition, interspecific competition can evidently also constrain the evolution of defence suppression when suppressors do not succeed in fully monopolising their feeding site (Glas et al., 2014). Herbivores may promote competing species not only through feeding, but also through oviposition. For example, A. thaliana leaves treated with egg extracts of the butterfly P. brassicae had suppressed JA-induced defence responses, resulting in increased performance of the larvae of their competitor Spodoptera littoralis (Bruessow et al., 2010). Interspecific competition can impose serious fitness costs on herbivores (Kaplan and Denno, 2007), and although some herbivores display behaviour (such as massive web production) associated with minimizing the ecological costs of suppressing plant defences (Sarmento et al., 2011b), such strategies can be costly physiological investments. Previous research suggests that web production by T. urticae spider mites shows a trade-off with reproduction (Tien et al., 2009), which would indicate a direct fitness cost of web production. In addition, like defence suppression, the web is a public good as it protects not only individual web builders but conspecifics too. Web builders could therefore also be subject to intraspecific cheaters that do not invest in web production but nevertheless benefit from its protection (Oku et al., 2009).
Given that the fitness benefits of defence suppression can vary with the ecological context, one may ask whether there is genetic variation for defence suppression within species. Kant et al. (2008) and Alba et al. (2015) obtained nearly isogenic lines from single populations of spider mites that were either suppressing or non-suppressing. Also, different populations of H. zea caterpillars were found to produce different amounts of glucose oxidase (Eichenseer et al., 1999). This suggests that genetic variation for defence suppression can be present at the species level. Hence, considering that defence suppression is potentially adaptive, what maintains this variation? First, it is likely that plants from different families differ in their molecular pathways of direct defence (Schulz, 1988). Therefore, generalist herbivores are expected to adapt to their local host plants (Agrawal, 2000), which maintains genetic variation among different herbivore populations on different host plant species. Second, defence suppression could show a trade-off with life-history traits such as oviposition and web production. In this case, the shape of the trade-off determines whether disruptive selection on defence suppression may maintain genetic variation. Third, varying ecological circumstances, such as the presence of competitors or predators, intraspecific cheaters, the relatedness of neighbours and food availability, can alter the direct fitness effects of defence suppression. Because herbivores typically cannot control such factors, it is likely that the benefits of defence suppression vary dynamically over time and space, allowing the maintenance of genetic variation within populations.
Finally, it is appealing to think of defence suppression as an adaptation of the herbivore. However, some studies suggest this may not always be the case. For example, mathematical models suggest that hosts can evolve not to mount an immune response upon infection with a pathogen if the pathogen reacts to the immune response with increased virulence (Restif, 2013). In a plant–herbivore context such ‘mafia behaviour’ (Soler et al., 1995) would mean that herbivores increase feeding upon experiencing induced plant defence. Although empirical studies of mafia behaviour are scarce (Ponton et al., 2006), the costs of mounting an induced defence response under certain circumstances can be higher than its benefits (Fagerström et al., 1987; Herms and Mattson, 1992). For example, when exposed to herbivores, N. attenuata plants typically initiate a JA-induced accumulation of nicotine, but the accumulation of nicotine is costly and decreases the competitive ability of the plant (Baldwin et al., 1998). Hence, when larvae of the nicotine-tolerant herbivore M. sexta feed on N. attenuata, the plant responds by downregulating the accumulation of nicotine (Kahl et al., 2000). Voelckel et al. (2001) argue that this response likely reflects an adaptation of the plant to shut down an inefficient and costly defence response, because using this energy to compete with conspecifics may be more rewarding. Furthermore, when these plants attract parasitoids of M. sexta, any nicotine present in the herbivore may decrease the efficiency of this indirect defence response. Viewed this way, not mounting a direct defence response may be beneficial for plants because it prevents intoxication of their bodyguards (Fordyce, 2001; Kumar et al., 2014). Indeed, whereas continued M. sexta feeding might downregulate nicotine accumulation in N. attenuata, the emission of volatile terpenoids that attract the parasitoids is upregulated (Kahl et al., 2000). These examples show that investigating defence suppression from the plant’s perspective can provide important new insights in the power balance between the plant and the herbivore. If defence suppression was solely an adaptation of the plant, there would not be a tragedy of the commons situation involved for the herbivore. We expect, however, that both plant and herbivore play an active role in this complex phenomenon, which would call for a coevolutionary approach to understand the evolution of defence suppression.
OUTLOOK
Plant defence suppression by herbivores is a largely unexplored phenomenon, but, together with induction, it may play a profound role in the plant-mediated indirect interactions that determine community structure in the phyllosphere. The vast majority of studies on defence suppression by plant eaters have focused on plant–pathogen interactions. These studies have predominantly focused on the mechanisms of defence suppression and the counter-adaptions of plants to undo such suppression. Although there may be many similarities in the mechanisms of defence suppression and the eco-evolutionary dynamics of traits underlying suppression, we do expect fundamental differences between the interactions of plants with immobile (pathogens) and mobile (herbivores) organisms. In this review we call attention to three main themes for future research.
How do herbivores suppress defences? Answering this question requires the transfer of molecular biology tools from phytopathology to the field of plant–herbivore interactions, especially to identify herbivore effectors and their in planta targets. This may deliver not only genetic markers that facilitate the study of their presence in (natural) populations, but also plant breeding targets to decrease the vulnerability of crops to key pest species that suppress plant defences, such as mites, aphids and whiteflies.
What are the consequences of suppression for community interactions? Answering this question requires long-term laboratory, greenhouse and field studies to assess the fitness costs and benefits in simple and more complex communities, not only for the herbivores, but also for the host plants on which they live. This knowledge may also have consequences for plant resistance breeding in conjunction with biological control strategies, e.g. when plant toxins decrease the beneficial effect of natural enemies more than is gained by decreasing herbivore performance directly.
Which conditions cause suppression to emerge, to persist or to disappear from populations? Answering this question requires ecological and evolutionary theory to make predictions on the invasion and population dynamics of herbivores with such traits, as well as a more detailed knowledge of the traits that allow either suppression or resistance to align these predictions with their dynamics under experimental and natural conditions.
ACKNOWLEDGEMENTS
This paper is dedicated to the loving memory of our dear friend and mentor Maurice Sabelis who passed away a week before the manuscript was submitted. M.R.K. was supported by the Netherlands Organization for Scientific Research NWO (Technology Foundation STW/VIDI 13492). F.L., B.K. and J.M.A. were supported by the Netherlands Organization for Scientific Research NWO (Earth & Life Sciences ALW/TOP 854.11.005). W.D. was supported by the Fund for Scientific Research Flanders (FWO). C.A.V. was supported by CONICYT. L.L. was supported by the China Scholarship Council (CSC). B.C.J.S. was supported by the Netherlands Organization for Scientific Research NWO (Earth & Life Sciences ALW/TTI Green Genetics 828.08.001). L.M.S.A. was supported by the Netherlands Organization for Scientific Research NWO (Earth & Life Sciences ALW 824.14.011. J.J.G. was supported by NWO (Technology Foundation STW 13550). M.W.S. was supported by the Royal Academy of Arts and Sciences (KNAW).
LITERATURE CITED
- Abe H, Tomitaka Y, Shimoda T, et al. 2012. Antagonistic plant defense system regulated by phytohormones assists interactions among vector insect, thrips and a tospovirus. Plant & Cell Physiology 53: 204–212. [DOI] [PubMed] [Google Scholar]
- Abramovitch RB, Anderson JC, Martin GB. 2006. Bacterial elicitation and evasion of plant innate immunity. Nature Reviews Molecular Cell Biology 7: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuña R, Padilla BE, Flórez-Ramos CP, et al. 2012. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proceedings of the National Academy of Sciences of the USA 109: 4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie B, Chico JM, Rubio-Somoza I, Solano R. 2007. Modulation of plant defenses by ethylene. Journal of Plant Growth Regulation 26: 160–177. [Google Scholar]
- Afshar K, Fikru-Dube F, Najafabadi HS, et al. 2013. Insights into the insect salivary gland proteome: diet-associated changes in caterpillar labial salivary proteins. Journal of Insect Physiology 59: 351–366. [DOI] [PubMed] [Google Scholar]
- Aggarwal R, Subramanyam S, Zhao C, et al. 2014. Avirulence effector discovery in a plant galling and plant parasitic arthropod, the hessian fly (Mayetiola destructor). PLoS One 9: e100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal AA. 2000. Host-range evolution: adaptation and trade-offs in fitness of mites on alternative hosts. Ecology 81: 500–508. [Google Scholar]
- Agrawal AA, Petschenka G, Bingham RA, Weber MG, Rasmann S. 2012. Toxic cardenolides: chemical ecology and coevolution of specialized plant–herbivore interactions. New Phytologist 194: 28–45. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Badenes-Pérez FR, Reichelt M, et al. 2011. Metabolic detoxification of capsaicin by UDP-glycosyltransferase in three Helicoverpa species. Archives of Insect Biochemistry and Physiology 78: 104–118. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Dermauw W, Wybouw N, Heckel DG, Van Leeuwen T. 2014. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochemistry and Molecular Biology 50: 43–57. [DOI] [PubMed] [Google Scholar]
- Akhtar TA, Matsuba Y, Schauvinhold I, et al. 2013. The tomato cis-prenyltransferase gene family. Plant Journal 73: 640–652. [DOI] [PubMed] [Google Scholar]
- Alba JM, Glas JJ, Schimmel BCJ, Kant MR. 2011. Avoidance and suppression of plant defenses by herbivores and pathogens. Journal of Plant Interactions 6: 221–227. [Google Scholar]
- Alba JM, Schimmel BCJ, Glas JJ, et al. 2015. Spider mites suppress tomato defences downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytologist 205: 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M. 2013. Peptides as triggers of plant defence. Journal of Experimental Botany 64: 5269–5279. [DOI] [PubMed] [Google Scholar]
- Alborn HT, Turlings TCJ, Jones TH, Stenhagen G, Loughrin JH, Tumlinson JH. 1997. An elicitor of plant volatiles from beet armyworm oral secretion. Science 9: 945–949. [Google Scholar]
- Alborn HT, Hansen TV, Jones TH, et al. 2007. Disulfooxy fatty acids from the American bird grasshopper Schistocerca americana, elicitors of plant volatiles. Proceedings of the National Academy of Sciences of the USA 104: 12976–12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC. 2004. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiology 135: 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Krasikov V, Allmann S, Rep M, Takken FL, Schuurink RC. 2010. Methyl salicylate production in tomato affects biotic interactions. Plant Journal 62: 124–134. [DOI] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, et al. 2010. Ethylene induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anten NPR, Pierik R. 2010. Moving resources away from the herbivore: regulation and adaptive significance. New Phytologist 188: 644–645. [DOI] [PubMed] [Google Scholar]
- Atamian HS, Chaudhary R, Cin VD, Bao E, Girke T, Kaloshian I. 2013. In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Molecular Plant-Microbe Interactions 26: 67–74. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. 2003. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Molecular Microbiology 49: 1537–1546. [DOI] [PubMed] [Google Scholar]
- Babst BA, Ferrieri RA, Thorpe MR, Orians CM. 2008. Lymantria dispar herbivory induces rapid changes in carbon transport and partitioning in Populus nigra. Entomologia Experimentalis et Applicata 128: 117–125. [Google Scholar]
- Balasundram N, Sundram K, Samman S. 2006. Phenolic compounds in plants and agri-industrial by-products: antioxidant activity, occurrence, and potential uses. Food Chemistry 99: 191-203. [Google Scholar]
- Baldwin IT. 1988. The alkaloid responses of wild tobacco to real and simulated herbivory. Oecologia 77: 378–381. [DOI] [PubMed] [Google Scholar]
- Baldwin IT. 1996. Methyl jasmonate-induced nicotine production in Nicotiana attenuata: inducing defenses in the field without wounding. Entomologia Experimentalis et Applicata 80: 213–220. [Google Scholar]
- Baldwin IT. 1998. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proceedings of the National Academy of Sciences of the USA 95: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Schultz JC. 1983. Rapid changes in tree leaf chemistry induced by damage: evidence for communication between plants. Science 221: 277–279. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Gorham D, Schmelz EA, Lewandowski CA, Lynds GY. 1998. Allocation of nitrogen to an inducible defense and seed production in Nicotiana attenuata. Oecologia 115: 541–552. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Halitschke R, Paschold A, von Dahl CC, Preston CA. 2006. Volatile signaling in plant-plant interactions: “Talking trees” in the genomics era. Science 311: 812–815. [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Constabel CP. 2011. Tannins in plant-herbivore interactions. Phytochemistry 72: 1551–1565. [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Jones CP, Yip L, Tran L, Constabel CP. 2007. Limited impact of elevated levels of polyphenol oxidase on tree-feeding caterpillars: assessing individual plant defenses with transgenic poplar. Oecologia 154: 129–140. [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Niewiadomski J, Kochmanski J. 2013. Importance of protein quality versus quantity in alternative host plants for a leaf-feeding insect. Oecologia 173: 1–12. [DOI] [PubMed] [Google Scholar]
- Barbosa P, Saunders JA, Waldvogel M. 1982. Plant-mediated variation in herbivore suitability and parasitoid fitness. In: Visser JH, Minks AK. eds. Proceedings of the 5th International Symposium on Insect-Plant Relationships. Wageningen: Pudoc, 63–71. [Google Scholar]
- Barth C, Jander G. 2006. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant Journal 46: 549–562. [DOI] [PubMed] [Google Scholar]
- Bateman KS, James MNG. 2011. Plant protein proteinase inhibitors: structure and mechanism of inhibition. Current Protein and Peptide Science 12: 340–347. [DOI] [PubMed] [Google Scholar]
- Bede JC, Musser RO, Felton GW, Korth KL. 2006. Caterpillar herbivory and salivary enzymes decrease transcript levels of Medicago truncatula genes encoding early enzymes in terpenoid biosynthesis. Plant Molecular Biology 60: 519–531. [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. 2007. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annual Review of Phytopathology 45: 399–436. [DOI] [PubMed] [Google Scholar]
- Beran F, Mewis I, Srinivasan R, et al. 2011. Male Phyllotreta striolata (F.) produce an aggregation pheromone: identification of male-specific compounds and interaction with host plant volatiles. Journal of Chemical Ecology 37: 85–97. [DOI] [PubMed] [Google Scholar]
- Beran F, Pauchet Y, Kunert G, et al. 2014. Phyllotreta striolata flea beetles use host plant defense compounds to create their own glucosinolate-myrosinase system. Proceedings of the National Academy of Sciences of the USA 111: 7349–7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum M. 1982. Coumarins and caterpillars: a case for coevolution. Evolution 37: 163–179. [DOI] [PubMed] [Google Scholar]
- Berenbaum M. 1987. Target site insensitivity in insect-plant interactions. In: Brattsten L, Ahmad S. eds. Molecular aspects of insect-plant associations . Springer US. [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. 2001. evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Bhuiyan NH, Selvaraj G, Wei Y, King J. 2009. Role of lignification in plant defense. Plant Signaling and Behavior 4: 158–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Mirabella R, Diergaarde PJ, et al. 2012. Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proceedings of the National Academy of Sciences of the USA 109: 20124–20129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatwright JL, Pajerowska-Mukhtar K. 2013. Salicylic acid: an old hormone up to new tricks. Molecular Plant Pathology 14: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Bonas U. 2010. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annual Review of Phytopathology 48: 419–436. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, VanDoorn A, Baldwin IT. 2011. Herbivore-associated elicitors: FAC signaling and metabolism. Trends in Plant Science 16: 294–299. [DOI] [PubMed] [Google Scholar]
- Bones AM, Rossiter JT. 1996. The myrosinase-glucosinolate system, its organization and biochemistry. Physiologia Plantarium 97: 194–208. [Google Scholar]
- Bos JIB, Prince D, Pitino M, et al. 2010. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach Aphid). PLoS Genetics 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouarab K, Melton R, Peart J, Baulcombe D, Osbourn A. 2002. A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418: 889–892. [DOI] [PubMed] [Google Scholar]
- Boubou A, Migeon A, Roderick GK, et al. 2012. Test of colonization scenarios reveals complex invasion history of the red tomato spider mite Tetranychus evansi. PloS One 7: e35601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattsten L. 1992. Metabolic defenses against plant allelochemicals. In: Rosenthal G, Berenbaum M. eds. Herbivores: their interactions with secondary plant metabolites . London: Academic Press, 175–242. [Google Scholar]
- Broadbent BA, Rhainds M, Shipp L, Murphy G, Wainman L. 2003. Pupation behaviour of westernflower thrips (Thysanoptera: Thripidae) on potted chrysanthemum. Canadian Entomologist 135: 741–744. [Google Scholar]
- Bryant JP, Chapin FS, Klein DR. 1983. Carbon nutrient balance of boreal plants in relation to vertebrate herbivory. Oikos 40: 357–368. [Google Scholar]
- Budd GE, Telford MJ. 2009. The origin and evolution of arthropods. Nature 457: 812–817. [DOI] [PubMed] [Google Scholar]
- Bruessow F, Gouhier-Garimont C, Buchala A, Metraux J-P, Reymond P. 2010. Insect eggs suppress plant defence against chewing herbivores. Plant Journal 62: 876–885. [DOI] [PubMed] [Google Scholar]
- Bruin J, Dicke M, Sabelis MW. 1992. Plants are better protected against spider-mites after exposure to volatiles from infested conspecifics. Experientia 48: 525–529. [Google Scholar]
- Buch F, Pauchet Y, Rott M, Mithöfer A. 2014. Characterization and heterologous expression of a PR-1 protein from traps of the carnivorous plant Nepenthes mirabilis. Phytochemistry 100: 43–50. [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. 2012. Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiology 160: 1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud MC, Dubreuil G, Quentin M, et al. 2008. Root-knot nematodes manipulate plant cell functions during a compatible interaction. Journal of Plant Physiology 165: 104–113. [DOI] [PubMed] [Google Scholar]
- Caldéron-Cortés N, Quesada M, Watanabe H, Cano-Camacho H, Oyama K. 2012. Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annual Review of Ecology, Evolution, and Systematics 43: 45–71. [Google Scholar]
- Campbell BC, Duffey SS. 1979. Tomatine and parasitic wasps: potential incompatibility of plant antibiosis with biological-control. Science 205: 700–702. [DOI] [PubMed] [Google Scholar]
- Caravas J, Friedrich M. 2010. Of mites and millipedes: recent progress in resolving the base of the arthropod tree. BioEssays 32: 488–495. [DOI] [PubMed] [Google Scholar]
- Cardoso MZ. 2008. Herbivore handling of a plant’s trichome: the case of Heliconius charithonia (L.) (Lepidoptera: Nymphalidae) and Passiflora lobata (Killip) Hutch. (Passifloraceae). Neotropical Entomology 37: 247–252. [DOI] [PubMed] [Google Scholar]
- Carlini CR, Grossi-de-Sa MF. 2002. Plant toxic proteins with insecticidal properties. A review on their potentialities as bioinsecticides. Toxicon 40: 1515–1539. [DOI] [PubMed] [Google Scholar]
- Carroll MJ, Schmelz EA, Meagher RL, Teal PE. 2006. Attraction of Spodoptera frugiperda larvae to volatiles from herbivore-damaged maize seedlings. Journal of Chemical Ecology 32: 1911–1924. [DOI] [PubMed] [Google Scholar]
- Castañera P, Steffens JC, Tingey WM. 1996. Biological performance of Colorado potato beetle larvae on potato genotypes with differing levels of polyphenol oxidase. Journal of Chemical Ecology 22: 91–101. [DOI] [PubMed] [Google Scholar]
- Cavallini C, Trettene M, Degan M, et al. 2010. Anti-angiogenic effects of two cystine-knot miniproteins from tomato fruit. British Journal of Pharmacology 162: 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA. 2005. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proceedings of the National Academy of Sciences of the USA 102: 19237–19242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gonzales-Vigil E, Wilkerson CG, Howe GA. 2007. Stability of plant defense proteins in the gut of insect herbivores. Plant Physiology 143: 1954–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhu YC, Whitworth RJ, Reese JC, Chen MS. 2013. Serine and cysteine protease-like genes in the genome of a gall midge and their interactions with host plant genotypes. Insect Biochemistry and Molecular Biology 43: 701–711. [DOI] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE. 2005. Ethylene signal transduction. Annals of Botany 95: 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, et al. 2014. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester KS. 1933. The problem of acquired physiological immunity in plants. Quarterly Review of Biology 8: 275–324. [Google Scholar]
- Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S. 2013. Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiology and Biochemistry 72: 1–20. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Choi J, Choi D, Lee S, Ryu C, Hwang I. 2011. Cytokinins and plant immunity: old foes or new friends? Trends in Plant Science 16: 388–394. [DOI] [PubMed] [Google Scholar]
- Choong MF. 1996. What makes a leaf tough and how this affects the pattern of Castanopsis fissa leaf consumption by caterpillars. Functional Ecology 10: 668–674. [Google Scholar]
- Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW. 2013. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proceedings of the National Academy of Sciences of the USA 110: 15728–15733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CW, Harvell CD. 1992. Inducible defenses and the allocation of resources: a minimal model. American Naturalist 139: 521–539. [Google Scholar]
- Coley PD, Bryant JP, Chapin FS. 1985. Resource availability and plant antiherbivore defense. Science 230: 895–899. [DOI] [PubMed] [Google Scholar]
- Coll M, Guershon M. 2002. Omnivory in terrestrial arthropods: mixing plant and prey diets. Annual Review of Entomology 47: 267–297. [DOI] [PubMed] [Google Scholar]
- Connor EF, Taverner MP. 1997. The evolution and adaptive significance of the leaf-mining habit. Oikos 79: 6–25. [Google Scholar]
- Consales F, Schweizer F, Erb M, et al. 2012. Insect oral secretions suppress wound-induced responses in Arabidopsis. Journal of Experimental Botany 63: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel CP, Barbehenn R. 2008. Defensive roles of polyphenol oxidase in plants. In: Schaller A. ed. Induced plant resistance to herbivory . Springer Science Business Media, 253–269. [Google Scholar]
- Constabel CP, Ryan CA. 1998. A survey of wound- and methyl jasmonate-induced leaf polyphenol oxidase in crop plants. Phytochemistry 47: 507–511. [Google Scholar]
- Constabel CP, Yip L, Patton JJ, Christopher ME. 2000. Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory. Plant Physiology 124: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Courdavault V, Papon N, Clastre M, Giglioli-Guivarc’h N, St-Pierre B, Burlat V. 2014. A look inside an alkaloid multisite plant: the Catharanthus logistics. Current Opinion in Plant Biology 19: 43–50. [DOI] [PubMed] [Google Scholar]
- Dąbrowska P, Shabab M, Brandt W, Vogel H, Boland W. 2011. Isomerization of the phytohormone precursor 12-oxophytodienoic acid (OPDA) in the insect gut. Journal of Biological Chemistry 286: 22348–22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla S, Swarts HGP, Koenderink JB, Dobler S. 2013. Amino acid substitutions of Na, K-ATPase conferring decreased sensitivity to cardenolides in insects compared to mammals. Insect Biochemistry and Molecular Biology 43: 1109–1115. [DOI] [PubMed] [Google Scholar]
- Van, Dam NM, Raaijmakers CE, Van der Putten WH. 2005. Root herbivory reduces growth and survival of the shoot feeding specialist Pieris rapae on Brassica nigra. Entomologia Experimentalis et Applicata 115: 161–170. [Google Scholar]
- Dangl JL, Jones JDG. 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Hiltpold I, Köllner TG, et al. 2009. Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proceedings of the National Academy of Sciences of the USA 106: 13213–13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney KJ, Higley LG. 2006. An insect countermeasure impacts plant physiology: midrib vein cutting, defoliation and leaf photosynthesis. Plant, Cell and Environment 29: 1245–1258. [DOI] [PubMed] [Google Scholar]
- DeLay B, Mamidala P, Wijeratne A, et al. 2012. Transcriptome analysis of the salivary glands of potato leafhopper, Empoasca fabae. Journal of Insect Physiology 58: 1626–1634. [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. 2011. Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF. 2012. SOS – too many signals for systemic acquired resistance? Trends in Plant Science 17: 538–545. [DOI] [PubMed] [Google Scholar]
- Denno RF, McClure MS, Ott JR. 1995. interspecific interactions in phytophagous insects: competition reexamined and resurrected. Annual Review of Entomology 40: 297–331 [Google Scholar]
- Dermauw W, Van Leeuwen T. 2014. The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochemistry and Molecular Biology 45: 89–110. [DOI] [PubMed] [Google Scholar]
- Dermauw W, Wybouw N, Rombauts S, et al. 2013a. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proceedings of the National Academy of Sciences of the USA 110: E113–E122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermauw W, Osborne EJ, Clark RM, Grbić M, Tirry L, Van Leeuwen T. 2013b. A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genomics 14: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S. 2012. Catch me if you can: bacterial effectors and plant targets . Trends in Plant Science 17: 644–655. [DOI] [PubMed] [Google Scholar]
- Despres L, David J, Gallet C. 2007. The evolutionary ecology of insect resistance to plant chemicals. Trends in Ecology & Evolution 22: 298–307. [DOI] [PubMed] [Google Scholar]
- Devonshire AL, Moores GD. 1982. A carboxylesterase with broad substrate specificity causes organophosphorus, carbamate and pyrethroid resistance in peach-potato aphids (Myzus persicae). Pesticide Biochemistry and Physiology 18: 235–246. [Google Scholar]
- Devonshire AL, Sawicki RM. 1979. Insecticide-resistant Myzus persicae as an example of evolution by gene duplication. Nature 280: 140–141. [Google Scholar]
- Dicke M, Sabelis MW. 1988. How plants obtain predatory mites as bodyguards. Netherlands Journal of Zoology 38: 148–165. [Google Scholar]
- Dicke M, VanBeek TA, Posthumus MA, Bendom N, VanBokhoven H, DeGroot AE. 1990. Isolation and identification of volatile kairomone that affects acarine predator-prey interactions - involvement of host plant in its production. Journal of Chemical Ecology 16: 381–396. [DOI] [PubMed] [Google Scholar]
- Diezel C, Von Dahl CC, Gaquerel E, Baldwin IT. 2009. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiology 150: 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh ST, Baldwin IT, Galis I. 2013. The HERBIVORE ELICITOR-REGULATED1 gene enhances abscisic acid levels and defenses against herbivores in Nicotiana attenuata plants. Plant Physiology 162: 2106–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobler S, Dalla S, Wagschal V, Agrawal AA. 2012. Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proceedings of the National Academy of Sciences of the USA 109: 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, et al. 2013. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogimont C, Chovelon V, Tual S, et al. 2008. Molecular diversity at the Vat/PM-W resistance locus in melon. In: Pitrat M. ed. Cucurbitaceae 2008. Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Avignon, France . Avignon: INRA, 219–228. [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, et al. 2007. MYC2 differentially modulates diverse jasmonate dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty HM, Selvendran RR, Bowles DJ. 1988. The wound response of tomato plants can be inhibited by aspirin and related hydroxy-benzoic acids. Physiological and Molecular Plant Pathology 33: 377–384. [Google Scholar]
- Douglas AE. 2006. Phloem-sap feeding by animals: problems and solutions. Journal of Experimental Botany 57: 747–754. [DOI] [PubMed] [Google Scholar]
- Du B, Zhang W, Liu B, et al. 2009. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proceedings of the National Academy of Sciences of the USA 106: 22163–22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Li X, Xue Q, Abo-El-Saad M, Xu O, Wu R. 1996. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nature Biotechnology 14: 444–498. [DOI] [PubMed] [Google Scholar]
- Ducluzeau A-L, Wamboldt Y, Elowsky CG, Mackenzie SA, Schuurink RC, Basset GJC. 2012. Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant Journal 69: 366–375. [DOI] [PubMed] [Google Scholar]
- Duffey SS. 1980. Sequestration of plant natural-products by insects. Annual Review of Entomology 25: 447–477. [Google Scholar]
- Dussourd DE, Denno RF. 1991. Deactivation of plant defense: correspondence between insect behaviour and secretory canal architecture. Ecology 72: 1383–1396. [Google Scholar]
- Eichenseer H, Mathews MC, Bi JL, Murphy JB, Felton GW. 1999. Salivary glucose oxidase: multifunctional roles for Helicoverpa zea? Archives of Insect Biochemistry and Physiology 42: 99–109. [DOI] [PubMed] [Google Scholar]
- Eichenseer H, Mathews MC, Powell JS, Felton GW. 2010. Survey of a salivary effector in caterpillars: glucose oxidase variation and correlation with host range. Journal of Chemical Ecology 36: 885–897. [DOI] [PubMed] [Google Scholar]
- Eigenbrode SD, Espelie KE. 1995. Effects of plant epicuticular lipids on insect herbivores. Annual Review of Entomology 49: 171–194. [Google Scholar]
- Eigenbrode SD, Shelton AM. 1990. Behavior of neonate diamondback moth larvae (Lepidoptera: Plutellidae) on glossy-leafed resistant Brassica oleracea L. Environmental Entomology 19: 1566–1571. [Google Scholar]
- Elbaz M, Halon E, Malka O, et al. 2012. Asymmetric adaptation to indolic and aliphatic glucosinolates in the B and Q sibling species of Bemisia tabaci (Hemiptera: Aleyrodidae). Molecular Ecology 21: 4533–4546. [DOI] [PubMed] [Google Scholar]
- Elzinga DA, De Vos M, Jander G. 2014. Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Molecular Plant-Microbe Interactions 27: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Moran NA. 2013. The gut microbiota of insects – diversity in structure and function. FEMS Microbiology Reviews 37: 699–735. [DOI] [PubMed] [Google Scholar]
- Engelberth J, Alborn HT, Schmelz EA, et al. 2004. Airborne signals prime plants against insect herbivore attack. Proceedings of the National Academy of Sciences of the USA 101: 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler HS, Spencer KC, Gilbert LE. 2000. Insect metabolism: preventing cyanide release from leaves. Nature 406: 144–145. [DOI] [PubMed] [Google Scholar]
- Erb M, Robert CAM, Hibbard BE, Turlings TCJ. 2011. Sequence of arrival determines plant-mediated interactions between herbivores. Journal of Ecology 99: 7–15. [Google Scholar]
- Erb M, Meldau S, Howe GA. 2012. Role of phytohormones in insect-specific plant reactions. Trends in Plant Science 17: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrick J, Behnke C, Czapla T, et al. 2002. Effects of a potato cysteine proteinase inhibitor on midgut proteolytic enzyme activity and growth of the southern corn rootworm, Diabrotica undecimpunctata howardi (Coleoptera: Chrysomelidae). Insect Biochemistry and Molecular Biology 32: 405–415. [DOI] [PubMed] [Google Scholar]
- Faeth SH. 1986. Indirect interactions between temporally separated herbivores mediated by the host plant. Ecology 67: 479–494. [Google Scholar]
- Fagan WF, Siemann E, Mitter C, et al. 2002. Nitrogen in insects: implications for trophic complexity and species diversification. American Naturalist 6: 784–802. [DOI] [PubMed] [Google Scholar]
- Fagerström T, Larsson S, Tenow O. 1987. On optimal defence in plants. Functional Ecology 1: 73–81. [Google Scholar]
- Fatouros NE, Broekgaarden C, Bukovinszkine’Kiss G, et al. 2008. Male-derived butterfly anti-aphrodisiac mediates induced indirect plant defense. Proceedings of the National Academy of Sciences of the USA 105: 10033–10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. 1990. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proceedings of the National Academy of Sciences of the USA 87: 7713–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeny P. 1976. Plant apparency and chemical defense. In: Wallace JW, Mansell RL. Biochemical interaction between plants and insects. Proceedings of the Fifteenth Annual Meeting of the Phytochemical Society of North America. New York: Plenum Press, 1–40. [Google Scholar]
- Felton GW. 2005. Indigestion is a plant’s best defense. Proceedings of the National Academy of Sciences of the USA 102: 18771–18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Tumlinson JH. 2008. Plant-insect dialogs: complex interactions at the plant-insect interface. Current Opinion in Plant Biology 11: 457–463. [DOI] [PubMed] [Google Scholar]
- Felton GW, Donato KK, Broadway RM, Duffey SS. 1992a. Impact of oxidized plant phenolics on the nutritional quality of dietary protein to noctuid herbivore, Spodoptera exigua. Journal of Insect Physiology 38: 277–285. [Google Scholar]
- Felton GW, Workman J, Duffey SS. 1992b. Avoidance of antinutritive plant defense: role of midgut pH in Colorado potato beetle. Journal of Chemical Ecology 18: 571–583. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieri AP, Agtuca B, Appel HM, Ferrieri RA, Schultz JC. 2013. Temporal changes in allocation and partitioning of new carbon as 11C elicited by simulated herbivory suggest that roots shape aboveground responses in Arabidopsis. Plant Physiology 161: 692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fescemyer HW, Sandoya GV, Gill TA, Ozkan S, Marden JH, Luthe DS. 2013. Maize toxin degrades peritrophic matrix proteins and stimulates compensatory transcriptome responses in fall armyworm midgut. Insect Biochemistry and Molecular Biology 43: 280–291. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. 2012. Insect CYP genes and P450 enzymes. In: Gilbert LI. ed. Insect molecular biology and biochemistry . London: Academic Press, Elsevier. [Google Scholar]
- Feyereisen R, Dermauw W, Van Leeuwen T. 2015. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pesticide Biochemistry and Physiology, in press. doi:10.1016/j.pestbp.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant RH. 2013. The molecular genetics of insecticide resistance. Genetics 194: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RC, Richter A, Wanek W, Mayer V. 2002. Plants feed ants: food bodies of myrmecophytic Piper and their significance for the interaction with Pheidole bicornis ants. Oecologia 133: 186–192. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, et al. 2009. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fordyce JA. 2001. The lethal plant defense paradox remains: inducible host-plant aristolochic acids and the growth and defense of the pipevine swallowtail. Entomologia Experimentalis et Applicata 100: 339–346. [Google Scholar]
- Fowler JH, Narvaez-Vasquez J, Aromdee DN, Pautot V, Holzer FM, Walling LL. 2009. Leucine aminopeptidase regulates defense and wound signaling in tomato downstream of jasmonic acid. Plant Cell 21: 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Pinto MES, Holloway DE, Bramley P. 2000. Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant Journal 24: 551–558. [DOI] [PubMed] [Google Scholar]
- Frehner M, Conn EE. 1987. The linamarin beta-glucosidase in Costa Rica wild lima beans (Phaseolus lunatus L) is apoplastic. Plant Physiology 84: 1296–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost CJ, Appel HM, Carlson JE, et al. 2007. Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecology Letters 10: 490–498. [DOI] [PubMed] [Google Scholar]
- Furch ACU, Hafke JB, Schulz A, Van Bel AJE. 2007. Ca2+-mediated remote control of reversible sieve tube occlusion in Vicia faba. Journal of Experimental Botany 58: 2827–2838. [DOI] [PubMed] [Google Scholar]
- Gaertner LS, Murray CL, Morris CE. 1998. Transepithelial transport of nicotine and vinblastine in isolated malpighian tubules of the tobacco hornworm (Manduca sexta) suggests a P-glycoprotein-like mechanism. Journal of Experimental Biology 201: 2637–2645. [DOI] [PubMed] [Google Scholar]
- Gaquerel E, Weinhold A, Baldwin IT. 2009. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VIII. An unbiased GCxGC-ToFMS analysis of the plant’s elicited volatile emissions. Plant Physiology 149: 1408–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston KJ, Reavey D, Valladares GR. 1991. Changes in feeding habits as caterpillar grow. Ecological Entomology 16: 339–344. [Google Scholar]
- Gatehouse AMR, Down RE, Powell KS, et al. 1996. Transgenic potato plants with enhanced resistance to the peach-potato aphid Myzus persicae. Entomologia Experimentalis et Applicata 79: 295–307. [Google Scholar]
- Gawehns F, Cornelissen BJC, Takken KLW. 2013. The potential of effector-target genes in breeding for plant innate immunity. Microbial Biotechnology 6: 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. 2007. The function of terpene natural products in the natural world. Nature Chemical Biology 3: 408–414. [DOI] [PubMed] [Google Scholar]
- Glas JJ, Schimmel BCJ, Alba JM, Escobar-Bravo R, Schuurink RC, Kant MR. 2012. Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. International Journal of Molecular Sciences 13: 17077–17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas JJ, Alba JM, Simoni S, et al. 2014. Defense suppression benefits herbivores that have a monopoly on their feeding site but can backfire within natural communities. BMC Biology 12: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender JL. 2008. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. Journal of Biological Chemistry 283: 16400–16407. [DOI] [PubMed] [Google Scholar]
- Glauser G, Dubugnon L, Mousavi SAR, Rudaz S, Wolfender JL, Farmer EE. 2009. Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. Journal of Biological Chemistry 284: 34506–34513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Gloss AD, Vassão DG, Hailey AL, et al. 2014. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Molecular Biology and Evolution 31: 2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez S, Steinbrenner AD, Osorio S, et al. 2012. From shoots to roots: transport and metabolic changes in tomato after simulated feeding by a specialist lepidopteran. Entomologia Experimentalis et Applicata 144: 101–111. [Google Scholar]
- Gonzales-Vigil E, Bianchetti CM, Phillips GN, Howe GA. 2011. Adaptive evolution of threonine deaminase in plant defense against insect herbivores. Proceedings of the National Academy of Sciences of the USA 108: 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. 2012. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proceedings of the National Academy of Sciences of the USA 109: 4674–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind G, Mittapalli O, Griebel T, Allmann S, Bocker S, Baldwin IT. 2010. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One 5: e8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozzo F, Faoro F. 2013. Systemic acquired resistance (50 years after discovery): moving from the lab to the field. Journal of Agricultural and Food Chemistry 61: 12473–12491. [DOI] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 14: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi DA, Engel MS. 2005. Evolution of the insects. Cambridge: Cambridge University Press. [Google Scholar]
- Gross N, Wasternack C, Köck M. 2004. Wound-induced RNase LE expression is jasmonate and systemin independent and occurs only locally in tomato (Lycopersicon esculentum cv. Lukullus). Phytochemistry 65: 1343–1350. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Holzer FM, Walling LL. 1999. Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato. European Journal of Biochemistry 263: 726–735. [DOI] [PubMed] [Google Scholar]
- Haegeman A, Mantelin S, Jones JT, Gheysen G. 2012. Functional roles of effectors of plant-parasitic nematodes. Gene 492: 19–31. [DOI] [PubMed] [Google Scholar]
- Hairston NG, Smith FE, Slobodkin LB. 1960. Community structure, population control, and competition. American Naturalist 94: 421–425. [Google Scholar]
- Halkier BA, Gershenzon J. 2006. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Hall DE, MacGregor KB, Nijsse J, Bown AW. 2004. Footsteps from insect larvae damage leaf surfaces and initiate rapid responses. European Journal of Plant Pathology 110: 441–447. [Google Scholar]
- Hamamouch N, Li CY, Seo PJ, Park CM, Davis EL. 2011. Expression of Arabidopsis pathogenesis-related genes during nematode infection. Molecular Plant Pathology 12: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD. 1964. The genetical evolution of social behaviour. Journal of Theoretical Biology 7: 1–16. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Du L, Naur P, et al. 2001. CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. Journal of Biological Chemistry 276: 24790–24796. [DOI] [PubMed] [Google Scholar]
- Hao P, Liu C, Wang Y, et al. 2008. Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiology 146: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare JD, Walling LL. 2006. Constitutive and jasmonate-inducible traits of Datura wrightii. Journal of Chemical Ecology 32: 29–47. [DOI] [PubMed] [Google Scholar]
- Harmel N, Letocart E, Cherqui A, et al. 2008. Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect Molecular Biology 17: 165–174. [DOI] [PubMed] [Google Scholar]
- Harris MO, Stuart JJ, Mohan M, Nair S, Lamb RJ, Rohfritsch O. 2003. Grasses and gall midges: plant defense and insect adaptation. Annual Review of Entomology 84: 549–577. [DOI] [PubMed] [Google Scholar]
- Harris MO, Freeman TP, Rohfritsch O, Anderson KG, Payne SA, Moore JA. 2006. Virulent hessian fly (Diptera: Cecidomyiidae) larvae induce a nutritive tissue during compatible interactions with wheat. Annals of the Entomological Society of America 99: 305–316. [Google Scholar]
- Harris MO, Freeman TP, Moore JA, et al. 2010. H-gene-mediated resistance to hessian fly exhibits features of penetration resistance to fungi. Phytopathology 100: 279–289. [DOI] [PubMed] [Google Scholar]
- Harrison RL, Bonning BC. 2010. Proteases as insecticidal agents. Toxins 2: 935–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S, Karban R. 1986. Effects of an early-season folivorous moth on the success of a later season-species, mediated by a change in the quality of the shared host, Lupinus arboreus Sims. Oecologia 69: 354–359. [DOI] [PubMed] [Google Scholar]
- Harrop TWR, Pearce SL, Daborn PJ, Batterham P. 2014. Whole-genome expression analysis in the third instar larval midgut of Drosophila melanogaster. G3: Genes. Genomes Genetics 4: 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M, Giri AP, Kaur H, Baldwin IT. 2010. Serine protease inhibitors specifically defend Solanum nigrum against generalist herbivores but do not Influence plant growth and development. Plant Cell 22: 4158–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. 2006. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575. [DOI] [PubMed] [Google Scholar]
- Heil M, Silva Bueno JC. 2007. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proceedings of the National Academy of Sciences of the USA 104: 5467–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Ton J. 2008. Long-distance signalling in plant defence. Trends in Plant Science 13: 264–272. [DOI] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ. 1992. The dilemma of plants: to grow or defend. Quarterly Review of Biology 67: 283–335. [Google Scholar]
- Hoballah ME, Gübitz T, Stuurman J, et al. 2007. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkison R, Ayasse M, Kalko EKV, et al. 2007. Chemical ecology of fruit bat foraging behavior in relation to the fruit odors of two species of paleotropical bat-dispersed figs (Ficus hispida and Ficus scortechinii). Journal of Chemical Ecology 33: 2097–2110. [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Bos JIB. 2011. Effector proteins that modulate plant-insect interactions. Current Opinion in Plant Biology 14: 422–428. [DOI] [PubMed] [Google Scholar]
- Hopkins RJ, Van Dam NM, Van Loon JJA. 2009. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annual Review of Entomology 54: 57–83. [DOI] [PubMed] [Google Scholar]
- van Houten YM, Glas JJ, Hoogerbrugge H, et al. 2013. Herbivory-associated degradation of tomato trichomes and its impact on biological control of Aculops lycopersici. Experimental and Applied Acarology 60: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA. 2004. Jasmonates as signals in the wound response. Journal of Plant Growth Regulation 23: 223–237. [Google Scholar]
- Howe GA, Jander G. 2008. Plant immunity to insect herbivores. Annual Review of Plant Biology 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. 1998. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271. [DOI] [PubMed] [Google Scholar]
- Huang J-S. 1986. Ultrastructure of bacterial penetration in plants. Annual Review of Phytopathology 24: 141–157. [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. 2006. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences of the USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Veyrat N, et al. 2013. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proceedings of the National Academy of Sciences of the USA 110: 5707–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Chadchawan S, Winge P, Thangstad OP, Bones AM. 2002. Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (Myrosinase) in Arabidopsis. Plant Physiology 128: 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Cox-Foster DL, Yang X, Ko WY, Cavener DR. 2007. Expansion and evolution of insect GMC oxidoreductases. BMC Evol Biol. 11:7: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar M, Doostdar H, Sonoda RM, Leibee GL, Mayer RT. 1998. Elicitors of plant defensive systems reduce insect densities and disease incidence. Journal of Chemical Ecology 24: 135–149. [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, et al. 2013. Biosynthesis of antinutritional alkaloids in Solanaceous crops is mediated by clustered genes. Science 341: 175–179. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Määttä K, Pirttilä, Törrönen R, Kärenlampi S, Hohtola A. 2002. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiology 130: 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G. 2012. Timely plant defenses protect against caterpillar herbivory. Proceedings of the National Academy of Sciences of the USA 109: 4343–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Pallini A, Venzon M, Sabelis MW. 1998. Behaviour and indirect interactions in food webs of plant-inhabiting arthropods. Experimental and Applied Acarology 22: 497–521. [Google Scholar]
- Jensen NB, Zagrobelny M, Hjernø K, et al. 2011. Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nature Communications 2: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji JH, Ding SW. 2001. The suppressor of transgene RNA silencing encoded by cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Molecular Plant-Microbe Interactions 14: 715–724. [DOI] [PubMed] [Google Scholar]
- Jongsma MA, Bolter C. 1997. The adaptation of insects to plant protease inhibitors. Journal of Insect Physiology 43: 885–895. [DOI] [PubMed] [Google Scholar]
- Jorda L, Vera P. 2000. Local and systemic induction of two defense-related subtilisin-like protease promoters in transgenic Arabidopsis plants. Luciferin induction of PR gene expression. Plant Physiology 124:1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, et al. 2000. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta 210: 336–342. [DOI] [PubMed] [Google Scholar]
- Kaloshian I, Walling LL. 2005. Hemipterans as plant pathogens. Annual Review of Phytopathology 43: 491–521. [DOI] [PubMed] [Google Scholar]
- Kamoun S. 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology 44: 41–60. [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, et al. 2007. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proceedings of the National Academy of Sciences of the USA 104: 12205–12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Ament K, Sabelis MW, Haring MA, Schuurink RC. 2004. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiology 135: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Sabelis MW, Haring MA, Schuurink RC. 2008. Intraspecific variation in a generalist herbivore accounts for induction and impact of host-plant defenses. Proceedings of the Royal Society B: Biological Sciences 275: 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Bleeker PM, Van Wijk M, Schuurink RC, Haring MA. 2009. Plant volatiles in defence. Advances in Botanical Research 51: 613–666. [Google Scholar]
- Kaplan I, Denno RF. 2007. Interspecific competition in phytophagous insects revisited: a quantitative assessment of competition theory. Ecology Letters 10: 977–994. [DOI] [PubMed] [Google Scholar]
- Kaplan I, Thaler JS. 2010. Plant resistance attenuates the consumptive and non-consumptive impacts of predators on prey. Oikos 119: 1105–1113. [Google Scholar]
- Karabourniotis G, Bornman JF. 1999. Penetration of UV-A, UV-B and blue light through the leaf trichome layers of two xeromorphic plants, olive and oak, measured by optical fibre microprobes. Physiologia Plantarum 105: 655–661. [Google Scholar]
- Karban R. 1989. Community organization of Erigeron glaucus folivores: effects of competition, predation, and host plant. Ecology 70: 1028–1039. [Google Scholar]
- Karban R, Agrawal AA. 2002. Herbivore offense. Annual Review of Ecology, Evolution, and Systematics 33: 641–664. [Google Scholar]
- Karban R, Maron J. 2002. The fitness consequences of interspecific eavesdropping between plants. Ecology 83: 1209–1213. [Google Scholar]
- Karban R, Shiojiri K, Huntzinger M, et al. 2006. Damage-induced resistance in sagebrush: volatiles are key to intra- and interplant communication. Ecology 87: 922–930. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95: 461–470. [DOI] [PubMed] [Google Scholar]
- Kauffman WC, Kennedy GG. 1989. Toxicity of allelochemicals from wild insect-resistant tomato Lycopersicon hirsutum f. glabratum to Campoletis sonorensis, a parasitoid of Heliothis zea. Journal of Chemical Ecology 15: 2051–2060. [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R. 2014. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26: 2285–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2009. Linking development to defense: auxin in plant–pathogen interactions. Trends in Plant Science 14: 373–382. [DOI] [PubMed] [Google Scholar]
- Kendrick MD, Chang C. 2008. Ethylene signaling: new levels of complexity and regulation. Current Opinion in Plant Biology 11: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C, Tierney K. 2013. Xenobiotic protection/resistance mechanisms in organisms. In: Laws EA. ed. Environmental toxicology . New York: Springer. [Google Scholar]
- Kessler A, Baldwin IT. 2001. Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144. [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. 2002. Plant responses to insect herbivory. The emerging molecular analysis. Annual Review of Plant Biology 53: 299–328. [DOI] [PubMed] [Google Scholar]
- Kessler A, Halitschke R, Baldwin IT. 2004. Silencing the jasmonate cascade: induced plant defenses and insect populations. Science 305: 665–668. [DOI] [PubMed] [Google Scholar]
- Kessler A, Halitschke R, Diezel C, Baldwin IT. 2006. Priming of plant defense responses in nature by airborne signaling between Artemisia tridentata and Nicotiana attenuata. Oecologia 148: 280–292. [DOI] [PubMed] [Google Scholar]
- Kessler D, Gase K, Baldwin IT. 2008. Field experiments with transformed plants reveal the sense of floral scents. Science 321: 1200–1202. [DOI] [PubMed] [Google Scholar]
- Kessler D, Bhattacharya S, Diezel C, et al. 2012a. Unpredictability of nectar nicotine promotes outcrossing by hummingbirds in Nicotiana attenuata. Plant Journal 71: 529–538. [DOI] [PubMed] [Google Scholar]
- Kessler D, Diezel C, Clark DG, Colquhoun TA, Baldwin IT. 2012b. Petunia flowers solve the defence/apparency dilemma of pollinator attraction by deploying complex floral blends. Ecology Letters 16: 299–306. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee BW, Schroeder FC, Jander G. 2008. Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant Journal 54: 1015–1026. [DOI] [PubMed] [Google Scholar]
- Kim J, Quaghebeur H, Felton GW. 2011. Reiterative and interruptive signaling in induced plant resistance to chewing insects. Phytochemistry 72: 1624–1634. [DOI] [PubMed] [Google Scholar]
- Kindt F, Joosten NN, Peters D, et al. 2003. Characterisation of the feeding behaviour of western flower thrips in terms of electrical penetration graph (EPG) waveforms. Journal of Insect Physiology 49: 183–191. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Kamei K, Taketani S, et al. 2010. Two chitinase-like proteins abundantly accumulated in latex of mulberry show insecticidal activity. BMC Biochemistry 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen JT, Eriksson R, Gershenzon J, Ståhl B. 2006. Diversity and distribution of floral scent. Botanical Review 72: 1–120. [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J. 2004. The sesquiterpene hydrocarbons of maize (Zea mays) form five groups with distinct developmental and organ-specific distributions. Phytochemistry 65: 1895–1902. [DOI] [PubMed] [Google Scholar]
- Komath SS, Kavitha M, Swamy MJ. 2006. Beyond carbohydrate binding: new directions in plant lectin research. Organic and Biomolecular Chemistry 4: 973–988. [DOI] [PubMed] [Google Scholar]
- Koo AJK, Gao X, Daniel Jones A, et al. 2009. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant Journal 59: 974–986. [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ. 2008. Cross talk in defense signaling. Plant Physiology 146: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Houshyani B, Achhami BB, et al. 2012. Herbivore-mediated effects of glucosinolates on different natural enemies of a specialist aphid. Journal of Chemical Ecology 38: 100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski SE, Plaisted RL, Steffens JC. 1993. Immunodetection of polyphenol oxidase in glandular trichomes of S. berthaultii, S. tuberosum and their hybrids . American Potato Journal 70: 185–199. [Google Scholar]
- Kramer KJ, Muthukrishnan S. 1997. Insect chitinases: molecular biology and potential use as biopesticides. Insect Biochemistry and Molecular Biology 27: 887–900. [DOI] [PubMed] [Google Scholar]
- Krimmel BA, Pearse IS. 2013. Sticky plant traps insects to enhance indirect defence. Ecology Letters 16: 219–224. [DOI] [PubMed] [Google Scholar]
- Kuhn J, Pettersson EM, Feld BK, et al. 2004. Selective transport systems mediate sequestration of plant glucosides in leaf beetles: a molecular basis for adaptation and evolution. Proceedings of the National Academy of Sciences of the USA 101: 13808–13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Steppuhn A, Baldwin IT. 2014. Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46’s role in a nicotine-mediated antipredator herbivore defense. Proceedings of the National Academy of Sciences of the USA 111: 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL. 1995. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiology 109: 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Rujan T, Martin W, Croteau R. 2000. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proceedings of the National Academy of Sciences of the USA 97: 13172–13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel D, Ober D. 2011. Evolutionary recruitment of a flavin-dependent monooxygenase for stabilization of sequestered pyrrolizidine alkaloids in arctiids. Phytochemistry 72: 1576–1584. [DOI] [PubMed] [Google Scholar]
- Larson KC, Whitham TG. 1991. Manipulation of food resources by a gall-forming aphid: the physiology of sink-source interactions. Oecologia 88: 15–21. [DOI] [PubMed] [Google Scholar]
- Lawrence SD, Novak NG. 2006. Expression of poplar chitinase in tomato leads to inhibition of development in Colorado potato beetle. Biotechnology Letters 28: 593–599. [DOI] [PubMed] [Google Scholar]
- Lawrence SD, Novak NG, Blackburn MB. 2011. Inhibition of proteinase inhibitor transcripts by Leptinotarsa decemlineata regurgitant in Solanum lycopersicum. Journal of Chemical Ecology 33: 1041–1048. [DOI] [PubMed] [Google Scholar]
- Lazzeri L, Curto G, Leoni O, Dallavalle E. 2004. Effects of glucosinolates and their enzymatic hydrolysis products via myrosinase on the root-knot nematode Meloidogyne incognita (Kofoid et White) Chitw. Journal of Agricultural and Food Chemistry 52: 6703–6707. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, Witters J, Nauen R, Duso C, Tirry L. 2010a. The control of eriophyoid mites: state of the art and future challenges. Experimental and Applied Acarology 51: 1–3. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. 2010b. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: a review. Insect Biochemistry and Molecular Biology 40: 563–572. [DOI] [PubMed] [Google Scholar]
- Leisinger A-K, Janzen DH, Hallwachs W, Igloi GL. 2013. Amino acid discrimination by the nuclear encoded mitochondrial arginyl-tRNA synthetase of the larva of a bruchid beetle (Caryedes brasiliensis) from northwestern Costa Rica. Insect Biochemistry and Molecular Biology 43: 1172–1180. [PubMed] [Google Scholar]
- Levin DA. 1976. Alkaloid-bearing plants: an ecogeographic perspective. American Naturalist 110: 261–284. [Google Scholar]
- Li C, Song X, Li G, Wang P. 2009. Midgut cysteine protease-inhibiting activity in Trichoplusia ni protects the peritrophic membrane from degradation by plant cysteine proteases. Insect Biochemistry and Molecular Biology 39: 726–734. [DOI] [PubMed] [Google Scholar]
- Li X, Schuler M, Berenbaum M. 2007. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annual Review of Entomology 52: 231–253. [DOI] [PubMed] [Google Scholar]
- Lindroth RL, Peterson SS. 1988. Effects of plant phenols on performance of southern armyworm larvae. Oecologia 75: 185–189. [DOI] [PubMed] [Google Scholar]
- Lison P, Rodrigo I, Conejero V. 2006. A novel function for the cathepsin D inhibitor in tomato. Plant Physiology 142: 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XM, Fritz AK, Reese JC, Wilde GE, Gill BS, Chen MS. 2005. H9, H10, and H11 compose a cluster of hessian fly-resistance genes in the distal gene-rich region of wheat chromosome 1AS. Theoretical and Applied Genetics 110: 1473–1480. [DOI] [PubMed] [Google Scholar]
- Liu X, Bai J, Huang J., et al. 2007. Gene expression of different wheat genotypes during attack by virulent and avirulent hessian fly (Mayetiola destructor) larvae. Journal of Chemical Ecology 33: 2171–2194. [DOI] [PubMed] [Google Scholar]
- Lomate PR, Jadhav BR, Giri AP, Hivrale VK. 2013. Alterations in the Helicoverpa armigera midgut digestive physiology after ingestion of pigeon pea inducible leucine aminopeptidase. PloS One 8: e74889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JD, Hamilton RS, Mitchell JL. 2007. Asymmetric competition via induced resistance: specialist herbivores indirectly suppress generalist preference and populations. Ecology 88: 1232–1240. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo MLR, de, Castro MM, das Graças Machado-Freire M. 2004. Mechanisms of the insecticidal action of TEL (Talisia esculenta lectin) against Callosobruchus maculatus (Coleoptera: Bruchidae). Archives of Insect Biochemistry and Physiology 56: 84–96. [DOI] [PubMed] [Google Scholar]
- Macintosh SC, Kishore GM, Perlak FJ, et al. 1990. Potentiation of Bacillus thuringiensis insecticidal activity by serine protease inhibitors. Journal Agricultural Food Chemistry 38: 1145–1152. [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL. 2002. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Mahanil S, Attajarusit J, Stout MJ, Thipyapong P. 2008. Overexpression of tomato polyphenol oxidase increases resistance to common cutworm. Plant Science 174: 456–466. [Google Scholar]
- Mainali RP. 2014. Biology and management of eggplant fruit and shoot borer, Leucinodes orbonalis Guenee (Lepidoptera: Pyralidae): a review. International Journal of Applied Sciences and Biotechnology 2: 18–28. [Google Scholar]
- Malone M, Alarcon JJ. 1995. Only xylem-borne factors can account for systemic wound signalling in the tomato plant. Planta 196: 740–746. [Google Scholar]
- Manton SM, Harding JP. 1964. Mandibular mechanisms and the evolution of arthropods. Philosophical Transactions of the Royal Society of London B 247: 1–183. [Google Scholar]
- Mao XB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY. 2013. Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Molecular Biology 83: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiacci L, Dicke M, Posthumus MA. 1995. beta-Glucosidase: an elicitor of herbivore-induced odor that attracts host-searching parasitic wasps. Proceedings of the National Academy of Sciences of the USA 92: 2036–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson WJ. 1980. Herbivory in relation to plant nitrogen content . Annual Review of Ecology and Systematics 11: 119–161. [Google Scholar]
- McNickle GG, Dybzinski R. 2013. Game theory and plant ecology. Ecology Letters 16: 545–555. [DOI] [PubMed] [Google Scholar]
- Van der Meer IM, Stam ME, Van, Tunen AJ, Mol JN, Stuitje AR. 1992. Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melangeli C, Rosenthal GA, Dalman DL. 1997. The biochemical basis for L-canavanine tolerance by the tobacco budworm Heliothis virescens (Noctuidae). Proceedings of the National Academy of Sciences of the USA 94: 2255–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Socha P, Meszaros P, et al. 2013. Glucan-rich diet is digested and taken up by the carnivorous sundew (Drosera rotundifolia L.): implication for a novel role of plant β-1,3-glucanases. Planta 238: 715–725. [DOI] [PubMed] [Google Scholar]
- Michiels K, Van Damme EJM, Smagghe G. 2010. Plant-insect interactions: what can we learn from plant lectins? Archives of Insect Biochemistry and Physiology 73: 193–212. [DOI] [PubMed] [Google Scholar]
- Miettinen K, Dong L, Navrot N, et al. 2014. The seco-iridoid pathway from Catharanthus roseus. Nature Communications 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles PW. 1999. Aphid saliva. Biological Reviews of the Cambridge Philosophical Society 74: 41–85. [Google Scholar]
- Milewski AV, Young TP, Madden D. 1991. Thorns as induced defense: experimental evidence. Oecologia 86: 70–75. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Hussey RS, Baum TJ, et al. 2013. Nematode effector proteins: an emerging paradigm of parasitism. New Phytologist 199: 879–894. [DOI] [PubMed] [Google Scholar]
- Mitteldorf J, Wilson DS. 2000. Population viscosity and the evolution of altruism. Journal of Theoretical Biology 204: 481–496. [DOI] [PubMed] [Google Scholar]
- De Moraes CM, Lewis WJ, Pare PW, Alborn HT, Tumlinson JH. 1998. Herbivore-infested plants selectively attract parasitoids. Nature 393: 570–573. [Google Scholar]
- Mou Z, Fan W, Dong X. 2003. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–44. [DOI] [PubMed] [Google Scholar]
- Mousavi SAR, Chauvin A, Pascaud F, Kellenberger S, Farmer E. 2013. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426. [DOI] [PubMed] [Google Scholar]
- Mouttet R, Kaplan I, Bearez P, Amiens-Desneux E, Desneux N. 2013. Spatiotemporal patterns of induced resistance and susceptibility linking diverse plant parasites. Oecologia 173: 1379–1386. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Nishimura MT, Dangl J. 2009. NPR1 in plant defense: it’s not over ‘til it’s turned over. Cell 137: 804–806. [DOI] [PubMed] [Google Scholar]
- Mumm R, Burow M, Bukovinszkine'kiss G, Kazantzidou E, Wittstock U, Dicke M, Gershenzon J. 2008. Formation of simple nitriles upon glucosinolate hydrolysis affects direct and indirect defense against the specialist herbivore, Pieris rapae. Journal of Chemical Ecology 34: 1311–1321. [DOI] [PubMed] [Google Scholar]
- Munnik T, Nielsen E. 2011. Green light for polyphosphoinositide signals in plants. Current Opinion in Plant Biology 14: 489–497. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C. 2006. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology 140: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, et al. 2002. Caterpillar saliva beats plant defences. Nature 416: 599–600. [DOI] [PubMed] [Google Scholar]
- Musser RO, Cipollini DF, Hum-Musser SM, Williams SA, Brown JK, Felton GW. 2005. Evidence that the caterpillar salivary enzyme glucose oxidase provides herbivore offense in solanaceous plants. Archives of Insect Biochemistry and Physiology 58: 128–137. [DOI] [PubMed] [Google Scholar]
- Mutti NS, Louis J, Pappan LK, et al. 2008. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proceedings of the National Academy of Sciences of the USA 105: 9965–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabity PD, Zavala JA, DeLucia EH. 2009. Indirect suppression of photosynthesis on individual leaves by arthropod herbivory. Annals of Botany 103: 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, et al. 2003. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant Journal 33: 887–898. [DOI] [PubMed] [Google Scholar]
- Newingham BA, Callaway RM, BassiriRad H. 2007. Allocating nitrogen away from a herbivore: a novel compensatory response to root herbivory. Oecologia 153: 913–920. [DOI] [PubMed] [Google Scholar]
- Nishida R. 2002. Sequestration of defensive substances from plants by Lepidoptera. Annual Review of Entomology 47: 57–92. [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY. 2011. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 108: 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty I, Yim JJ, Schmelz EA, Schroeder FC. 2011. Synthesis of caeliferins, elicitors of plant immune responses: accessing lipophilic natural products via cross metathesis. Organic Letters 13: 5900–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi T. 2005. Indirect interaction webs: herbivore-induced indirect effects through trait change in plants. Annual Review of Ecology, Evolution and Systematics 36: 81–105. [Google Scholar]
- Oku K, Magalhães S, Dicke M. 2009. The presence of webbing affects the oviposition rate of two-spotted spider mites, Tetranychus urticae (Acari: Tetranychidae). Experimental and Applied Acarology 49: 167–172. [DOI] [PubMed] [Google Scholar]
- Opitz SW, Müller C. 2009. Plant chemistry and insect sequestration. Chemoecology 19: 117–154. [Google Scholar]
- Oppel CB, Dussourd DE, Garimella U. 2009. Visualizing a plant defense and insect counterploy: alkaloid distribution in Lobelia leaves trenched by a plusiine caterpillar. Journal of Chemical Ecology 35: 625–634. [DOI] [PubMed] [Google Scholar]
- Orians CM, Pomerleau J, Ricco R. 2000. Vascular architecture generates fine scale variation in systemic induction of proteinase inhibitors in tomato. Journal of Chemical Ecology 26: 471–485. [Google Scholar]
- Osborn RW, De Samblanx GW, Thevissen K, et al. 2009. Isolation and characterisation of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Letters 368: 257–262. [DOI] [PubMed] [Google Scholar]
- Park YL, Lee JH. 2002. Leaf cell and tissue damage of cucumber caused by twospotted spider mite (Acari: Tetranychidae). Journal of Economic Entomology 95: 952–957. [DOI] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. 2007. Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant Journal 51: 79–91. [DOI] [PubMed] [Google Scholar]
- Paszota P, Escalante-Perez M, Thomsen LR, et al. 2014. Secreted major Venus flytrap chitinase enables digestion of arthropod prey. Biochimica et Biophysica Acta 1844: 374–383. [DOI] [PubMed] [Google Scholar]
- Pauchet Y, Heckel DG. 2013. The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacteria through horizontal gene transfer. Proceedings of the Royal Society of London B: Biological Sciences 280: 20131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Paz Celorio-Mancera M, Wheat CW, Vogel H, Söderlind L, Janz N, Nylin S. 2013. Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Molecular Ecology 22: 4884–4895. [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. 1991. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897. [DOI] [PubMed] [Google Scholar]
- Pearse IS, Harris DJ, Karban R, Sih A. 2013. Predicting novel herbivore-plant interactions. Oikos 122: 1554–1564. [Google Scholar]
- Pechan T, Cohen A, Williams WP, Luthe DS. 2002. Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proceedings of the National Academy of Sciences of the USA 99: 13319–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer M, Felton GW. 2009. Do caterpillars secrete “oral secretions”? Journal of Chemical Ecology 35: 326–335. [DOI] [PubMed] [Google Scholar]
- Peiffer M, Tooker JF, Luthe DS, Felton GW. 2009. Plants on early alert: glandular trichomes as sensors for insect herbivores. New Phytologist 184: 644–656. [DOI] [PubMed] [Google Scholar]
- Pemberton RW, Lee JH. 1996. The influence of extrafloral nectaries on parasitism of an insect herbivore. American Journal of Botany 83: 1187–1194. [Google Scholar]
- Pentzold S, Zagrobelny M, Rook F, Bak S. 2013. How insects overcome two-component plant chemical defence: plant β-glucosidases as the main target for herbivore adaptation. Biological Reviews 89: 531–551. [DOI] [PubMed] [Google Scholar]
- Pentzold S, Zagrobelny M, Roelsgaard PS, Møller BL, Bak S. 2014. The multiple strategies of an insect herbivore to overcome plant cyanogenic glucoside defence. PLoS One 9: e91337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LE, Cribb BW, Brewer PB, et al. 2013. Generalist insects behave in a jasmonate-dependent manner on their host plants, leaving induced areas quickly and staying longer on distant parts. Proceedings of the Royal Society B: Biological Sciences 280: 20122646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschenka G, Pick C, Wagschal V, Dobler S. 2013. Functional evidence for physiological mechanisms to circumvent neurotoxicity of cardenolides in an adapted and a non-adapted hawk-moth species. Proceedings of the Royal Society B: Biological Sciences 280: 20123089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Dicke M. 2007. Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends in Plant Science 12: 564–569. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, León-Reyes A, Van der, Ent S, Van Wees SCM. 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5: 308–316. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Pinto MFS, Fensterseifer ICM, Migliolo L, et al. 2012. Identification and structural characterization of novel cyclotide with activity against an insect pest of sugar cane. Journal of Biological Chemistry 287: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Hogenhout SA. 2013. Aphid protein effectors promote aphid colonization in a plant species-specific manner. Molecular Plant-Microbe Interactions 26: 130–139. [DOI] [PubMed] [Google Scholar]
- Van Poecke RMP, Dicke M. 2002. Induced parasitoid attraction by Arabidopsis thaliana: involvement of the octadecanoid and the salicylic acid pathway. Journal of Experimental Botany 53: 1793–1799. [DOI] [PubMed] [Google Scholar]
- Van Poecke RMP, Posthumus MA, Dicke M. 2001. Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: chemical, behavioral and gene-expression analysis. Journal of Chemical Ecology 27: 1911–1928. [DOI] [PubMed] [Google Scholar]
- Poelman EH, Broekgaarden C, Van Loon JJA, Dicke M. 2008a. Early season herbivore differentially affects plant defence responses to subsequently colonizing herbivores and their abundance in the field. Molecular Ecology 17: 3352–3365. [DOI] [PubMed] [Google Scholar]
- Poelman EH, Galiart R, Raaijmakers CE, van, Loon JJA, van Dam NM. 2008b. Performance of specialist and generalist herbivores feeding on cabbage cultivars is not explained by glucosinolate profiles. Entomologia Experimentalis et Applicata 127: 218–228. [Google Scholar]
- Poelman EH, Zheng SJ, Zhang Z, Heemskerk NM, Cortesero AM, Dicke M. 2011. Parasitoid-specific induction of plant responses to parasitized herbivores affects colonization by subsequent herbivores. Proceedings of the National Academy of Sciences of the USA 108: 19647–19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelman EH, Bruinsma M, Zhu F, et al. 2012. Hyperparasitoids use herbivore-induced plant volatiles to locate their parasitoid host. PLoS Biology 10: e1001435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton F, Biron DG, Moore J, Møller AP, Thomas F. 2006. Facultative virulence: a strategy to manipulate host behaviour? Behavioural Processes 72: 1–5. [DOI] [PubMed] [Google Scholar]
- Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. 2011. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proceedings of the National Academy of Sciences of the USA 108: 10127–10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott C, McLoughlin S, Shunqing W, Friis EM. 2012. Trichomes on the leaves of Anomozamites villosus sp. nov. (Bennettitales) from the Daohugou beds (Middle Jurassic), Inner Mongolia, China: mechanical defence against herbivorous arthropods. Review of Paleobotany and Palynology 169: 48–60. [Google Scholar]
- Pourcel L, Routaboul J-M, Cheynier V, Lepiniec L, Debeaujon I. 2007. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends in Plant Science 12: 29–36. [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PW, Bouton CE, Gross P, McPheron BA, Thompson JN, Weis AE. 1980. Interactions among three trophic levels: influence of plants on interactions between insect herbivores and natural enemies. Annual Review of Ecology and Systematics 11: 41–65. [Google Scholar]
- Prokopy RJ, Roitberg BD. 2001. Joining and avoidance behavior in nonsocial insects. Annual Review of Entomology 46: 631–665. [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, et al. 2011. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR. 2009. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes and Development 23: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quideau S, Deffieux D, Douat-Casassus C, Pouységu L. 2011. Plant polyphenols: chemical properties, biological activities, and synthesis. Angewandte Chemie International Edition 50: 586–621. [DOI] [PubMed] [Google Scholar]
- Ragusa S, Tsolakis H. 2000. Notes on the adaptation of some phytophagous and predacious mites to various ecological parameters in the Mediterranean countries. Web Ecology 1: 35–47. [Google Scholar]
- Rahfeld P, Kirsch R, Kugel S, et al. 2014. Independently recruited oxidases from the glucose-methanol-choline oxidoreductase family enabled chemical defences in leaf beetle larvae (subtribe Chrysomelina) to evolve. Proceedings of the Royal Society B: Biological Sciences 281: 20140842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin DJ, Bargum K, Kokko H. 2007. The tragedy of the commons in evolutionary biology. Trends in Ecology & Evolution 22: 643–651. [DOI] [PubMed] [Google Scholar]
- Rasmann S, Köllner TG, Degenhardt J, et al. 2005. Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434: 732–737. [DOI] [PubMed] [Google Scholar]
- Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J. 2002. Disarming the mustard oil bomb. Proceedings of the National Academy of Sciences of the USA 99: 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. 2014. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research 42: D503–D509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M. 2005. Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiology Letters 253: 19–27. [DOI] [PubMed] [Google Scholar]
- Restif O. 2013. An offer you cannot refuse: down-regulation of immunity in response to a pathogen’s retaliation threat. Journal of Evolutionary Biology 26: 2021–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. 2000. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhainds M, Eveleigh E, Francis B, Silk P. 2011. Factors affecting oral regurgitation by larval spruce budworm. Entomologia Experimentalis et Applicata 140: 254–261. [Google Scholar]
- Rhodes JD, Thain JF, Wildon DC. 1999. Evidence for physically distinct systemic signalling pathways in the wounded tomato plant. Annals of Botany 84: 109–116. [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG. 2011. Hormone crosstalk in plant disease and defense: more than just JASMONATE-SALICYLATE antagonism. Annual Review of Phytopathology 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Rodrigues D, Maia PHS, Trigo JR. 2010. Sabotaging behaviour and minimal latex of Asclepias curassavica incur no cost for larvae of the southern monarch butterfly Danaus erippus. Ecological Entomology 35: 504–513. [Google Scholar]
- Rodriguez PA, Bos JIB. 2013. Toward understanding the role of aphid effectors in plant infestation. Molecular Plant-Microbe Interactions Journal 26: 25–30. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Saona C, Thaler JS. 2005. The jasmonate pathway alters herbivore feeding behaviour: consequences for plant defences. Entomologia Experimentalis et Applicata 115: 125–134. [Google Scholar]
- Rodriguez-Saona C, Crafts-Brandner SJ, Cañas LA. 2003. Volatile emissions triggered by multiple herbivore damage: beet armyworm and whitefly feeding on cotton plants. Journal of Chemical Ecology 29: 2539–2550. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Saona C, Musser R, Vogel H, Hum-Musser S, Thaler JS. 2010. Molecular, biochemical, and organismal analyses of tomato plants simultaneously attacked by herbivores from two feeding guilds. Journal of Chemical Ecology 36: 1043–57. [DOI] [PubMed] [Google Scholar]
- Rooney HC, Van’t Klooster JW, van der, Hoorn RA, et al. 2005. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786. [DOI] [PubMed] [Google Scholar]
- Rosenthal G, Dahlman D, Janzen D. 1976. A novel means for dealing with L-canavanine, a toxic metabolite. Science 192: 256–258. [DOI] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. 1998. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proceedings of the National Academy of Sciences of the USA 95: 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenich H, Boshoven JC, Thomma BPHJ. 2014. Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Current Opinion in Plant Biology 20: 96–103. [DOI] [PubMed] [Google Scholar]
- Runyon J, Mescher MC, De Moraes CM. 2006. Volatile chemical cues guide host location and host selection by parasitic plants. Science 313: 1964–1967. [DOI] [PubMed] [Google Scholar]
- Ryan CA. 1990. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annual Review of Phytopathology 28: 425–449. [Google Scholar]
- Ryan CA. 2000. The systemin signaling pathway: differential activation of plant defensive genes. Biochimica et Biophysica Acta 1477: 112–121. [DOI] [PubMed] [Google Scholar]
- Sabelis MW, Janssen A, Kant MR. 2001. The enemy of my enemy is my ally. Science 291: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Sano H, Seo S, Orudgev E, Youssefian S, Ishizuka K, Ohashi Y. 1994. Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proceedings of the National Academy of Sciences of the USA 91: 10556–10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria ME, Cambra I, Martinez M, et al. 2012. Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae. PLoS One 7: e43011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir-Mir M, Mett A, Belausov E, et al. 2008. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiology 148: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento RA, Lemos F, Bleeker PM, et al. 2011a. A herbivore that manipulates plant defence. Ecology Letters 14: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento RA, Lemos F, Dias CR, et al. 2011b. A herbivorous mite down-regulates plant defence and produces web to exclude competitors. PLoS One 6: e23757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Alba JM, Sabelis MW. 2014. Testing for reproductive interference in the population dynamics of two congeneric species of herbivorous mites. Heredity 113: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvion N, Charles H, Febvay G, Rahbé Y. 2004. Effects of jackbean lectin (ConA) on the feeding behavior and kinetics of intoxication of the pea aphid, Acyrthosiphon pisum. Entomologia Experimentalis et Applicata 110: 31–44. [Google Scholar]
- Savchenko T, Pearse IS, Ignatia L, Karban R, Dehesh K. 2013. Insect herbivores selectively suppress the HPL branch of the oxylipin pathway in host plants. Plant Journal 73: 653–662. [DOI] [PubMed] [Google Scholar]
- Schaeffer A, Bronner R, Benveniste P, Schaller H. 2001. The ratio of campesterol to sitosterol which modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2-1. Plant Journal 25: 605–615. [DOI] [PubMed] [Google Scholar]
- Schäfer M, Fischer C, Meldau S, Seebald E, Oelmüller, Baldwin IT. 2011. Lipase activity in insect oral secretions mediates defense responses in Arabidopsis. Plant Physiology 156: 1520–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CCN, Haring MA, Schuurink RC. 2007. Tomato linalool synthase is induced in glandular trichomes by jasmonic acid. Plant Molecular Biology 64: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CCN, Takken FLW. 2014. Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology 52: 551–581. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. 2005. Systemic signaling in the wound response. Current Opinion in Plant Biology 8: 369–377. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, et al. 2009. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proceedings of the National Academy of Sciences of the USA 106: 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Preston CA, Baldwin IT. 2000. Eating the evidence? Manduca sexta larvae can not disrupt specific jasmonate induction in Nicotiana attenuata by rapid consumption. Planta 210: 343–346. [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH, Teal PEA. 2009. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proceedings of the National Academy of Sciences of the USA 106: 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Carroll MJ, Alborn HT, Ali JG, Teal PEA. 2012. An amino acid substitution inhibits specialist herbivore production of an antagonist effector and recovers insect-induced plant defenses. Plant Physiology 160: 1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Baldwin IT. 2006. Systemin in Solanum nigrum. The tomato-homologous polypeptide does not mediate direct defense responses. Plant Physiology 142: 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Held M, Turlings TCJ, Gershenzon J, Degenhardt J. 2006. The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proceedings of the National Academy of Sciences of the USA 103: 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RW, Grogan RG. 1977. Tomato leaf trichomes, a habitat for resident populations of Pseudomonas tomato. Phytopathology 67: 898–902. [Google Scholar]
- Schramm K, Vassão DG, Reichelt M, Gershenzon J, Wittstock U. 2012. Metabolism of glucosinolate-derived isothiocyanates to glutathione conjugates in generalist lepidopteran herbivores. Insect Biochemistry and Molecular Biology 42: 174–182. [DOI] [PubMed] [Google Scholar]
- Schuler MA. 2012. Insect P450s: mounted for battle in their war against toxins. Molecular Ecology 21: 4157–4159. [DOI] [PubMed] [Google Scholar]
- Schulz JC. 1988. Many factors influence the evolution of herbivore diets, but plant chemistry is central. Ecology 69: 896–897. [Google Scholar]
- Schuman MC, Barthel K, Baldwin IT. 2012. Herbivory-induced volatiles function as defenses increasing fitness of the native plant Nicotiana attenuata in nature. eLife 1: e00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. 2006. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proceedings of the National Academy of Sciences of the USA 103: 12935–12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzberg EG, Böröczky K, Tumlinson JH. 2011. Pea aphids, Acyrthosiphon pisum, suppress induced plant volatiles in broad bean, Vicia faba. Journal of Chemical Ecology 37: 1055–1062. [DOI] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, et al. 2013. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmler W. 1989. Insect endocytobiosis: morphology, physiology, genetics, evolution. Boca Raton: CRC Press. [Google Scholar]
- Scranton MA, Fowler JH, Girke T, Walling LL. 2013. Microarray analysis of tomato’s early and late wound response reveals new regulatory targets for leucine aminopeptidase A. PloS One 8: e77889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehlmeyer S, Wang L, Langel D, et al. 2010. Flavin-dependent monooxygenases as a detoxification mechanism in insects: new insights from the arctiids (Lepidoptera). PLoS One 5: e10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels J, Mathys J, De Coninck BMA, Cammue BPA, De, Bolle MFC. 2008. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiology and Biochemistry 46: 941–950. [DOI] [PubMed] [Google Scholar]
- Shabab M, Khan SA, Vogel H, Heckel DG, Boland W. 2014. OPDA isomerase GST16 is involved in phytohormone detoxification and insect development. FEBS Journal 281: 2769–2783. [DOI] [PubMed] [Google Scholar]
- Shade RE, Schroeder HE, Pueyo JJ, et al. 1994. Transgenic pea seeds expressing the alpha-amylase inhibitor of the common bean are resistant to bruchid beetles. Nature Biotechnology 12: 793–796. [Google Scholar]
- Shepherd RW, Bass WT, Houtz RL, Wagner GJ. 2005. Phylloplanins of tobacco are defensive proteins deployed on aerial surfaces by short glandular trichomes. Plant Cell 17: 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau YS, Horng SB, Chen CS, et al. 2006. Structural analysis of the unique insecticidal activity of novel mungbean defensin VrD1 reveals possibility of homoplasy evolution between plant defensins and scorpion neurotoxins. Journal of Molecular Recognition 19: 441–450. [DOI] [PubMed] [Google Scholar]
- Shiojiri K, Takabayashi J, Yano S, Takafuji A. 2002. Oviposition preferences of herbivores are affected by tritrophic interaction webs. Ecology Letters 5: 186–192. [Google Scholar]
- Shiraishi T, Oku H, Yamashita M, Ouchi S. 1978. Elicitor and suppressor of pisatin induction in spore germination fluid of pea pathogen, Mycosphaerella pinodes. Annals of the Phytopathologicial Society of Japan 44: 659–665. [Google Scholar]
- Shroff R, Vergara F, Muck A, et al. 2008. Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proceedings of the National Academy of Sciences of the USA 105: 6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukle RH, Subramanyam S, Salzmann KA, Williams CE. 2010. Ultrastructural changes in the midguts of hessian fly larvae feeding on resistant wheat. Journal of Insect Physiology 56: 754–760. [DOI] [PubMed] [Google Scholar]
- Shulaev V, Silverman P, Raskin I. 1997. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385: 718–721. [Google Scholar]
- Sikkema J, De Bont JAM, Poolman B. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiological Reviews 59: 201–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AT, Gurr GM. 2005. Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agricultural and Forest Entomology 7: 265–276. [Google Scholar]
- Simmons AT, Gurr GM, McGrath D, Martin PM, Nicol HI. 2004. Entrapment of Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) on glandular trichomes of Lycopersicon species. Australian Journal of Entomology 43: 196–200. [Google Scholar]
- Siritunga D, Arias-Garzon D, White W, Sayre RT. 2004. Over-expression of hydroxynitrile lyase in transgenic cassava roots accelerates cyanogenesis and food detoxification. Plant Biotechnology Journal 2: 37–43. [DOI] [PubMed] [Google Scholar]
- Smith AB, Daly NL, Craik DJ. 2011. Cyclotides: a patent review. Expert Opinion on Therapeutic Patents 21: 1657–1672. [DOI] [PubMed] [Google Scholar]
- Soler M, Soler JJ, Martinez JG, Møller AP. 1995. Magpie host manipulation by great spotted cuckoos: evidence for an avian mafia? Evolution 49: 770–775. [DOI] [PubMed] [Google Scholar]
- Soler R, Bezemer TM, Van der Putten WH, Vet LEM, Harvey JA. 2005. Root herbivore effects on above-ground herbivore, parasitoid and hyperparasitoid performance via changes in plant quality. Journal of Animal Ecology 74: 1121–1130. [Google Scholar]
- Soler R, Badenes-Perez FR, Broekgaarden C, et al. 2012. Plant-mediated facilitation between a leaf-feeding and a phloem-feeding insect in a brassicaceous plant: from insect performance to gene transcription. Functional Ecology 26: 156–166. [Google Scholar]
- Soler R, Erb M, Kaplan I. 2013. Long distance root-shoot signalling in plant-insect community interactions. Trends in Plant Science 18: 149–156. [DOI] [PubMed] [Google Scholar]
- Sorensen J, Dearing MD. 2006. Efflux transporters as a novel herbivore countermechanism to plant chemical defenses. Journal of Chemical Ecology 32: 1181–1196. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Loake GJ. 2011. Redox-based protein modifications: the missing link in plant immune signalling. Current Opinion in Plant Biology 14: 358–364. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, et al. 2003. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. 2009. Proteasome-mediated turnover of the transcription co-activator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamp N. 2003. Out of the quagmire of plant defense hypotheses. Quarterly Review of Biology 78: 23–55. [DOI] [PubMed] [Google Scholar]
- Stauber EJ, Kuczka P, van Ohlen M, et al. 2012. Turning the ‘mustard oil bomb’ into a ‘cyanide bomb’: aromatic glucosinolate metabolism in a specialist insect herbivore. PLoS One 7: e35545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauer MJ, Burns AE, Hall A, Riegler M, Taylor GS. 2014. Nutritional enhancement of leaves by a psyllid through senescence-like processes: insect manipulation or plant defence? Oecologia 176: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A., Baldwin IT. 2007. Resistance management in a native plant: nicotine prevents herbivores from compensating for plant proteinase inhibitors. Ecology Letters 10: 499–511. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. 2001. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proceedings of the National Academy of Sciences of the USA 98: 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stireman JO, III, Cipollini D. 2008. Stealth tactics of galling parasites and their potential indirect effects. New Phytologist 178: 462–465. [DOI] [PubMed] [Google Scholar]
- Stone GN, Schonrogge K. 2003. The adaptive significance of insect gall morphology. Trends in Ecology & Evolution 18: 512–522. [Google Scholar]
- Stork W, Diezel C, Halitschke R, Gális I, Baldwin IT. 2009. An ecological analysis of the herbivory-elicited JA burst and its metabolism: plant memory processes and predictions of the moving target model. PLoS One 4: e4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Thomson JG, Wang Y. 2009. Plant defensins defense, development and application. Plant Signaling & Behavior 4: 1010–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout MJ. 2013. Reevaluating the conceptual framework for applied research on host-plant resistance. Insect Science 20: 263–272. [DOI] [PubMed] [Google Scholar]
- Stout MJ, Workman KV, Bostock RM, Duffey SS. 1998. Specificity of induced resistance in the tomato, Lycopersicon esculentum. Oecologia 113: 74–81. [DOI] [PubMed] [Google Scholar]
- Stout MJ, Fidantsef AL, Duffey SS, Bostock RM. 1999. Signal interactions in pathogen and insect attack: systemic plant-mediated interactions between pathogens and herbivores of the tomato, Lycopersicum esculentum. Physiological and Molecular Plant Pathology 54: 115–130. [Google Scholar]
- Stowe KA, Marquis RJ, Hochwender CG, Simms EL. 2000. The evolutionary ecology of tolerance to consumer damage. Annual Review of Ecology and Systematics 31: 565–595. [Google Scholar]
- Strauss AS, Peters S, Boland W, Burse A. 2013. ABC transporter functions as a pacemaker for sequestration of plant glucosides in leaf beetles. eLife 2: e01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JJ, Chen MS, Shukle R, Harris MO. 2012. Gall midges (hessian flies) as plant pathogens. Annual Review of Phytopathology 50: 339–57. [DOI] [PubMed] [Google Scholar]
- Su YL, Li JM, Li M, et al. 2012. Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS One 7: e39303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Oliver KM, Xie W, Wu Q, Wang S, Zhang Y. 2015. The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defenses in tomato. Functional Ecology, in press. doi: 10.1111/1365-2435.12405. [Google Scholar]
- Sugimoto K, Matsui K, Iijima Y, et al. 2014. Intake and transformation to a glycoside of (Z)-3-hexenol from infested neighbors reveals a mode of plant odor reception and defense. Proceedings of the National Academy of Sciences of the USA 111: 7144–7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. 2011. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proceedings of the National Academy of Sciences of the USA 108: E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Li CP, Poulton JE. 1992. Tissue and subcellular localization of enzymes catabolizing (R)-amygdalin in mature Prunus serotina seeds. Plant Physiology 100: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerwska-Muhkar K, et al. 2008. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 2005. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis . Plant Physiology 139: 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Feyereisen R. 1996. Molecular biology and evolution of resistance of toxicants. Molecular Biology and Evolution 13: 719–734. [DOI] [PubMed] [Google Scholar]
- Terra WR, Ferreira C. 1994. Insect digestive enzymes: properties, compartmentalization and function. Comparative Biochemistry and Physiology B 109: 1–62. [Google Scholar]
- Thaler JS. 1999. Jasmonate-inducible plant defences cause increased parasitism of herbivores. Nature 399: 686–688. [Google Scholar]
- Thaler JS, Stout MJ, Karban R, Duffey SS. 1996. Exogenous jasmonates simulate insect wounding in tomato plants (Lycopersicon esculentum) in the laboratory and field. Journal of Chemical Ecology 22: 1767–1781. [DOI] [PubMed] [Google Scholar]
- Thaler JS, Fidantsef AL, Duffey SS, Bostock RM. 1999. Trade-offs in plant defense against pathogens and herbivores: a field demonstration of chemical elicitors of induced resistance. Journal of Chemical Ecology 25: 1597–1607. [Google Scholar]
- Thaler JS, Stout MJ, Karban R, Duffey SS. 2001. Jasmonate-mediated induced plant resistance affects a community of herbivores. Ecological Entomology 26: 312–324. [Google Scholar]
- Thaler JS, Karban R, Ullman DE, Boege K, Bostock RM. 2002. Crosstalk between jasmonate and salicylate plant defense pathways: effects on several plant parasites. Oecologia 131: 227–235. [DOI] [PubMed] [Google Scholar]
- Thaler JS, Agrawal AA, Halitschke R. 2010. Salicylate-mediated interactions between pathogens and herbivores. Ecology 91: 1075–1082. [DOI] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. 2012. Evolution of jasmonate and salicylate signal crosstalk. Trends in Plant Science 17: 260–270. [DOI] [PubMed] [Google Scholar]
- Thayer SS, Conn EE. 1981. Subcellular localization of dhurrin beta-glucosidase and hydroxynitrile lyase in the mesophyll cells of Sorghum leaf blades. Plant Physiology 67: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, et al. 2007. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Camme BPA, Thevissen K. 2002. Plant defensins. Planta 216: 193–202. [DOI] [PubMed] [Google Scholar]
- Tian D, Peiffer M, Shoemaker E, et al. 2012. Salivary glucose oxidase from caterpillars mediates the induction of rapid and delayed-induced defenses in the tomato plant. PLoS One. 7: e36168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Huitema E, da Cunha L, Torto-Alalibo T, Kamoun S. 2004. A kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. Journal of Biological Chemistry 279: 26370–26377. [DOI] [PubMed] [Google Scholar]
- Tien NSH, Sabelis MW, Egas M. 2009. Heritability of defence and life-history traits in the two-spotted spider mite. Evolutionary Ecology Research 11: 1271–1281. [Google Scholar]
- Tissier A. 2012. Glandular trichomes: what comes after expressed sequence tags? Plant Journal 70: 51–68. [DOI] [PubMed] [Google Scholar]
- Torres-Vera R, García JM, Pozo MJ, López-Ráez JA. 2013. Do strigolactones contribute to plant defence? Molecular Plant Pathology 15: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooker JF, De Moraes CM. 2007. Jasmonate, salicylate, and benzoate in insect eggs. Journal of Chemical Ecology 33: 331–43. [DOI] [PubMed] [Google Scholar]
- Tooker JF, Rohr JR, Abrahamson WG, De Moraes CM. 2008. Gall insects can avoid and alter indirect plant defenses. New Phytologist 178: 657–671. [DOI] [PubMed] [Google Scholar]
- Tran LT, Taylor JS, Constabel CP. 2012. The polyphenol oxidase gene family in land plants: lineage-specific duplication and expansion. BMC Genomics 13: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traugott MS, Stamp NE. 1997. Effects of chlorogenic acid- and tomatine-fed caterpillars on performance of an insect predator. Oecologia 109: 265–272. [DOI] [PubMed] [Google Scholar]
- Traw MB, Dawson TE. 2002. Differential induction of trichomes by three herbivores of black mustard. Oecologia 131: 526–532. [DOI] [PubMed] [Google Scholar]
- Truitt CL, Wei HX, Pare PW. 2004. A plasma membrane protein from Zea mays binds with the herbivore elicitor volicitin. Plant Cell 16: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlings TCJ, Tumlinson JH, Eller FJ, Lewis WJ. 1991. Larval-damaged plants - source of volatile synomones that guide the parasitoid Cotesia marginiventris to the microhabitat of its hosts. Entomologia Experimentalis et Applicata 58: 75–82 [Google Scholar]
- Vadassery J, Reichelt M, Mithöfer A. 2012. Direct proof of ingested food regurgitation by Spodoptera littoralis caterpillars during feeding on Arabidopsis. Journal of Chemical Ecology 38: 865–872. [DOI] [PubMed] [Google Scholar]
- Valette C, Andary C, Geiger JP, Sarah JL, Nicole M. 1998. Histochemical and cytochemical investigations of phenols in roots of banana infected by the burrowing nematode Radopholus similis. Phytopathology 88: 1141–1148. [DOI] [PubMed] [Google Scholar]
- VanDoorn A, Bonaventure G, Schmidt DD, Baldwin IT. 2011. Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum: COI1 and JAR4 play overlapping yet distinct roles. New Phytologist 190: 640–652. [DOI] [PubMed] [Google Scholar]
- VanDoorn A, de Vries M, Kant MR, Schuurink RC. 2015. Whiteflies glycosylate salicylic acid and secrete the conjugate via their honeydew. Journal Chemical Ecology 41: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, et al. 2013. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. 1984. Biosynthesis of jasmonic acid by several plant species. Plant Physiology 75: 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervuori L, Pasonen HL, Lyytikainen-Saarenmaa P. 2008. Density and composition of an insect population in a field trial of chitinase transgenic and wild-type silver birch (Betula pendula) clones. Environmental Entomology 37: 1582–1591. [DOI] [PubMed] [Google Scholar]
- Vihervuori L, Lyytikainen-Saarenmaa P, Lu J, Pasonen HL. 2013. Effects on lepidopteran herbivores of feeding on leaves of transgenic birch (Betula pendula) expressing the sugar beet chitinase IV gene. European Journal of Entomology 110: 253–262. [Google Scholar]
- Vijaysegaran S, Walter GH, Drew RAI. 1997. Mouthpart structure, feeding mechanisms, and natural food sources of adult Bactrocera (Diptera: Tephritidae). Annals of the Entomological Society of America 90: 184–201. [Google Scholar]
- Vilanova C, Marco G, Domínguez-Escribà L, et al. 2012. Bacteria from acidic to strongly alkaline insect midguts: potential sources of extreme cellulolytic enzymes. Biomass and Bioenergy 45: 288–294. [Google Scholar]
- Vinson SB. 1976. Host selection by insect parasitoids. Annual Review of Entomology 21: 109–133. [Google Scholar]
- Viswanathan DV, Thaler JS. 2004. Plant vascular architecture and within-plant spatial patterns in resource quality following herbivory. Journal of Chemical Ecology 30: 531–543. [DOI] [PubMed] [Google Scholar]
- Viswanathan DV, Narwani AJT, Thaler JS. 2005. Specificity in induced plant responses shapes patterns of herbivore occurrence on Solanum dulcamara. Ecology 86: 886–896. [Google Scholar]
- De Vleesschauwer D, Gheysen G, Höfte M. 2013. Hormone defense networking in rice: tales from a different world. Trends in Plant Science 18: 555–565. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey MA, Klessig DF. 2009. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology 47: 177–206. [DOI] [PubMed] [Google Scholar]
- Voegele RT, Mendgen K. 2003. Rust haustoria: nutrient uptake and beyond. New Phytologist 159: 93–100. [DOI] [PubMed] [Google Scholar]
- Voelckel C, Schittko U, Baldwin IT. 2001. Herbivore-induced ethylene burst reduces fitness cost of jasmonate- and oral secretion-induced defenses in Nicotiana attenuata. Oecologia 127: 274–280. [DOI] [PubMed] [Google Scholar]
- De Vos M, van Zaanen W, Koornneef A, et al. 2006. Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiology 142: 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GJ, Wang E, Shepherd RW. 2004. New approaches for studying and exploiting an old protuberance, the plant trichome. Annals of Botany 93: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. 2000. The myriad plant responses to herbivores. Journal of Plant Growth Regulation 19: 195–216. [DOI] [PubMed] [Google Scholar]
- Walling LL. 2009. Adaptive defense responses to pathogens and pests. Advances in Botanical Research 51: 551–612. [Google Scholar]
- Walter DE. 1996. Living on leaves: mites, tomenta and leaf domatia. Annual Review of Entomology 41: 101–114. [DOI] [PubMed] [Google Scholar]
- Wang L, Beuerle T, Timbilla J, Ober D. 2012. Independent recruitment of a flavin-dependent monooxygenase for safe accumulation of sequestered pyrrolizidine alkaloids in grasshoppers and moths. PLoS One 7: e31796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Constabel CP. 2004. Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220: 87–96. [DOI] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. 2002. Ethylene biosynthesis and signaling networks. Plant Cell 14: S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany . Annals of Botany 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JL. 1984. Cyanide intoxication and its mechanism of antagonism. Annual Review of Pharmacology and Toxicology 24: 451–481. [DOI] [PubMed] [Google Scholar]
- Weech MH, Chapleau M, Pan L, Ide C, Bede JC. 2008. Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. Journal of Experimental Botany 59: 2437–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FM, et al. 2013. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342: 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, Lewsey MG, Murphy AM, et al. 2014. Interference with jasmonic acid-regulated gene expression is a general property of viral suppressors of RNA silencing but only partly explains virus-induced changes in plant-aphid interactions. Journal of General Virology 95: 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–571. [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, Van Bel AJE. 2007. Molecular sabotage of plant defense by aphid saliva. Proceedings of the National Academy of Sciences of the USA 104: 10536–10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Furch ACU, Zimmermann MR. 2013. How phloem-feeding insects face the challenge of phloem-located defenses. Frontiers in Plant Science 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M, Schmeller T, Latz-Brüning B. 1998. Modes of action of allelochemical alkaloids: interaction with neuroreceptors, DNA, and other molecular targets. Journal of Chemical Ecology 24: 1881–1937. [Google Scholar]
- Wise MJ, Kieffer DL, Abrahamson WG. 2006. Costs and benefits of gregarious feeding in the meadow spittlebug Philaenus spumarius. Ecological Entomology 31: 548–555. [Google Scholar]
- Wittstock U, Agerbirk N, Stauber EJ, et al. 2004. Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proceedings of the National Academy of Sciences of the USA 101: 4859–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzgall P, Stelinski L, Gut L, Thomson D. 2008. Codling moth management and chemical ecology. Annual Review of Entomology 53: 503–522. [DOI] [PubMed] [Google Scholar]
- Wouters FC, Reichelt M, Glauser G, et al. 2014. Reglucosylation of the benzoxazinoid DIMBOA with inversion of stereochemical configuration is a detoxification strategy in lepidopteran herbivores. Angewandte Chemie 126: 11502–11506. [DOI] [PubMed] [Google Scholar]
- Wright GA, Lutmerding A, Dudareva N, Smith BH. 2005. Intensity and the ratios of compounds in the scent of snapdragon flowers affect scent discrimination by honeybees (Apis mellifera). Journal of Comparative Physiology A 191: 105–114. [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. 2010. New insights into plants responses to the attack from insect herbivores. Annual Review of Genetics 44: 1–24. [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, et al. 2007. Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liu X, Zhang S, Zhu Y, Whitworth J, Chen M. 2008. Differential responses of wheat inhibitor-like genes to hessian fly, Mayetiola destructor, attacks during compatible and incompatible interactions. Journal of Chemical Ecology 34: 1005–1012. [DOI] [PubMed] [Google Scholar]
- Wu S, Peiffer M, Luthe DS, Felton GW. 2012. ATP hydrolyzing salivary enzymes of caterpillars suppress plant defenses. PLoS One 7: e41947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw N, Balabanidou V, Ballhorn DJ, et al. 2012. A horizontally transferred cyanase gene in the spider mite Tetranychus urticae is involved in cyanate metabolism and is differentially expressed upon host plant change. Insect Biochemistry and Molecular Biology 42: 881–889. [DOI] [PubMed] [Google Scholar]
- Wybouw N, Dermauw W, Tirry L, et al. 2014. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife 3: e02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A. 2011. Endogenous peptide elicitors in higher plants. Current Opinion in Plant Biology 14: 351–357. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. 2010. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Barona G, Ryan CA, Pearce G, et al. 2011. GmPep914, an eight-amino acid peptide isolated from soybean leaves, activates defense-related genes. Plant Physiology 156: 932–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Dong X. 2014. Perception of the plant immune signal salicylic acid. Current Opinion in Plant Biology 20: 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Borrego E, Kolomiets MV. 2013. Jasmonate biosynthesis, perception and function in plant development and stress responses. In: Baez RV. ed. Lipid metabolism. InTech, 393–442. [Google Scholar]
- Yang D-L, Yao J, Mei C-S, et al. 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences of the USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZW, Duan XD, Jin S, et al. 2013. Regurgitant derived from the tea geometrid Ectropis obliqua suppresses wound-induced polyphenol oxidases activity in tea plants. Journal of Chemical Ecology 39: 744–751. [DOI] [PubMed] [Google Scholar]
- Yeh KW, Lin ML, Tuan SJ, Chen YM, Lin CY, Kao SS. 1997. Sweet potato (Ipomoea batatas) trypsin inhibitors expressed in transgenic tobacco plants confer resistance against Spodoptera litura. Plant Cell Reports 16: 696–699. [DOI] [PubMed] [Google Scholar]
- Yoshinaga N, Alborn HT, Nakanishi T, et al. 2010. Fatty acid-amino acid conjugates diversification in lepidopteran caterpillars. Journal of Chemical Ecology 36: 319–325. [DOI] [PubMed] [Google Scholar]
- Yu H, Kowalski SP, Steffens JC. 1992. Comparison of polyphenol oxidase expression in glandular trichomes of Solanum and Lycopersicon species. Plant Physiology 100: 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrobelny M, Møller BL. 2011. Cyanogenic glucosides in the biological warfare between plants and insects: the burnet moth-birdsfoot trefoil model system. Phytochemistry 72: 1585–1592. [DOI] [PubMed] [Google Scholar]
- Van Zandt PA, Agrawal AA. 2004. Community-wide impacts of herbivore-induced plant responses in milkweed (Asclepias syriaca). Ecology 85: 2616–2629. [Google Scholar]
- Zangerl AR, Rutledge CE. 1996. The probability of attack and patterns of constitutive and induced defense: a test of optimal defense theory. American Naturalist 147: 599–608. [Google Scholar]
- Zarate SI, Kempema LA, Walling LL. 2007. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiology 143: 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebelo SA, Maffei ME. 2012. The ventral eversible gland (VEG) of Spodoptera littoralis triggers early responses to herbivory in Arabidopsis thaliana. Arthropod-Plant Interactions 6: 543–551. [Google Scholar]
- Zebelo SA, Maffei ME. 2015. Role of early signalling events in plant–insect interactions. Journal of Experimental Botany 66: 435–448. [DOI] [PubMed] [Google Scholar]
- Zhao YF, Thilmony R, Bender CL, Schaller A, He SY, Howe GA. 2003. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant Journal 36: 485–499. [DOI] [PubMed] [Google Scholar]
- Zhang PJ, Zheng SJ, van Loon JJA, et al. 2009. Whiteflies interfere with indirect plant defense against spider mites in Lima bean. Proceedings of the National Academy of Sciences of the USA 106: 21202–21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhu X, Huang F, et al. 2011. Suppression of jasmonic acid-dependent defense in cotton plant by the mealybug Phenacoccus solenopsis. PLoS One 6: e22378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PJ, Broekgaarden C, Zheng S-J, et al. 2013a. Jasmonate and ethylene signaling mediate whitefly-induced interference with indirect plant defense in Arabidopsis thaliana. New Phytologist 197: 1291–1299. [DOI] [PubMed] [Google Scholar]
- Zhang PJ, Xu C-X, Zhang J-M, et al. 2013b. Phloem-feeding whiteflies can fool their host plants, but not their parasitoids. Functional Ecology 27: 1304–1312. [Google Scholar]
- Zhang T, Luan J, Qi J, et al. 2012. Begomovirus–whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Molecular Ecology 21: 1294–1304. [DOI] [PubMed] [Google Scholar]
- Zhen Y, Aardema ML, Medina EM, Schumer M, Andolfatto P. 2012. Parallel molecular evolution in an herbivore community. Science 337: 1634–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu S, Chen X, et al. 2014. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant Journal 77: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Oger PM, Schrammeijer B, Hooykaas PJJ, Farrand SK, Winans SC. 2000. The bases of crown gall tumorigenesis. Journal of Bacteriology 182: 3885–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, et al. 2011. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K, Zeng R. 2015. Insect response to plant defensive protease inhibitors. Annual Review of Entomology 60: 233–252. [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Luthe DS, Felton GW. 2008. Arthropod-inducible proteins: broad spectrum defenses against multiple herbivores. Plant Physiology 146: 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]