Abstract
Kernel row number (KRN) is an important component of yield during the domestication and improvement of maize and controlled by quantitative trait loci (QTL). Here, we fine-mapped a major KRN QTL, KRN4, which can enhance grain productivity by increasing KRN per ear. We found that a ~3-Kb intergenic region about 60 Kb downstream from the SBP-box gene Unbranched3 (UB3) was responsible for quantitative variation in KRN by regulating the level of UB3 expression. Within the 3-Kb region, the 1.2-Kb Presence-Absence variant was found to be strongly associated with quantitative variation in KRN in diverse maize inbred lines, and our results suggest that this 1.2-Kb transposon-containing insertion is likely responsible for increased KRN. A previously identified A/G SNP (S35, also known as Ser220Asn) in UB3 was also found to be significantly associated with KRN in our association-mapping panel. Although no visible genetic effect of S35 alone could be detected in our linkage mapping population, it was found to genetically interact with the 1.2-Kb PAV to modulate KRN. The KRN4 was under strong selection during maize domestication and the favorable allele for the 1.2-Kb PAV and S35 has been significantly enriched in modern maize improvement process. The favorable haplotype (Hap1) of 1.2-Kb-PAV-S35 was selected during temperate maize improvement, but is still rare in tropical and subtropical maize germplasm. The dissection of the KRN4 locus improves our understanding of the genetic basis of quantitative variation in complex traits in maize.
Author Summary
Maize (Zea mays L.) is one of the world's most important sources of calories for humans. With an expanding global population, the demands for maize-derived food, feed, and fuel are rapidly increasing. To meet these needs, geneticists and breeders are facing the challenge of enhancing grain yield through genetic improvement of maize germplasm. Understanding the genetic basis of grain yield is necessary to guide breeding efforts towards the development of high-yielding hybrids. Kernel row number (KRN) in maize is one of the most important yield components and a significant breeding target. Over the last few decades, many genes that determine inflorescence development and architecture have been identified and characterized. The formation of kernel rows is an integral part of the development of the female inflorescence in maize. Nevertheless, the genetic basis and molecular regulation of quantitative variation in KRN is poorly understood. This study provides experimental evidence for the hypothesis that variation in intergenic regions can regulate quantitative variation of important grain yield-related traits, and also provides tools for improving KRN in maize.
Introduction
Understanding the genetic and molecular basis of grain yield is necessary to guide breeding efforts towards the development of high-yielding maize hybrids. Kernel row number (KRN) in maize is one of the most important yield components and a significant breeding target. During the domestication of maize, KRN underwent a dramatic change from two rows in teosinte to more than eight rows in modern maize [1]. A number of quantitative trait loci (QTL) have been reported [2–3] to control quantitative variation in KRN. However, the genetic and molecular mechanisms of these KRN QTL are unknown.
Switching from vegetative to reproductive development turns axillary meristems (AMs) into ear inflorescence meristems (IMs) [4]. The IMs then elongate and produce spikelet-pair meristems (SPMs). Each SPM makes two spikelet meristems (SMs), which then give rise to floral meristems (FMs) that form kernels after fertilization [4]. The initial number of SPMs on the female inflorescence meristem determines the number of kernel rows on the maize ear, while the meristematic activity of IMs determines the potential number of kernels in each kernel row. The initial number of SPMs is correlated with the size of the inflorescence meristem, which provides space for the development of SPMs. The CLAVATA-WUSCHEL (CLV-WUS) feedback-signalling loop regulates IM size by restricting stem cell proliferation and maintaining meristem activity. Recently, several genes in the CLV-WUS feedback loop, including thick tassel dwarf1 (td1) [5], fasciated ear2 (fea2) [6–7], and COMPACT PLANT2 (CT2) [8], were isolated in maize. Additionally, the RAMOSA genes [9], Corngrass1 (Cg1) [10], tasselsheath4 (tsh4) [11], FLORICAULA/LEAFY (ZFL1 and ZFL2) [12], unbranched2 (ub2) and ub3 [13] and others, all affect ear morphology by regulating the development of SPMs and SMs. However, these genes were originally isolated through genetic assays of inflorescence mutants, the mechanisms of them to affect quantitative variation of ear-related traits remain unknown, except for fea2 and ub3 [7, 13]. Thus, the genetic basis and molecular regulation of quantitative variation in KRN deserves further study.
Previously, a major KRN QTL, KRN4, with a large additive effect was identified by combining linkage and association mapping [2–3]. We found that the associated SNPs within KRN4 constitute a linkage disequilibrium block (Chr4:198.9Mb–199.9Mb) in our association mapping panel (S1 Fig). In the present study, we isolated KRN4 by positional cloning and analysed the putative causal variant using maize mutants, gene expression, and association mapping. We then examined changes in the allelic composition of populations for the causal variant during the domestication and improvement of maize. Finally, we assessed the utility of KRN4 for maize breeding by allele substitution using marker-assisted selection.
Results
Positional cloning of KRN4
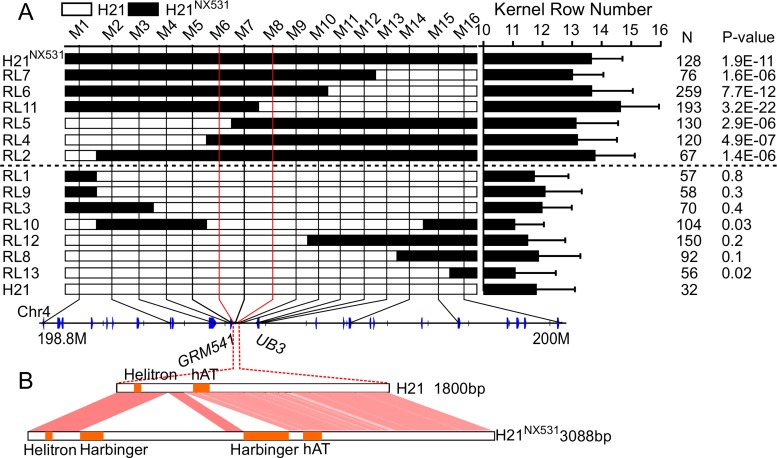
To fine-map KRN4, a near isogenic line (H21NX531) containing the QTL was developed. In comparison with H21, H21NX531 exhibited similar plant appearance (Fig 1A). The KRN (P-value = 5.87 E-07), ear diameter (P-value = 0.0017), cob diameter (P-value = 0.0075), kernel number (P-value = 8.70 E-05), and grain yield (P-value = 7.47 E-05) were significantly increased in H21NX531 (Table 1 and Fig 1B). However, 100-kernel weight of H21NX531 did not differ from that of H21 (Table 1). To understand the developmental basis of the increase in KRN, we measured the inflorescence meristem size of the 2-mm immature ear. The diameter of ear IM in H21NX531 is significantly larger (P-value = 5.2 E-04) than that of H21 in the developing female inflorescence (Fig 1C and 1D). Next, to fine map KRN4, a total of 31 recombinants representing 13 distinct crossover events were found in over 10,000 F2 individuals derived from the cross H21×H21NX531. We compared the KRN of H21 with homozygous recombinant lines derived from the 13 representative recombinants, and found that the homozygous recombinant lines (RL2, RL4, RL5, RL6, RL7, and RL11) carrying the H21NX531 genomic segment between marker M6 and M8 displayed higher KRN (more than 13 rows, P-value < 1.0 E-05, Student’s t-test) than H21 (11.8 ± 1.3), while the other homozygous recombinant lines carrying the H21 genomic segment exhibited almost the same KRN as H21 (Fig 2A). To exclude the effect of residual genetic background, we also compared the KRN of offspring individuals derived from each of the 13 heterozygous recombinants in four environments. We found that only when the offspring populations were segregated with KRN4 H21 and KRN4 NX531 in M6-M8 marker interval (RL6-RL10, S1 Dataset), the KRN of those individuals with the homozygous H21NX531 genotype in the M6-M8 marker interval were significantly higher than that of individuals with the homozygous H21 genotype (P-value < 0.01, S1 Dataset, Student’s t-test). Therefore, we could narrow the genomic location of KRN4 down to a 3-Kb intergenic region flanked by M6 and M8 markers (Fig 2A and S1 Dataset), which is located ~60 Kb downstream from an SBP-box gene UB3 [13] and ~300 bp upstream of a gene of unknown function, GRMZM2G001541 (Fig 2A). The genomic region between marker M6 and M8 was defined as KRN4. In comparison with H21, two regions totaling 1.2 Kb in length (the 1.2-Kb PAV) each containing a fragment of the harbinger transposable element are present in H21NX531 (Fig 2B and S2 Dataset). Several SNPs and small indels are also present in this region (Fig 2B and S2 Dataset). Therefore, sequence differences within the 3-Kb genomic region between H21 and H21NX531 could be the potential causative sites for KRN4 to control KRN variation.
Fig 1. The plant and inflorescence performance of H21 and H21NX531.
A) The plant performance of H21 (right) and H21NX531 (left). B) The mature ears of H21 and H21NX531. C) Diameter of inflorescence meristems in H21 and H21NX531, micrograph of apical 2-mm immature ears of H21 (left) and H21NX531 (right), white bracketed lines represent inflorescence meristem diameters. Bar = 200 μm. D) The statistical analysis of inflorescence meristem diameters between H21 and H21NX531. *** P < 0.001. N: 8/7 for H21 and H21NX531.
Table 1. Pleiotropic effects of KRN4.
| Trait | H21 | H21NX531 | P-value | N a |
|---|---|---|---|---|
| Kernel row number | 11.1 ± 1.2 | 13.3 ± 1.2 | 5.85 E-07 | 17/24 |
| Ear diameter (cm) | 4 ± 0.3 | 4.4 ± 0.3 | 0.0017 | 17/24 |
| Cob diameter (mm) | 29 ± 1.5 | 30.8 ± 2.5 | 0.0075 | 17/24 |
| Kernel number per ear | 309.6 ± 41.7 | 366.3 ± 32.5 | 8.70 E-05 | 17/24 |
| Kernel yield per ear (g) | 64.8 ± 11.1 | 81.1 ± 10.4 | 7.47 E-05 | 15/23 |
| Tassel branch number | 8.7 ± 3.6 | 10.2 ± 4 | 0.21 | 17/24 |
| Ear length (cm) | 12.5 ± 1.3 | 11.8 ± 1.2 | 0.07 | 17/24 |
| Kernel number per row | 28.2 ± 2.6 | 27.4 ± 2.6 | 0.31 | 17/24 |
| 100-kernel weight (g) | 12.4 ± 1.5 | 12.8 ± 1.6 | 0.49 | 17/24 |
aN, sample size, H21/H21NX531
Fig 2. Fine mapping of KRN4.
A) The graphical genotypes of homozygous recombinants (HR). The white boxes in the graphical genotype represent the genomic segments from H21, the black boxes represent genomic segments from H21NX531. A progeny test was conducted to examine whether the KRN of HRs were significantly higher than that of H21. N: the total number of HR phenotyped in four environments (S1 Dataset). P-value: Student’s t-test of the difference in KRN between HRs and H21. The axis represents the physical map of KRN4, and the blue box represents genes within KRN4. B) Nucleotide sequence differences in the KRN4 region between H21 and H21NX531. The orange solid boxes represent the locations of transposable elements in KRN4.The shadowed regions represent homologous KRN4 sequence between H21 and H21NX531 (S2 Dataset).
Expression analysis of UB3 and GRMZM2G001541
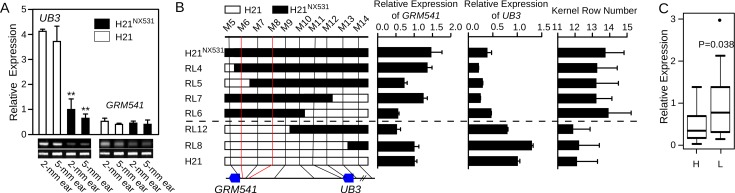
We first examined the expression atlas for UB3 and GRMZM2G001541. The expression data were obtained from qteller (http://www.qteller.com/) and MaizeGDB (http://www.maizegdb.org/). We found both UB3 and GRMZM2G001541 exhibited similar mRNA expression patterns and accumulated in developing ears and tassels (S2 Fig). They also express in the non-reproductive tissues such as leaf, internode etc. (S2 Fig). However, in the immature ear at spikelet-pair meristems (2-mm ear) and spikelet meristems (5-mm ear) differentiation stages, only UB3 exhibited differential expression between H21 and H21NX531, with an expression level almost threefold higher in H21 than in H21NX531 (Fig 3A). Differential expression of UB3 was also observed in stems, roots, and leaves (S3A Fig). However, in 5-mm tassel and 10-mm tassel, expression of UB3 did not show an obvious decrease in H21NX531 relative to H21 (S3A Fig), which might explain why tassel branch number did not differ between H21 and H21NX531 (Table 1). To explore the relationship between expression of UB3 and KRN4, we analysed the expression of UB3 and GRMZM2G001541 in immature ears of six homozygous recombinant lines (RL4, RL5, RL6, RL7, RL8, and RL12) and two parental lines (H21 and H21NX531), and found that RL4, RL5, RL6, and RL7, which carry the KRN4 NX531 allele, showed lower expression of UB3 and higher KRN, while the lines RL8 and RL12, which carry the KRN4 H21 allele, showed higher expression of UB3 and correspondingly lower KRN (Fig 3B). In contrast, the expression of GRMZM2G001541 in the lines with the KRN4 NX531 allele was similar to that in lines with the KRN4 H21 allele (P-value = 0.42) (Fig 3B). Therefore, the expression of UB3 is regulated by KRN4, shows a strong negative correlation with KRN (Fig 3B). We further divided these 38 diverse maize inbred lines into two groups: Group L carrying the KRN4 H21 allele (N = 26) and Group H carrying the KRN4 NX531 allele (N = 12), according to their genotypes for the 1.2-Kb PAV of KRN4 (S1 Table). By examining UB3 expression at the 2-mm ear stage, we found that the expression of UB3 in Group L lines was significantly higher than that in Group H lines (P-value = 0.038, Student’s t-test, Fig 3C), and KRN in these 38 inbred lines was again negatively correlated with the expression level of UB3 (r = -0.35, P-value = 0.037, Pearson’s correlation coefficient, S3B Fig).
Fig 3. Analysis of UB3 and GRMZM2G001541 expression.
A) Expression patterns of UB3 and GRMZM2G001541 (GRM541) in immature ears of H21 and H21NX531, **: P-value < 0.01. B) Analysis of UB3 and GRMZM2G001541 (GRM541) expression in recombinant lines in immature 2-mm ear. The white boxes in the graphical genotype represent the genomic segment from H21, and the black boxes represent the genomic segment from H21NX531. C) Expression of UB3 in immature 2-mm ear in 38 diverse inbred lines. H represents lines with the H21NX531genotype at the KRN4 locus (N = 12), and L represents lines with the H21genotype at the KRN4 locus (N = 26).
DNA sequence variation and putative causal polymorphic sites in KRN4 and UB3
We sequenced KRN4 (~3 Kb, between marker M6 and M8) and UB3 genic region (~4 Kb, including promoter to 3′-UTR but not first intron) in our association mapping panel (S3 Dataset) [3, 14], and identified 69 and 46 polymorphic sites, respectively, with Minor Allele Frequency (MAF) ≥ 0.05 (S4 Fig). Association analysis using the MLM K + Q model [15–16] revealed that four sites were associated with KRN at P-value <1.0 E-04 (Table 2), including one A/G SNP in the third exon of UB3 (S35, P = 3.81E-08, N = 428), one G/A SNP in the 3'-UTR region of UB3 (S45, P-value = 7.35 E-05, N = 384), one ~700 bp insertion/deletion (S23, P-value = 6.69 E-05, N = 416) in the promoter region of UB3, and the 1.2-Kb PAV in KRN4 (P-value = 7.28 E-06, N = 428) (Table 2). The four sites could be classified into three LD groups at R2 > 0.4: group 1 including S23, group 2 including S35 and S45, and group 3 including the 1.2-Kb PAV (S4 Fig). Conditional association analysis was then conducted using these four sites as covariates under an MLM K + Q model, to determine whether these sites were independent or not. When S35 was conditioned, neither S45 nor S23 were significantly associated with KRN (P-value 0.49 and 0.41, S2 Table), but the 1.2-Kb PAV was found to be weakly associated with KRN (P-value = 0.03, S2 Table). The signals for association of S35 and the 1.2-Kb PAV with KRN were only slightly decreased when conditioned by any one of S23 and S45 (S2 Table). Finally, when conditioned on the 1.2-Kb PAV, the other variants were also still significantly associated with KRN (S2 Table). Hence, the association of the 1.2-Kb PAV with KRN might be independent of S23 and S45 but partially related to S35, and the association of S23 and S45 with KRN might depend on that of S35. The dependence of S45 on S35 might be due to its high linkage disequilibrium with S35; thus, S35 could actually represent the association of S45 with KRN, while S23 might not, because of the weak linkage disequilibrium between S23 and S35 (R2 = 0.21).
Table 2. The four polymorphisms in KRN4 and UB3 associated with KRN under the MLM K + Q model.
| Site | Location | Allelea | Frequency a | P-value |
|---|---|---|---|---|
| S23 | Promoter of UB3 | 700/170/0-bp Insertion | 86/86/243 | 6.69 E-05 |
| S35 | Exon of UB3 | A/G | 59/369 | 3.81 E-08 |
| S45 | 3' UTR of UB3 | G/A | 104/280 | 7.35 E-05 |
| 1.2-Kb PAV | KRN4 | 1.2-Kb Presence/Absence | 153/257 | 7.28 E-06 |
a The alleles represent ‘desirable allele/undesirable allele’.
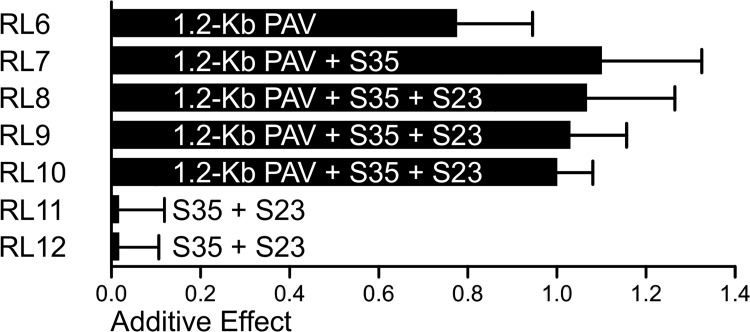
To further determine the relationship between the 1.2-Kb PAV, S35, and S23, the segregating populations derived from selfing the heterozygous recombinants RL6-RL12 were used to evaluate the additive effects of these three tightly linked loci. The 1.2-Kb PAV showed a large additive effect (0.78) in RL6 offspring segregating population, while the additive effect of S35 and S23 were zero in RL11-RL12 (Fig 4). However, combination of the 1.2-Kb PAV + S35 (RL7) or the 1.2-Kb PAV + S35 + S23 (RL8-RL10) had an additive effect more than 1.07 rows, almost 40% higher than that of the 1.2-Kb PAV alone in RL6 (Fig 4). These two kinds of combinations exhibited a similar additive effect, which suggests that the increased additive effect was caused mainly by S35 or polymorphisms tagged by S35. Therefore, the 1.2-Kb PAV or a locus near 1.2-Kb PAV that genetically interacts with a locus tagged by S35, and their interaction, might strongly promote the additive effect on KRN (Fig 4). We next constructed haplotypes using 1.2-Kb PAV and S35 (1.2-Kb-PAV-S35) and found that they showed stronger association with KRN (P-value = 2.41 E-09, N = 428, MLM K + Q) than did each individual locus, when comparing the high-KRN haplotype against the low-KRN haplotype using the MLM K + Q model. In the association mapping panel, a total of four haplotypes (Hap1-Hap4) were observed for the 1.2-Kb-PAV-S35 (Table 3). Lines with Hap1 exhibited higher KRN than lines with the other three haplotypes, and lines containing Hap2 to Hap4 did not significantly differ from each other in KRN (Table 3).
Fig 4. The additive effects of 1.2-Kb PAV and S35 estimated in RLs.
The heterozygous recombination lines RL6-RL12 were selfed to generate segregating populations for either 1.2-Kb PAV, S35, and S23, or all of them. The average additive effects estimated in four environments for each RL were treated as the genetic effect of the segregating site in the RL.
Table 3. KRN and frequencies of haplotypes between 1.2-Kb PAV and S35.
| Haplotype | 1.2-Kb PAV | S35 | KRN | Frequency in Maize | ||
|---|---|---|---|---|---|---|
| All Maize a | TST b | TEMP c | ||||
| Hap1 | 1.2-Kb Presence | A | 14.5 ± 1.9 | 12.8% | 2.5% | 21.1% |
| Hap2 | 1.2-Kb Presence | G | 13.2 ± 1.3 | 23.3% | 36.5% | 12.6% |
| Hap3 | 1.2-Kb Absence | A | 13.4 ± 0.5 | 0.4% | 0.0% | 0.8% |
| Hap4 | 1.2-Kb Absence | G | 13.0 ± 1.5 | 63.5% | 61.0% | 65.4% |
a The sample size for all maize is 428
b TST: Tropical and SubTropical maize germplasm, sample size: 234
c TEMP: Temperate maize germplasm, sample size: 194.
Analysis of the molecular evolution of KRN4
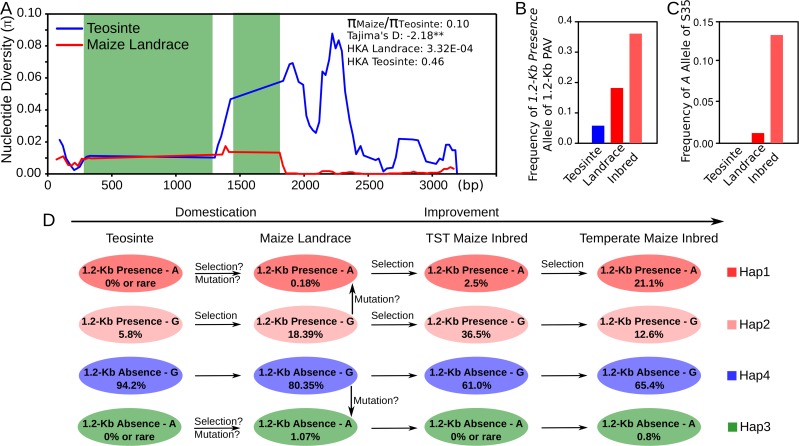
A total of 29 maize wild relatives Z. mays subsp. parviglumis teosinte accessions and 36 diverse maize landraces were employed to estimate the selection pressure during maize domestication (S4 Dataset). The genomic sequence of KRN4 was sequenced in them. Then three expectations of past selection were assessed. First, we compared the nucleotide diversity (π) of KRN4 between teosintes and maize landraces. We found KRN4 had undergone strong reduction in nucleotide diversity from teosintes to maize landraces with πmaize/πteosinte = 0.10, indicating that only 10% nucleotide diversity in teosintes was retained in maize landraces (Fig 5A). Second, a significantly negative Tajima’s D-statistic (-2.18, P-value < 0.01, length of tested region = 3,144 bp, number of sites = 1,722, Fig 5A) of KRN4 was acquired in maize landraces which suggested a recent selection in the KRN4 region. Furthermore, the Hudson–Kreitman–Aguade (HKA) test was applied to assesses the ratio of diversity in maize landrace to divergence from an outgroup (Z. diploperennis) for KRN4 relative to four neutral genes. KRN4 in landrace showed significant selection based on HKA test result (P-value = 3.32E-04, length of tested region = 3,144 bp, number of sites = 1,722, Fig 5A and S3 Table), but KRN4 in teosinte doesn’t (P-value = 0.46, length of tested region = 3,300 bp, number of sites = 1,642, Fig 5A and S3 Table). These results revealed that KRN4 was under strong selection during domestication from teosinte to maize, similar to tga1 promoter and tb1upstream region [17–18]. However, different from tga1 and tb1 loci [17–18], no fixed difference between teosintes and maize landraces could be observed in KRN4.
Fig 5. The evidence of significant selection in KRN4 during maize domestication and improvement.
A) Nucleotide diversities (π) within ~3 Kb of KRN4 for maize landrace (red line) and teosinte (blue line). The green shades represent the 1.2-Kb PAV region. **: P-value < 0.01. B) Allele frequencies of 1.2-Kb Presence of 1.2-Kb PAV in teosinte, maize landrace and inbred lines. C) Allele frequencies of A of S35 in teoisnte, maize landrace and inbred lines. D) A putative evolutionary pattern of 1.2-Kb PAV and S35 during maize domestication and improvement. The circles in colors represent the four haplotypes between 1.2-Kb PAV and S35. The genotypes of haplotypes in 1.2-Kb PAV and S35 are showed in the circles. The number inside the circle is the frequency of the haplotype. TST maize: Tropical and SubTropical maize germplasms. All of the steps marked as “Selection” are of significantly frequency change, P-value < 0.001 (χ2 test based on the frequencies).
To explore the evolution of 1.2-Kb-PAV and S35 loci, we genotyped them in 120 teosinte accessions, 280 maize landraces (S5 Dataset) and 428 maize inbred lines, respectively. In teosinte, the frequencies of favorable alleles for 1.2-Kb PAV (1.2-Kb Presence allele) and S35 (A allele) were 5.8% and 0% (Fig 5B and 5C). The 1.2-Kb Presence allele had a higher frequency (9.6%) in Z. parviglumis but rare in Z. mexicana (2.4%), implying that the favorable allele of 1.2-Kb PAV in modern maize was probably selected from Z. parviglumis (S5 Dataset). In maize landrace, the frequencies of favorable alleles for 1.2-Kb PAV and S35 were increased to 18.6% and 1.25%, respectively (Fig 5B and 5C). During modern maize improvement, they were enriched to 36.1% and 13.2% (Fig 5B and 5C), and the R2 of them in the association mapping panel were 5.0% and 12.2%. The favorable haplotype of 1.2-Kb-PAV-S35, Hap1 was not detected in teosinte accessions (Fig 5D), and the frequency of Hap1 in maize inbred lines increased to 12.8% (N = 428, Fig 5D), but differed dramatically between temperate (21.1%, N = 234, Fig 5D) and TST (tropical and subtropical, 2.5%, N = 194, Fig 5D) maize inbred lines. The unequal distribution of Hap1 in different subpopulations suggests that favorable Hap1 has been selected to increase grain yields by increasing the number of kernel rows in temperate germplasm. Based on these results, we proposed an evolutionary pattern of 1.2-Kb PAV and S35 during maize domestication and improvement (Fig 5D). Hap2 of 1.2-Kb-PAV-S35, which harbors the 1.2-Kb Presence allele, was selected and enriched from teosinte to landrace and then to tropical and subtropical maize inbred lines (Fig 5D). The favorable Hap1 allele might have been selected from teosinte or could have arisen by mutation at S35 after domestication (Fig 5D). However, the intensive selection on Hap1 only occurred during temperate maize inbred lines improvement (Fig 5D).
UB3 regulates inflorescence meristem development
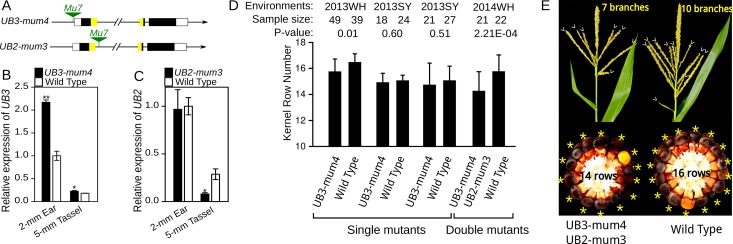
UB3 is an ortholog of OsSPL14, which is responsible for IPA1 (ideal plant architecture 1) and WFP (WEALTHY FARMER’S PANICLE) in rice (S5 Fig) [19–20], and is also homologous with UB2. Recent study has revealed that ub2 and ub3 knock-out mutants exhibit increase in maize KRN [13]. Two novel Mutator-mediated mutants, UB3-mum4, with a Mu7 insertion in the promoter region of UB3, and UB2-mum3, with a Mu7 insertion in the first intron of UB2 (Fig 6A), were obtained from Maize Stock Center. UB3 expression in 2-mm immature ears and 5-mm tassels of the UB3-mum4 line was significantly higher than that in the wild type (WT) (Fig 6B). Similarly, a previous study has identified that a Mu transposon insertion in 5’UTR of P1 gene increases P1 expression in maize [21]. UB2 expression in 2-mm immature ears of the UB2-mum3 line did not differ significantly from WT (Fig 6C), but ~14% of UB2-mum3 transcripts contained an extra 295-bp fragment composed of a 145-bp intron sequence flanking Mu7 insertion sites and a 150-bp terminal inverted repeat of Mu7 (S6 Fig). The 295-bp fragment was inserted into the SBP-box domain-encoding sequence and might result in loss of function of the alternatively spliced transcript. We developed segregating populations to evaluate the influence in KRN by the Mu7 insertion in UB3-mum4 and UB2-mum3. Each single mutant did not show an obvious change in KRN or ear diameter (Fig 6D and S4 and S5 Tables), only UB3-mum4 showed a slight but significant decrease in KRN in 2013 Wuhan environment (P-value = 0.01, Fig 6D and S4 Table). Interestingly, double mutants of UB3-mum4 and UB2-mum3 showed a significant decrease in KRN (P-value = 2.21 E-04) and ear diameter (P-value = 2.90 E-05) relative to WT (Fig 5D and 5E and S6 Table). In addition, UB3-mum4 and double mutant also showed a slight but significant reduction in tassel branch number relative to wild types (Fig 6E and S6 Table).
Fig 6. Expression analysis and phenotypic characterization of UB3-mum4 and UB2-mum3 mutants.
A) The insertion site of Mutator in UB3-mum4 and UB2-mum3 mutants. B and C) Expression level of UB3 (B) and UB2 (C) in mutant and wild type. ** P < 0.01, * P < 0.05. D) KRN performance single and double mutants of UB3-mum4/UB2-mum3 and wild type; WH: Wuhan; SY: Sanya. E) Tassel and ear of wild type and double mutant of UB3-mum4/UB2-mum3. Detailed information regarding phenotypes is presented in S3–S6 Tables.
The potential for use of KRN4 in maize improvement
The introgression of the 1.2-Kb PAV from NX531 into H21 results in significant enlargement of the inflorescence meristem in the immature ear of H21NX531 (Table 1 and Fig 1C and 1D). The enlarged diameter of the inflorescence meristem provides a larger space to support the larger number of spikelet-pair meristems generated. Accompanying the increase in KRN in H21NX531, kernel number per ear also significantly increased, but 100-kernel weight was not affected, and so the grain yield of H21NX531 was markedly enhanced (Table 1). The enhanced yield resulting from the increased KRN with unaltered kernel weight may only apply to the specific genetic backgrounds or growth conditions. Then, we anticipate that selection for the favorable allele at KRN4 will contribute positively to maize productivity. To test this hypothesis, we used marker-assisted selection to introgress the 1.2-Kb Presence alleles from two inbred lines carrying the 1.2-Kb Presence alleles, TY6 and Qi205, into W138 and Mo17 carrying the 1.2-Kb Absence alleles. To minimize the influence of genetic background, heterozygotes at the 1.2-Kb PAV in BC3F1 were selfed to develop a segregating population, and then two homozygous genotype subgroups (1.2-Kb Presence subgroup, 1.2-Kb Absence subgroup) were identified in each segregating population for KRN evaluation to maximum randomize genetic background. We found that mean of KRN of the 1.2-Kb Presence subgroup was almost 2 rows higher than that of the 1.2-Kb Absence subgroup, indicating that the introgression of the superior alleles could increase KRN of recurrent parents (S7 Table).
Discussion
UB3 is distally regulated by KRN4 and controls kernel row number in maize
In this study, we fined mapping a major KRN QTL, KRN4, and suggested the 3-Kb intergenic region that includes a 1.2-Kb PAV ~60 Kb downstream of UB3 is the causation underlying the major KRN QTL. Expression analysis in immature ear indicated that the expression difference of UB3 between H21 and H21NX531, and also among diverse inbred lines, was highly correlated with variation in KRN4. Further, the weak mutants of UB3-mum4 and UB2-mum3 used in this study demonstrated that elevation of UB3 expression reduces the KRN and ear diameter, which is consistent with previous characterized ub3 and ub2 knock-out mutations which cause KRN increase and ear diameter enlargement [13]. The elevation of UB3 expression in UB3-mum4/UB2-mum3 may reduce the inflorescence meristem size of the developing ear, resulting in formation of less spikelet-paired meristems (SPMs), and then decreased number of kernel rows and ear diameter. This hypothesis can be supported in H21 and H21NX531, where the higher UB3 expression in H21 is correlated with smaller inflorescence meristem size and less SPMs formation than H21NX531, and also is consistent with ub3 knock-out mutants with enlargement in inflorescence meristem size [13]. However, we observed that an increase of UB3 expression in UB3-mum4 slightly reduces the tassel branch number, which is inconsistent with the results of ub3 knock-out mutants, which show highly suppressed tassel branch [13]. These observations imply that the allele effect on tassel branch number of UB3-mum4 used in this study is different from previous identified ub3 knock-out mutants. The ortholog of UB3 and UB2 in rice, OsSPL14, negatively regulates axillary bud outgrowth to repress shoot tillering, but positively regulates the number of panicle branches by enhancing meristematic activity and cell proliferation [19–20, 22–24]. Unlike OsSPL14, UB3 and UB2 exhibit redundant biological functions on negative regulation of KRN, a kind of short branch in maize ear. It seems like that UB3 and UB2 evolved from a common ancestral gene with OsSPL14 and retained similar biological functions, but may act in opposite ways. Therefore, we suggest that KRN4 controls the natural variation of KRN by acting as a distal regulator of UB3 expression and UB3 negatively regulates KRN in maize.
Previous study revealed that ub3 shows more severe phenotype than ub2 [13]. The UB3 locus is also a KRN and tassel branch number QTLs hotspot detected by many studies [2–3], and UB3 is found to be the causative gene underlying a major KRN QTL, KRN4, in this study. However, the natural variation in UB2 locus has not been found to be associated with inflorescence traits in maize [2–3]. So, alterations in UB3 by mutations or natural variation are more likely to cause the response on inflorescence traits than UB2. In addition, the expression differences of UB3 was not in developing tassels, consistent with ear traits being modulated and tassel traits not. Thus, KRN4 may not be responsible for the TBN QTLs at this locus, which is consistent with previous suggestion that KRN and TBN are controlled by different polymorphisms of UB3 [13].
The association analysis of KRN4 revealed that only the 1.2-Kb PAV containing TE fragments was significantly associated with KRN in diverse inbred lines. Hence, variation in KRN between H21 and H21NX531 due to UB3 expression is possibly caused by the 1.2-Kb PAV. This kind of distal regulation of gene expression being responsible for variation in important traits has been previously described in maize, and two different mechanisms may account for it. First, like tb1, Vgt1, ZmCCT, and prol1.1, the causal sequences (commonly transposon derived sequences) act as enhancers to regulate gene expression level or pattern in cis [18, 25–28]. In a second mechanism, non-coding tandem repeat sequences located ∼100 kb upstream of b1 express dsRNA, which mediates trans-communication between alleles to establish paramutation [29]. KRN4 may interact with the UB3 regulatory region in cis to promote expression of UB3, or the transposon fragments in KRN4 may express small RNAs and affect UB3 expression by an epigenetic regulation mediated by small RNAs. These assumptions are yet to be investigated.
KRN4 and UB3 might genetically interact to regulate KRN
In addition to 1.2-Kb PAV, an A/G SNP designated as S35 that is significantly associated with KRN was also detected in our association mapping panel. Located in an exon of UB3, this is the same as the Ser220Asn polymorphism mentioned by Chuck et al. [13]. S35 showed stronger association with KRN and had better support in conditional analysis than did the 1.2-Kb PAV. However, unlike the 1.2-Kb PAV, in the recombinant lines of the fine mapping population, the introgression of A (or Asn220) from H21NX531 to replace the G (or Ser220) in H21 did not result in increased KRN in RL12. Further, when S35 was segregating in RL11-RL12, no significant additive effect was observed. But the additive effects of 1.2-Kb PAV could be promoted 40% by S35 in the background of 1.2-Kb PAV, implying a positive genetic interaction between them and a larger genetic effect due to their combination. This hypothesis is supported by the stronger association of KRN with the creating haplotype 1.2-Kb-PAV-S35 than with either of the individual loci. We propose that a change in UB3 protein function due to S35 made UB3 more efficient in modulating inflorescence development. Although S35 alone or other polymorphisms in linkage disequilibrium with KRN4 did not display apparent genetic effects in H21, S35 might still affect the biological function of UB3 in KRN formation in another genetic background. Therefore, the 1.2-Kb-PAV-S35 combination could represent the high- and low-KRN haplotypes for KRN4 among these diverse inbred lines, and Hap1 was the most favorable haplotype for KRN.
KRN4 was a selection target during modern maize domestication and improvement
Domestication leads to the loss of genetic diversity throughout the genome, or in specific regions, and desirable alleles for important traits have been selected and enriched [17–18, 30]. For KRN4, the nucleotide diversity in maize landrace is markedly reduced relative to that in teosinte. The strong selection signal was also observed by Tajima’s D test and HKA test. Accompanying the selection on KRN4, the 1.2-Kb Presence allele was continuously enriched during maize domestication and improvement for desirable alleles of KRN4. Its frequency was increased more than twofold from teosinte to maize landrace, and was further doubled from landrace to modern inbred line. In the corresponding processes, the mean values of allele frequency at four neutral genes (adh1, adh2, fus6 and te1) were small changed, just 0.37 fold change from teosinte to landrace, and 0.15 fold change from landrace to modern inbred line for low frequent allele, respectively. Additionally, the favourable allele of KRN4 was enriched rather than was fixed in modern maize lines, which is different from the case of tga1 ant tb1, indicating that KRN4 may be not the critical locus that determines the transition from 2 rows in teosinte to more than 4 rows in modern maize. This was further supported by the fact that neither KRN4 nor UB3 is located within domestication-associated QTL [30]. However, the favourable A allele of S35 in UB3 is not detected in teosinte and has low frequency in maize landraces, indicating that it might have emerged during the post-domestication improvement of modern maize. Because of the larger genetic effect exhibited by the interaction between 1.2-Kb Presence allele of KRN4 and A allele of S35, Hap1 was likely the selection target in modern temperate maize improvement, and the frequency of Hap1 increased more than 7 folds from tropical to temperate maize. Meanwhile, the frequency of A allele of S35 is enriched in temperate maize, but the 1.2-Kb Presence allele shows similar frequency between tropical and temperate maize. The decrease of selection pressure on KRN4 during temperate maize breeding might be caused by the selection on the other KRN loci or the diverse breeding objectives.
Despite the continued improvement during breeding program, the favourable Hap1 is still absent in most modern maize inbred lines that are included in our association mapping panel. For the TST lines in our association mapping panel, Hap1 was still a rare haplotype. Thus KRN4 and UB3 could be subjected to more intense selection by molecular breeding to improve yield by increasing number of kernel rows in maize ear. In conclusion, the dissection of KRN4 in our study not only extends our knowledge about the genetic and molecular mechanisms of important traits in maize, but also provides diagnostic and germplasm tools for improving maize KRN and grain yields.
Materials and Methods
Association analysis
A subset of an association mapping panel with 368 diverse inbred lines was genotyped with 500K SNP markers [31]. KRN of these 368 lines was evaluated in five environments and reported in previous study, including Ya'an (30°N, 103°E), Sanya (18°N, 109°E), and Kunming (25°N, 102°E) in 2009, and Wuhan (30°N, 114°E) and Kunming (25°N, 102°E) in 2010 [3]. The best linear unbiased prediction (BLUP) of KRN was estimated using a linear mixed model in SAS software (SAS Institute Inc., 2001) by previous study [3, 32]. The association of KRN4 with KRN (BLUP data) [3] was established using Tassel v3.0 with a mixed linear model (MLM) approach considering varietal relatedness (K) and population structure (Q) (MLM K + Q) [3, 15–16]. The linkage disequilibrium among associated SNPs was estimated using Haploview v4.1 [33].
Fine mapping of KRN4
A near-isogenic line, H21NX531, that incorporates the KRN4 QTL for kernel row number (Chr4:198.9Mb-199.9Mb, B73 RefGen V2, S1 Fig), was developed by four cycles of backcrossing (BC) followed by two cycles of selfing, using H21 as the recurrent parent and NX531 as the donor of the favorable allele. Over 10,000 F2 individuals derived from the H21×H21NX531 cross were genotyped with markers flanking KRN4 and 14 newly developed markers (Primers were listed in S6 Dataset) within the QTL interval to identify the recombinants. The heterozygous recombinants were self-crossed to segregate the homozygous recombinant (HR) and non-recombinant (HNR) progeny pairs from each recombinant derived family. The HR and NHR progeny pairs were phenotyped at Wuhan (30°N, 114°E) and Sanya (18°N, 109°E) in 2013 (S1 Dataset), with two replications under a randomized block design for each. And the HRs and HNRs were self-crossed to generate homozygous progeny lines for replicated testing at Wuhan and Baoding (38°N, 115°E) in 2014 (S1 Dataset) with two replications under a randomized block design for each. The substitution mapping procedure widely used in fine mapping [34] was employed by examining the KRN differences between HRs and H21, also between HRs and HNRs progeny pairs from each recombinant derived family, using Student’s t-test with significant threshold P-value < 0.01.
Expression analysis
To identify candidate genes for the KRN4 QTL, analysis of the expression of genes in the relevant interval was performed on developing ears and tassels from H21, H21NX531, recombinant lines, and 38 diverse inbred maize lines (S1 Table) using Quantitative PCR (qPCR). Total RNA was extracted using TRIzol Reagent (Life Technologies, Invitrogen, Carlsbad, CA, USA). Total RNAs of H21 and H21NX531 lines were extracted from roots, leaves, stems, immature 5-mm tassel (5-mm tassel, 6-leaf stage with branch meristem initiation), immature 10-mm tassel (10-mm tassel, 10-leaf stage with branches), immature ear stage 1 (2-mm ear, 10-leaf stage with Inflorescence meristems IMs and spikelet-pair meristems SPMs), and immature ear stage 2 (5-mm ear, 12-leaf stage with IM, SPM, and spikelet-meristems SM). Total RNAs of 38 diverse maize inbred lines were extracted from immature ears at the S1 stage (S1 Table). Total RNAs of UB3-mum4 and UB2-mum3 lines were extracted from immature 5-mm tassel and 2-mm ear, respectively. DNase I (TaKaRa Biotech, Dalian, China) was used to remove genomic DNA contamination. An oligo(dT) primer and M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) were used to synthesize first-strand cDNAs. A SYBR Green RT-PCR kit (Bio-Rad, Hercules CA, USA) was used to perform qPCR with gene-specific primers (S7 Dataset). Expression levels were normalized using beta-actin (NM_001155179) as an endogenous control. The expression data for UB3 and GRMZM2G001541 in B73 were downloaded from the qTeller website (www.qteller.com) and MaizeGDB website (www.maizegdb.org).
Mutant analysis
Two Mutator-mediated insertion mutants were obtained from the Maize Genetics Cooperation Stock Center at the University of Illinois, Champaign-Urbana. According to information from the Maize Stock Center [35], UB3-mum4 (UFMu-06293) has Mutator (Mu) inserted upstream of UB3, and UB2-mum3 (UFMu-06514) has Mu inserted into the first intron of UB2. The insertion site of Mu was detected by PCR with gene-specific primers and TIR6 primers designed from the TIR sequence of Mu (S3 Dataset). To characterize the phenotypic effects of the mutants and eliminate the influence of the other Mu insertion, UB3-mum4 and UB2-mum3 were backcrossed with its parent W22, and self-crossed to develop the F2 segregating populations. In each segregating population, wild types (+/+) and homozygous mutants (-/-) were identified by genotyping (Primers used are listed in S3 Dataset) and were phenotyped. The UB3-mum4 and UB2-mum3 were crossed to develop double mutant, which was also crossed with W22, and self-crossed to develop F2 segregating populations. In the segregating populations, double mutant and wild type individuals were genotyped and phenotyped. Student's t-test was used to evaluate the phenotypic differences between wild types and mutants.
Analysis of nucleotide diversity and molecular evolution
To discover DNA sequence variation and putative causal polymorphisms in UB3 and KRN4, gene-specific primers (S3 Dataset) were designed to amplify UB3 and KRN4 in the association mapping panel [3, 14]. We genotyped 428 inbred lines using the 1.2-Kb PAV in KRN4 as a marker, and sequenced about 4.0 Kb of DNA from 5'-upstream of UB3 to its 3'-UTR and ~3 Kb containing KRN4 in 110 or 428 inbred lines of the AM panel, respectively (the line number of the lines that were sequenced is listed in the S3 Dataset). SNPs and indels with MAF > 0.05 were used to estimate pairwise LD and to evaluate the association between polymorphic sites and KRN under the MLM K+ Q model [15–16]. Conditional analysis was conducted using the associated sites as covariates under an MLM K + Q model in Tassel v3.0. The MLM K + Q model was also used for haplotype-based association analysis. The selfed progeny of heterozygous recombinants RL6-RL12 which are segregating at 1.2-Kb PAV, S35 and S23 were employed to evaluate the genetic effect of 1.2-Kb PAV, S35 and S23 (S1 Dataset). The individuals, which harbored homozygous alleles of the three sites, were used to estimate the additive effects of in each segregating population (S1 Dataset).
The selection pressure on KRN4 during the domestication and improvement of maize was estimated using 36 randomly selected landraces (S4 Dataset) from 280 diverse maize landrace collections (S5 Dataset) [36] and 29 Z. mays subsp. parviglumis teosinte (S4 Dataset) from 120 teosinte accessions (S5 Dataset). The KRN4 genomic region was amplified and sequenced using primers listed in S3 Dataset. Nucleotide diversity (π) and Tajima’s D were estimated using DnaSP ver. 5.0 [37]. The 1.2-Kb PAV was treated as single PAV when estimating the nucleotide diversity (π). Four neutral loci (adh1, adh2, fus6 and te1) [38–41] were used as controls for the HKA test [42] using Zea diploperennis as the outgroup. The overall HKA P-value was obtained by summing the individual χ2 values of the four control genes. Another 88 teosinte accessions (including 35 Z. mays subsp. parviglumis and 54 Z. mays subsp. mexicana accessions, S5 Dataset) and 244 maize landraces were genotyped by a PCR marker for the 1.2-Kb PAV and a KASP marker (http://www.kbioscience.co.uk/) for S35, to estimate their frequency in teosinte accessions and maize landraces (Primers are listed in S3 Dataset). All of the sequences have been deposited in NCBI Genebank KT928654—KT931615.
Phylogenetic tree of SBP-box proteins in six plant species
A total of 130 SBP-box genes were predicted in six plant species, including 16 SBP-box genes from Arabidopsis, 18 from Brachypodium, 18 from sorghum, 19 from rice, 20 from foxtail millet, and 29 from maize [43–48], and used for phylogenetic analysis.
Supporting Information
An LD heatmap was constructed using pairwise R2 of the nine KRN-associated SNPs in 368 inbred lines. The X axis represents genomic locations of SNP and Y axis represents -log10(P-observed). The three red points indicate the SNPs most highly associated with KRN, and the dotted line indicates a SNP located in UB3. The horizontal lines represent–log10(0.05/N) and–log10(1/N).
(TIF)
The expression data is obtained from qTeller (www.qteller.com) and MaizeGDB (www.maizegdb.org). Expression pattern of UB3 observed from qteller (A) and MaizeGDB (B). Expression pattern of GRMZM2G001541 observed from qteller (C) and MaizeGDB (D).
(TIF)
A) 5-mm tassel: 6-leaf stage, with BM initiating; 10-mm tassel: 10-leaf stage, with BM; 2-mm ear: 10-leaf stage, with IM and SPM; 5-mm ear: 12-leaf stage, with IM, SPM, and SM. B) The correlation between expression of UB3 and KRN in these 38 inbred lines.
(TIF)
All polymorphisms with a minor allele frequency (MAF) >5% were used to calculate the pairwise linkage disequilibrium (LD). The four polymorphisms most significantly associated with KRN are indicated. In the gene structure of UB3, the blue boxes represent the transposon fragments inserted in the promoter region (S23), the white boxes represent the UTR regions, the black boxes and the yellow boxes represent exons, and the yellow boxes also represent the SBP-box domain.
(TIF)
The legend indicates the scale of branch lengths. Different colors represent the 14 different subfamilies of SBP-box genes.
(PDF)
A) Detection of alternative spliced transcripts of UB2 in UB2-mum3. The MuIS-Primer (S7 Dataset) was used to amplify the cDNA sequence of UB2 flanking the Mu7 insertion site. In the 2-mm ear sample of UB2-mum3, a larger band than the predicted transcript was observed. B) A diagram of the sequence composition of the alternatively spliced transcript of UB2. A 145-bp segment originating from the intron flanking the Mu7 insertion site and a 150-bp segment consist of the terminal Mu7 inverted repeat.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOC)
(DOC)
(DOCX)
(DOC)
(XLS)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
We are grateful to Dr. David Jackson (Cold Spring Harbor Laboratory) for critically reviewing the manuscript. We are grateful to Dr. Feng Tian (China Agricultural University) for his helpful comments and Dr. Xiaohong Yang (China Agricultural University) for her kindly offering maize landrace DNA samples. We gratefully thank Dr. Nathan M. Springer and the three anonymous reviewers for their valuable suggestions.
Data Availability
All relevant data are within the paper and its Supporting Information files. All sequence data of the resequence of teosinte, landrace and maize are available from the nucleotide database of NCBI KT928654–KT931615 (https://www.ncbi.nlm.nih.gov/nucleotide/).
Funding Statement
This work was supported by the National Basic Research Program of China (2014CB138200, 2009CB118400); from the Ministry of Science and Technology of the People´s Republic of China(http://www.most.gov.cn/); received by ZZ. This work was also supported by the National Natural Science Foundation of China (91335110); from The National Natural Science Foundation of China (http://www.nsfc.gov.cn/); received by ZZ. This work was also supported by the Science Foundation of the Ministry of Agriculture of China (2011ZX08009-001); from the Ministry of Agriculture of the People´s Republic of China(http://www.moa.gov.cn/); received by ZZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Doebley J. The genetics of maize evolution. Ann Rev Genet. 2004; 38: 37–59. [DOI] [PubMed] [Google Scholar]
- 2. Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 2011; 7:e1002383 10.1371/journal.pgen.1002383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lei L, Yanfang D, Dongao H, Man W, Shen X, Bing Y, et al. Genetic architecture of maize kernel row number and whole genome prediction. Theor Appl Genet. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vollbrecht E, Schmidt RJ. Development of the inflorescences. In: Bennetzen, JL Hake, SC, editors. Handbook of Maize: Its Biology, eds New York: Springer; 2009. pp.; 13–40. [Google Scholar]
- 5. Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2005; 132: 1235–45. [DOI] [PubMed] [Google Scholar]
- 6. Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001; 15: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bommert P, Nagasawa NS, Jackson D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet. 2013; 45(3): 334–337. 10.1038/ng.2534 [DOI] [PubMed] [Google Scholar]
- 8. Bommert P, Je BI, Goldshmidt A, Jackson D. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature. 2013; 502: 555–558. 10.1038/nature12583 [DOI] [PubMed] [Google Scholar]
- 9. McSteen P. Branching out: the ramosa pathway and the evolution of grass inflorescence morphology. Plant Cell. 2006; 18(3): 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007; 39(4): 544–549. [DOI] [PubMed] [Google Scholar]
- 11. Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010; 137: 1243–1250. 10.1242/dev.048348 [DOI] [PubMed] [Google Scholar]
- 12. Bomblies K1, Doebley JF. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics. 2006; 172: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chuck GS, Brown PJ, Meeley R, Hake S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci USA. 2014; 111(52): 18775–18780. 10.1073/pnas.1407401112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang X, Gao S, Xu S, Zhang Z, Prasanna B M, Li L, et al. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breed. 2011; 28: 511–526. [Google Scholar]
- 15. Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006; 38: 203–208. [DOI] [PubMed] [Google Scholar]
- 16. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007; 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- 17. Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, et al. The origin of the naked grains of maize. Nature. 2005; 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1 . Nat Genet. 2011; 43: 1160–1163. 10.1038/ng.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010; 42: 541–545. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- 20. Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. 2010; Nat Genet. 42: 545–549. 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- 21. Robbins ML, Sekhon RS, Meeley R, Chopra S. A Mutator transposon insertion is associated with ectopic expression of a tandemly repeated multicopy Myb gene pericarp color1 of maize. Genetics. 2008; 178: 1859–1874.A Mutator transposon insertion is associated with ectopic expression of a tandemly repeated multicopy Myb gene 10.1534/genetics.107.082503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, et al. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell. 2013; 25: 3743–3759. 10.1105/tpc.113.113639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, et al. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003; 33: 513–520. [DOI] [PubMed] [Google Scholar]
- 24. Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009; 41: 494–497. 10.1038/ng.352 [DOI] [PubMed] [Google Scholar]
- 25. Salvi S, Sponza G, Morgante M, Tomes D, Niu X, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA. 2007; 104: 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hung HY, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA. 2012; 109: E1913–1921. 10.1073/pnas.1203189109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA. 2013; 110: 16969–16974. 10.1073/pnas.1310949110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wills DM, Whipple CJ, Takuno S, Kursel LE, Shannon LM, et al. From Many, One: Genetic Control of Prolificacy during Maize Domestication. PLoS Genet. 2013; 9(6): e1003604 10.1371/journal.pgen.1003604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arteaga-Vazquez M, Sidorenko L, Rabanal FA, Shrivistava R, Nobuta K, Green PJ, et al. RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc Natl Acad Sci U S A. 2010; 107(29): 12986–91. 10.1073/pnas.1007972107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006; 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- 31. Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013; 45: 43–50. 10.1038/ng.2484 [DOI] [PubMed] [Google Scholar]
- 32. SAS Institute 2001. SAS/STAT User’s Guide v. 8.2. SAS Institute, Cary, N.C., [Google Scholar]
- 33. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21: 263–5. [DOI] [PubMed] [Google Scholar]
- 34. Paterson AH, DeVerna JW, Lanini B, Tanksley SD. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics. 1990; 124:735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, et al. Steady-state transposon mutagenesis in inbred maize. Plant J. 2005; 44: 52–61. [DOI] [PubMed] [Google Scholar]
- 36. Wen W, Franco J, Chavez-Tovar VH, Yan J, Taba S. Genetic characterization of a core set of a tropical maize race Tuxpeño for further use in maize improvement. PLoS One. 2012;7(3):e32626 10.1371/journal.pone.0032626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 38. Eyre Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci U S A. 1998; 95: 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci U S A. 2001;98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tenaillon MI, U’Ren J, Tenaillon O, Gaut BS. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol Biol Evol. 2004; 21: 1214–25. [DOI] [PubMed] [Google Scholar]
- 41. White SE, Doebley JF. The molecular evolution of terminal ear1, a regulatory gene in the genus Zea . Genetics. 1999; 153: 1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hudson RR, And MK, Aguadé M. A Test of Neutral Molecular Evolution Based on Nucleotide Data. Genetics. 1987; 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Z, Wang X, Gu S, Hu Z, Xu H, Xu C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene. 2008; 407: 1–11. [DOI] [PubMed] [Google Scholar]
- 44. Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, et al. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature. 2010; 463: 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- 45. Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009; 457: 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- 46. Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science. 2002; 296: 92–100. [DOI] [PubMed] [Google Scholar]
- 47. Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 2012; 30: 549–554. 10.1038/nbt.2195 [DOI] [PubMed] [Google Scholar]
- 48. Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009; 326: 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An LD heatmap was constructed using pairwise R2 of the nine KRN-associated SNPs in 368 inbred lines. The X axis represents genomic locations of SNP and Y axis represents -log10(P-observed). The three red points indicate the SNPs most highly associated with KRN, and the dotted line indicates a SNP located in UB3. The horizontal lines represent–log10(0.05/N) and–log10(1/N).
(TIF)
The expression data is obtained from qTeller (www.qteller.com) and MaizeGDB (www.maizegdb.org). Expression pattern of UB3 observed from qteller (A) and MaizeGDB (B). Expression pattern of GRMZM2G001541 observed from qteller (C) and MaizeGDB (D).
(TIF)
A) 5-mm tassel: 6-leaf stage, with BM initiating; 10-mm tassel: 10-leaf stage, with BM; 2-mm ear: 10-leaf stage, with IM and SPM; 5-mm ear: 12-leaf stage, with IM, SPM, and SM. B) The correlation between expression of UB3 and KRN in these 38 inbred lines.
(TIF)
All polymorphisms with a minor allele frequency (MAF) >5% were used to calculate the pairwise linkage disequilibrium (LD). The four polymorphisms most significantly associated with KRN are indicated. In the gene structure of UB3, the blue boxes represent the transposon fragments inserted in the promoter region (S23), the white boxes represent the UTR regions, the black boxes and the yellow boxes represent exons, and the yellow boxes also represent the SBP-box domain.
(TIF)
The legend indicates the scale of branch lengths. Different colors represent the 14 different subfamilies of SBP-box genes.
(PDF)
A) Detection of alternative spliced transcripts of UB2 in UB2-mum3. The MuIS-Primer (S7 Dataset) was used to amplify the cDNA sequence of UB2 flanking the Mu7 insertion site. In the 2-mm ear sample of UB2-mum3, a larger band than the predicted transcript was observed. B) A diagram of the sequence composition of the alternatively spliced transcript of UB2. A 145-bp segment originating from the intron flanking the Mu7 insertion site and a 150-bp segment consist of the terminal Mu7 inverted repeat.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOC)
(DOC)
(DOCX)
(DOC)
(XLS)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All sequence data of the resequence of teosinte, landrace and maize are available from the nucleotide database of NCBI KT928654–KT931615 (https://www.ncbi.nlm.nih.gov/nucleotide/).