Abstract
Gene expression programs undergo constant regulation to quickly adjust to environmental stimuli that alter the physiological status of the cell, like cellular stress or infection. Gene expression is tightly regulated by multi-layered regulatory elements acting in both cis and trans. Post-transcriptional regulation of the 3′ untranslated region (3′ UTR) is a powerful regulatory process that determines the rate of protein translation from messenger RNA. Regulatory elements targeting the 3′ UTR comprise microRNAs, RNA-binding proteins and long non-coding RNAs, which dramatically alter the immune response. Here we provide an overview of our current understanding of post-transcriptional regulation of immune gene expression. The focus of this review will be on regulatory elements that target the 3′ UTR. We will delineate how the synergistic or antagonistic interactions of post-transcriptional regulators determine gene expression levels, and how dysregulation of 3′ UTR-mediated post-transcriptional control associates with human diseases.
Introduction
The human genome contains approximately 20,000–25,000 protein-coding genes, yet they comprise only 1–2 % of the genome. Recent data suggest the large proportion of the genome is also transcribed into noncoding RNA that potentially regulate cellular processes (1, 2). Precise control of gene expression is vital for the host to maintain homeostasis as well as to mount a rapid and effective immune response during infection. This becomes critical for genes associated with immune responses as they not only have to be rapidly induced in response to cellular stress, infection and inflammatory stimuli, but also quickly turned off to limit undesirable immune-pathology caused by persistent immune activation. For such precise control the cellular machinery has evolved regulators at several stages from transcription to translation fine-tuning gene expression. These include structural and chemical modifications of chromosomal DNA, transcriptional regulation, post-transcriptional control of messenger RNA (mRNA), varying translational efficiency and protein turnover. These mechanisms in concert determine the spatio-temporal control of genes to elicit an optimal immune response.
Messenger RNA is composed of a protein-coding region, and 5´ and 3 untranslated regions (UTRs). The 3′ UTR is variable in sequence and size; it spans between the stop codon and the poly(A) tail. Importantly, the 3′ UTR sequence harbors several regulatory motifs that determine mRNA turnover, stability and localization, and thus governs many aspects of post-transcriptional gene regulation. Over the past decade, it has become increasingly apparent that these regulatory motifs are critical in modulating the immune responses (3–5).
Immune genes are particularly good models to study post-transcriptional gene regulation since an adequate, properly-dosed immune response depends on their rapid but transient expression. The importance of post-transcriptional regulatory elements in these processes is evident from the evolution of multiple instability motifs and polymorphisms in the 3′ UTR of immune genes associated with pathogen pressure. Interleukins, interferons and chemokines encode mRNA instability motifs such as adenylate uridylate (AU)-rich elements (AREs), constitutive decay elements (CDEs) and stem loops, which are targeted by specific RNA binding proteins to destabilize the mRNA (6). These genes also harbor microRNA (miRNA) recognition elements (MREs), which fine-tune immune responses through more sequence-specific binding to the 3′ UTR (7). Recent evidence suggests that motifs in RNA secondary and tertiary structures interact with post-transcriptional regulators to dictate the transcript stability of immune genes. Dysregulated gene expression resulting from polymorphisms in 3′ UTR sequences or their interacting regulatory proteins have been associated with diseases, including cancer, infection and autoimmune disorders (5, 8, 9). Thus, it is clear that the “language” of the 3′ UTR needs to be decoded to comprehend and potentially engineer ideal immune responses.
This review will provide an integrated overview of the mechanisms and components that act in an additive, synergistic and/or antagonistic manner through interaction with the 3′ UTR of mRNA, hence defining the “post-transcriptional regulome” of the 3′ UTR for control of immune gene expression and its implications for immune-mediated diseases.
Immune regulation by microRNAs
Major players of the post-transcriptional regulation are endogenously encoded miRNAs. These small (20–25 nt) non-coding, single-stranded RNAs were first discovered in Caenorhabditis elegans in 1993 (10) and are distributed widely in eukaryotes. Since then the potential regulatory role for this class of small RNAs has become increasingly appreciated (11). Canonical miRNA binding to target mRNA is determined by the ‘seed region’ at the miRNA 5′ end (nucleotides 2 to 7/8), which perfectly matches the MRE in the 3′ UTR of target mRNA (12). A more recent study also demonstrated a mechanism of non-canonical miRNA-mRNA interaction that does not require perfect base pairing within the seed region, but depends instead on G-bulge sites within target mRNA (13).
During miRNA-dependent gene silencing a polyprotein complex, the miRNA-induced silencing complex (miRISC), is recruited by a miRNA to target mRNA. Two distinct mechanisms for miRISC-mediated silencing have been documented. Initially, binding of the miRISC to target mRNA was thought only to interfere with translation and protein synthesis by inhibiting ribosome assembly, interfering with translational initiation factors, or by blocking translation post-initiation. However, subsequent studies identified a major contribution of miRISC to mRNA deadenylation and degradation (14, 15).
Micro RNAs are important regulators of immune responses and are involved in nearly all aspects of the immune system, ranging from immune cell ontogeny to innate and adaptive immunity against infections. Chen and colleagues identified miR-181, miR-223 and miR-142 as modifiers of hematopoietic lineage differentiation (16). Furthermore, the crucial role of miRNAs in immune cell development has been demonstrated wherein T cell lineage specific deletion of Dicer, an essential enzyme for miRNA processing, results in impaired T cell development and a dysregulated CD4+ T cell cytokine signature (17, 18). Likewise, differentiation into B1 cells is controlled by miR-150, which is required to downregulate c-Myb expression (19).
Of several miRNAs key to modulating adaptive immune responses, miR-155 is one of the most prominent. SHIP1, a major regulator of the biology of various hematopoietic cells, is targeted by miR-155 through its 3′ UTR, with impacts on immune cell physiology, malignancies and autoimmune disorders (20, 21). The Bradley and Rajewsky groups also demonstrated that microRNA-155-deficient mice present impaired B and T cell immunity, caused by diminished activation of T cells through DCs, impaired germinal center responses due to decreased TNF levels in GC B cells, and increased c-Maf expression skewing T cell differentiation towards a Th2 phenotype (22, 23). miR-155 expression driven by Foxp3 is crucial for developing thymic regulatory T (Treg) cells, as it limits SOCS1 protein expression and thus indirectly increases sensitivity to IL-2 signaling required for Treg expansion (24). Gracias and colleagues have also found that miR-155 induced during primary CD8+ T cell activation renders the cells resistant to the antiproliferative effects of type I IFN, thus enabling establishment of effector memory. For a more comprehensive overview of literature on miR-155, its functions in immune cell biology and implications for autoimmunity please refer to these reviews (25, 26).
Various other studies reviewed by Baumjohann & Ansel highlight specific mechanisms of miRNA-mediated regulation of CD4+ T cell differentiation and plasticity (27). MicroRNA-182, which is induced in CD4+ T cells after stimulation with IL-2 regulates Foxo1 to promote clonal expansion (28). A study by Li et al. identified that miR-181a fine-tunes T cell sensitivity and selection during thymic development (29). Finally, miRNA targeting extends to effector cytokines such as interferon-gamma (IFNγ, IFNG), which harbors a conserved miR-29 MRE in its 3′ UTR. Several studies examined the role of miR-29 in regulating IFNG expression and identified miR-29 directly affecting IFNG mRNA stability or indirectly through targeting of TBET and EOMES mRNA (30, 31).
MicroRNAs are also important regulators of innate immune sensing pathways as initially shown by Baltimore’s group (32). They identified that miR-146 acts as a negative regulator of TLR4 signaling by targeting TLR adapters, TRAF6 and IRAK1, upon induction via its NF-κB-dependent promoter. Thus, miR-146 plays an important role in preventing excessive antimicrobial inflammatory responses. In line with these findings, miR-146a-/- mice develop spontaneous inflammation, which progresses with age and leads to the development of myeloid malignancies (33, 34). TLR signaling is also regulated by the microRNAs let-7i, miR-145, miR-155, and miR-346, which target receptors or downstream adapter molecules; of these, miR-155 correlates directly while let-7i correlates inversely with TLR signaling and immune response (35, 36). Two other studies show a role of miR-223 in granulocyte development and function, where miR-223 deficient mice displayed increased granulocyte numbers, hypersensitivity to stimulation and suppressed neutrophil activation (37, 38).
Sensing of pathogen-derived components by endosomal and cytosolic pattern recognition receptors induces innate immune responses. Expression of type I and type III IFNs is a hallmark of early innate immune response against viral infection. A variety of miRNAs regulate IFN-mediated immune responses by targeting IFN transcripts, the type I IFN receptor and/or downstream transcription factors (5). We have recently discovered that infection with hepatitis C virus induces expression of miR-208b and miR-499a-5p that target IFNL2 and IFNL3 genes (39). We have also found that these miRNAs target IFNAR1 mRNA and thus control responses to type I IFN (unpublished observations). Interestingly, while most miRNAs are endogenously encoded by the host genome, some viruses are known to encode their own viral miRNAs, which are predominantly involved in manipulation of the host immune responses (40, 41).
Since miRNAs fine-tune immune responses through control of immune gene expression, a dysregulated miRNA expression has been linked to autoimmune diseases. One of the best-studied miRNAs in this context is miR-146, whose expression is decreased in systemic lupus erythematosus (SLE) patients, leading to elevated levels of type I IFN, a key characteristic of SLE (42). In contrast, rheumatoid arthritis (RA) patients present higher miR-146 expression which in turn downregulates pro-inflammatory cytokines such as TNF-α and IL-17 (43). Other miRNA signatures distinguish the two diseases: miR-155 and miR-15a are increased in SLE mouse models and affect regulatory T cell activity and production of anti-dsDNA antibodies by B cells (43). miR-155 is also increased in RA patients alongside miR-132 and miR-16 (44). Ectopic expression of the miR-17-92 cluster in the lymphocyte compartment of mice results in lymphoproliferative disease and autoimmunity (45). A recent study shows that in addition to miR-146, miR-155 is also involved in regulation of chronic inflammation (21). Several excellent reviews discuss the roles of miRNAs in immune regulation and immune responses in more (5, 46–48) depth.
Altogether, these examples demonstrate the role of miRNAs as critical modulators of the immune system, controlling expression of genes involved in immune cell ontogeny, innate and adaptive immune responses, as well as autoimmunity. Future investigations into post-transcriptional regulation through miRNAs will help to identify novel miRNA targets and mechanisms of immune regulation.
Immune regulation by RNA-binding proteins
The 3′ UTRs harbor sequence or structural motifs that serve as recognition sites for RNA-binding proteins (RBPs) that can affect mRNA stability. The best-characterized RBP recognition motif is the adenylate-uridylate (AU)-rich element (ARE), which is found in 8–10 % of the human transcriptome. AREs can range from 40 to 150 nt in length and characteristically contain at least one AUUUA pentamer flanked by AU-rich sequence stretches (49–51). Almost three decades ago, the ARE motifs were first identified in human and mouse TNF genes (52). Shaw and Kamen then provided the first direct evidence that AREs influence mRNA stability by introducing an AU-rich sequence from the human GMCSF gene into the 3′ UTR of the rabbit beta-globin gene, which resulted in drastic decay of beta-globin mRNA (53). Several subsequent studies identified AREs in proto-oncogenes, transcription factors, interferons and cytokines, suggesting a major contribution of ARE-mediated decay to those genes (54).
Various motifs that facilitate interaction of RBPs with mRNAs have been identified including, but not limited to, CU-rich elements (CREs), GU-rich elements (GREs), repetitive C-rich sequences (CRSes), and constitutive decay element (CDEs), all of which determine mRNA stability (6). Classically, RBP-mediated decay begins with binding of a RBP to its respective target mRNA leading to recruitment of deadenylases to remove the poly(A) tail followed by 3′ to 5′ exonucleolytic mRNA degradation, a process called ARE-mediated decay (AMD) when it occurs via an ARE motif. RBPs have also been shown to mediate 5′ decapping and 5′ to 3′ mRNA degradation, and conversely can also enhance mRNA stability by protecting it from other decay proteins.
One of the best-studied RBPs targeting AREs is tristetraprolin (TTP). Mouse and human TTP were first described in the early 1990s, as proteins encoded by the Zfp36/ZFP36 gene (55). TTP and similar RBPs contain two tandem repeats of zinc finger motifs that enable direct interaction with AREs within the 3′ UTR of target mRNAs. TTP was first shown to act by recruiting the exosome to ARE-mRNAs to cause 3′ to 5′ exonucleolytic mRNA decay (56). However, Lykke-Andersen and Wagner observed that TTP also interacts with enzymes involved in mRNA decapping and 5′ to 3′ exonuclease activity, suggesting additional mechanisms of TTP-mediated mRNA decay (57). More recently, another study demonstrated that TTP also interacts with the CCR4-CAF1-NOT deadenylation complex, possibly facilitating poly(A) tail deadenylation (58). The importance of TTP in gene regulation is evident as TTP-/- mice develop a severe systemic inflammatory syndrome with patchy alopecia, dermatitis, erosive arthritis, cachexia, conjunctivitis, myeloid hyperplasia, glomerular mesangial thickening, and antinuclear antibodies within 8 weeks after birth (59). Post-transcriptional regulation by TTP is important in controlling the expression of various cytokine genes, especially inflammatory cytokines like TNF, GMCSF, IFNG, IL10, IL12, IL17, IL23, CCL3, and CXCL1 (60–69). Furthermore, linkage of a polymorphism in the ZFP36 gene with development of rheumatoid arthritis (RA) has been reported (70). However, the genetic risk score for this polymorphism in RA is low and this phenotype occurs only in a subset of analyzed individuals and appears to be specific to their ethnicity. The same class of zinc finger proteins includes the TTP paralogs butyrate response factors 1 and 2 (BRF1 and BRF2). Of these, BRF1 has been shown with a functional cDNA library cloning approach to stabilize IL3, IL6, GMCSF, and TNF transcripts in an ARE-dependent manner (71, 72).
KH-type splicing regulatory protein (KSRP) is another RBP important for immune regulation, especially in controlling cytokine expression. Similar to TTP, KSRP participates in exosome-mediated mRNA decay of immune mediators like TNF, CXCL2, CXCL3, IL2, IL6, and IL8 (56, 73, 74). KSRP also controls Ifna4 and Ifnb expression such that its absence increases resistance to viral infection due to rescue of IFN levels (75). Besides its role in post-transcriptional control of immune genes, KSRP participates in miRNA maturation and processing (76) by forming a complex with Drosha and Dicer, and interacting with the terminal loop of target precursor miRNA.
Three zinc-finger RBPs have been recently identified which destabilize immune genes. Interaction of the zinc finger protein roquin-1 (RC3H1) with the 3′ UTR of Icos mRNA limits Icos expression in T cells (77, 78). Srivastava et al. identified a novel role for roquin in miRNA homeostasis through direct interaction with Argonaute2 (AGO2), a central component of the miRISC. They also showed that roquin controls levels of miR146a, a miRNA targeting Icos mRNA, which may explain another way in which roquin modulates Icos (79). Mutation of Rc3h1 in sanroque mice results in an autoimmune disorder caused by accumulation of lymphocytes (80) and increased TNF-α levels (81). The roquin-1 paralog roquin-2 (Rc3h2) has a redundant role in Icos mRNA downregulation (82, 83). Similar to roquin and TTP, regulatory RNase 1 (regnase-1)-deficient mice develop a systemic inflammatory phenotype, which is caused by increased production of IL-6 and IL-12p40 (84). Furthermore, regnase-1 constitutively regulates expression of c-Rel, Ox40, and Il2 in naïve T cells through 3′ UTR targeting and cleavage of mRNA (85). Upon triggering of the T cell receptor, the Malt1 paracaspase cleaves regnase-1 to relieve suppression of its targeted genes and thus allow timely effector T cell activation and expansion (85, 86). Interestingly, although regnase-1 and roquin share a specific set of target mRNAs, a recent study demonstrated differential mRNA specificity between the two which depends on subcellular localization and translational status of the respective target mRNA (87).
The human antigen R (HuR) identified in 1996 (88) is a ubiquitously expressed member of the embryonic lethal abnormal vision (ELAV)-like protein family that harbors three RNA-binding domains that interact with AREs. In contrast to the destabilizing RBPs mentioned above, HuR overexpression stabilizes many ARE-containing mRNA transcripts (89, 90), including TNF (91, 92), suggesting competition between RBPs for mRNA binding and a dynamic interplay of post-transcriptional regulatory elements. For example, Ogilvie and coworkers proposed a model of direct competition for IL2 mRNA binding by HuR and TTP. Binding of HuR to the mRNA prevents TTP-mediated recruitment of the exosome to the transcript (93). Furthermore, dysregulation of the HuR-TTP equilibrium has been shown to drive colon carcinogenesis, in which loss of TTP expression and simultaneous elevated expression of HuR greatly increases tumorigenic COX-2 expression (94). In recent years, however, it has been found that HuR can also destabilize mRNA mainly through concerted actions with the miRISC (95). Thus the effect of HuR on mRNA stability depends on the composite context of regulatory elements on the 3′ UTR. Likewise, AU-rich element-binding protein 1 (AUF1) can either stabilize or destabilize immune gene transcripts in a cell-type dependent manner that is not completely understood. AUF1 controls post-transcriptional regulation of numerous immune mediators like IL-3, IL-10, TNF-α, and GM-CSF (96–100), as well as the miRNA processing machinery by targeting and repressing DICER1 (101). A recent study established that AUF1 interacts with the miRISC through AGO2 in either a cooperative or reciprocal manner depending on which target mRNA is associated, which could partly explain context-dependent differential effects of AUF1 (102).
Some RBPs have been shown to interact with each other or compete for binding to the same recognition element, thereby having synergistic or antagonistic effects on stability of targeted mRNA (6). For instance, roquin and regnase-1 cooperate to regulate Th17 cell differentiation and expression of Th17 cell-promoting factors like IL-6 and ICOS (86). The dynamics of such RBP interactions and their effects on gene expression are determined by tissue- or cell-type-specific expression of respective RBPs as well as by external stimuli that alter abundance or activity of certain RBPs. Notably, TTP levels and its extensive regulatory activity are altered by external stimuli. To start with, the p38 mitogen-activated protein kinase (MAPK) pathway is a critical regulator of TTP expression and activity (103, 104). MK-2 mediated phosphorylation of TTP which occurs after cytokine exposure and other stress-inducing external stimuli leads to binding of 14-3-3 proteins, exclusion of TTP from stress granules, and thereby loss of TTP-dependent suppression of gene expression (105, 106). Fine-tuning of TTP activity through MK2-mediated phosphorylation stabilizes Tnf mRNA as it decreases TTP affinity to the ARE and its ability to compete with Tnf-stabilizing HuR for the same AREs (107).
The role of RBPs as immune regulators needs to be further explored in order to identify additional RBPs involved in immune regulation, delineate RBP mechanisms of mRNA degradation/stabilization, and identify novel RBP-mRNA interaction motifs.
Genetic variation within the 3′ UTR modulates post-transcriptional regulation in disease
Single-nucleotide polymorphisms (SNPs) of a gene are frequently associated with human diseases (39, 108–114). A single nucleotide change in the 3′ UTR can result in dysregulated post-transcriptional regulation. First, alterations in MRE sequences can change the affinity of miRNA-mRNA interaction. Functional polymorphisms located directly within destabilization motifs have been reported for MREs. These SNPs disrupt the interaction sites for miRNA binding, which usually leads to stabilization of the mRNA transcript and increased protein levels. Such miRNA target site polymorphisms have been linked to immune-associated diseases like cancer (113).
Genetic variation within the HLA-C gene associates with control of HIV infection (115) and HLA-C mRNA levels as well as cell surface expression (116). We revealed a causal relation between variations within the 3′ UTR of HLA-C mRNA and its effect on HIV control (117). Variation of the HLA-C 3′ UTR determines binding by miR-148a, directly affecting HLA-C expression and control of HIV. The SNP (rs10889677) in the IL23R 3′ UTR associates with inflammatory bowel diseases (IBD), caused by disruption of the MRE for let-7e and let-7f resulting in increased IL-23R expression (118).
Single nucleotide polymorphisms can also change the secondary and tertiary structure of the mRNA by changing base pair complementarity affecting stem-loop formation (119). Since 3′ UTR stem-loops act as scaffolds for RBP-mRNA interaction, such changes may disrupt interaction sites for RBPs and affect mRNA stability (120). We identified a functional SNP, rs4803217, within the 3′ UTR of IFNL3 previously tagged as one of the strongest predictors of natural and therapy-induced HCV clearance. This SNP dictates the turnover of IFNL3 mRNA by influencing the extent of both AMD and miRNA-mediated decay (39). We have identified the HCV-induced miRNAs dictating IFNL3 mRNA instability (see above) and that rs4803217 allows escape of miRNA-mediated decay to increase IFNL3 mRNA expression. However, how the 3′ UTR SNP influences IFNL3 AMD and what regulatory components are involved are still unclear.
Taken together, genetic variation within the 3′ UTR of immune genes is a strong determinant of immune response. Sequence variations can disrupt binding sites for miRNAs and/or RBPs, altering their ability to regulate transcripts (as summarized in Figure 1). New sequencing technologies have advanced the investigations to understand these interactions and mechanisms behind the many disease polymorphisms in non-coding regions.
Figure 1. The post-transcriptional regulome of the 3′ UTR.
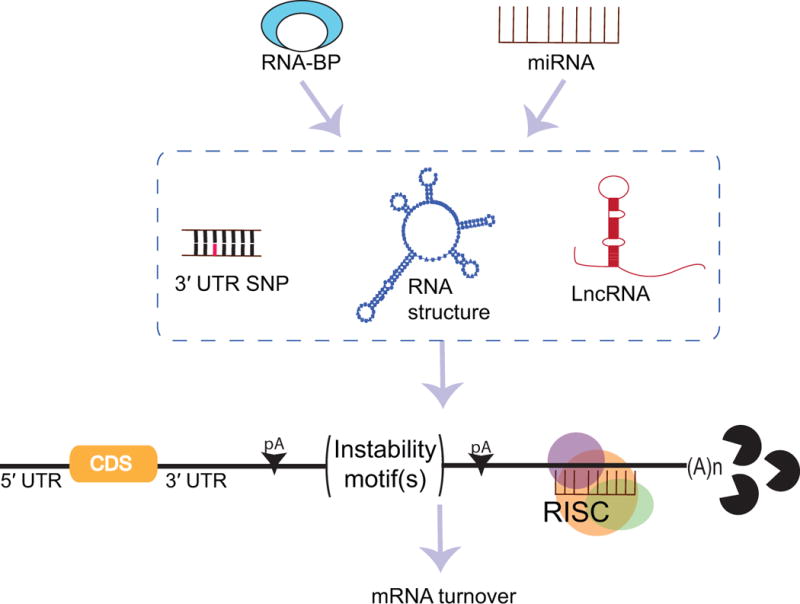
mRNA stability and gene expression is dictated by various post-transcriptional modulators interacting with the 3′-untranslated region (UTR). microRNAs (miRNAs) recruit the RNA-induced silencing complex (RISC) to specific target regions leading to ribonuclease-mediated mRNA decay. Similarly, instability motifs located within the 3′ UTR are targeted by RNA-binding proteins (RBPs), resulting in rapid poly(A) tail deadenylation and mRNA degradation, or in mRNA stabilization, respectively. Single nucleotide polymorphisms (SNPs) in the 3′ UTR disrupt the nucleotide complementarity needed for miRNA-mRNA interaction hence altering the binding capacity of miRNAs. In a similar fashion, SNPs can change the overall mRNA structure or particular instability motifs required for efficient RBP-mRNA interaction. Long non-coding RNAs (lncRNAs) are overarching modifiers of miRNA and RBP activity through sequestration, thereby suppressing their function. Finally, shortening of the 3′ UTR through usage of alternative polyadenylation sites (pA) affects overall mRNA stability by decreasing the number of potential interaction sites/motifs for the previously mentioned post-transcriptional modulators. These regulatory elements acting in sync define the post-transcriptional regulome of the 3′ UTR and ultimately dictate mRNA turnover and expression of a given gene.
Polyadenylation and 3′ UTR shortening dictate mRNA function and turnover
Alternative polyadenylation (APA) presents a powerful mechanism to modulate the strength of 3′ UTR-mediated regulation. Several polyadenylation sites can be found in a single 3′ UTR in a majority of human genes, giving rise to different isoforms of a specific gene transcript (121, 122). APA alters the length of the 3′ UTR, which may affect mRNA stability, localization and/or translation of the isoforms (123). Messenger RNAs with shorter 3′ UTRs are generally more stable than those with longer 3′ UTRs as they encode fewer regulatory elements allowing them to escape regulation (124). Regulation of APA of an mRNA transcript thereby controls mRNA stability and protein levels.
Alternative polyadenylation has been shown to be involved in B lymphocyte differentiation and switch from membrane-bound to secreted IgM. B cell protein levels of the essential polyadenylation factor CSTF2 affect alternative processing of IgM mRNA through differential usage of poly(A) sites (125). Low levels of CSTF2 in early-stage B cells promotes cleavage at the distal IgM poly(A) site leading to expression of membrane-bound IgM. During B cell maturation, increasing concentrations of CSTF2 skews cleavage site selection towards a more proximal intronic poly(A) site resulting in expression of secreted IgM. A similar role for APA and involvement of CSTF2 has also been shown in effector T cell maturation (126). Naïve T cells express two mRNA isoforms of the transcription factor NFATc at low levels. However, after differentiation into effector T lymphocytes and second exposure to antigens, T cells rapidly start expressing high amounts of a third, shorter NFATc isoform. This change is caused by increased CSTF2 levels, which lead to usage of a proximal alternative polyadenylation site in the NFATc mRNA.
Alternative polyandenylation can also precipitate profound immune dysfunction and pathology. Genetic variation within the IRF5 gene has been attributed to increased risk of SLE development (111, 127). One of three SNPs identified by Graham et al. affects IRF-5 protein levels and associates with high risk for SLE. This SNP creates an APA signal, hence increasing the stability of IRF5 mRNA by truncating the 3′ UTR (128). Similarly, 3′ UTR variants have been discovered in a diverse subset of other genes such as TREX1, TLR7, CD247 that also associate with SLE risk through screening of SLE patient cohorts (129–131). A mutation in the single poly(A) site of FOXP3 mRNA has been associated with development of immune dysfunction, polyendocrinopathy, enteropathy, X-linked (IPEX) (109). Here, a single A to G mutation in the hexameric polyadenylation signal results in impaired polyadenylation, failure to terminate transcription and decreased FOXP3 expression. In the case of proto-oncogenes, 3′ UTR shortening through APA is associated with development of cancer (132).
Thus, APA presents an efficient and powerful mechanism by which mRNA stability can be transiently increased within short time frame through shortening of its 3′ UTR. However, we still do not know how often APA is used to regulate gene expression in immune cells. Advancement in deep sequencing technologies will determine the extent of APA in immune genes under steady state and activated conditions.
Long non-coding RNAs are sinks for post-transcriptional regulators
Recently discovered long non-coding RNAs (lncRNAs) are generally classified as non-coding RNAs with a size of more than 200 nucleotides. The role of lncRNAs as overarching modifiers of gene expression became more evident in the last decade as thousands of novel lncRNA transcripts were identified by improved sequencing technology (133). lncRNAs interact with mRNAs, miRNAs and RBPs to modulate mRNA splicing, mRNA stability and translation, thus adding complexity to the post-transcriptional regulatory mechanisms determining gene expression and protein output. lncRNAs compete with miRNAs for binding to target mRNA, thus masking miRNA binding sites and repressing miRISC-mediated mRNA decay (134). Similarly, lncRNAs also interfere with miRNA-mediated gene regulation by acting as sponges to sequester miRNAs (135). Furthermore, lncRNAs have also been shown to alter mRNA stability by recruitment of RBPs to the 3′ UTR (136). Several outstanding reviews discuss the contribution of lncRNAs to the overall post-transcriptional regulatory machinery in more detail (137, 138).
Post-transcriptional regulome – interaction between regulators of the post-transcriptional machinery dictates gene expression
Rather than considering each of the above-mentioned regulatory elements individually, we propose a more dynamic model of post-transcriptional gene regulation, including synergistic or antagonistic interaction of several of these regulators. Jing et al. were first to show that miRNAs and RBPs can mediate mRNA degradation in a cooperative manner (139). While this study uncovered that TTP-mediated TNF mRNA decay requires both miR-16 processing by Dicer as well as presence of Argonaute, direct binding of miR-16 to TTP was not found. The authors suggested an indirect interaction of miR-16 with TTP through association and complex formation with Argonaute family members (139). Later, miR-221 was found to associate with TTP to facilitate TNF mRNA decay (140). Furthermore, we have shown that IFNL3 mRNA stability is determined by cooperative actions of miRNAs and RBPs targeting AREs in the IFNL3 3′ UTR (39). On the other hand, miRNAs can also directly compete with RBPs for 3′ UTR binding. For example, the seed region of miR-466l is complementary to the pentameric AUUUA sequence of the ARE. In the presence of miR-4661, IL10 mRNA and protein increase (141), as miR-466l competes with TTP for the AUUUA motif and prevents TTP-mediated degradation. miR-29 has also been identified as a stabilizer of tumor suppressor A20 mRNA through interaction with other post-transcriptional elements where it acts as a RNA decoy for the RBP HuR, thus preventing A20 mRNA decay (142). Additionally, we have uncovered a similar mechanism by which miR-29 stabilizes gene expression of IFNG through competitive binding to RBP recognition elements. To study the simultaneous effects of miR-29 and AREs on IFNG mRNA stability, we retained RNA structural integrity by using constructs containing the complete 3′ UTR sequence. We observed that miR-29 stabilizes IFNG expression in presence of the tristetraprolin (TTP) complex (unpublished observation). The AU-rich elements (AREs) targeted by TTP for mRNA decay are in close proximity to the miR-29 binding site in the secondary mRNA structure. We propose that this prevents the recruitment of GW182 so that miRISC cannot degrade the transcript. Although miR-29 has been shown enhance degradation of IFNG (31), these studies used partial UTR sequences lacking the AREs and thus may overlook crucial interactions between miR-29 and TTP. Thus, overall miR-29 acts as a stabilizer of IFNG mRNA through its antagonism of ARE mediated decay (AMD).
Another prominent example of interaction between miRNAs and RBPs in gene expression control comprises the Lin28/let-7 axis and its role in hematopoiesis and cancer. The RBPs Lin28a and Lin28b bind to pre-let-7 to inhibit its processing by Drosha and Dicer (143). Lin28a/b also facilitate oligo-uridylation of pre-let-7 by TUT4, leading further to its decay (144). Yuan and colleagues highlighted the importance of Lin28/let7 balance in the developing immune system when they found Lin28b expressed in fetal but absent in adult lymphocyte progenitor cells, and that Lin28b in these cells inversely correlated with expression levels of let-7 family members. Ectopic expression of Lin28 in bone-marrow hematopoietic stem/progenitor cells represses let-7 miRNAs and allows multi-lineage reconstitution (145).
Many examples given in previous sections of this review combine diverse simultaneous post-transcriptional regulatory elements, which can either compete or cooperate. Given this ample evidence that the interplay of multiple factors can result in a vastly different phenotype from the actions of individual components, integrating these regulatory pathways whilst studying these regulations will provide a complete picture of the 3′ UTR “regulome” of the gene.
Conclusions
Post-transcriptional regulation through the 3′ UTR is a potent mechanism to control development and homeostasis of the immune system and to quickly adjust expression of immune genes upon stimulation by cell-intrinsic or -extrinsic cues. In this review we discussed different mechanisms by which post-transcriptional regulation through the 3′ UTR of mRNA transcripts occurs. miRISC and AMD are the most powerful regulatory elements dictating mRNA stability and/or degradation. However, alteration of 3′ UTR sequence, structure and length by SNPs and alternative polyadenylation add an additional level of complexity to the actions of miRISC and AMD. Furthermore, lncRNAs interact directly with mRNA, miRNA and RBPs to modify their individual activity as an additional layer of control (summarized in Fig. 1).
Not surprisingly, a number of immune-mediated diseases are associated with genetic variation within 3′ UTR elements of immune genes (Figure 2). Identification of novel SNPs within the 3′ UTR allowed deeper insights into the linkage between post-transcriptional dysregulation and human disease. Such variations can affect mRNA localization, stability and translation efficiency, all of which ultimately dictate expression of a given gene. Future investigations should be aimed at the identification of novel post-transcriptional regulatory elements, their specific mode(s) of action, and a detailed understanding of how multiple elements act in synergy and/or antagonism. Deep-sequencing approaches and mass spectrometry will help to uncover novel RNA-RBP interactions and provide mechanistic explanations for genetic data. Furthermore, RNA-seq-based approaches will unravel novel miRNAs and lncRNAs and their possible target sites within specific 3′ UTRs. A recently published study by Zhao et al. presents an excellent example of how novel sequencing approaches massively improve identification of novel cis-acting regulatory elements in the 3′ UTR and how genetic variation within these elements affects mRNA stability (146).
Figure 2. Post-transcriptional regulation of the 3′ UTR and its implications for immune disorders.
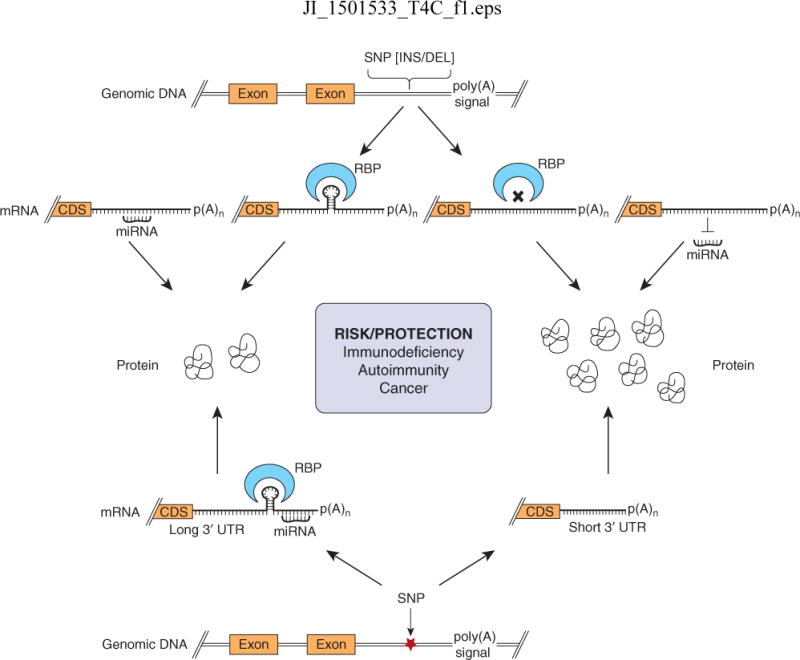
Insertion (INS) or deletion (DEL) mutations in the 3′ UTR of the gene could create or destroy binding sites for regulators such as RNA binding proteins (RBPs) and microRNAs (miRNAs). This could lead increased or decreased mRNA stability and protein levels. Likewise, single nucleotide polymorphisms (SNPs) in the 3′ UTR can create new alternative polyadenylation signals or destroy poly(A) signals altering the length of the 3′ UTR affecting the stability of mRNA. Elimination of interaction motifs/sequences for RBPs and miRNAs leads to increased mRNA stability and protein levels. These SNPs affect post-transcriptional control of genes involved in innate and adaptive immunity. Hence, carriers of the risk or protective alleles can be more likely to develop or resist immune disorders like immunodeficiency, autoimmunity or cancer.
A better understanding of the complex interplay of regulatory elements interacting with the 3′ UTR will help to link immune disorders to dysregulation of post-transcriptional mechanisms. In addition, it may pave the way for development of novel therapeutic approaches and the design of genotype-specific, tailored treatment for immune diseases.
Acknowledgments
The authors thank Abigail P. Jarret, Chrissie Lim, Adelle P. McFarland, Snehal S. Ozarkar and Justin A. Roby for critical reading of the manuscript. The authors apologize to all whose work could not be cited due to space limitations.
Funding: This work was supported in part by 1R01AI108765 (R.S.).
References
- 1.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium EP. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 4.Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol. 2014;15:492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- 5.Savan R. Post-transcriptional regulation of interferons and their signaling pathways. J Interferon Cytokine Res. 2014;34:318–329. doi: 10.1089/jir.2013.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov P, Anderson P. Post-transcriptional regulatory networks in immunity. Immunol Rev. 2013;253:253–272. doi: 10.1111/imr.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 8.Jia J, Yao P, Arif A, Fox PL. Regulation and dysregulation of 3′UTR-mediated translational control. Current opinion in genetics & development. 2013;23:29–34. doi: 10.1016/j.gde.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J Interferon Cytokine Res. 2005;25:1–10. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nature structural & molecular biology. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 16.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 17.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 20.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, Dongen S van, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 24.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng RX, Pan HF, Qin WZ, Chen GM, Ye DQ. Role of microRNA-155 in autoimmunity. Cytokine Growth Factor Rev. 2011;22:141–147. doi: 10.1016/j.cytogfr.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 27.Baumjohann DK, Ansel M. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 29.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 32.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao JL, Rao DS, Boldin MP, Taganov KD, O’Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cellular & molecular immunology. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 37.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 38.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 39.McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro Ba, Delker Da, Hagedorn CH, Carrington M, Gale M, et al. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nature Immunology. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 41.David M. Interferons and microRNAs. J Interferon Cytokine Res. 2010;30:825–828. doi: 10.1089/jir.2010.0080. [DOI] [PubMed] [Google Scholar]
- 42.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, Vries N de, Tak PP, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis and rheumatism. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 43.Ceribelli A, Satoh M, Chan EK. MicroRNAs and autoimmunity. Curr Opin Immunol. 2012;24:686–691. doi: 10.1016/j.coi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis research & therapy. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen CZ, Schaffert S, Fragoso R, Loh C. Regulation of immune responses and tolerance: the microRNA perspective. Immunol Rev. 2013;253:112–128. doi: 10.1111/imr.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 48.Schwerk J, Jarret AP, Joslyn RC, Savan R. Landscape of post-transcriptional gene regulation during hepatitis C virus infection. Curr Opin Virol. 2015;12:75–84. doi: 10.1016/j.coviro.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asson-Batres MA, Spurgeon SL, Diaz J, DeLoughery TG, Bagby GC., Jr Evolutionary conservation of the AU-rich 3′ untranslated region of messenger RNA. Proc Natl Acad Sci U S A. 1994;91:1318–1322. doi: 10.1073/pnas.91.4.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillis P, Malter JS. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991;266:3172–3177. [PubMed] [Google Scholar]
- 52.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 54.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends in biochemical sciences. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 55.Taylor GA, Lai WS, Oakey RJ, Seldin MF, Shows TB, Eddy RL, Jr, Blackshear PJ. The human TTP protein: sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Res. 1991;19:3454. doi: 10.1093/nar/19.12.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 57.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & development. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 60.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 61.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 62.Datta S, Biswas R, Novotny M, Herjan PG, Pavicic T, Jr, Mandal P, Hamilton TA. Tristetraprolin regulates CXCL1 (KC) mRNA stability. Journal of immunology. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 63.Gu L, Ning H, Qian X, Huang Q, Hou R, Almourani R, Fu M, Blackshear PJ, Liu J. Suppression of IL-12 production by tristetraprolin through blocking NF-kcyB nuclear translocation. Journal of immunology. 2013;191:3922–3930. doi: 10.4049/jimmunol.1300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang JG, Amar MJ, Remaley AT, Kwon J, Blackshear PJ, Wang PY, Hwang PM. Zinc finger protein tristetraprolin interacts with CCL3 mRNA and regulates tissue inflammation. Journal of immunology. 2011;187:2696–2701. doi: 10.4049/jimmunol.1101149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee HH, Yoon NA, Vo MT, Kim CW, Woo JM, Cha HJ, Cho YW, Lee BJ, Cho WJ, Park JW. Tristetraprolin down-regulates IL-17 through mRNA destabilization. FEBS Lett. 2012;586:41–46. doi: 10.1016/j.febslet.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 66.Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, Van Simaeys G, Blackshear PJ, Leo O, Goriely S. Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J Exp Med. 2013;210:1675–1684. doi: 10.1084/jem.20120707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogilvie RL, Sternjohn JR, Rattenbacher B, Vlasova IA, Williams DA, Hau HH, Blackshear PJ, Bohjanen PR. Tristetraprolin mediates interferon-gamma mRNA decay. J Biol Chem. 2009;284:11216–11223. doi: 10.1074/jbc.M901229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian X, Ning H, Zhang J, Hoft DF, Stumpo DJ, Blackshear PJ, Liu J. Posttranscriptional regulation of IL-23 expression by IFN-gamma through tristetraprolin. Journal of immunology. 2011;186:6454–6464. doi: 10.4049/jimmunol.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carrick DM, Chulada P, Donn R, Fabris M, McNicholl J, Whitworth W, Blackshear PJ. Genetic variations in ZFP36 and their possible relationship to autoimmune diseases. J Autoimmun. 2006;26:182–196. doi: 10.1016/j.jaut.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 71.Stoecklin G, Stoeckle P, Lu M, Muehlemann O, Moroni C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001;7:1578–1588. [PMC free article] [PubMed] [Google Scholar]
- 72.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Molecular and cellular biology. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 75.Lin WJ, Zheng X, Lin CC, Tsao J, Zhu X, Cody JJ, Coleman JM, Gherzi R, Luo M, Townes TM, et al. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Molecular and cellular biology. 2011;31:3196–3207. doi: 10.1128/MCB.05073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, Kremmer E, Wang X, Heissmeyer V. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- 78.Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 79.Srivastava M, Duan G, Kershaw NJ, Athanasopoulos V, Yeo JH, Ose T, Hu D, Brown SH, Jergic S, Patel HR, et al. Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis. Nat Commun. 2015;6:6253. doi: 10.1038/ncomms7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 81.Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 82.Pratama A, Ramiscal RR, Silva DG, Das SK, Athanasopoulos V, Fitch J, Botelho NK, Chang PP, Hu X, Hogan JJ, et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 83.Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, Zoller J, Warth SC, Hoefig KP, Lohs C, et al. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity. 2013;38:655–668. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 84.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 85.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 86.Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 87.Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, et al. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell. 2015;161:1058–1073. doi: 10.1016/j.cell.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 88.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 89.Brennan CM, Steitz JA. HuR and mRNA stability. Cellular and molecular life sciences: CMLS. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Di Marco S, Hel Z, Lachance C, Furneaux H, Radzioch D. Polymorphism in the 3′-untranslated region of TNFalpha mRNA impairs binding of the post-transcriptional regulatory protein HuR to TNFalpha mRNA. Nucleic Acids Res. 2001;29:863–871. doi: 10.1093/nar/29.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Molecular and cellular biology. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. Journal of immunology. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 94.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Current protein & peptide science. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brewer G, Saccani S, Sarkar S, Lewis A, Pestka S. Increased interleukin-10 mRNA stability in melanoma cells is associated with decreased levels of A + U-rich element binding factor AUF1. J Interferon Cytokine Res. 2003;23:553–564. doi: 10.1089/107999003322485053. [DOI] [PubMed] [Google Scholar]
- 97.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes & development. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Molecular and cellular biology. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Molecular and cellular biology. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abdelmohsen K, Tominaga-Yamanaka K, Srikantan S, Yoon JH, Kang MJ, Gorospe M. RNA-binding protein AUF1 represses Dicer expression. Nucleic Acids Res. 2012;40:11531–11544. doi: 10.1093/nar/gks930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu X, Chesoni S, Rondeau G, Tempesta C, Patel R, Charles S, Daginawala N, Zucconi BE, Kishor A, Xu G, et al. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carballo E, Cao H, Lai WS, Kennington EA, Campbell D, Blackshear PJ. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J Biol Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Molecular and cellular biology. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J Biol Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 107.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS genetics. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D, Torkvist L, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS genetics. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 110.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Escribano Gonzalez MF, Argentine et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 112.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, KZhang C, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends in genetics: TIG. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 114.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 115.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thomas R, Apps R, Qi Y, Gao X, Male V, O’HUigin C, O’Connor G, Ge D, Fellay J, Martin JN, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, Martin MP, Hunt P, Deeks SG, Telenti A, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011:1–5. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zwiers A, Kraal L, van de Pouw Kraan TC, Wurdinger T, Bouma G, Kraal G. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. Journal of immunology. 2012;188:1573–1577. doi: 10.4049/jimmunol.1101494. [DOI] [PubMed] [Google Scholar]
- 119.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS genetics. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 121.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature reviews. Genetics. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 124.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 126.Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10:261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- 127.Sigurdsson S, Nordmark G, Goring HH, Lindroos K, Wiman AC, Sturfelt G, Jonsen A, Rantapaa-Dahlqvist S, Moller B, Kere J, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. American journal of human genetics. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shen N, Fu Q, Deng Y, Qian X, Zhao J, Kaufman KM, Wu YL, Yu CY, Tang Y, Chen JY, et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010;107:15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsuzaka K, Fukuhara I, Setoyama Y, Yoshimoto K, Suzuki K, Abe T, Takeuchi T. TCR zeta mRNA with an alternatively spliced 3′-untranslated region detected in systemic lupus erythematosus patients leads to the down-regulation of TCR zeta and TCR/CD3 complex. Journal of immunology. 2003;171:2496–2503. doi: 10.4049/jimmunol.171.5.2496. [DOI] [PubMed] [Google Scholar]
- 131.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, Silva U de, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 132.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Faghihi MA, Zhang M, Huang J, Modarresi F, Brug MP, Van der Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol. 2014;26:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 140.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem. 2010;285:20940–20951. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ma F, Liu X, Li D, Wang P, Li N, Lu L, Cao X. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. Journal of immunology. 2010;184:6053–6059. doi: 10.4049/jimmunol.0902308. [DOI] [PubMed] [Google Scholar]
- 142.Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, London CA, Kraybill W, Perrotti D, Croce CM, et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal. 2013;6:ra63. doi: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 145.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. Massively parallel functional annotation of 3′ untranslated regions. Nature biotechnology. 2014;32:387–391. doi: 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]


