Abstract
The lysine (K)-specific demethylase (LSD1) family of histone demethylases regulates chromatin structure and the transcriptional potential of genes. LSD1 is frequently deregulated in tumors, and depletion of LSD1 family members causes developmental defects. Here, we report that reductions in the expression of the Pumilio (PUM) translational repressor complex enhanced phenotypes due to dLsd1 depletion in Drosophila. We show that the PUM complex is a target of LSD1 regulation in fly and mammalian cells and that its expression is inversely correlated with LSD1 levels in human bladder carcinoma. Unexpectedly, we find that PUM posttranscriptionally regulates LSD1 family protein levels in flies and human cells, indicating the existence of feedback loops between the LSD1 family and the PUM complex. Our results highlight a new posttranscriptional mechanism regulating LSD1 activity and suggest that the feedback loop between the LSD1 family and the PUM complex may be functionally important during development and in human malignancies.
INTRODUCTION
Chromatin has a fundamental role in regulating the transcriptional potential of each gene within the genome. The basic unit of chromatin is the nucleosome, which consists of DNA wrapped around a histone octamer. Dynamic posttranslational modifications of histones influence the accessibility of chromatin and the expression of transcripts (1). Aberrant patterns of chromatin modifications are strongly associated with a wide variety of human diseases, including cancer (2). One such modification, the methylation of lysine residues on histone tails, is instrumental in regulating gene transcription and is required for development and tissue differentiation (3). Histone methylation is dynamically controlled by the antagonistic activity of histone methyltransferases and demethylases.
To date, two families of histone lysine demethylases have been identified, the Jumonji domain-containing (JmjC) protein and the lysine (K)-specific demethylase 1 (KDM1) protein. The human KDM1 family of histone demethylases, LSD1 (KDM1A) and LSD2 (KDM1B), catalyze the demethylation of mono- and dimethyl marks of lysines 4 (K4) and 9 (K9) of histone H3 (4–7). This dual activity enables LSD1 to regulate both the repression and activation of genes. When associated with the REST corepressor (coREST) or the Mi-2/nucleosome remodeling and deacetylase (NuRD) complexes, LSD1 promotes gene silencing by removing activating methyl marks from H3K4 (8). In contrast, when LSD1 interacts with the androgen or estrogen receptor, it promotes transcriptional activation by demethylating the repressive H3K9me2 (dimethylation of histone H3 at K9) histone modification (2); these findings suggest that LSD1 has a context-dependent effect on transcription.
The KDM1 family of proteins has important roles during development. Mutation of the gene for the sole member of the family in Drosophila, Lsd1 [dLsd1; also known as Su(var)3-3], results in tissue-specific phenotypes, including wing defects and germ line abnormalities (9). dLsd1 mutant flies have an abnormal number of germ line stem cells and follicle cells due to incorrect differentiation, resulting in rudimental ovaries and sterility (9–11). In mammalian cells, both LSD1 and LSD2 (LSD1/2) are required for cellular differentiation (12–14) and DNA imprinting (15).
Aberrant LSD1 levels have also been identified in a number of human malignancies ranging from prostate cancer and bladder carcinoma to acute myeloid leukemia (8). However, very little is known about how the levels of the LSD1 family are regulated in vivo and about how changes in the levels of LSD1/2 contribute to developmental processes and human diseases, such as cancer.
To identify novel modulators of LSD1 activity in vivo, we screened for modifiers of dLsd1-RNA interference (RNAi) wing phenotypes in Drosophila. Among the hits of the screen, we found that RNAi of two components of the Pumilio translation repressor complex, Pumilio (Pum) and brain tumor (Brat), significantly enhanced dLsd1-RNAi phenotypes.
The Pumilio posttranscriptional repressor complex in Drosophila is comprised of two RNA-binding proteins, Pum and Nanos (Nos), and a trim-like protein, Brat. The Pum complex binds to a Nanos regulatory element (NRE) (UGUAXAUA) in the 3′ untranslated region (UTR) of its substrates and represses their translation (16–18). In mammalian systems, two homologs of the PUM (19, 20) and three of the NANOS (21) genes have been identified. The PUM complex mediates translational silencing via a number of complex mechanisms including deadenylation (22), mRNA decapping (23), and microRNA (miRNA) recruitment (24–26) or by blocking translation initiation (27, 28). PUM regulation is conserved throughout evolution, and PUM homologs have essential roles in regulating cell cycle progression, stress responses, and differentiation (29–31). In addition to these roles, the PUM complex has an important evolutionarily conserved function in maintaining stem cell pluripotency (32–34) and tissue specification (35).
Here, we provide genetic evidence for a strong synergistic interaction between dLsd1 and the Pumilio complex during Drosophila development. We show that dLsd1 directly binds to the nanos and pumilio loci in Drosophila and that this binding is conserved in mouse and human cells. In addition, we find that disruption of LSD1 and LSD2 function using small-molecule inhibitors or short hairpin RNA (shRNA) in human cell lines induces expression of NANOS 1 and 3 (NANOS1/3). Consistent with these results, quantitative expression experiments confirm that the relative mRNA levels of the PUM complex (NANOS 1, PUM 1, and PUM 2) are significantly lower in bladder tumors expressing high levels of LSD1 than in normal bladder tissue samples.
Unexpectedly, we identify a novel feedback mechanism in which the Pumilio complex directly represses the translation potential of dLsd1 in Drosophila and LSD2 in human cells by binding to NRE motifs in their 3′ UTRs.
In summary, our work has led to the discovery of a complex and functionally important interaction between the LSD1 family of histone demethylases and the Pumilio posttranscriptional repressor complex.
MATERIALS AND METHODS
Fly strains and genetic analysis.
The following stocks were obtained from Bloomington Stock center: Pum01688, Brat1, Engrailed-GAL4 GFP, and Actin5C-GAL4 genotypes. Transgenic upstream activation sequence (UAS)-RNAi lines were obtained from the Transgenic RNAi Project (TRiP) collection at Harvard Medical School (accession numbers JF02267, HMS05078, and JF02931) and from the Vienna Drosophila Research Center (VDRC) (accession numbers 25218/GD, 45815/GD, 105054/KK, and 108900/KK). w1118 flies were used as a wild-type control in all experiments. A complete list of the fly stocks used in this study is shown in Table S1 in the supplemental material. Flies were grown on standard Drosophila medium and maintained at 25°C.
The effect on the ectopic-wing-vein phenotype was studied by crossing females heterozygous for Engrailed-GAL4 and Engrailed-GAL4; UAS-Lsd1 RNAi with males carrying UAS-RNAi for Pumilio, Brat, and Nanos. The effect was quantified in females by counting the number of wings with ectopic veins.
To assess the viability, five females and two males were crossed at 25°C, and the number of progeny of each genotype was counted for 7 days.
Cells and cloning.
The cells used in the study reported in this paper are Drosophila S2 cells, mouse 32D cells, and human breast cancer cell lines, MCF7 and MDA-MB-231. S2 cells were grown in Schneider's Drosophila medium supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin. MCF7 cells were grown in Dulbecco's modified Eagle medium with nutrient mixture F-12 (DMEM–F-12) and supplemented with 10% FCS, penicillin-streptomycin, and 0.01 mg/ml insulin. MDA-MB-231 cells were grown in DMEM–F-12 and supplemented with 10% FCS and penicillin-streptomycin. Luciferase reporter constructs were cloned by inserting XbaI- and NheI-digested PCR fragments into the XbaI site of the pGL3 plasmid. The pGL3 and pGL4 plasmids were purchased from Promega (catalog numbers E1751 and E6881). Site-directed mutagenesis was conducted using Stratagene Pfu Turbo as per the manufacturer's instructions. Primer sequences can be found in the supplemental material.
RNAi in Drosophila S2 cells.
Double-stranded RNA (dsRNA) for RNAi experiments was generated using a RiboMax large-scale RNA production system (Promega) according to the manufacturer's instructions. Drosophila S2 cells were incubated with 25 to 50 μg of dsRNA for 4 days as previously described (36). All RNAi experiments described in this paper were conducted in triplicate, and data represent averages and standard deviations.
TAP tag RNA pulldown assays.
Wild-type (w1118), nos-GAL4/UAS-TAP-PumHD, and nos-GAL4/UAS-TAP-PABP adult females were grown on standard cornmeal-molasses fly food. The adults were then frozen in liquid nitrogen and stored at −80°C. Adult flies (2.5 g) were suspended in 15 ml of buffer 1 (20 mM Tris-HCl, pH 8, 150 mM NaCl, 10 mM EDTA, 0.2% Nonidet P-40, 0.02 mg/ml heparin, 1.5 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 μg of leupeptin, 20 units/ml DNase I, 10 units/ml RNasin) and ground to a powder with a pestle. The powder was then subjected to Dounce homogenization and centrifuged twice at 10,000 × g for 10 min. Cleared extracts were then incubated with 500 μl of slurry (50%, vol/vol) of IgG-agarose beads at 4°C for 90 min. The beads were then washed once in buffer 1 for 15 min before being washed three more times in buffer 2 (20 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.01% Nonidet P-40, 1 mM DTT, 10 units/ml RNasin). Tandem affinity purification (TAP)-tagged proteins were removed from the beads by incubation with 150 units of AcTEV protease (Invitrogen) for 2 h. RNA was then isolated using TRIzol reagent (Invitrogen), followed by RNeasy (Qiagen) purification as per the manufacturer's instructions (31). Each RNA TAP tag pulldown was conducted in triplicate, and averages and standard deviations from real-time quantitative PCR (RT-qPCR) experiments are reported in this paper.
RT-qPCR.
Total RNA from human cell lines was purified using an RNeasy extraction kit (Qiagen). Reverse transcription was performed using a TaqMan reverse transcription kit (PE Applied Biosystems) according to the manufacturer's specifications. RT-PCR was performed for 50 cycles using an ABI Prism 7900 HD Sequence Detection system. mRNA levels were measured using SYBR green detection chemistry (Applied Biosystems).
Total RNA from Drosophila was extracted using 500 μl of TRIzol reagent (Invitrogen). RNA was treated with a DNA-free DNA Removal kit (Ambion) for 30 min at 37°C as described by the manufacturer. cDNA was synthesized from 1 μg of the total RNA preparation using TaqMan reverse transcription reagents (PE Applied Biosystems) according to the manufacturer's specifications. Real-time PCR analyses were performed using a CFX Connect real-time PCR detection system (Bio-Rad) and FastStart Universal SYBR green master (Rox) mix (Roche Applied Science).
Quantification was performed using the comparative threshold cycle (ΔCT) method as described by the manufacturer. Tubulin, Actin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase gene), RpL32 (RP49), and Rsp26 were used as controls for normalization. Gene-specific primer sequences are included in the supplemental material. All RT-qPCR experiments were conducted in biological triplicates and technical duplicates. Graphs representing RT-qPCR data contain averages and standard deviations. Use of human normal and cancer tissues in this study was approved by the Cambridgeshire Local Research Ethics Committee (Ref 03/018), and the biorepository is supported by National Institute for Health Research funding and by the Cambridge Biomedical Research Centre.
Transfections.
Drosophila S2 cells were transfected for 48 h using X-tremeGENE HP transfection reagent (Roche) in accordance with the manufacturer's instructions. Human cells were transfected for 48 h with X-treme GENE transfection reagent (Roche) and Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. All transfection experiments were conducted in biological triplicates.
Antibodies.
Antibodies used in this study include antitubulin (E7 [Developmental Studies Hybridoma Bank] and clone DM1A [Sigma]), anti-Pumilio 1 (anti-PUM1) (A300 201A; Bethyl), anti-Pumilio 2 (anti-PUM2) (A300-202A; Bethyl), anti-dLsd1 (9), anti-LSD1 (ab17721; Abcam) anti-LSD2 (ab52001; Abcam), and IgG from rabbit serum (I5006; Sigma).
Drug treatments.
MCF7 cells were treated with tranylcypromine (TCP) dissolved in dimethyl sulfoxide (DMSO) at the concentrations outlined in Results. Treatment with DMSO alone was used to normalize the gene expression changes. Cells were treated with the drug for 24 h before being lysed, and the effect on gene expression was analyzed using RT-qPCR. Specific primer sequences can be found in the supplemental material.
Luciferase assays.
S2 Drosophila cells were transfected in 12-well plates with 100 ng of pGL4 and 150 ng of the pGL3-dLsd1-3′ UTR or dLsd1-3′ UTR-NRE mut (NRE mut). For LSD1, LSD2, and LSD2-NRE-mut luciferase experiments, MDA-MB-231 cells were transfected in 12-well plates with 150 ng of pGL3-LSD1 (or variations thereof). Unless otherwise stated, luciferase levels were measured 48 h posttransfection (data are expressed as means ± standard errors [SE]; n = 3). Luciferase readings were taken using a Dual-Luciferase reporter assay system (Promega) as per the manufacturer's instructions. All luciferase assays were conducted in biological triplicate and technical duplicate. Luciferase readings are reported as averages and standard deviations of these measurements.
Lentiviral shRNA.
The DNA preparation, transfections, and virus preparation methods have been published elsewhere (37). LKO.1 shRNA vectors targeting the human PUM complex were as follows: for PUM 1, TRCN0000147347 (sh15), TRCN0000148785 (sh16), TRCN0000148491 (sh17), TRCN0000148263 (sh18), and TRCN0000146945 (sh19); for PUM 2 (GenBank accession number NM_015317), TRCN0000061858 (sh1), TRCN0000061859 (sh2), TRCN0000061860 (sh3), TRCN0000061861 (sh4), and TRCN0000061862 (sh5); for LSD1 (KDM1A), TRCN0000046068 (shRNA1), TRCN0000046069 (shRNA2), TRCN0000046070 (shRNA3), TRCN0000046071 (shRNA4), and TRCN0000046072 (shRNA5); for LSD2 (KDM1B), TRCN0000046073 (shRNA1), TRCN0000046074 (shRNA2), TRCN0000046075 (shRNA3), TRCN0000046076 (shRNA4), and TRCN0000046077 (shRNA5). The shRNAs were obtained from the RNAi Consortium (Boston, MA). The effect of each shRNA was tested in triplicate, and representative samples are displayed.
Chromatin immunoprecipitations (ChIP).
Drosophila adult ovaries and wing discs from third-instar larvae were dissected in phosphate-buffered saline (PBS) and resuspended in buffer A (60 mM KCl, 15 mM NaCl, 4 mM NaCl, 15 mM HEPES [pH 7.6], 1 mM EDTA, 0.5 mM EGTA, 0.5% Triton X-100, 0.5 mM DTT, protease inhibitors). After being homogenized and fixed in 1% formaldehyde for 15 min at room temperature, samples were resuspended in lysis buffer (140 mM NaCl, 15 mM HEPES pH 7.6, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.5 mM DTT, 0.1% sodium deoxycholate, 0.05% SDS) and lysed by sonication. The lysates were cleared by centrifugation, preabsorbed by incubation with protein G- and A-Sepharose beads (GE Healthcare), and incubated overnight at 4°C with 10 μl of anti-dLsd1 (described in reference 9) or in preimmune serum. Antibody complexes were recovered with a mixture of protein A- and G-Sepharose. After extensive washes, immunocomplexes were eluted from the beads and cross-links were reversed. The DNA was recovered by phenol-chloroform extraction and ethanol precipitation. DNA was resuspended in 150 μl of water, and 7.5 μl was used for real-time qPCRs.
S2 and MCF7 cells were fixed in 1% formaldehyde for 15 min at room temperature; the reaction was stopped by addition of glycine, and cells were washed in PBS and harvested in SDS buffer (100 mM NaCl, 50 mM Tris-HCl, pH 8, 5 mM EDTA, pH 8.0, 0.5% SDS, protease inhibitors). Following centrifugation cells were resuspended in immunoprecipitation (IP) buffer (1 volume of SDS buffer to 0.5 volume of Triton dilution buffer), and chromatin was sonicated to an average size of 750 bp. The samples were then processed for ChIP as described above. For the ChIP experiments in MCF7 cells, lysates were incubated overnight at 4°C with 1 μg of IgG and 3 μg of anti-LSD2 antibody.
The databases analyzed for LSD1 binding to the Pum/PUM complex promoters and a description of how the LSD1 ChIP from mouse 32D cells and MCF7 cells were conducted are reported elsewhere (14, 38).
Primer sequences are included in the supplemental material.
RNA pulldown assays.
RNA affinity isolations were performed as described previously (29). HeLa cells were grown to 90% confluence and then washed in 1× PBS and collected by centrifugation at 2,000 × g for 10 min. Cells were then lysed in polysome lysis buffer (10 mM HEPES-KOH, pH 7, 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% Nonidet P-40, 2 mM DTT, 0.2 mg/ml heparin, 50 units/ml RNase OUT [Invitrogen], 50 units/ml SUPERase IN, and 1× complete protease inhibitor tablet [Roche]). This lysate was then spun three times at 14,000 × g for 10 min, and any cellular debris was removed. Aliquots were then flash frozen and stored at −80°C. Fifty microliters of protein G or protein A beads (Amersham) was equilibrated in NT2 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40, 5% bovine serum albumin [BSA; Sigma], 0.02 mg/ml heparin). Twenty micrograms of Pum 1 antibody and 50 μg of Pum 2 were then coupled to the beads and incubated for 12 h at 4°C. The beads were then washed three times in NT2 buffer. Twenty milligrams of lysate was then added to the bead-coupled antibody and mixed for 6 h at 4°C. The beads were then washed four times in NT2 buffer, and RNP was eluted in SDS-EDTA buffer (50 mM Tris, pH 8, 100 mM NaCl, 10 mM EDTA, 1% SDS). Purified RNA from these pulldown experiments was analyzed using RT-qPCR. Each RNA pulldown was done in duplicate, and the RT-qPCR of each sample was analyzed in triplicate. RT-qPCR data from these experiments are displayed averages and standard deviations.
RESULTS
dLsd1 and the Pumilio complex genetically interact in Drosophila.
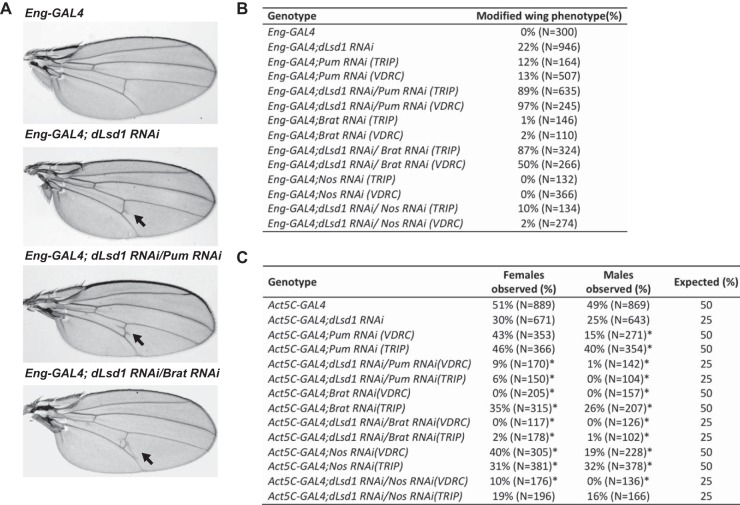
To identify novel regulators of dLsd1 activity, we conducted a dLsd1-RNAi screen in the Drosophila wing. For screening, we depleted dLsd1 in the posterior compartment of the developing Drosophila wing with UAS-RNAi constructs driven by the Engrailed-GAL4 driver (Eng-GAL4). Reduced dLsd1 levels in the wing resulted in a reproducible extra-vein phenotype in 22% of the flies (Fig. 1A and B). We then tested RNAi lines from the TRiP (Transgenic RNAi Project) collection for candidate genes that could modify the frequency of the dLsd1 RNAi-induced phenotype (unpublished data). This analysis identified components of the Pumilio translational repressor complex (Pumilio and brain tumor) as strong enhancers of dLsd1 RNAi phenotypes (Fig. 1A and B). Codepletion of dLsd1 with either Pum or Brat increased the frequency of the appearance of ectopic wing veins from 22% to 89% and 87%, respectively (Fig. 1B). To exclude possible off-target effects of our RNAi lines, we acquired alternative RNAi lines from the Vienna Drosophila RNAi Center (VDRC). Using VDRC RNAi lines of Brat and Pum, we obtained comparable enhancement of the dLsd1-RNAi phenotype to that seen in the lines from the TRiP collection (Fig. 1B). Importantly, RNAi of Brat or Pum alone only rarely produced extra wing vein material (Brat, 0%; Pum, 12%) (Fig. 1B). Depletion of Nanos in dLsd1-RNAi wings did not enhance the phenotype (Fig. 1B). This result is consistent with the fact that the Nanos transcript is mainly expressed in early embryo and in germ line cells and is very weakly expressed in wing discs (39, 40; also data not shown). We next tested the effect of crossing mutant hypomorphic alleles of Pumilio and Brat (Pumilio01688 and Brat1) to dLsd1ΔN (dLsd1 encoding a sequence with an N-terminal deletion) mutants. These hemizygous flies (Pumilio01688/dLsd1ΔN and Brat1; dLsd1ΔN) displayed a moderate enhancement of the dLsd1 ectopic-vein phenotype, in agreement with our RNAi experiments (data not shown). These results show that dLsd1 and components of the Pumilio complex (Pumilio and Brat) act synergistically during wing vein development.
FIG 1.
dLsd1 genetically interacts with the Pumilio complex in Drosophila. (A) Pumilio and Brat depletion results in an enhancement of the dLsd1-RNAi phenotype in the wings. Shown are images of wings expressing the Engrailed-GAL4 driver alone (Eng-GAL4) or dLsd1 RNAi constructs alone (Eng-GAL4; dLsd1 RNAi), dLsd1 and Pumilio RNAi constructs (Eng-GAL4; dLsd1 RNAi/Pum RNAi), or dLsd1 and Brat RNAi constructs (Eng-GAL4; dLsd1 RNAi/Brat RNAi). (B) Table showing the enhancement of the ectopic-vein phenotype penetrance by codepletion of dLsd1 and Pumilio or Brat in the posterior part of the wing using the Eng-GAL4 driver. Nanos depletion does not enhance the extra-wing-vein phenotype observed upon dLsd1 depletion. N, number of wings counted. (C) Codepletion of dLsd1 and Pumilio or Brat by RNAi using the Act5C-GAL4 driver results in synthetic lethality. The table shows the percentage of viable females and males observed for each genotype indicated. N, the number of flies counted. *, P value < 0.001 (binomial test).
To verify this genetic interaction in another dLsd1-mediated phenotype, we tested the effect of cumulative loss of dLsd1 and components of the Pumilio complex on viability. Lowering dLsd1 levels by RNAi using a ubiquitous driver (Act5C-GAL4) did not affect viability. Individually reducing the levels of Pumilio, Brat, or Nanos only subtly changed viability (Fig. 1C). However, codepletion of dLsd1 and components of the PUM complex produced a strong synthetic lethality, and interestingly this effect was significantly stronger in males than in females (Fig. 1C). Our results show that dLsd1 genetically interacts with components of the Pumilio complex and suggest that dLsd1 and the Pumilio complex act synergistically during Drosophila development.
dLsd1 regulates the expression of Nanos and Brat in Drosophila.
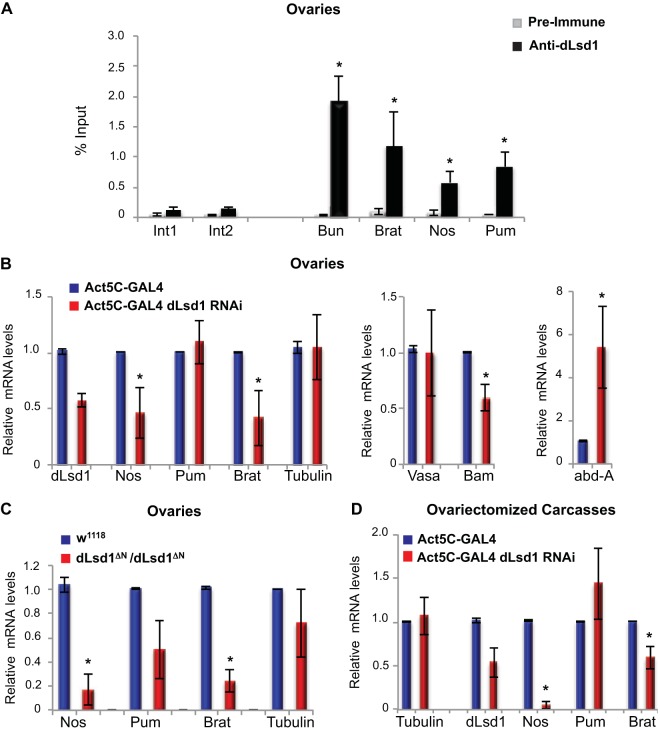
To determine the molecular mechanisms underlying the genetic interaction between dLsd1 and the Pumilio complex, we investigated whether the components of the Pumilio complex were direct transcriptional targets of dLsd1. To do this, we conducted dLsd1 ChIP from Drosophila adult ovaries and third-instar larva wing discs. As shown in Fig. 2A, the promoters of genes encoding all three components of the Pumilio complex (Pum, Nos, and Brat) are directly bound by dLsd1, akin to the previously characterized dLsd1 target, Bunched (Bun). Two intergenic regions, Int1 and Int2, were used as negative controls and were not bound by dLsd1. A similar result was obtained in third-instar larva wing discs (data not shown). As a secondary assay, we performed dLsd1 ChIP experiments in Drosophila S2 cells and found that dLsd1 is bound to the promoters of the Pum complex (data not shown). Importantly, the dLsd1 ChIP signal is dramatically reduced in S2 cells depleted of dLsd1 using dsRNA (data not shown). These ChIP results indicate that the regulatory regions of the components of the Pumilio posttranscriptional repressor complex are directly bound by dLsd1.
FIG 2.
dLsd1 binds to the regulatory regions of the Pumilio complex genes and regulates the transcription of Nos and Brat. (A) dLsd1 binds to Nanos, Pumilio, and Brat regulatory regions. ChIP analysis of dLsd1 binding was performed using an anti-dLsd1-specific antibody in wild-type (w1118) ovaries. Preimmune serum was used as a control for specificity. Int1 and Int2 are intergenic regions used as negative controls, and Bun is a previously identified dLSD1 target used as a positive control. ChIP data are the result of three independent immunoprecipitations. The y axis represents enrichment as percent input. (B) Depletion of dLsd1 results in downregulation of Nos and Brat expression in ovaries. RT-qPCR analysis of the expression of Pum, Nos, Brat, Vasa, Bam, and abd-A in ovaries depleted of dLsd1 using an Act5C driver. Tubulin was used as a control. The expression level was normalized against an Act5C wild-type control, and RP49 and RpL32 transcripts were used as a reference. (C) RT-qPCR analysis of the expression of Nanos, Pumilio, Brat, and Tubulin in w1118 (wild-type) and dLsd1 ΔN homozygous mutant ovaries. The expression level was normalized to that of wild-type ovaries. (D) RT-qPCR analysis of the expression of Nanos, Pumilio, and Brat in ovariectomized carcasses depleted of dLsd1 using an Act5C-GAL4 driver. Act5C-GAL4 was used as a wild-type control. dLsd1 depletion was verified using dLsd1-specific primers, and the levels of the housekeeping gene Tubulin were monitored as a control. All experiments were performed at least in triplicate, and error bars indicate standard errors of the means. A Welch two-sample t test was performed to indicate significance (*, P < 0.05).
To determine the role of dLsd1 in regulating the expression of the Pumilio complex, we compared the ovaries of dLsd1ΔN homozygous mutants and dLsd1-RNAi flies to those of wild-type animals. Surprisingly, loss of dLsd1 expression resulted in reduced levels of Nos and Brat mRNAs, while Pum mRNAs levels remained unchanged (Fig. 2B and C). In agreement with previous reports (9, 10), we found that the mRNA levels of dLsd1 target genes Bam and abd-A were changed upon dLsd1 loss. The expression of the germ line-specific gene Vasa remained unaffected (Fig. 2B). To determine that these changes were not due to aberrant developmental programs in the ovary, we examined the expression of the Pumilio complex upon dLsd1 depletion in ovariectomized carcasses. In these samples, we also observed reduced expression of Nos and Brat (Fig. 2D), supporting a role for dLsd1 in promoting Pum complex expression. These findings are consistent with previous work showing that dLsd1 can either repress or activate gene expression, depending on its association with specific protein complexes (8).
Pumilio posttranscriptionally regulates dLsd1 levels.
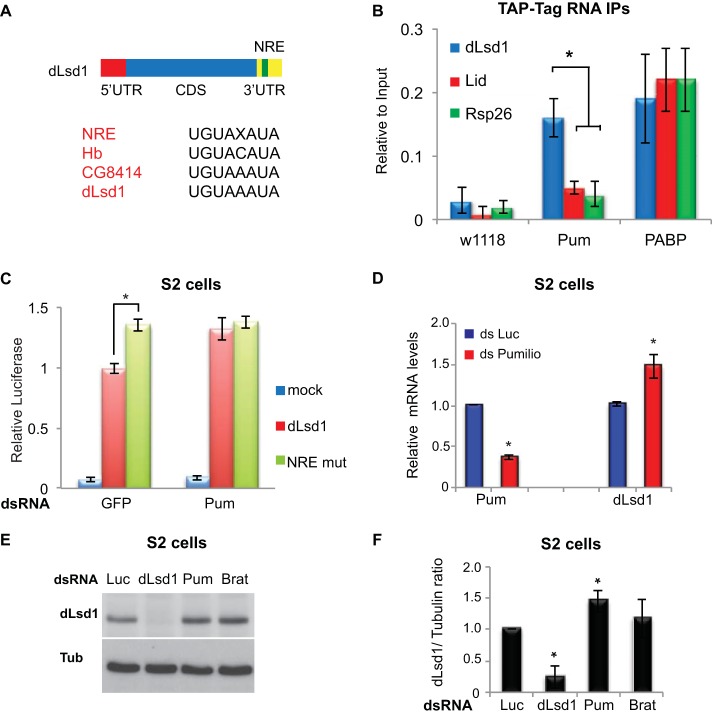
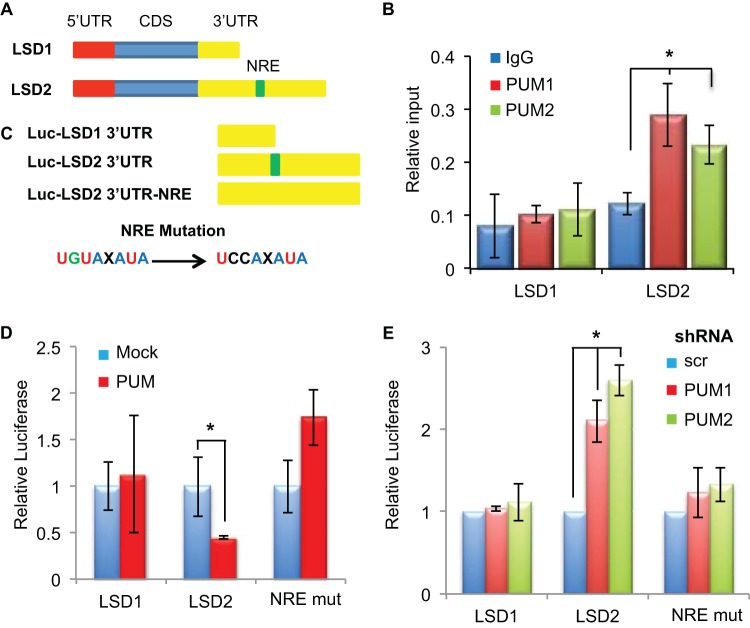
The Pumilio complex regulates transcript stability and translation potential of its target mRNAs by binding a Nanos regulatory element (NRE) (UGUAXAUA) within the 3′ UTR. We next investigated whether the Drosophila Pumilio complex was able to regulate dLsd1 levels. To do this, we examined the transcripts of dLsd1 and other Drosophila chromatin-related modifiers (Lid) for putative Nanos regulatory elements (NREs). This analysis revealed a putative NRE sequence within the 3′ UTR of Drosophila dLsd1, similar to findings in previously characterized Pum substrates (Hunchback [Hb] and CG8414) (Fig. 3A). To determine if the Drosophila dLsd1 transcript is a posttranscriptional target of the Pumilio complex, we conducted RNA immunoprecipitation (RIP) experiments with flies expressing TAP-tagged Pumilio (Pum) or TAP-tagged poly(A)-binding protein (PABP) (positive control) and wild-type (w1118) flies (negative control). RNA enriched from each of these TAP tag immunoprecipitations was assayed using RT-qPCR. From this analysis, we found enrichment for the dLsd1 transcript in the TAP-tagged Pum purified samples compared to transcript levels of control RNAs, which do not contain NRE motifs (lid and rsp26) (Fig. 3B). To verify the functionality of the putative NRE motif, we cloned the 3′ UTR of dLsd1 downstream of a luciferase reporter construct. Using site-directed mutagenesis, we mutated the putative NRE element UGUAAAUA to an inactive UCCAAAUA sequence and measured the effect on luciferase production in Drosophila S2 cells. Mutation of the NRE motif or depletion of Pum by dsRNA increased the levels of luciferase (Fig. 3C). Loss of Pum function did not modify luciferase production from the NRE mutant plasmid, suggesting that the dLsd1 transcript contains only one functional NRE (Fig. 3C).
FIG 3.
The Pumilio complex regulates dLsd1 levels in Drosophila. (A) Schematics of the dLsd1 coding sequences of the 5′ UTR and 3′ UTR regions, including the position of the NRE within dLsd1. Sequence alignment of the Nanos regulatory element (NRE), the characterized NREs in Pum substrates Hunchback (Hb)and CG8414, and the putative NRE in the dLsd1 3′ UTR (dLsd1) is shown. (B) RNA immunoprecipitation using TAP-tagged purification from wild-type flies (w1118) and flies expressing TAP-tagged Pumilio (Pum) or TAP-tagged poly(A)-binding protein (PABP). The RNAs from these purifications were analyzed using RT-qPCR for Rsp26 (negative control), dLsd1, and Lid. (C) Relative luciferase expression from luciferase constructs containing the dLsd1 3′ UTR (dLsd1) or the dLsd1 3′ UTR containing a disrupted Nanos regulatory element (NRE mut) from S2 cells treated with dsRNA targeting green fluorescent protein (GFP) or Pumilio (Pum). (D) Relative dLsd1 mRNA levels upon Pumilio depletion by double-stranded RNA in S2 cells. Double-stranded RNA against luciferase (Luc) was used as a control. (E) Western blots of dLsd1 (dLsd1) and tubulin (Tub) from Drosophila S2 cells treated with dsRNA targeting a control sequence (Luc), dLsd1, Pum, and Brat. (F) Quantification of dLsd1 protein levels relative to tubulin levels from three independent Western blot analyses performed in S2 cells upon depletion of Pumilio or Brat. All experiments were performed at least in triplicate, and error bars indicate standard errors of the means. A Student's t test was performed to determine significance (*, P < 0.05).
To determine how Pumilio regulation affected dLsd1 mRNA, we assayed dLsd1 mRNA levels in Drosophila S2 cells depleted of Pumilio by dsRNA. As shown in Fig. 3D, we detected a small but statistically significant increase in dLsd1 mRNA levels upon Pumilio depletion. A greater increase was observed in Drosophila ovariectomized carcasses upon depletion of Pum, Brat, and Nos using the Act5C-GAL4 ubiquitous driver (data not shown). To determine if Pumilio regulation also affected dLsd1 protein production, we performed Western blot analysis using an antibody directed against dLsd1 in S2 cells depleted of Pumilio and Brat by dsRNA. In agreement with the qPCR analysis, reducing Pumilio complex function resulted in a small increase in dLsd1 protein levels (Fig. 3E). A quantification of dLsd1 protein levels relative to the level of the tubulin control from three independent experiments indicated that this increase in dLsd1 upon Pum silencing is statistically significant. We observed a tendency toward an increase when we depleted Brat; however, this increase was not statistically significant (Fig. 3F). Taken together, these results suggest that, in Drosophila, the dLsd1 transcriptional program and the Pumilio posttranscriptional repressor complex are functionally linked and important for developmental processes.
LSD1 regulates the expression of the Pumilio complex in mammalian cells.
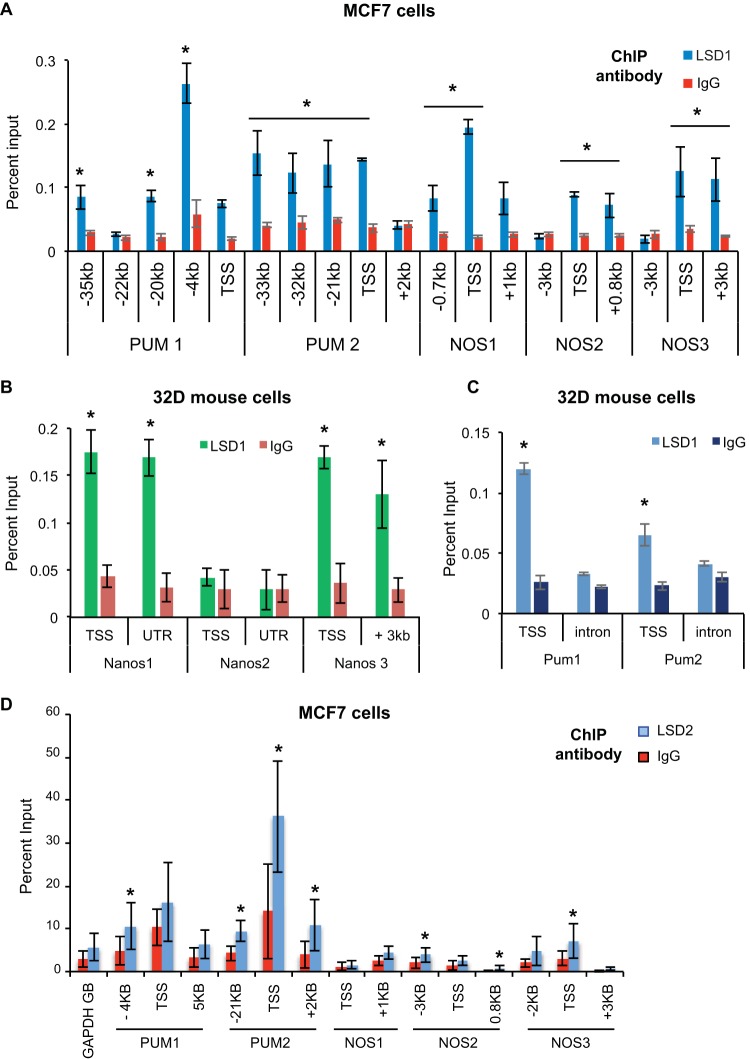
To establish whether LSD1 regulation of the Pumilio complex is conserved in mammalian systems, we examined published LSD1 data sets derived from ChIP with microarray technology (ChIP-chip) of mouse 32D (14) and human K562 cells (38, 41). From these analyses, we found that the promoters and distal regulatory elements of the PUM complex (Pum1/PUM1, Pum2/PUM2, Nanos1/NANOS1, NANOS2, and Nanos3/NANOS3) appeared to be occupied by LSD1. The Nanos2 gene in murine 32D cells showed no enrichment for LSD1 binding.
To test whether the NANOS and PUM genes are bound by LSD1, we conducted LSD1 ChIP and quantitative RT-PCR from mouse 32D cells and from human MCF7 breast cancer cells; LSD1 ChIP from these cells confirmed that all of the NANOS (NOS1 to NOS3) and PUMILIO (PUM1 and PUM2) homolog genes are bound by LSD1 around the transcription start site (TSS) in human cells (Fig. 4A). In mouse 32D cells, all the Pum complex genes are bound by LSD1, except for Nanos2 (Fig. 4B and C). To then verify if the other member of the KDM1 family can also bind to NANOS and PUM genes, we analyzed a recently published LSD2 data set derived from ChIP with DNA sequencing (ChIP-Seq) (42). This analysis showed binding of LSD2 to NANOS and PUM genes in HepG2 cells (42). To confirm that this binding occurs in MCF7 cells, we performed ChIP-qPCR analysis. As shown in Fig. 4D, LSD2 can be found on the NANOS and PUM genes.
FIG 4.
LSD1 and LSD2 bind to the Pumilio complex genes in mammalian cells. (A) ChIP analysis of LSD1 binding to the NANOS1 to NANOS3 (NOS1 to NOS3) and PUM1 and PUM2 loci in human MCF7 breast cancer cells (*, P < 0.01). (B) Relative binding of LSD1 to the Nanos1, Nanos2, and Nanos3 loci in mouse 32D cells was measured by ChIP followed by RT-qPCR. IgG was used as a negative control (*, P < 0.01). (C) Relative binding of LSD1 to the Pum1 and Pum2 loci in mouse 32D cells was measured by ChIP followed by RT-qPCR. IgG was used as a negative control (*, P < 0.01). (D) Relative binding of LSD2 to the PUM1, PUM2, NOS1, NOS2, and NOS3 loci in MCF7 cells was measured by ChIP followed by RT-qPCR. IgG was used as a negative control. *, P < 0.05 (Welch two-sample t test). GB, gene body.
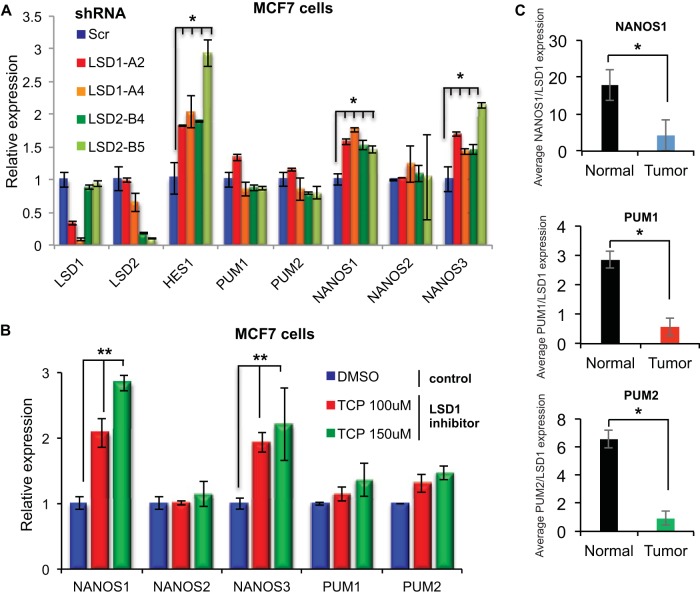
We next investigated the functional consequences of LSD1 and LSD2 binding. To do this, we depleted LSD1, LSD2, or scrambled control sequences (Scr) using shRNA from MCF7 and MDA-MB-231 breast cancer cell lines (Fig. 5A and data not shown). Loss of either LSD1 or LSD2 activity induced NANOS1 and NANOS3 expression to levels similar to the level of the validated LSD1 target gene, HES1 (43) (Fig. 5A), but did not affect the expression of PUM1 and PUM2. To determine if the lack of regulation of PUM1 and PUM2 was due to a redundant function of LSD1 and LSD2, we measured gene expression changes in the PUM complex in human breast cancer cells treated with the LSD1 and LSD2 inhibitor tranylcypromine (TCP) or in cells depleted of both LSD family members. Cells treated with TCP at working concentrations (100 μM and 150 μM) and cells codepleted for LSD1 and LSD2 showed significant induction in the expression of NANOS1 and NANOS3 (Fig. 5B and data not shown) compared to levels in DMSO-treated cells. Codepletion of LSD1 and LSD2 did not significantly increase NANOS expression compared to the level with single-LSD silencing (data not shown). The PUM genes were not significantly changed by TCP treatment or by codepletion of LSD1 and LSD2. These results implicated both LSD1 and LSD2 as important in controlling NANOS levels. These findings are in agreement with previous work, which identified fluctuations in NANOS expression as important for stabilizing the PUM complex (17) and promoting PUM function (44). This suggested that although the binding of the LSD1 family of histone demethylases to the promoters of PUM complex components is conserved throughout evolution, in mammalian cells LSD1 and LSD2 repress rather than promote the expression of the PUM complex.
FIG 5.
LSD1 regulates NANOS1 and NANOS3 expression in human cells. (A) Relative mRNA levels of the PUM complex component genes (PUM1 and PUM2 and NANOS1, -2, and -3) and bona fide LSD1 target (HES1) from MCF7 cells infected with two different shRNAs targeting LSD1 (LSD1-A2/A4), LSD2 (LSD2-B4/B5), and a scrambled control (Scr). (B) Relative RT-qPCR levels of the mammalian PUM complex member genes (PUM1, PUM2, NANOS1, NANOS2, and NANOS3) from MCF7 cells treated with DMSO or the LSD1 inhibitor TCP. (C) Relative LSD1, NANOS1, PUM1, and PUM2 expression in human normal bladder and tumor samples measured by RT-qPCR. GAPDH transcripts were used as a reference. Error bars indicate standard deviations. *, P < 0.05.
We next sought to examine how this regulation was changed in a human disease setting as we recently showed that NANOS1 misregulation was important in tumors lacking a functional Retinoblastoma 1 gene (44). Elevated LSD1 levels have been demonstrated to promote the oncogenic potential of cells, and the overexpression of LSD1 has been found in numerous human tumors (45–47). LSD1 levels have been shown to be particularly important determinants in bladder cancer growth and patient outcome (48). To investigate how LSD1 levels in human tumors correlates with the expression of the PUM complex components, we conducted RT-qPCR from normal human bladder tissues (samples BN11B, BN25A, BN26A, BN2B, and BN4A) and bladder carcinomas (samples BT48, BT53, BT69, BT74, BT120, BT128, BT129, and BT139). The expression levels of NANOS2 and NANOS3 were below the threshold of detection and were thus excluded from further studies. Tumor samples with high levels of LSD1 express lower levels of NANOS1, PUM1, and PUM2 than normal bladder tissue (data not shown). Accordingly, Fig. 5C shows that the ratios between the average expression levels of NANOS1, PUM1, and PUM2 and the average expression levels of LSD1 are reduced in tumor versus normal samples. These data are in agreement with our findings from tissue culture cell lines and suggest that LSD1 represses the expression of the PUM complex and, in particular, NANOS1 in bladder tumors.
PUM posttranscriptionally regulates LSD2 but not LSD1 in human cells.
We next sought to test whether the posttranscriptional repression of the LSD1 family by the PUM complex is conserved throughout evolution. To do this, we used a bioinformatics approach to search the human LSD1 and LSD2 3′ UTRs for NRE (Nanos regulatory element) motifs. This analysis revealed a putative NRE in the 3′ UTR of LSD2 but not LSD1 (Fig. 6A) and is in agreement with previous work, which identified LSD2 as a target of PUM1 and PUM2 in HeLa cells (29). To examine the PUM-mediated regulation of LSD2, we conducted RNA immunoprecipitation (RIP) experiments from MCF7 cells, using antibodies specific to PUM1 and PUM2. RT-qPCR analysis of the RNA purified from these RIP experiments demonstrated that LSD2 but not LSD1 is bound by both PUM1 and PUM2 in these cells (Fig. 6B). To characterize the NRE motif responsible for this regulation, the 3′ UTRs of both LSD1 and LSD2 were cloned downstream of a luciferase reporter gene. The putative NRE within the 3′ UTR of LSD2 was mutated to an inactive UCCAXAUA sequence (Fig. 6C). These constructs were then transfected into MCF7 cells either overexpressing PUM1 or depleted of PUM1 or PUM2 function using a shRNA (data not shown). Elevated PUM1 levels repressed the luciferase levels of the LSD2 3′ UTR but not LSD1 or the LSD2-NRE mutant (Fig. 6D). Depletion of the either PUM protein produced the converse effect on LSD2 luciferase levels (Fig. 6E), implicating PUM as a direct regulator of LSD2. These findings establish LSD2 as a target of PUM posttranscriptional regulation via a Pumilio regulatory element within its 3′ UTR.
FIG 6.
PUM regulates LSD2 but not LSD1 translation. (A) Schematics of the LSD1 and LSD2 coding sequences and 5′ UTR and 3′ UTR regions, including the position of the NRE within LSD2. (B) Relative binding of PUM1 and PUM2 proteins to LSD1 and LSD2 mRNAs from PUM RNA immunoprecipitations measured by RT-qPCR. Error bars indicate standard deviations. *, P < 0.05 (C) Schematic representation of luciferase constructs containing the 3′ UTRs of LSD1, LSD2, and LSD2 containing a mutation within its NRE sequence (3′ UTR-NRE). (D) Relative luciferase expression from luciferase constructs containing the LSD1 and LSD2 3′ UTR sequences in MCF7 cells transfected with PUM1 overexpression constructs. Error bars indicate standard deviations. *, P < 0.05. (E) Relative luciferase expression from LSD1 and LSD2 3′ UTRs from MCF7 cells infected with shRNAs targeting PUM1, PUM2, and a scrambled control (Scr). Error bars indicate standard deviations. *, P < 0.05.
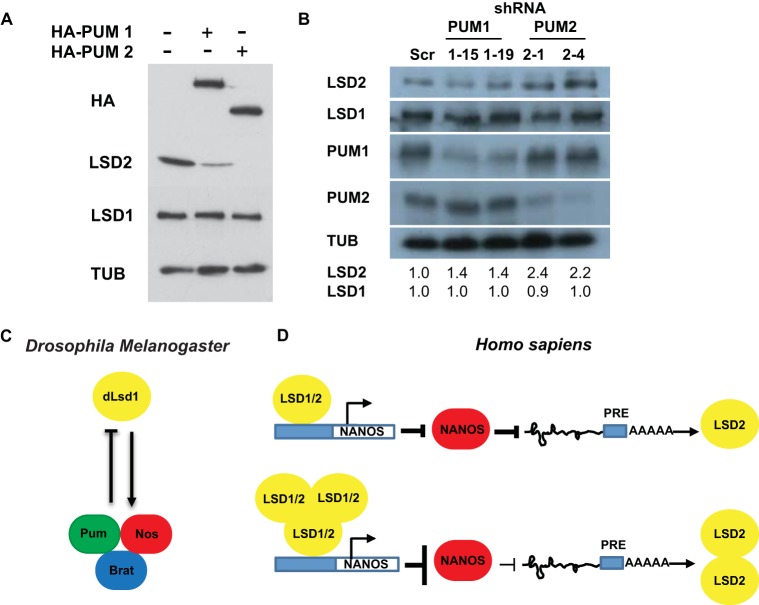
To assess the role of PUM regulation of endogenous LSD2 in cells, we transfected MCF7 cells with plasmids containing PUM1 or PUM2 and assayed LSD1 and LSD2 levels by Western blotting. The exogenous source of the PUM complex was sufficient to dramatically reduce LSD2 but not LSD1 protein levels (Fig. 7A). Conversely, we find that depletion of PUM2 using shRNA resulted in an increase in the protein levels of LSD2 (Fig. 7B) but did not affect the mRNA levels of either LSD1 or LSD2 (data not shown). These results confirm that LSD2 is a substrate for PUM posttranscriptional regulation in human cells and suggest that PUM acts to inhibit the translation of LSD2 rather than promoting the degradation of the LSD2 transcript.
FIG 7.
Schematic representation of the interplay between the LSD1 family and the Pumilio complex. (A) Western blots of hemagglutinin (HA), LSD1, LSD2, and tubulin (TUB) from MCF7 cells transfected with HA-PUM expression constructs. (B) Western blots of LSD1, LSD2, PUM1, PUM2, and TUB from MCF7 cells infected with shRNAs targeting PUM1 (lanes 1-15 and 1-19), PUM2 (lanes 2-1 and 2-4), and a scrambled shRNA sequence (lane Scr). Relative protein level changes of LSD1 and LSD2 in cells depleted of PUM by shRNA are shown. (C) Schematic representation of the feedback loop between dLsd1 and the Pumilio complex in Drosophila melanogaster. (D) Schematic representation of the feedback loop between LSD1, LSD2, and the Pumilio complex in Homo sapiens.
DISCUSSION
In this study, we have identified a novel regulatory mechanism between the KDM1 family of histone demethylases and the PUM posttranscriptional repressor complex. Specifically, we find that the components of the PUM complex are directly bound by LSD1 in flies, mice, and humans. In addition, we have discovered a conserved regulatory feedback loop between the PUM complex and members of the LSD1 family. We propose that LSD1 regulates the expression of the PUM complex and that PUM posttranscriptionally fine-tunes the translation of dLsd1 and LSD2. Importantly, our studies suggest that this interplay is physiologically relevant as Pumilio and dLsd1 have synergistic roles during Drosophila development.
In support of this hypothesis, we find that the concomitant depletion of dLsd1 and components of the Pumilio complex in Drosophila results in synthetic lethality. In addition, we found a strong enhancement of the dLsd1-RNAi wing phenotype when we codepleted components of the Pumilio complex specifically in the wings. These findings suggest that dLsd1 and Pum act synergistically to regulate cell fate and cell survival decisions during Drosophila development and are in agreement with previous findings in Caenorhabditis elegans (49). The site-specific effect on wing formation might be dependent on gradients of signaling molecules and/or transcription factors and highlights the importance of studying this interplay in vivo.
To determine the molecular link between LSD1 and the Pumilio complex, we began by testing whether LSD1 can modulate Pumilio complex expression. By conducting LSD1 ChIP from Drosophila and multiple mammalian cell lines, we found that LSD1 is bound to the promoters of all of the components of the PUM complex. Although the binding of LSD1 to these promoters is conserved throughout evolution, LSD1's function in regulating PUM complex expression appears different in Drosophila and humans. Specifically, we find that LSD1 acts as a transcriptional repressor of the NANOS genes in the human cells tested. In contrast, our results in Drosophila suggest that dLsd1 functions to promote rather than repress Nanos and Brat expression. We cannot exclude the possibility that in complex tissues in Drosophila, dLsd1 depletion indirectly causes Nanos and Brat downregulation by altering tissue differentiation or by changing the expression of Pumilio complex transcriptional regulators. However, a dual role for LSD1 in controlling gene expression is consistent with previous studies showing that LSD1 can associate with both repressive (e.g., coREST) and activating (e.g., Androgen receptor) cofactors to modulate gene expression (8). Intriguingly, some of the genes of the Pumilio complex, which are bound by LSD1, show only minor expression changes upon LSD1/2 depletion or inhibition (PUM1 and PUM2). The results from double depletion of LSD1 and LSD2 seem to exclude the possibility that LSD1 and LSD2 functions in the regulation of Pum complex genes are redundant. Another possibility would be that LSD1 catalytic activity at these genes is blocked by specific cofactors or by the presence of acetylated histones, as previously observed in embryonic stem cells (12). Therefore, LSD1 may be bound to the actively transcribed Pumilio genes and be able to prime them for repression rather than directly contribute to their transcriptional potential. Our results highlight the importance of LSD1 in modulating the expression of the PUM complex but also suggest that this regulation is very context dependent.
In addition, we also identified a conserved feedback mechanism between the PUM complex and LSD1 family members. Our results demonstrate that the PUM complex directly targets the dLsd1/LSD2 transcripts and prevents their translation. It does this by binding to a NANOS regulatory element (NRE) within each of the 3′ UTRs. Using luciferase reporter constructs, we have characterized a functional NRE motif within dLsd1 and LSD2. Consistently, we have found that dLsd1 and LSD2 protein levels are sensitive to Pumilio manipulation. Our results show that PUM does not affect LSD2 mRNA levels, suggesting that PUM acts to directly inhibit the translation of LSD2 rather than by promoting the degradation of the LSD2 transcript. We propose that the regulation of LSD2 and dLsd1 by the Pumilio complex represents a novel posttranscriptional regulatory mechanism to control the expression of these genes in specific cell types. The lack of PUM-binding sites in the shortened 3′ UTR of LSD1 could allow for other ways to regulate LSD1, thus differentiating LSD1 and LSD2. For example, previous studies have identified additional posttranscriptional mechanisms, such as miRNA 137 (miR-137) (50), in constraining LSD1 levels. These results suggest that in human cells, the translation of both KDM1 family members is tightly regulated by different components of the posttranscriptional network.
Taken together, our results suggest that dLsd1 and the Pumilio complex function with a built-in feedback loop which is important for tissue homeostasis during Drosophila development (Fig. 7C). Coupling Pum and dLsd1 expression may be a safeguard mechanism to control cell fates in a number of developmental contexts. Consistently, we have shown that concomitant depletion of dLsd1 and the Pumilio complex results in defects in wing vein determination. This interplay may also be important for oogenesis. In the Drosophila ovary, two independent Pumilio complexes function to regulate the balance between germ line stem cell (GSC) self-renewal and differentiation into cystoblasts during oogenesis (51). The Nanos-Pumilio translational repressor complex is expressed in GSCs and promotes GSC self-renewal. In cystoblasts, a Brat-Pumilio complex functions to support differentiation. Importantly, dLsd1 mutant ovaries have an increased number of GSC-like cells and display a stem cell tumor phenotype (10). We propose that misregulation of Nos, Brat, and Bam in dLsd1 mutant ovaries may contribute to the failure of stem cells to correctly differentiate into cystoblasts. These partially differentiated precursor cells accumulate in the ovary and generate stem cell tumors associated with dLsd1 loss.
In mammalian systems, the family of LSD1 demethylases and the PUM complex act in an antagonistic manner (Fig. 7D). These findings have important ramifications for tumors as LSD1 amplifications have been identified in a number of different tumor types. In tumors that overexpress LSD1, such as bladder carcinoma, one might predict that LSD1 represses the expression of the PUM complex. Consistently, in our analysis of bladder carcinoma tumors, we find significantly lower levels of NANOS1 and reduced levels of both PUM homologs (PUM1 and PUM2). Diminishing the levels of the PUM complex is likely to promote the translation of NRE-containing transcripts, including LSD2, and may contribute to the cellular changes associated with LSD1 amplifications. In support of this hypothesis, the overexpression of miRNAs targeting the PUM complex has been linked to aberrant expression of PUM substrates and to the progression of non-small-cell lung tumors (52).
Based on our results, we propose that the LSD1 family of histone demethylases and the PUM posttranscriptional repressor complex are functionally linked in multiple organisms. These proteins are involved in intricate feedback loops during specific developmental contexts, which are likely to have important implications for stem cell biology and human cancers. These findings provide unexpected insights into the physiological consequences of altering epigenetic and posttranscriptional regulatory pathways and open the road to a detailed study of their impact on the balance between stem cell renewal and differentiation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing the transgenic RNAi fly stocks used in this study. We thank Didier Trouche and members of the LBCMCP and of the Dyson laboratory for technical assistance and helpful discussion.
This work was supported by National Institutes of Public Health Service grants R01 CA163698 and R01 CA64402 to N.J.D. and by the Fondation pour la Recherche Médicale (FRM AJE201109), the Agence Nationale de la Recherche (ANR 2012-CHEX-0002-01), a Marie Curie Career Integration Grant (PCIG13-GA-2013-618272), and the Région Midi-Pyrénées (reference 13051317) to L.D.S. N.J.D. is the James and Shirley Curvey MGH Research Scholar. J.M.J.L. is supported by a postdoctoral fellowship from the Association de la Recherche sur le Cancer.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00755-15.
REFERENCES
- 1.Chi P, Allis CD, Wang GG. 2010. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen MT, Helin K. 2010. Histone demethylases in development and disease. Trends Cell Biol 20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439. [DOI] [PubMed] [Google Scholar]
- 6.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG. 2010. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell 39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Essen D, Zhu Y, Saccani S. 2010. A feed-forward circuit controlling inducible NF-κB target gene activation by promoter histone demethylation. Mol Cell 39:750–760. doi: 10.1016/j.molcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Amente S, Lania L, Majello B. 2013. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta 1829:981–986. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. 2007. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol 17:808–812. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eliazer S, Shalaby NA, Buszczak M. 2011. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc Natl Acad Sci U S A 108:7064–7069. doi: 10.1073/pnas.1015874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MC, Spradling AC. 2014. The progenitor state is maintained by lysine-specific demethylase 1-mediated epigenetic plasticity during Drosophila follicle cell development. Genes Dev 28:2739–2749. doi: 10.1101/gad.252692.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. 2011. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 14.Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O'Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. 2013. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 16.Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol 1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 17.Sonoda J, Wharton RP. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev 13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonoda J, Wharton RP. 2001. Drosophila brain tumor is a translational repressor. Genes Dev 15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spassov DS, Jurecic R. 2002. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene 299:195–204. doi: 10.1016/S0378-1119(02)01060-0. [DOI] [PubMed] [Google Scholar]
- 20.Zamore PD, Williamson JR, Lehmann R. 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 21.Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. 2003. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol 213:120–126. [DOI] [PubMed] [Google Scholar]
- 22.Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. 2012. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blewett NH, Goldstrohm AC. 2012. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol 32:4181–4194. doi: 10.1128/MCB.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. 2010. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 25.Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. 2012. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev 26:356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolde MJ, Saka N, Reinert KL, Slack FJ. 2007. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′ UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol 305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. 2012. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol 19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Q, Padmanabhan K, Richter JD. 2010. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA 16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerber AP, Herschlag D, Brown PO. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci U S A 103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H. 2012. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr Biol 22:420–425. doi: 10.1016/j.cub.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai F, Singh A, King ML. 2012. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development 139:1476–1486. doi: 10.1242/dev.079608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. 2003. Conserved role of nanos proteins in germ cell development. Science 301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 35.Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. 1999. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell 99:271–281. doi: 10.1016/S0092-8674(00)81658-X. [DOI] [PubMed] [Google Scholar]
- 36.Dimova DK, Stevaux O, Frolov MV, Dyson NJ. 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearlberg J, Degot S, Endege W, Park J, Davies J, Gelfand E, Sawyer J, Conery A, Doench J, Li W, Gonzalez L, Boyce FM, Brizuela L, Labaer J, Grueneberg D, Harlow E. 2005. Screens using RNAi and cDNA expression as surrogates for genetics in mammalian tissue culture cells. Cold Spring Harbor Symp Quant Biol 70:449–459. doi: 10.1101/sqb.2005.70.047. [DOI] [PubMed] [Google Scholar]
- 38.Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. 2013. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Lehmann R. 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66:637–647. doi: 10.1016/0092-8674(91)90110-K. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Lin H. 2004. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- 41.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, Durham T, Zhang X, Donaghey J, Epstein CB, Regev A, Bernstein BE. 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaoka K, Hino S, Sakamoto A, Anan K, Takase R, Umehara T, Yokoyama S, Sasaki Y, Nakao M. 2015. Lysine-specific demethylase 2 suppresses lipid influx and metabolism in hepatic cells. Mol Cell Biol 35:1068–1080. doi: 10.1128/MCB.01404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, Mostoslavsky R, Gygi SP, Gill G, Dyson NJ, Naar AM. 2011. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 42:689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miles WO, Korenjak M, Griffiths LM, Dyer MA, Provero P, Dyson NJ. 2014. Post-transcriptional gene expression control by NANOS is up-regulated and functionally important in pRb-deficient cells. EMBO J 33:2201–2215. doi: 10.15252/embj.201488057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schule R, Buettner R. 2006. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res 66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 46.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. 2010. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 47.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, Metzger E, Schule R, Eggert A, Buettner R, Kirfel J. 2009. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res 69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 48.Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R. 2011. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer 128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 49.Katz DJ, Edwards TM, Reinke V, Kelly WG. 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Althoff K, Beckers A, Odersky A, Mestdagh P, Koster J, Bray IM, Bryan K, Vandesompele J, Speleman F, Stallings RL, Schramm A, Eggert A, Sprussel A, Schulte JH. 2013. MiR-137 functions as a tumor suppressor in neuroblastoma by downregulating KDM1A. Int J Cancer 133:1064–1073. doi: 10.1002/ijc.28091. [DOI] [PubMed] [Google Scholar]
- 51.Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. 2011. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell 20:72–83. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, Condorelli G, Banfi S, Verde P. 2015. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene 34:3240–3250. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.