Abstract
Spinal cord injury interrupts descending motor tracts and creates persistent functional deficits due to the absence of spontaneous axon regeneration. Of descending pathways, the corticospinal tract (CST) is thought to be the most critical for voluntary function in primates. Even with multiple tracer injections and genetic tools, the CST is visualized to only a minor degree in experimental studies. Here, we identify and validate the mu-crystallin (crym) gene as a high-fidelity marker of the CST. In transgenic mice expressing green fluorescent protein (GFP) under crym regulatory elements (crym-GFP), comprehensive and near complete CST labeling is achieved throughout the spinal cord. Bilateral pyramidotomy eliminated the 17,000 GFP-positive CST axons that were reproducibly labeled in brainstem from the spinal cord. We show that CST tracing with crym-GFP is 10-fold more efficient than tracing with biotinylated dextran amine (BDA). Using crym-GFP, we reevaluated the CST in mice lacking nogo receptor 1 (NgR1), a protein implicated in limiting neural repair. The number and trajectory of CST axons in ngr1−/− mice without injury was indistinguishable from ngr1+/+ mice. After dorsal hemisection in the midthoracic cord, CST axons did not significantly regenerate in ngr1+/+ mice, but an average of 162 of the 6000 labeled thoracic CST axons (2.68%) regenerated >100 μm past the lesion site in crym-GFP ngr1−/− mice. Although traditional BDA tracing cannot reliably visualize regenerating ngr1−/− CST axons, their regenerative course is clear with crym-GFP. Therefore the crym-GFP transgenic mouse is a useful tool for studies of CST anatomy in experimental studies of motor pathways.
SIGNIFICANCE STATEMENT Axon regeneration fails in the adult CNS, resulting in permanent functional deficits. Traditionally, inefficient extrinsic tracers such a biotinylated dextran amine (BDA) are used to label regenerating fibers after therapeutic intervention. We introduce crym-green fluorescent protein (GFP) transgenic mice as a comprehensive and specific tool with which to study the primary descending motor tract, the corticospinal tract (CST). CST labeling with crym-GFP is 10 times more efficient compared with BDA. The enhanced sensitivity afforded by crym-GFP revealed significant CST regeneration in NgR1 knock-out mice. Therefore, crym-GFP can be used as a standardized tool for future CST spinal cord injury studies.
Keywords: corticospinal tract, nogo receptor, regeneration, spinal cord injury, transgene
Introduction
Spinal cord injury (SCI) results in permanent disability due to the limited growth capacity of adult CNS axons. In part, cell-intrinsic programs limit axonal regeneration (Liu et al., 2011). In addition, the adult CNS is an inhibitory environment due to the expression of chondroitin sulfate proteoglycans (Yiu and He, 2006) and myelin-associated inhibitors (NogoA, myelin-associated glycoprotein, oligodendrocyte myelin glycoprotein; Akbik et al., 2012). These molecules restrict axon growth by signaling through neuron-specific Nogo Receptors 1 and 3 (NgR1, (Fournier et al., 2001; Dickendesher et al., 2012), PirB (Atwal et al., 2008), PTPsigma (Shen et al., 2009; Fry et al., 2010), leukocyte common antigen-related phosphatase (Fisher et al., 2011), and the sphingolipid receptor S1PR2 (Kempf et al., 2014). Receptor antagonism via either genetic perturbation (Kim et al., 2003; Simonen et al., 2003; Kim et al., 2004; Dimou et al., 2006; Cafferty et al., 2010; Dickendesher et al., 2012; Bartus et al., 2014) or pharmacological treatment (Schnell and Schwab, 1990; Bradbury et al., 2002; GrandPré et al., 2002; Liebscher et al., 2005; Wang et al., 2006; Wang et al., 2011; Lang et al., 2015) results in enhanced (yet incomplete) recovery of motor function after experimental SCI (Basso et al., 1995; Basso et al., 2006). In general, the anatomical substrate supporting functional recovery remains unclear and correlative. In contrast to universally adopted SCI behavioral assessments such as BMS and BBB, methods to label motor tracts such as the corticospinal tract (CST) vary widely (Steward et al., 2008b; Lee et al., 2010; Geoffroy et al., 2015). Therefore, important anatomical data can be lost, overinterpreted, and difficult to reproduce (Kim et al., 2003; Simonen et al., 2003; Woolf, 2003; Zheng et al., 2005; Cafferty et al., 2007a; Steward et al., 2007; Steward et al., 2008a; Steward et al., 2012).
Traditionally, CST axon regeneration and plasticity is assessed after injection of anterograde tracers such as biotinylated dextran amine (BDA) into motor cortex (Tuszynski and Steward, 2012). Cortical BDA injection is convenient because only layer V corticospinal motor neurons (CSMNs) project to spinal cord. Multiple standardized stereotaxic injection sites targeting the fore and hindlimb motor cortex are used routinely to maximize CSMN labeling. Despite these measures, CST axon labeling varies greatly between laboratories and as much as 70-fold within groups (Steward et al., 2008b).
In addition to inconsistent partial labeling with BDA, extrinsic tracer injections further complicate SCI experiments through additional surgeries with unintended damage. To limit these issues, genetic approaches have begun to emerge to more comprehensively and exclusively label CSMNs. One report describes a double transgenic thy1-STOP-YFP × Emx-Cre mouse in which yellow fluorescent protein (YFP) is driven via cre-recombinase-mediated excision of a transcriptional stop site under the control of thy1 elements (Bareyre et al., 2005). In this line, 11,000 YFP+ CST axons are labeled in the cervical cord. However, the utility of this approach is limited by the stochastic nature of thy1 promoter elements (Feng et al., 2000; Willenberg and Steward, 2015) and the need for multiple transgenes. A mouse in which a single transgene drives a CST-specific reporter would facilitate breeding with multiple genetically modified models for SCI studies. Here, we introduce the mu-crystallin green fluorescent protein (crym-GFP) transgenic mouse to study the intact and lesioned adult CST. Crym-GFP mice express soluble GFP under the mu-crystallin promoter and demonstrate comprehensive and near specific CST labeling. Furthermore, we demonstrate the utility of the crym-GFP line by crossing it with NgR1 knock-out mice and reassessing CST regeneration after dorsal hemisection (DhX). The increased sensitivity afforded by the crym-GFP line reveals significant CST regeneration in ngr1−/− compared with ngr1+/+ mice, an anatomical finding not observed in previous studies using BDA (Kim et al., 2004). We propose that the crym-GFP mice will streamline and standardize SCI experiments that assess CST regeneration as a primary outcome.
Materials and Methods
Mice.
BAC transgenic Tg(crym-EGFP)GF82Gsat mice (hereafter referred to as crym-GFP mice) were procured from the GENSAT project [stock number 012003-UCD, The Gene Expression Nervous System Atlas (GENSAT) Project, National Institutes of Health–National Institute of Neurological Disorders and Stroke Contracts N01NS02331 and HHSN271200723701C to The Rockefeller University, New York) and back crossed with C57BL/6 mice for 9 generations. Crym-GFP mice were then crossed with ngr1−/− mice as described previously (Kim et al., 2004), subsequent crym-GFP ngr1+/− mice were crossed with nontransgenic ngr1+/− mice to create either crym-GFP ngr1+/+ or crym ngr1−/− mouse lines. To minimize the number of animals used while maintaining enough rigor to achieve our scientific objectives (Festing and Altman, 2002), we used freely available power analysis (Hedwig.mgh.harvard.edu/sample_size/js/js_parallel_quant.html) to estimate sample sizes. The experiments are powered at 90% based on the number of animals in each group, the SD as determined by our previous BMS data (Cafferty et al., 2010), and a significance level of 0.05. Before behavioral assessment and surgery, mice were randomized and split into cages of three. These blocks were maintained unless evidence of fighting was apparent, upon which aggressors were singly housed.
Surgery.
All procedures and postoperative care were performed in accordance with the guidelines of the Institutional Animal Use and Care Committee at Yale University.
Retrograde fast blue and anterograde BDA tracing.
To complete retrograde fast blue (FB) and anterograde BDA labeling of CSMNs, adult (7–9 weeks of age) male crym-GFP ngr1+/+ (n = 6) and crym-GFP ngr1−/−(n = 6) mice were anesthetized with ketamine (100 mg/kg) and xylazine (15 mg/kg) and placed in a stereotaxic frame (Stoelting). An incision was made over the cervical enlargement and the C5–C8 vertebrae were revealed by blunt dissection of overlying muscle. A hemi-laminectomy was performed to expose the underlying C5–C8 spinal cord and a small incision was made in the dura mater. The tip of a pulled glass capillary tube attached to a Micro4 infusion device (World Precision Instruments) was slowly inserted stereotaxically to a depth of 500 μm into the C5 level of the spinal cord and ∼500 μm lateral from the midline. Thirty seconds after introduction of the capillary tube 75 nl of a 0.1% solution of FB (Polysciences) was infused into the spinal cord over 2 min. The tip was left in situ for an additional 30 s before removal. This procedure was completed three additional times at C6, C7, and C8, resulting in a total infusion of 300 nl of FB. Anterograde CSMN labeling with BDA was completed as described previously (Kim et al., 2003; Kim et al., 2004; Cafferty et al., 2007b; Cafferty et al., 2010). Briefly, burr holes were made over the sensorimotor cortex and 5 microinfusions of 75 nl of a 10% solution of BDA were made to a depth of 0.7 mm (coordinates, +1 mm to −1 mm posterior to bregma and 0.5–1.5 mm lateral to bregma) using a pulled glass capillary tube attached to a Micro4 infusion device to deliver a total volume of 375 nl of BDA. Muscle was sutured with Vicryl and skin with monofilament suture. Two weeks after external tracer injections, mice were perfused with 4% paraformaldehyde and the tissue was postfixed overnight at 4°C and embedded in 10% gelatin for immunohistochemical processing. An investigator blinded to genotype completed all surgical procedures.
DhX and anterograde CSMN tracing.
Adult (7–9 weeks of age) female crym-GFP ngr1+/+ (n = 11) and crym-GFP ngr1−/− (n = 15) mice were anesthetized with ketamine (100 mg/kg) and xylazine (15 mg/kg) and an incision made over their thoracic spinal cord. A laminectomy was performed to expose the dorsal portion of spinal cord corresponding to the T6 and T7 levels. The dura mater was pierced and the spinal cord exposed and a pledget of gelfoam soaked in 1% lidocaine was placed on the exposed cord for 1 min before lesion. A DhX lesion was performed at T6 with a 30 gauge needle and a pair of microscissors to a depth of 1.0 mm to completely sever the dorsal and dorsolateral CSTs. The overlying muscle was sutured with Vicryl and skin layer with monofilament suture. Four weeks after SCI, mice received unilateral cortical micro infusion (Micro4; World Precision Instruments) with BDA (10,000 mol/wt; Life Technologies) to anterogradely label the CST as described above. Six weeks after DhX, mice were perfused with 4% paraformaldehyde and the tissue was postfixed overnight at 4°C and embedded in 10% gelatin for immunohistochemical processing. An investigator blinded to genotype completed all surgical procedures.
Bilateral pyramidotomy.
To complete bilateral pyramidotomy (bPyX), adult wild-type crym-GFP mice (n = 6) were anesthetized with ketamine (100 mg/kg) and xylazine (15 mg/kg) and placed in a supine position, an incision was made to the left of the trachea, and blunt dissection exposed the occipital bone at the base of the skull. The occipital bone was removed on either side of the basilar artery with blunt Dumont #2 forceps to expose the medullary pyramids. The dura mater was pierced with a 30 gauge needle and resected. The pyramids were transected bilaterally with fine Dumont #5 forceps to a depth of 0.25 mm or just exposed for sham lesion. No internal sutures were made and skin was closed with monofilament suture. Four weeks after bPyX, mice were perfused with 4% paraformaldehyde. Tissue was postfixed overnight at 4°C and embedded in 10% gelatin for immunohistochemical processing.
Behavioral analysis.
Mice that underwent DhX lesions were assessed using the Basso Mouse Score (BMS) (Basso et al., 2006). Data are presented as average BMS ± SEM. Data were analyzed via repeated-measures ANOVA with Bonferonni correction for multiple comparisons. Post hoc analysis was completed for statistically significant differences comparing BMS score at each time point between genotypes with ANOVA. Two investigators blinded to genotype completed BMS scoring.
Histology.
Mice were killed with an overdose of ketamine (100 mg/kg) and xylazine (15 mg/kg) and were transcardially perfused with 0.9% NaCl (normal saline) followed by 4% paraformaldehyde in PBS. Brains and spinal cords were dissected, postfixed in 4% paraformaldehyde overnight at 4°C, and subsequently embedded in 10% gelatin (Sigma-Aldrich) dissolved in water for vibratome sectioning. Transverse sections (35–40 μm) of cervical and lumbar spinal cord (C6–C7), sagittal and horizontal sections of thoracic spinal cord (T4–T8), and coronal sections of brain and brainstem were processed for BDA with streptavidin-conjugated secondary antibodies (Life Technologies) and tyramide signal amplification (PerkinElmer). Immunofluorescence used antibodies directed against green fluorescent protein (GFP, 1:5000; Life Technologies), Crym (1:500; Abcam), and GFAP (1:10,000; DAKO) with Alexa Fluor 488 and Alexa Fluor 594 (1:500; Life Technologies). Direct application of Nissl-568 (Life Technologies)-labeled neuronal somata in cortex. An investigator blinded to genotype completed all immunohistochemical procedures.
Quantification: percentage of cortical layer V neuronal somata crym-GFP+ and crym-GFP+FB+.
Mice that underwent unilateral intraspinal FB infusion were prepared for immunohistochemical analysis and the number of crym-GFP+ and crym-GFP+FB+ CSMNs was determined. Then, 35 μm sections through forelimb primary motor cortex from ngr1+/+ (n = 6) and ngr1−/− (n = 6) mice were processed for GFP immunohistochemistry and counterstained with Nissl-568. FB emits fluorescence at 420 nm under 365 nm illumination and therefore does not require additional methods for detection. Layer V of cortex was identified via depth from pial surface and density of large pyramidal shaped somata. The number of Nissl+, crym-GFP+Nissl+, and crym-GFP+FB+ neurons were counted in a 250 × 250 μm box superimposed upon five randomly selected sections from each mouse and photomicrographs taken under epifluorescent illumination at 20× magnification (Leica Microsystems). Data are presented as average percentage of the number of Nissl+ somata that were crym-GFP+ and the average number of FB+ somata that were crym-GFP+ ± SEM and analyzed by Student's t test with Bonferonni correction.
Number of CST axons in spinal white matter.
Mice that underwent sham lesion in the DhX cohort that received intracortical BDA infusion were prepared for immunohistochemical analysis to determine the absolute number of CST axons that were crym-GFP+ and BDA+ in the brainstem and dorsal, dorsolateral, and ventral CST in the cervical, thoracic, and lumbar spinal cord. Then, 35 μm transverse sections from brainstem and spinal cord from ngr1+/+ (n = 6) and ngr1−/− (n = 6) mice were processed for GFP immunohistochemistry and BDA detection. Photomicrographs were taken at 63× under oil illumination (Zeiss Imager Z1) from five randomly selected sections from each location from each animal. The absolute number crym-GFP+ and BDA+ axons were counted for the dorsolateral and ventral CSTs. For brainstem and dorsal CST, low-power photomicrographs were taken at 10× to determine the area occupied by the pyramidal tract and the CST in the ventral dorsal columns, respectively. A 25 × 25 μm grid was then overlaid on the 63× photomicrographs and the crym-GFP+ or BDA+ axons in five randomly selected boxes from within this grid were counted. The average number of axons from these boxes was then scaled up to determine the total number of axons labeled. Data are presented as the average number of crym-GFP+ or BDA+ axonal profiles ± SEM and analyzed by ANOVA with Bonferonni correction.
Density of crym-GFP+ and BDA+ axons in spinal gray matter.
Densitometric analysis of crym-GFP+ and BDA+ axons terminating in the cervical spinal cord was completed in ImageJ version 1.45s as described previously (Cafferty and Strittmatter, 2006). Briefly, labeled axons were selected by thresholding and the average fiber length was measured using the skeletonize function in five sections per animal. Data are presented as the average length of crym-GFP+ and BDA+ axons in mm2 ± SEM and analyzed by ANOVA with Bonferonni correction.
Number of regenerating crym-GFP+ CST axons.
To quantify the number of regenerating crym-GFP+ CST axons after DhX, we cut 35 μm sagittal sections of thoracic spinal cord through the lesion site and completed GFP immunohistochemistry. All sections with evidence of the fasciculated dorsal CST rostral to the lesion site were included in our analysis (5–6 sections/animal). The crym-GFP+ axons at the lesion epicenter and at 50, 100, 250, 500, 750, 1000, 1250, 1500, 1750, 2000, 2250, and 2500 μm caudal to the lesion site were counted in each sagittal section. The sum of crym-GFP+ CST axons in five sagittal sections was divided by the average number of crym-GFP+ CST axons in the dorsal CST at the mid-thoracic spinal level in intact wild-type crym-GFP mice to derive an axon index value (Lee et al., 2010; Blackmore et al., 2012; Geoffroy et al., 2015; Wang et al., 2015). Data are shown as the average number of regenerating crym-GFP+ axons after T6 DhX per animal indexed to the total number of crym-GFP± axons in intact crym-GFP mice ± SEM/genotype and analyzed with two-way ANOVA with repeated measures with Bonferonni correction for multiple comparisons with post hoc t test. An investigator blinded to genotype completed all immunohistochemical analyses.
Results
Crym-GFP transgenic mice reveal detailed wiring of the CST
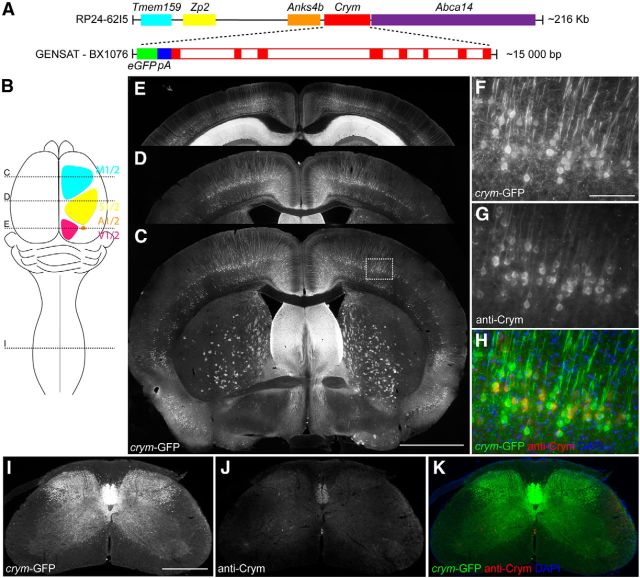
The GENSAT project provides a compendium of mouse lines in which bacterial artificial chromosome (BAC) transgenes have been introduced to express fluorescent reporters under the control of the promoter and regulatory elements of specific genes of interest. These lines facilitate the detailed study of the temporal and spatial expression of specific genes of interest. We browsed their transgenic catalog (Gong et al., 2003; Heintz, 2004) in search of a mouse line that would label CSMNs, their axons, and their terminals with high efficiency. We found that the GENSAT clone BX1076 fulfilled these criteria. GENSAT used the BAC clone RP24-62I5 containing 216,104 base pairs of mouse chromosome 7 (Fig. 1A) to make GENSAT BAC clone BX1076 (Fig. 1A); that is, ∼15 kilobases containing the crym promoter, the eight exons, and seven introns of the crym gene. The sequence for green fluorescent protein (GFP) with its own start codon followed by a polyadenylation signal was inserted upstream of the ATG of the first exon of the crym gene (Fig. 1A) allowing for the expression of soluble GFP under the crym promoter. We obtained the BAC-BX1076 transgenic mouse from GENSAT (Gong et al., 2003) herein referred to as the crym-GFP mouse. Crym is a NADP-regulated thyroid hormone binding protein (Vié et al., 1997; Hallen et al., 2011) and lens protein (Kim et al., 1992) that is also enriched in layer V neurons that project to the spinal cord (Arlotta et al., 2005). Other than its use as a marker of CSMNs in cortex (Yasvoina et al., 2013), its functional role remains unknown. Examination of crym knock-out mice revealed no observable anatomical or functional differences (data not shown). Gross examination of the GFP expression in the CNS of crym-GFP mice (localization schematized in Fig. 1B) confirmed that crym-GFP is expressed in neuronal somata in cortical layer V projection neurons in primary and secondary motor cortex (Fig. 1C,F–H), sensory cortex (Fig. 1D), and auditory and visual cortex (Fig. 1E). In addition, crym-GFP can be seen in the septal nuclei (Fig. 1C,D), striatum (Fig. 1C), and hippocampus (Fig. 1E). In the spinal cord, the CST is robustly labeled with crym-GFP, revealing the dense innervation pattern of the CST into the gray matter from axons exiting the dorsal, dorsolateral, and ventral white matter (Fig. 1I). Next, we investigated whether GFP transgenic labeling under the control of the crym promoter was faithful to endogenous Crym expression. Immunostaining of brain and spinal cord sections using an antibody to Crym revealed complete overlap between endogenous Crym and transgenic GFP in layer V neurons of cortex (Fig. 1F–H) and in the spinal cord (Fig. 1I–K). Endogenous Crym expression is weak and especially difficult to detect in the finer CST axon terminals in spinal cord, suggesting that Crym functions within the neuronal somata (Fig. 1J); however, crym-GFP transgenic mice express soluble GFP that is not restricted to any cellular compartment and therefore fills somata, axons, and terminals, thereby underscoring its utility as a comprehensive intrinsic CST marker (Fig. 1I–K).
Figure 1.
BAC transgenic Tg(Crym-EGFP)GF82Gsat expresses soluble GFP in adult brain and spinal cord. Schematic (A) shows a linear representation of the BAC clone RP24-62I5, which contains Tmem159 (cyan), Zp2 (yellow), Anks4b (orange), Crym (red), and Abca14 (purple). Enlarged sequence shows inferred schematic of GENSAT clone BX1076, which contains EGFP (green) with its own PolyA (blue) 5′ to the start codon of Crym (red solid boxes are exons, open boxes are introns). BX1076 was injected into fertilized ova to create Tg(crym-EGFP)GF82 (crym-GFP) mice. Schematic (B) shows location of primary and secondary motor (cyan), sensory (yellow), auditory (orange), and visual cortex (magenta). Stippled lines labeled C, D, and I in schematic B indicate location of photomicrographs through the transverse plane of the brain (C–E) and cervical spinal cord (I–K) of an adult wild-type crym-GFP mouse. Intense GFP labeling is observed in layer V of motor (C, ∼0.86 mm anterior to bregma), sensory (D, ∼0.86 posterior to bregma), auditory and visual cortex (E, ∼3.5 mm posterior to bregma). GFP is additionally localized in a diffuse pattern in the striatum (C) and in the CA1 and CA2 regions of the hippocampus (E). High-power photomicrographs (F–H) show crym-GFP expression in pyramidal neurons in layer V of primary motor cortex. Antibodies to Crym label all crym-GFP+ pyramidal neurons (G, H). Low-power photomicrographs (I–K) of a transverse section of C7 spinal cord show robust crym-GFP expression in the dorsal, lateral, and ventral funiculi (I) and throughout dorsal and intermediate gray matter. Low levels of Crym protein are observed in the ventral dorsal column (J), which overlaps 100% with crym-GFP (K). Scale bars: C, 1 mm; F, 100 μm; I, 500 μm.
Crym-GFP comprehensively labels the CST throughout the spinal axis
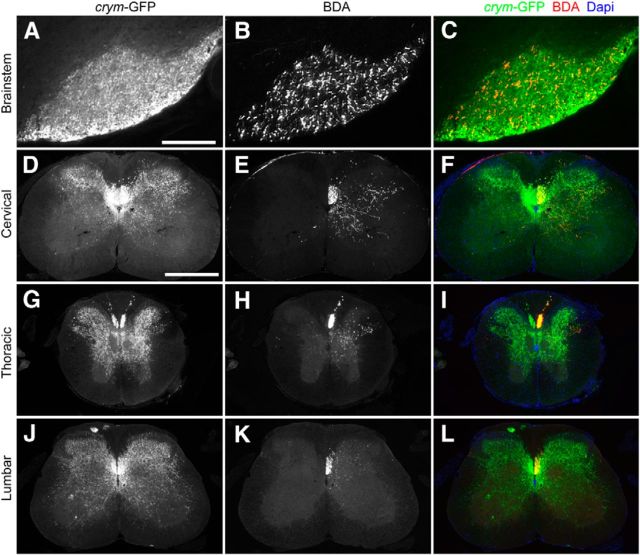
We sought to examine the utility of crym-GFP mice to intrinsically label the CST for spinal cord injury studies. To this end, we completed bPyX in crym-GFP mice to singularly axotomize the CST bilaterally in its entirety. Intact crym-GFP mice show robust labeling of CST axons in the dorsal CST (dCST) the dorsolateral CST (dlCST), and ventral CST (vCST) in the cervical (Fig. 2A) and lumbar (Fig. 2B) spinal cord. In addition, GFP labeled CST terminals can be seen densely innervating spinal gray matter in transverse sections of cervical (Fig. 2A) and lumbar (Fig. 2B) cord, and in sagittal (Fig. 2C,D) and horizontal (Fig. 2E,F) sections of thoracic cord. After bPyX, crym-GFP is almost entirely eliminated from cervical (Fig. 3A), thoracic (Fig. 3C–F), and lumbar spinal cord (Fig. 3B). A few spinal interneurons can be seen along the midline; however, they are easily distinguishable from the axon tracts and complex CST innervation pattern (Fig. 2). These data clearly demonstrate that crym-GFP is a comprehensive and robust label of the CST and also reveal the full extent of its complex and dense terminal arborization in both dorsal and ventral spinal gray matter.
Figure 2.
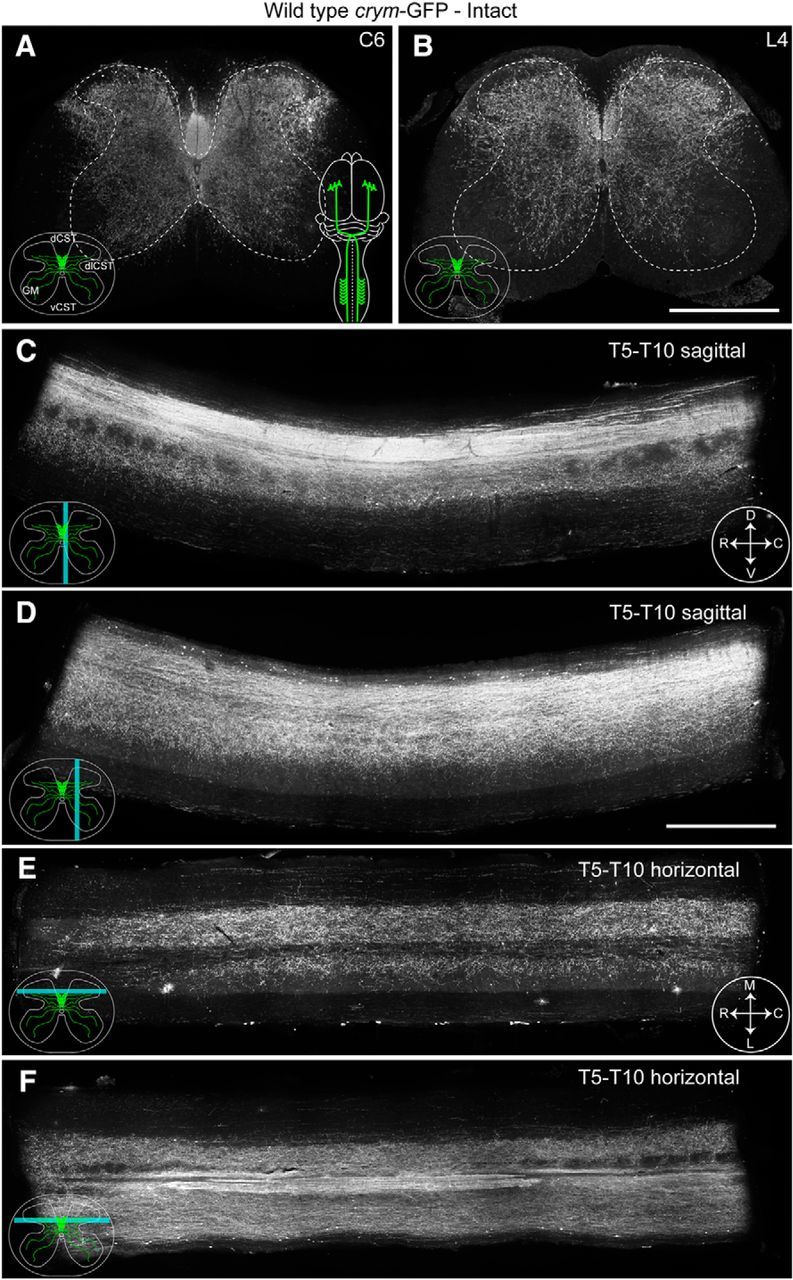
Intact crym-GFP transgenic mice illustrate comprehensive labeling of the CST. Low-power photomicrographs of transverse sections of cervical (C6, A) and lumbar (L4, B) spinal cord from an intact (color schematic in A shows a bilaterally intact CST in green) adult wild-type crym-GFP mouse show intrinsic GFP expression in the dorsal (dCST), dorsolateral (dlCST), and ventral corticospinal (vCST) tracts and throughout spinal gray matter (GM). Inset schematics (A) reflect intact terminals. Spinal gray and white matter are delineated with a white stippled line (A, B). Sagittal sections through the midline (C, cyan line depicts relative location of section, compass reflects orientation of section: D, dorsal; V, ventral; R, rostral; C, caudal) and mediolateral (D) thoracic cord show intense GFP labeling in the main dorsal CST and GFP+ terminals entering spinal gray matter. Horizontal thoracic sections through the upper (E, cyan line depicts relative location of section, compass reflects orientation of section: M, medial; L, lateral; R, rostral; C, caudal) and lower dorsal horn (F) show GFP+ CST terminals densely localized throughout gray matter. Scale bars: B, 500 μm; D, 1 mm.
Figure 3.
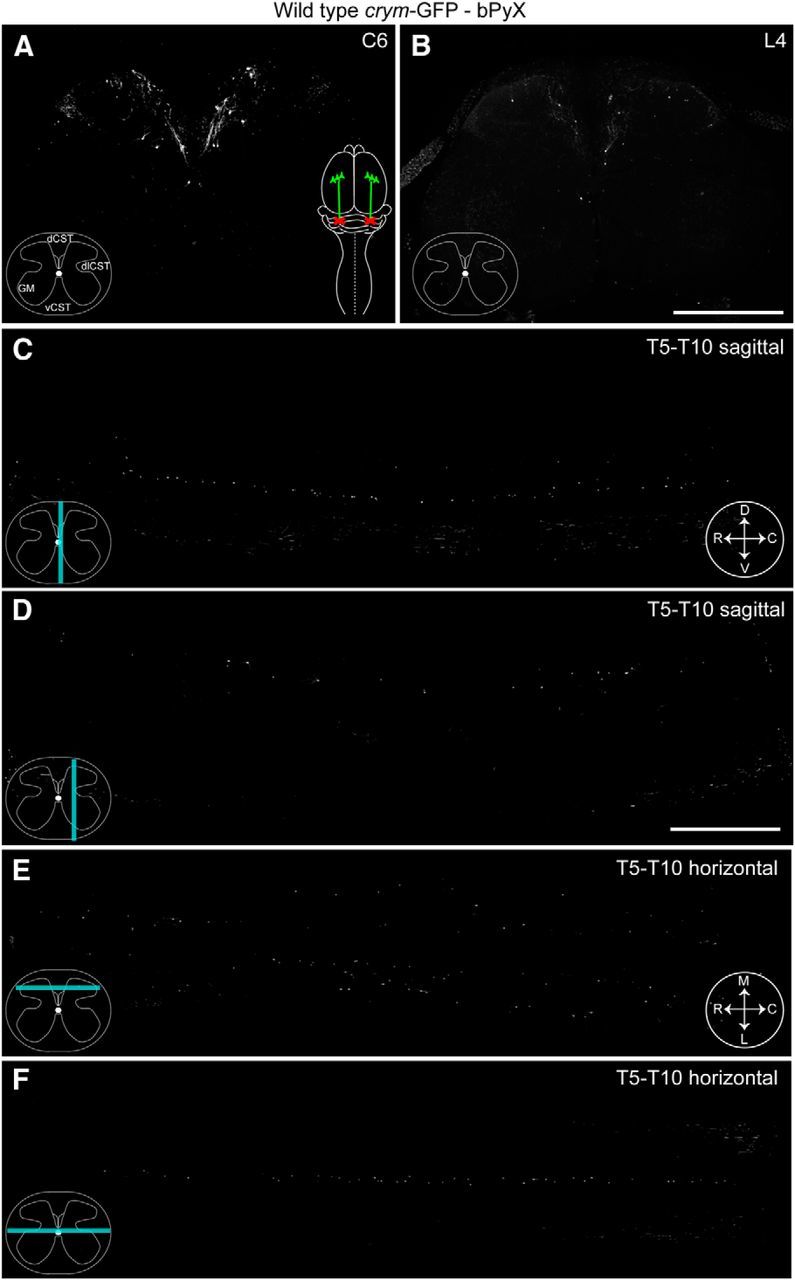
bPyX results in complete loss of spinal GFP labeling in crym-GFP mice. Low-power photomicrographs of transverse sections of cervical (C6, A) and lumbar (L4, B) spinal cord after bPyX [color schematic in A shows bilateral axotomy (red crosses) of the CST (green) in the medullary pyramids] in an adult wild-type crym-GFP mouse show a complete loss of GFP in the dCST, dlCST, vCST, and throughout spinal gray matter. Inset schematic (A) reflects lesioned terminals. Midline (C, cyan line depicts relative location of section, compass reflects orientation of section: D, dorsal; V, ventral; R, rostral; C, caudal) and mediolateral (D) sagittal sections show a complete loss of GFP labeling in white and gray matter after bPyX. Horizontal sections through the upper (E, cyan line depicts relative location of section, compass reflects orientation of section: M, medial; L, lateral; R, rostral; C, caudal) and lower dorsal horn also show a complete loss of GFP+ CST axons and terminals after bPyX. Few sporadic GFP+ cells can be seen scattered in gray matter in transverse (A, B), sagittal (C, D) and horizontal (E, F) sections. Scale bars: B, 500 μm; D, 1 mm.
Crym-GFP+ CSMNs exclusively terminate in spinal cord in ngr1+/+ and ngr1−/− mice
The sensitivity of CST labeling afforded by crym-GFP transgenic mice allows for comprehensive localization of fine structures that are typical of newly sprouted or regenerating axons (Steward et al., 2003; Tuszynski and Steward, 2012). Previous studies have shown that mice null mutant for NgR1 exhibit plasticity of uninjured CSMNs after unilateral pyramidotomy (uPyX) (Cafferty and Strittmatter, 2006) and with experience-dependent plasticity in visual cortex (McGee et al., 2005) and somatosensory cortex (Akbik et al., 2013). Strikingly, however, no CST axon regeneration was observed after DhX in ngr1−/− mice (Kim et al., 2004). Together, these data suggest that either developmental differences in the wiring of ngr1−/− mice account for the anatomical changes observed after uPyX and during activity-dependent plasticity or the methodology used to label regenerating CST axons after DhX in ngr1−/− mice was insufficient to detect regenerating CST axons. To explore these possibilities, we completed a comprehensive evaluation of the CST in ngr1+/+ crym-GFP and ngr1−/− crym-GFP mice (see Figs. 4, 5, 6, 7) and assessed CST regeneration in both lines of mice after DhX SCI (see Figs. 8, 9, 10, 11, 12).
Figure 4.
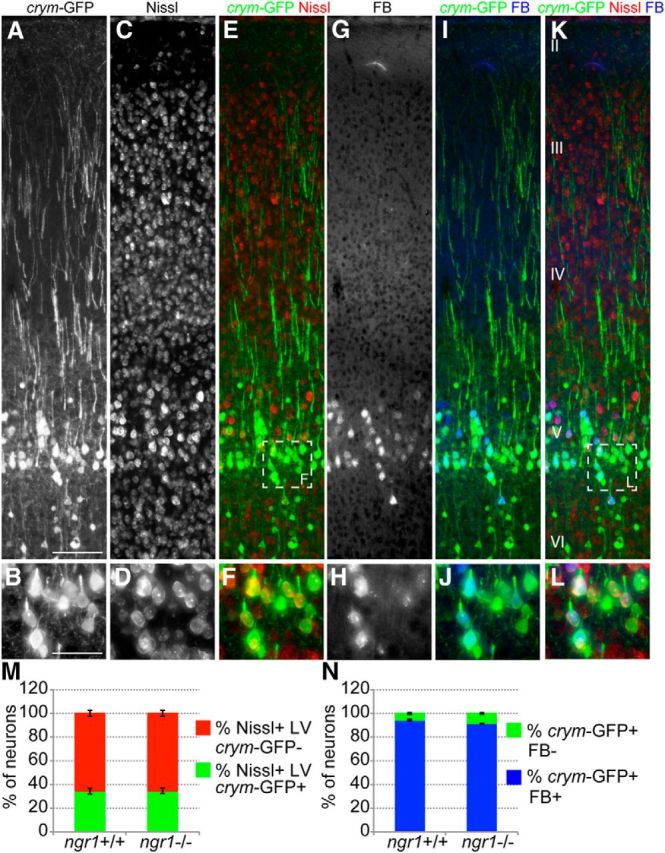
Crym-GFP is expressed in cortical layer V projection neurons. Photomicrographs (A, B) show robust GFP expression in layer V (cortical layers shown in K) cortical neuron somata, axons and primary dendrites (B, D, F are high-power insets from box shown in E). GFP+ somata are exclusively neuronal as 100% of GFP+ cells are Nissl+ (C–F). Of all Nissl+ cells in layer V (LV), 34.44 ± 2.4 (average number of Nissl+GFP+ somata ± SEM) were GFP+ in ngr1+/+ and 34.93 ± 2.4 were GFP+ in ngr1−/− mice (M). There was no significant difference between genotypes (Student's t test). Unilateral injection of the retrograde tracer FB into the cervical spinal cord resulted in dense labeling of layer V pyramidal neuron somata (G–L) in contralateral cortex 2 weeks after injection (H, J, L are high-power insets from box shown in K). Of all FB+ neurons, 94.69 ± 0.6 were GFP+ in ngr1+/+ and 94.23 ± 0.4 were GFP+ in ngr1−/− mice (N). There was no significant difference between genotypes (Student's t test). Scale bars: A, 100 μm; B, 50 μm.
Figure 5.
Intrinsic GFP labeling of the CST in crym-GFP transgenic mice is superior to extrinsic labeling with anterogradely transported BDA. Low-power photomicrographs of brainstem (A–C), cervical (C8, D–F), thoracic (T8, G–I), and lumbar (L4, J–L) spinal cord show robust intrinsic GFP+ CST axons in the medullary pyramid (A) dorsal, dorsolateral and ventral spinal white and gray matter (D, G, J). In contrast, fewer CST axons and terminals are BDA+ in the pyramids (B) and spinal cords (E, H, K) compared with GFP+-labeled axons and terminals in crym-GFP mice (see overlays C, F, I, L; GFP, green; BDA, red; DAPI, blue). Scale bars in A and D, 500 μm.
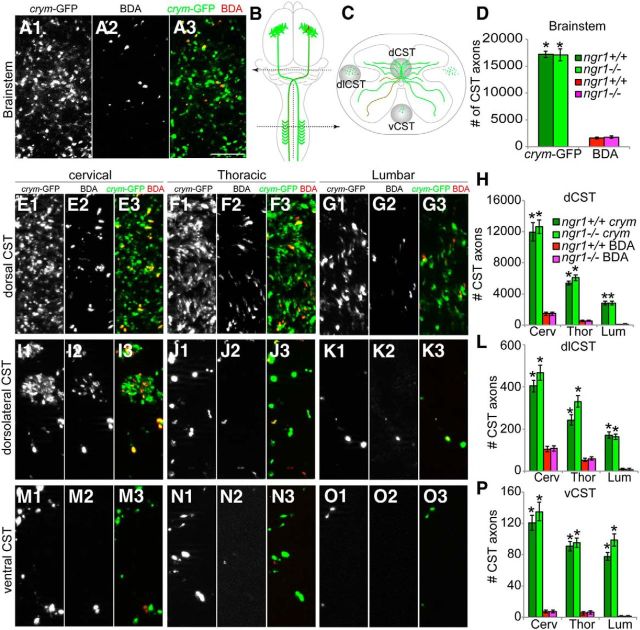
Figure 6.
Intrinsic GFP labeling of the CST in crym-GFP transgenic mice is superior to extrinsic labeling with anterogradely transported BDA. Single-channel high-power photomicrographs show crym-GFP expression (A1) and BDA tracing (A2) of CST axons (overlay A3; crym-GFP, green; BDA, red) in the medullary pyramid of a wild-type crym-GFP mouse. Sections (A1–A3) were assessed and quantification completed on CST axons in the brainstem rostral to the pyramidal decussation, shown in schematic B. Subsequent evaluation of the number of CST axons entering the cervical, thoracic, and lumbar spinal cord were parsed into three groups according to the their white matter location (schematic C) in the dCST, dlCST, or vCST. Significantly more CST axons were labeled with GFP compared with BDA in both ngr1+/+ and ngr1−/− mice (D, *F(3, 99) = 966.652, p < 0.0005, one-way ANOVA with Bonferonni post hoc comparisons). There was no significant difference in the number of crym-GFP+ axons or BDA+ axons between genotypes. Data are shown as average number of labeled axons ± SEM. Single-channel high-power photomicrographs show crym-GFP expression (E1, F1, G1) and BDA tracing (E2, F2, G2) of CST axons in the dCST in transverse sections of cervical (E1–E3), thoracic (F1–F3), and lumbar (G1–G3) spinal cord of a wild-type crym-GFP mouse. Significantly more CST axons were labeled with GFP compared with BDA (E3, F3, G3, overlays; crym-GFP, green; BDA, red) at all spinal levels in both ngr1+/+ and ngr1−/− mice (H, *F(11, 273) = 381.683, p < 0.0005, one-way ANOVA with Bonferonni post hoc comparisons, data are shown as average number of labeled axons ± SEM). There was no significant difference in the number of crym-GFP labeled axons or BDA+ axons between genotypes at any spinal level. Data are shown as average number of labeled axons ± SEM. Single channel high-power photomicrographs show crym-GFP expression (I1, J1, K1) and BDA tracing (I2, J2, K2) of CST axons in the dlCST in transverse sections of cervical (I1–I3), thoracic (J1–J3), and lumbar (K1–K3) spinal cord of a wild-type crym-GFP mouse. Significantly more CST axons were labeled with GFP compared with BDA (I3, J3, K3, overlays; crym-GFP, green; BDA, red) at all spinal levels in both ngr1+/+ and ngr1−/− mice (L, *F(11, 318) = 292.458, p < 0.0005, one-way ANOVA with Bonferonni post hoc comparisons, data are shown as average number of labeled axons ± SEM). There was no significant difference in the number of crym-GFP labeled axons or BDA+ axons between genotypes at any spinal level. Single-channel high-power photomicrographs show crym-GFP expression (M1, N1, O1) and BDA tracing (M2, N2, O2) of CST axons in the vCST in transverse sections of cervical (M1–M3), thoracic (N1–N3), and lumbar (O1–O3) spinal cord of a wild-type crym-GFP mouse. Significantly more CST axons were labeled with GFP compared with BDA (M3, N3, O3, overlays; crym-GFP, green; BDA, red) at all spinal levels in both ngr1+/+ and ngr1−/− mice (P, *F(11, 313) = 282.994, p < 0.0005, one-way ANOVA with Bonferonni post hoc comparisons, data are shown as average number of labeled axons ± SEM). There was no significant difference in the number of crym-GFP-labeled axons or BDA+ axons between genotypes at any spinal level. Scale bar in A3, 10 μm.
Figure 7.
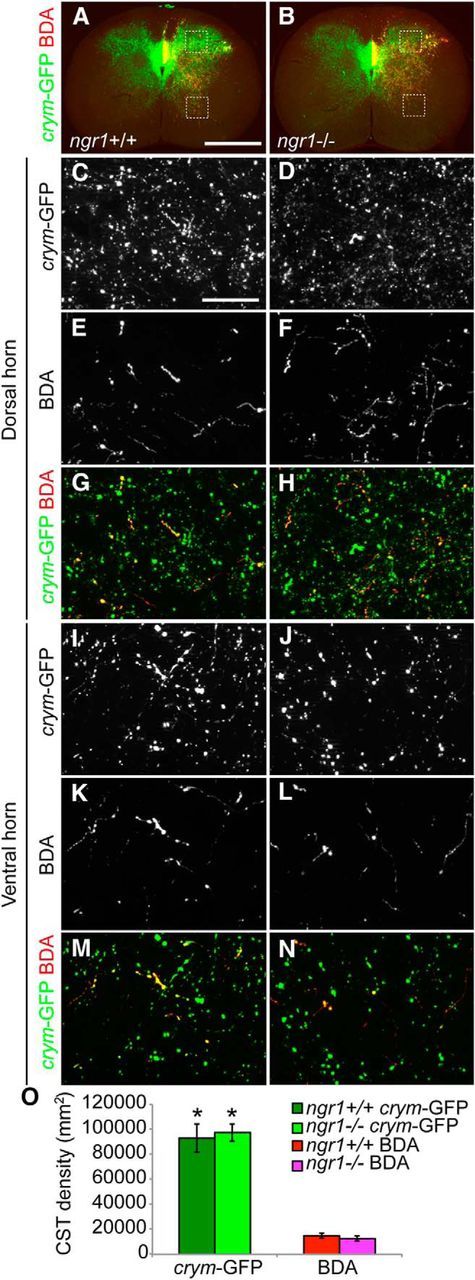
Intrinsic GFP labeling of gray matter CST terminals in crym-GFP transgenic mice is superior to extrinsic labeling with anterogradely transported BDA. Low-power photomicrographs of transverse sections of intact C8 spinal cord from adult ngr1+/+ (A) and ngr1−/− (B) mice show dense GFP+ CST fibers terminating throughout gray matter (A, B, green). BDA+ fibers can be seen exiting the dorsal columns and innervating a more sparse area of gray matter (A, B, red). High-power photomicrographs of the dorsal (C–H) and ventral (I–N) horns show significantly denser GFP+ CST axon terminals (C, D, I, J) compared with BDA+ CST axon terminals (E, F, K, L). Multichannel overlays (G, H, M, N; GFP, green; BDA, red) illustrate the superior efficiency of crym-GFP compared with anterograde BDA tracing. Densitometric analysis showed that significantly more GFP+ CST fibers terminated in intact cervical spinal gray matter compared with BDA+ CST fibers (O, *F(3, 96) = 231.293, p < 0.0005, one-way ANOVA with Bonferonni post hoc comparisons, data are shown as the average density of CST occupation per square millimeter for each label ± SEM). There was no significant difference in the density of crym-GFP-labeled terminals or BDA+ terminals between genotypes. Scale bars: A, 500 μm; C, 10 μm.
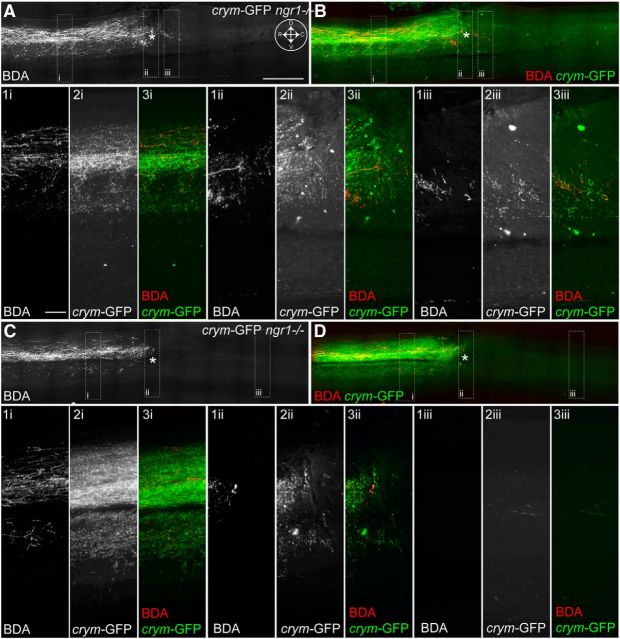
Figure 8.
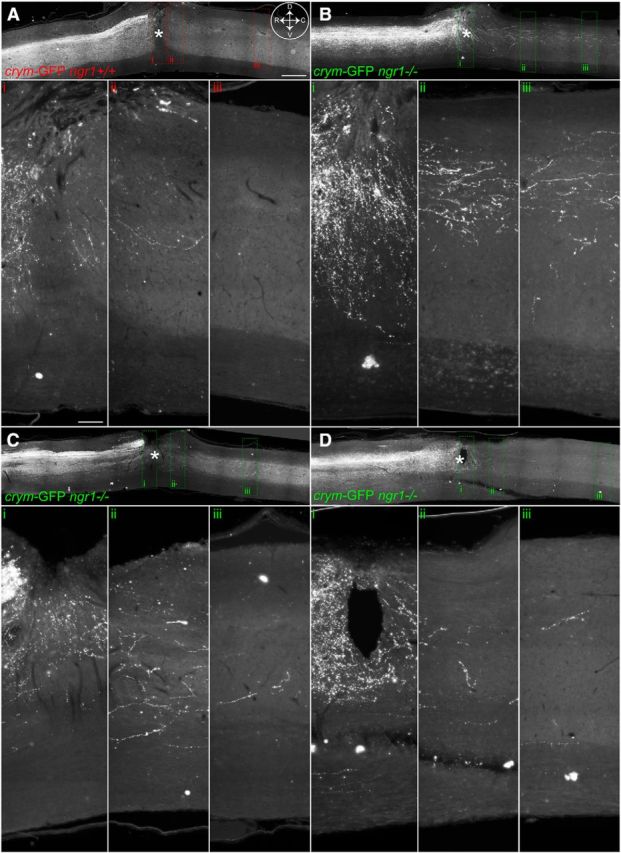
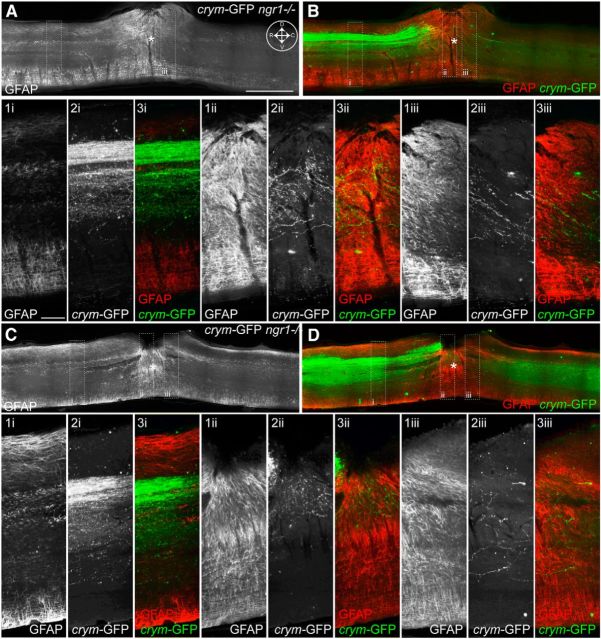
Comprehensive CST labeling in crym-GFP mice reveal axon regeneration of axotomized CST neurons after DhX SCI in ngr1+/+ and ngr1−/− mice. Low-power photomicrographs of mid-sagittal sections from ngr1+/+ (A) and ngr1−/− (B–D) show intense GFP+ labeling of the CST rostral to the lesion site (inset compass in A shows orientation; D, dorsal; V, ventral; R, rostral; C, caudal). The epicenter of the lesion is marked with an asterisk. In both ngr1+/+ (A) and ngr1−/− (B–D) mice GFP+ CST axons can be seen caudal to the lesion site. High-power photomicrographs of inset boxes labeled i–iii clearly show GFP+ CST axons regenerating into the lesion site in ngr1+/+ mice (Ai–Aiii). Significantly more GFP+ CST axons can be seen crossing the lesion site and entering the distal spinal cord in ngr1−/− mice (Bi–Biii, Ci–Ciii, Di–Diii). Scale bars: A, 500 μm; Ai, 150 μm.
Figure 9.
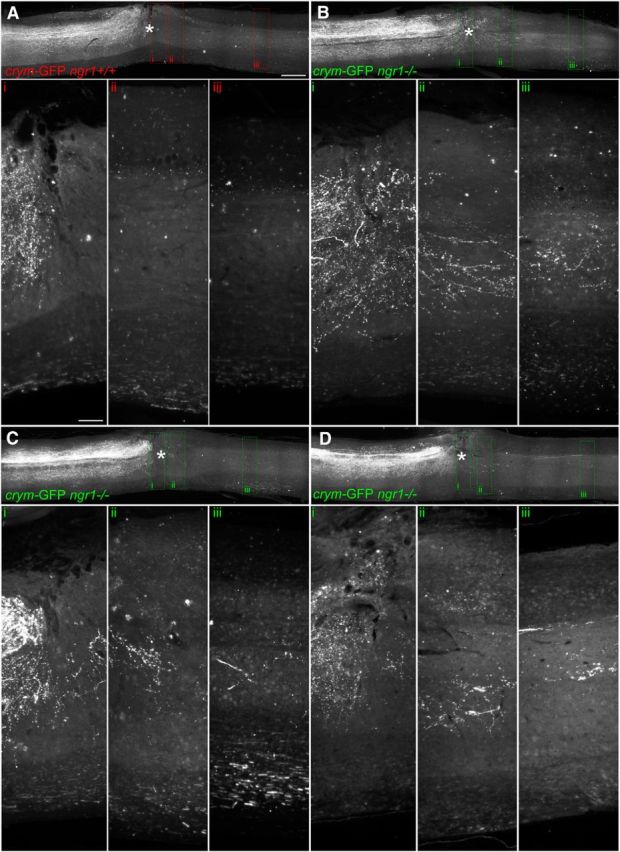
Additional examples of CST regeneration in ngr1+/+ and ngr1−/− after DhX. The epicenter of the lesion is marked with an asterisk. In both ngr1+/+ (A) and ngr1−/− (B–D) mice, GFP+ CST axons can be seen caudal to the lesion site. High-power photomicrographs of inset boxes labeled i–iii clearly show GFP+ CST axons regenerating into the lesion site in ngr1+/+ mice (Ai–Aiii). Significantly more GFP+ CST axons can be seen crossing the lesion site and entering the distal spinal cord in ngr1−/− mice (Bi–Biii, Ci–Ciii, Di–Diii). Scale bars: A, 500 μm; Ai, 150 μm.
Figure 10.
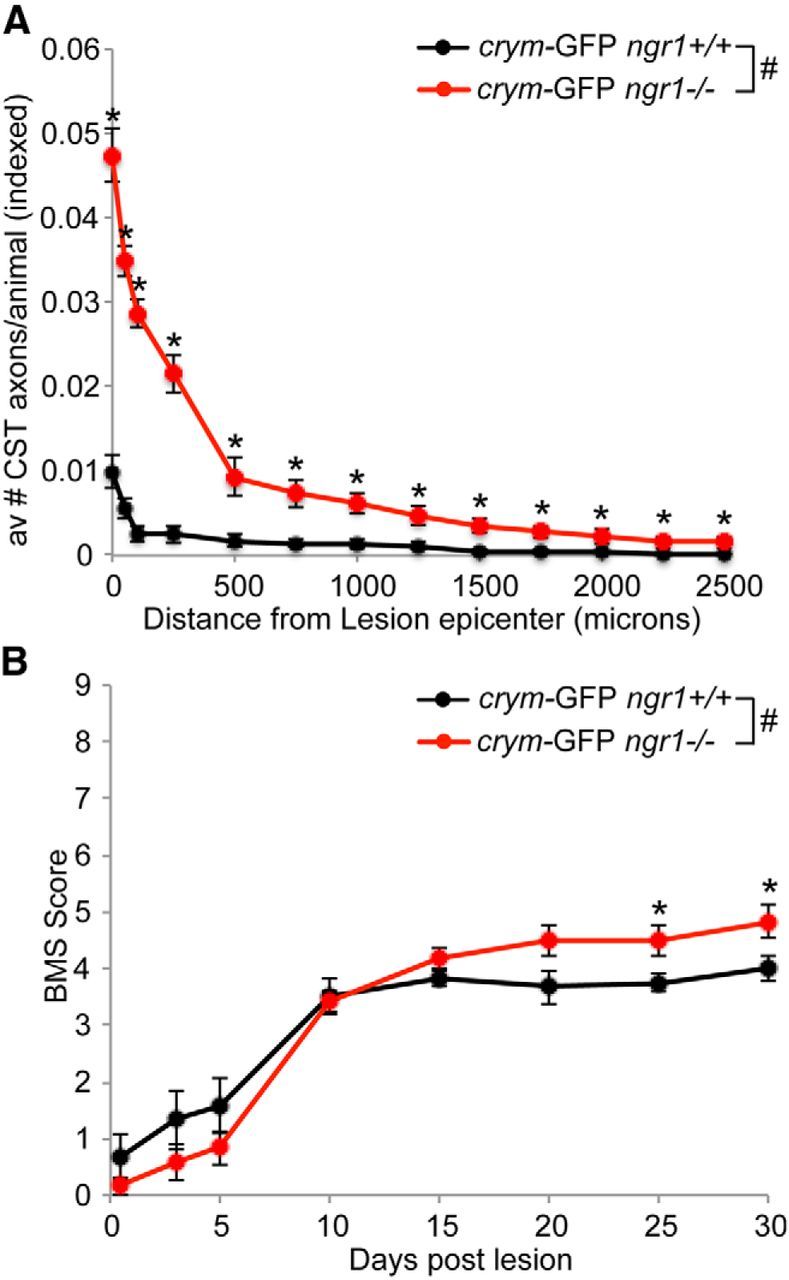
Ngr1−/− mice show significantly more CST regeneration and functional recovery after DhX compared with ngr1+/+ mice. The number of GFP+ CST axons were counted in five medial sagittal sections of ngr1+/+ and ngr1−/− mice and normalized to the number of CST axons present in the dorsal columns of intact crym-GFP mice. Significantly more GFP+ CST axons were observed caudal to the lesion site in ngr1−/− compared with ngr1+/+ mice (A, #F(1, 10) = 11.694, p < 0.01 two-way repeated-measures ANOVA with Bonferonni correction for multiple comparisons, with post hoc t test, *p < 0.05). Lesioned mice underwent behavioral assessment in the open field using the BMS. Acutely after DhX both ngr1+/+ and ngr1−/− mice exhibited flaccid paralysis. By 30 d after lesion, ngr1−/− mice recovered significant function compared with ngr1+/+ mice (B, #F(1, 5) = 588.193, p < 0.05 two-way repeated-measures ANOVA with Bonferonni correction for multiple comparisons with post hoc ANOVA, *p < 0.05).
Figure 11.
Comprehensive CST labeling in crym-GFP mice reveal substantially more regenerating CST axons compared with extrinsic BDA tracing. Low-power photomicrographs of midsagittal sections from two crym-GFP ngr1−/− mice (A, C) show BDA+ CST axons rostral to a DhX lesion site (lesion epicenter is marked with an asterisk, inset compass in A shows orientation; D, dorsal; V, ventral; R, rostral; C, caudal). High-power photomicrographs from stippled box insets show dense BDA labeling in the dorsal column projection (A1i, C1i), few BDA+ axons entering the rostral lesion site and (A1ii, C1ii), and virtually no BDA+ axons caudal to the lesion site (A1iii, C1iii). However, viewing sections under GFP illumination revealed robust detection of crym-GFP+ CST axons both rostral and caudal to the lesion site (B, D; crym-GFP, green; BDA, red). High-power photomicrographs show considerably more crym-GFP+ CST axons rostral (A2i, C2i), within (A2ii, C2ii), and caudal to the lesion site (A2iii, C2iii) compared with BDA+ axons (A3i–A3iii and C3i–C3iii, overlays; crym-GFP, green; BDA, red). Scale bars: A, 1 mm; A1i, 100 μm.
Figure 12.
Comprehensive CST labeling in crym-GFP transgenic mice reveal extensive CST regeneration within the lesion site. Low-power photomicrographs of midsagittal sections from two crym-GFP ngr1−/− mice (A, C) show GFAP immunoreactivity (GFAP-IR) and reveal extensive astrogliosis at the DhX site (lesion epicenter is marked with an asterisk, inset compass in A shows orientation: D, dorsal; V, ventral; R, rostral; C, caudal). High-power photomicrographs from stippled box insets show GFAP-IR of quiescent astrocytes rostral to the lesion (A1i, C1i), dense hypertrophy of astrocytes in the lesion epicenter (A1ii, C1ii), and a decreasing hypertrophic profile caudal to the lesion site (A1iii, C1iii). Viewing sections under GFP illumination revealed robust detection of crym-GFP+ CST axons both rostral and caudal to the lesion site (B, D; crym-GFP, green; GFAP, red). High-power photomicrographs show extensive labeling of crym-GFP+ CST axons rostral to the lesion site (A2i, C2i), and regenerating into (A2ii, C2ii) and caudal to the lesion site (A2iii, C2iii) through areas of intense GFAP-IR (A3i-iii, C3i-iii, overlays; crym-GFP, green; GFAP, red). Scale bars: A, 1 mm; A1i, 100 μm.
We examined crym-GFP labeling in transverse sections of motor cortex (Fig. 4A, B) in intact ngr1+/+ and ngr1−/− mice. Costaining for Nissl (Fig. 4C–F) revealed that crym-GFP labeled 34.4 ± 2.4% of layer V pyramidal neurons in ngr1+/+ mice and 34.9 ± 2.4% of layer V neurons in ngr1−/− mice (Fig. 4M). Because only a subset of layer V neurons were crym-GFP+, we sought to determine whether this subset was projecting to either the spinal cord, intracortically across the corpus callosum, or to brainstem nuclei. To this end, we injected the retrograde tracer FB into the cervical enlargement to label CSMNs that project to the spinal cord. FB can be seen accumulating in a subset of layer V neurons in forelimb motor cortex 2 weeks after intraspinal injection (Fig. 4G–L). Colabeling sections with GFP revealed that 94.7 ± 0.6% and 94.2 ± 0.4% of FB+ cells were also GFP+ in crym-GFP ngr1+/+ and crym-GFP ngr1−/− mice, respectively (Fig. 4N). These data confirm that crym-GFP is specifically labeling spinally projecting layer V neurons in motor cortex and that there is no significant difference in the number or location of spinally projecting CSMNs in ngr1−/− compared with ngr1+/+ mice.
CST fascicule labeling is superior in crym-GFP mice compared with anterograde BDA transport
We next sought to examine CST axon number and spinal terminal density in crym-GFP ngr1+/+ mice and determine whether the CST is miswired in crym-GFP ngr1−/− mice. Traditionally, the CST has been visualized in the spinal cord after injection of BDA into multiple sites in motor cortex (Tuszynski and Steward, 2012). Using this method, normal CST wiring was observed in ngr1−/− mice previously (Kim et al., 2004; Cafferty and Strittmatter, 2006). We sought to determine whether crym-GFP labeling of the CST was more efficient than BDA tracing and thus able to distinguish any potential differences in CST anatomy between ngr1+/+ and ngr1−/− mice. Transverse sections through the brainstem (Fig. 5A–C), cervical (Fig. 5D–F), thoracic (Fig. 5G–I), and lumbar (Fig. 5J–L) spinal cord 2 weeks after intracortical BDA delivery revealed that, qualitatively, BDA labeled a fraction of the CST compared with crym-GFP at all levels of the neuroaxis.
Close inspection of BDA+ and crym-GFP+ CST axon profiles in the pyramidal tract rostral to the decussation in the brainstem (Fig. 6A,B) showed that, on average, 17205 ± 593 and 17107 ± 1078 CST axons were GFP+ in crym-GFP ngr1+/+ and crym-GFP ngr1−/− mice, respectively (Fig. 6D). However, intracortical BDA injection (Fig. 6A) labeled only 1634 ± 167 and 1787 ± 204 CST axons in crym-GFP ngr1+/+ and crym-GFP ngr1−/− mice, respectively, significantly less than the number of GFP+ axons (Fig. 6D; *p < 0.0001, ANOVA). These data show that crym-GFP is labeling 10 times the number of CST axons that are commonly labeled with intracortical BDA injection. To determine whether this trend was consistent throughout the neuroaxis, we examined the number of GFP+ CST axon profiles and BDA+ profiles in the dCST (Fig. 6C,E–H), dlCST (Fig. 6C,I–L), and vCST (Fig. 6C,M–P) in the cervical (Fig. 6E,I,M), thoracic (Fig. 6F,J,N), and lumbar (Fig. 6G,K,O) spinal cord in intact crym-GFP ngr1+/+ and crym-GFP ngr1−/− mice 2 weeks after intracortical BDA injection. As expected, the average number of crym-GFP+ and BDA+ CST profiles decreased caudally as axons terminated in spinal gray matter (Fig. 6H,L,P). However, significantly more GFP+ CST axon profiles were observed in all white matter locations at all spinal levels compared with the number of BDA+ axonal profiles (Fig. 6H,L,P; *p < 0.001, ANOVA), maintaining at least a 10:1 CST labeling superiority of crym-GFP over BDA. Despite the difference in the efficiency between crym-GFP and BDA in labeling the CST, there was no significant difference in the number of GFP+ CST axon profiles or BDA+ profiles between genotypes, confirming normal CST fasciculation in ngr1−/− mice.
Crym-GFP reveals extensive CST terminal innervation in spinal gray matter and normal CST wiring in ngr1−/− mice
Because the number of CST axons labeled in crym-GFP was 10-fold higher than commonly observed with BDA tracing, we sought to explore the CST termination pattern in spinal gray matter (Fig. 7A,B) and also to determine whether spinal CST innervation was aberrant in ngr1−/− mice. Densitometric analysis (Fig. 7O) was completed on high-power photomicrographs of transverse sections of crym-GFP (Fig. 7C,D,G–J,M,N) and BDA (Fig. 7E–H, K–N)-labeled axons in the cervical spinal dorsal (Fig. 7C–H) and ventral horns (Fig. 7I–N). Consistent with the white matter quantification, crym-GFP labels 10 times more axons in spinal gray matter than BDA (Fig. 7O). No difference in gray matter innervation was observed between crym-GFP ngr1+/+ and crym-GFP and ngr1−/− mice (Fig. 7O), indicating that there are no developmental wiring defects in the CST of ngr1−/− mice.
Comprehensive CST labeling reveals CST regeneration in ngr1−/− mice
CSMNs express NgR1 (McGee and Strittmatter, 2003; Zheng et al., 2005), so CST axons are restricted from spontaneous plasticity and regeneration by myelin-associated inhibitors expressed in the intact and degenerating CNS (Schwab and Strittmatter, 2014). Previously, we have shown that mice null for NgR1 display enhanced plasticity of the processes of CSMNs in the adult cortex (Lee et al., 2004) and spinal cord (Cafferty and Strittmatter, 2006). Furthermore, pharmacological inhibition of NgR1 activation resulted in increased CST regeneration after experimental SCI (Li et al., 2004a; Wang et al., 2006; Wang et al., 2011). However, despite increased regeneration of rubrospinal axons and increased sprouting of raphespinal axons, no CST regeneration was observed after DhX in ngr1−/− mice (Kim et al., 2004). With the significantly enhanced CST labeling efficiency afforded by crym-GFP mice over BDA labeling and the lack of developmental CST wiring defects observed in crym-GFP ngr1−/− mice, we decided to revisit whether bona fide CST regeneration occurs in ngr1−/− mice after DhX.
Crym-GFP ngr1+/+ mice (n = 11) and crym-GFP ngr1−/− mice (n = 15) received DhX SCI at T6 for anatomical and behavioral analyses. Six weeks after injury, no residual degenerating crym-GFP axons were seen caudal to the lesion site in either genotype (Figs. 8, 9). GFP+ CST axons can be seen regenerating up to the lesion site in crym-GFP ngr1+/+ mice, with very few, if any, regenerating CST axons observed caudal to the lesion site (Figs. 8A, 9A). However, crym-GFP ngr1−/− mice exhibit significant regeneration of crym-GFP CST axons through and caudal to the lesion site (Figs. 8B–D, 9B–D). Figures 8 and 9 exhibit comparable spinal sections from six independent examples that demonstrate CST regeneration in ngr1−/− mice is robust and reproducible. Crym-GFP+ regenerating axons were quantified at several distances caudal to the lesion epicenter (Fig. 10A). Ngr1−/− mice exhibited significantly greater CST regeneration compared with ngr1+/+ mice (Fig. 10A; #p < 0.01, two-way ANOVA with repeated measures). Ngr1−/− mice also exhibited enhanced locomotor recovery in the open field (Fig. 10B; #p < 0.05, two-way ANOVA with repeated measures), consistent with our previous data (Kim et al., 2004).
To determine whether detection sensitivity was wholly responsible for the previously observed lack of CST regeneration in ngr1−/− mice, crym-GFP mice that received DhX also received intracortical BDA injections 2 weeks before killing. Midsagittal sections through the lesion site revealed BDA+ CST axons rostral to the lesion site in the main dorsal CST fascicule and terminating in gray matter (Fig. 11A,C). Few BDA+ CST axons can be seen regenerating through and past the lesion site (Fig. 11A1ii, C1ii,A1iii,C1iii). Costaining the same sections with for GFP (Fig. 11B,D) revealed densely labeled crym-GFP+ CST axons regenerating into and immediately caudal to the lesion site (Fig. 11A2ii–3ii, C2ii–C3ii) and evidence of crym-GFP+ CST axons more distally from the lesion site (Fig. 11A2iii–A3iii,C2iii–C3iii). These data further suggest that inefficient BDA labeling obscures the comprehensive CST regenerative phenotype in ngr1−/− mice.
The significantly enhanced capacity of crym-GFP to localize the CST allows for more detailed investigation of how plastic or regenerating CST axons interact with the intact or lesioned CNS environment. To explore how crym-GFP+ CST axons respond to the glial scar after DhX, we costained sagittal sections through the lesion site from crym-GFP ngr1−/− mice with GFAP (Fig. 12A–D). GFAP staining clearly delineates the lesion site in crym-GFP ngr1−/− mice (Fig. 12A,A1ii,C,C1ii) in a manner indistinguishable from previous studies (Cafferty et al., 2007b; Cafferty et al., 2010; Geoffroy et al., 2015). Strikingly, significant numbers of GFP+ CST axons can be seen growing into and beyond scar regions intensely stained with GFAP (Fig. 12B, B3ii, B3iii,D,D3ii,D3iii), thereby confirming bona fide CST axon regeneration. This is consistent with the hypothesis that eliminating NgR1 desensitizes axons to the inhibitory effects of degenerating CNS myelin (Fournier et al., 2001; Kim et al., 2004; Cafferty et al., 2010) and CSPGs (Dickendesher et al., 2012) deposited in and around the glial scar. In sum, these data indicate that, by labeling the CST in a comprehensive manner using the crym-GFP system, we are able to observe significant CST regeneration in ngr1−/− mice after DhX and furthermore support the utility of this model when assessing the CST regenerative capacity of potential therapeutic interventions, thereby minimizing anatomical variations within experiments and between laboratories.
Discussion
To date, comprehensive assessment of axon regeneration after SCI has been fraught by the need to use inefficient and inconsistent extrinsic labeling methodologies (Tuszynski and Steward, 2012). Here, we introduce a model in which the CST is comprehensively and reliably genetically labeled via expression of a fluorescent reporter controlled by the crym promoter. In this study, we: (1) show that crym-GFP mice label 10 times the number of CST axons at all levels of the neuroaxis compared with the anterograde CST tracer BDA, (2) reveal the comprehensive spinal gray matter termination pattern of the CST, and (3) demonstrate the streamlined utility of this transgenic line for in vivo SCI experiments for crossing onto knock-out strains. In addition, by completing DhX SCI on crym-GFP ngr1+/+ and crym-GFP ngr1−/− mice, we can draw several conclusions regarding the capacity of the CST to mount a regenerative response in the presence and absence of NgR1. First, ngr1−/− mice have indistinguishable CST white and gray matter wiring compared with ngr1+/+ mice. Second, crym-GFP ngr1+/+ reveal that the CSMNs mount a modest regenerative response after DhX, with a small number of CST axons regenerating up to and into the lesion site. Third, crym-GFP ngr1−/− mice show significant CST axon regeneration after DhX compared with ngr1+/+ mice, providing further evidence that NgR1 activation is a significant abortive signal for regeneration of adult CST axons.
Inefficiency of extrinsic axon tracing methodology
Abortive axon regeneration in the adult CNS remains a significant barrier to functional SCI recovery (Filli and Schwab, 2012). To test the functional efficacy of proregeneration therapeutics in vivo, investigators routinely deliver subcomplete lesions to the dorsal spinal cord and monitor functional recovery over time. These data are then correlated with anatomy in fixed tissue sections at the end of the experiment. This approach is preferred from a functional perspective as these lesions are moderately easy to complete; interrupt a number of crucial descending spinal motor tracts, including the CST; and result in a reproducible loss of motor function that can be evaluated using standardized tests (Basso et al., 1995; Basso et al., 2006). However, assessing axon regeneration in these models is complicated because it requires discrete delivery of anterograde tracers to axotomized somata. BDA is commonly used for this purpose via cortical microinjections into layer V of M1. Although BDA is absorbed and transported by a number of CSMNs, its delivery requires an additional surgery and multiple injection sites and therefore routinely results in varied, inconsistent, and incomplete filling of regenerating axons in experimental animals within and between groups (Steward et al., 2008b).
Transgenic labeling of the CST with crym-GFP is superior to BDA
We sought to identify an alternative method for labeling CSMNs that would obviate the need for inefficient extrinsic tracer delivery and more comprehensively label the CST. To this end, we explored the GENSAT catalog and found the crym BAC transgenic line that appeared to express GFP specifically in CSMNs. Close investigation revealed robust and comprehensive GFP expression in CSMNs, their axons, and their terminals. We injected BDA into M1 of crym-GFP mice to compare directly the efficiency of these two approaches to label the CST. We found that crym-GFP mice label 10 times the number of CST axons in spinal white matter and CST terminals in spinal gray matter compared with BDA (Figs. 6, 7). Previous studies have also reported the benefits of using transgenic fluorescent reporter lines to study CST anatomy; however, none has directly compared labeling efficiency directly with BDA (Bareyre et al., 2005; Carter et al., 2008). The YFP-H line expresses YFP under the thy1 promoter and therefore labels, not only CSMNs, but also other classes of cortical, subcortical, and spinal neurons (Carter et al., 2008). This line has myriad benefits for studying anatomy ex vivo (Chung and Deisseroth, 2013) and in vivo (Holtmaat et al., 2012; Akbik et al., 2013); however, the stochastic nature of YFP expression (Feng et al., 2000) renders it inappropriate for CST SCI studies (Carter et al., 2008). A more refined approach to labeling the CST was achieved by Bareyre et al. (2005), who developed a double transgenic line by crossing thy1-STOP-YFP mice with Emx1-cre mice. Progeny of this cross (CST-YFP mice) show robust expression of YFP in CSMNs. The number of YFP-labeled CST axons in the dCST, dlCST, and vCST in the cervical, thoracic, and lumbar spinal cord in CST-YFP mice were comparable to our counts in the crym-GFP line. However, despite equivalent labeling of CST axons in the spinal cord, >90% of layer V pyramidal neurons were labeled in the CST-YFP line; however, only 30% of layer V neurons were labeled in our crym-GFP line. This suggests that YFP expression in CST-YFP mice is not restricted to spinally projecting CSMNs, but also labels layer V neurons, sending projections intracortically and corticofugally. Our data show that crym-GFP exclusively labels spinally projecting CSMNs because ∼100% of CSMNs retrogradely traced via intraspinal delivery of FB were crym-GFP+ (Fig. 4). Furthermore, differential gene expression analysis comparing corticospinal, cortico–cortico, and corticofugal CSMNs revealed that crym was enriched in the corticospinal class (Arlotta et al., 2005), thus confirming crym as an ideal candidate to target reporter expression to the CST.
CST axon regeneration in ngr1+/+ and ngr1−/− mice
The single transgenic approach presented by crym-GFP line mice allows for streamlined breeding compared with double transgenic CST-YFP mice onto existing knock-out and knock-in lines. We crossed crym-GFP mice with ngr1−/− mice to revisit CST axon regeneration in these knock-out mice. Abundant genetic reports support a critical role for NgR1 in transducing inhibitory signals in the intact and lesioned CNS that prevent axon regeneration (Li et al., 2005), sprouting (Cafferty and Strittmatter, 2006; Geoffroy et al., 2015; Siegel et al., 2015), and plasticity (Lee et al., 2004; McGee et al., 2005; Akbik et al., 2013). Furthermore, pharmacological inhibition of NgR1 signaling also results in significant CST axon regeneration (Li et al., 2004b; Wang et al., 2006; Wang et al., 2011). Therefore, it was puzzling that we did not detect CST axon regeneration after DhX in our previous study (Kim et al., 2004) despite the emergence of locomotor recovery. The discrepancy in axon regeneration phenotype between constitutive knock-out and pharmacological inhibition of NgR1 may have arisen due aberrant CST wiring in ngr1−/− compared with ngr1+/+ mice or the incomplete tracing of regenerating CST axons with BDA. We addressed both of these possibilities using crym-GFP line mice. By crossing ngr1−/− with crym-GFP, we were able to complete careful anatomical analysis of crym-GFP+ and BDA+ CST axons in the same animals in both crym-GFP ngr1+/+ and crym-GFP ngr1−/− mice. Despite crym-GFP labeling being 10 times more sensitive than BDA tracing, we did not find a difference in the number of crym-GFP+ or BDA+ CST axons in white matter (Fig. 6) or terminals in gray matter (Fig. 7) between ngr1+/+ and ngr1−/− mice. Therefore, using a more sensitive measure of CST wiring, we can confirm our earlier report that constitutive ngr1−/− mice have normal CST patterning (Kim et al., 2004; Cafferty and Strittmatter, 2006). Next, we sought to determine whether the significantly enhanced sensitivity of CST labeling afforded by crym-GFP would yield a deeper insight into the regenerative capacity of axotomized CST axons. Indeed, inspection of sagittal sections through the lesion site 6 weeks after DhX in crym-GFP ngr1+/+ mice showed that large numbers of GFP+ CST axons had regenerated up to the lesion site, with a few axons observed caudal to the lesion epicenter (Figs. 8A,9A). Significantly more GFP+ CST axons were observed regenerating caudal to the lesion site in crym-GFP ngr1−/− mice (Figs. 8B–D,9B–D). A percentage of regenerating GFP+ CST axons were also BDA+ (Fig. 11); no axons were BDA+ and GFP−. The majority of GFP+-regenerating CST axons in crym-GFP ngr1−/− mice were BDA−, further underscoring the need for more comprehensive CST labeling techniques to subvert underinterpretation or overinterpretation of regeneration data. From these data, we conclude that the CST mounts a significant regenerative response in constitutive ngr1−/− mice.
In summary, incomplete CST labeling may in part be responsible for the difficulty in reproducing SCI experiments (Steward et al., 2012). Our current data demonstrate that BDA labels ∼10% of a mixed population of corticospinal, cortico–cortico, and corticofugal CSMN axons. This paltry percentage allows for significant variation in region and density of layer V neurons in M1 that are labeled between animals, surgical days, surgeons, and laboratories. Comprehensive, robust, and invariant CST labeling in transgenic crym-GFP mice allows for the standardization of anatomical analyses of CST regeneration after SCI and provides a framework with which to study the molecular mechanisms that restrict and enhance CST axon plasticity, sprouting, and regeneration.
Footnotes
This work was supported by the National Institutes of Health (Grant R01NS080388 to S.M.S.) and the Falk Medical Research Trust (S.M.S.).
S.M.S. is a cofounder of Axerion Therapeutics, seeking to develop NgR- and PrP-based therapeutics. The remaining authors declare no competing financial interests.
References
- Akbik FV, Bhagat SM, Patel PR, Cafferty WB, Strittmatter SM. Anatomical plasticity of adult brain is titrated by Nogo Receptor 1. Neuron. 2013;77:859–866. doi: 10.1016/j.neuron.2012.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbik F, Cafferty WB, Strittmatter SM. Myelin associated inhibitors: a link between injury-induced and experience-dependent plasticity. Exp Neurol. 2012;235:43–52. doi: 10.1016/j.expneurol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Atwal JK, Pinkston-Gosse J, Syken J, Stawicki S, Wu Y, Shatz C, Tessier-Lavigne M. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- Bareyre FM, Kerschensteiner M, Misgeld T, Sanes JR. Transgenic labeling of the corticospinal tract for monitoring axonal responses to spinal cord injury. Nat Med. 2005;11:1355–1360. doi: 10.1038/nm1331. [DOI] [PubMed] [Google Scholar]
- Bartus K, James ND, Didangelos A, Bosch KD, Verhaagen J, Yáñez-Muñoz RJ, Rogers JH, Schneider BL, Muir EM, Bradbury EJ. Large-scale chondroitin sulfate proteoglycan digestion with chondroitinase gene therapy leads to reduced pathology and modulates macrophage phenotype following spinal cord contusion injury. J Neurosci. 2014;34:4822–4836. doi: 10.1523/JNEUROSCI.4369-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso DM, Beattie MS, Bresnahan JC. A sensitive and reliable locomotor rating scale for open field testing in rats. J Neurotrauma. 1995;12:1–21. doi: 10.1089/neu.1995.12.1. [DOI] [PubMed] [Google Scholar]
- Basso DM, Fisher LC, Anderson AJ, Jakeman LB, McTigue DM, Popovich PG. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J Neurotrauma. 2006;23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- Blackmore MG, Wang Z, Lerch JK, Motti D, Zhang YP, Shields CB, Lee JK, Goldberg JL, Lemmon VP, Bixby JL. Kruppel-like Factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc Natl Acad Sci U S A. 2012;109:7517–7522. doi: 10.1073/pnas.1120684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Cafferty WB, Strittmatter SM. The Nogo-Nogo receptor pathway limits a spectrum of adult CNS axonal growth. J Neurosci. 2006;26:12242–12250. doi: 10.1523/JNEUROSCI.3827-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty WB, Kim JE, Lee JK, Strittmatter SM. Response to correspondence: Kim et al., “axon regeneration in young adult mice lacking Nogo-A/B.”. Neuron. 2007a;38:187–199. doi: 10.1016/j.neuron.2007.04.005. Neuron 54:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty WB, Yang SH, Duffy PJ, Li S, Strittmatter SM. Functional axonal regeneration through astrocytic scar genetically modified to digest chondroitin sulfate proteoglycans. J Neurosci. 2007b;27:2176–2185. doi: 10.1523/JNEUROSCI.5176-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty WB, Duffy P, Huebner E, Strittmatter SM. MAG and OMgp synergize with Nogo-A to restrict axonal growth and neurological recovery after spinal cord trauma. J Neurosci. 2010;30:6825–6837. doi: 10.1523/JNEUROSCI.6239-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter LM, Starkey ML, Akrimi SF, Davies M, McMahon SB, Bradbury EJ. The yellow fluorescent protein (YFP-H) mouse reveals neuroprotection as a novel mechanism underlying chondroitinase ABC-mediated repair after spinal cord injury. J Neurosci. 2008;28:14107–14120. doi: 10.1523/JNEUROSCI.2217-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Dickendesher TL, Baldwin KT, Mironova YA, Koriyama Y, Raiker SJ, Askew KL, Wood A, Geoffroy CG, Zheng B, Liepmann CD, Katagiri Y, Benowitz LI, Geller HM, Giger RJ. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat Neurosci. 2012;15:703–712. doi: 10.1038/nn.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou L, Schnell L, Montani L, Duncan C, Simonen M, Schneider R, Liebscher T, Gullo M, Schwab ME. Nogo-A-deficient mice reveal strain-dependent differences in axonal regeneration. J Neurosci. 2006;26:5591–5603. doi: 10.1523/JNEUROSCI.1103-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/S0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Festing MF, Altman DG. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 2002;43:244–258. doi: 10.1093/ilar.43.4.244. [DOI] [PubMed] [Google Scholar]
- Filli L, Schwab ME. The rocky road to translation in spinal cord repair. Ann Neurol. 2012;72:491–501. doi: 10.1002/ana.23630. [DOI] [PubMed] [Google Scholar]
- Fisher D, Xing B, Dill J, Li H, Hoang HH, Zhao Z, Yang XL, Bachoo R, Cannon S, Longo FM, Sheng M, Silver J, Li S. Leukocyte common antigen-related phosphatase is a functional receptor for chondroitin sulfate proteoglycan axon growth inhibitors. J Neurosci. 2011;31:14051–14066. doi: 10.1523/JNEUROSCI.1737-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPré T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- Fry EJ, Chagnon MJ, López-Vales R, Tremblay ML, David S. Corticospinal tract regeneration after spinal cord injury in receptor protein tyrosine phosphatase sigma deficient mice. Glia. 2010;58:423–433. doi: 10.1002/glia.20934. [DOI] [PubMed] [Google Scholar]
- Geoffroy CG, Lorenzana AO, Kwan JP, Lin K, Ghassemi O, Ma A, Xu N, Creger D, Liu K, He Z, Zheng B. Effects of PTEN and Nogo codeletion on corticospinal axon sprouting and regeneration in mice. J Neurosci. 2015;35:6413–6428. doi: 10.1523/JNEUROSCI.4013-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- GrandPré T, Li S, Strittmatter SM. Nogo-66 receptor antagonist peptide promotes axonal regeneration. Nature. 2002;417:547–551. doi: 10.1038/417547a. [DOI] [PubMed] [Google Scholar]
- Hallen A, Cooper AJ, Jamie JF, Haynes PA, Willows RD. Mammalian forebrain ketimine reductase identified as mu-crystallin; potential regulation by thyroid hormones. J Neurochem. 2011;118:379–387. doi: 10.1111/j.1471-4159.2011.07220.x. [DOI] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, de Paola V, Wilbrecht L, Trachtenberg JT, Svoboda K, Portera-Cailliau C. Imaging neocortical neurons through a chronic cranial window. Cold Spring Harb Protoc. 2012;2012:694–701. doi: 10.1101/pdb.prot069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf A, Tews B, Arzt ME, Weinmann O, Obermair FJ, Pernet V, Zagrebelsky M, Delekate A, Iobbi C, Zemmar A, Ristic Z, Gullo M, Spies P, Dodd D, Gygax D, Korte M, Schwab ME. The sphingolipid receptor S1PR2 is a receptor for Nogo-a repressing synaptic plasticity. PLoS Biol. 2014;12:e1001763. doi: 10.1371/journal.pbio.1001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Li S, GrandPré T, Qiu D, Strittmatter SM. Axon regeneration in young adult mice lacking Nogo-A/B. Neuron. 2003;38:187–199. doi: 10.1016/S0896-6273(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Kim JE, Liu BP, Park JH, Strittmatter SM. Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron. 2004;44:439–451. doi: 10.1016/j.neuron.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Kim RY, Gasser R, Wistow GJ. mu-crystallin is a mammalian homologue of Agrobacterium ornithine cyclodeaminase and is expressed in human retina. Proc Natl Acad Sci U S A. 1992;89:9292–9296. doi: 10.1073/pnas.89.19.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BT, Cregg JM, DePaul MA, Tran AP, Xu K, Dyck SM, Madalena KM, Brown BP, Weng YL, Li S, Karimi-Abdolrezaee S, Busch SA, Shen Y, Silver J. Modulation of the proteoglycan receptor PTPsigma promotes recovery after spinal cord injury. Nature. 2015;518:404–408. doi: 10.1038/nature13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Kim JE, Sivula M, Strittmatter SM. Nogo receptor antagonism promotes stroke recovery by enhancing axonal plasticity. J Neurosci. 2004;24:6209–6217. doi: 10.1523/JNEUROSCI.1643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Geoffroy CG, Chan AF, Tolentino KE, Crawford MJ, Leal MA, Kang B, Zheng B. Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66:663–670. doi: 10.1016/j.neuron.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu BP, Budel S, Li M, Ji B, Walus L, Li W, Jirik A, Rabacchi S, Choi E, Worley D, Sah DW, Pepinsky B, Lee D, Relton J, Strittmatter SM. Blockade of nogo-66, myelin-associated glycoprotein, and oligodendrocyte myelin glycoprotein by soluble nogo-66 receptor promotes axonal sprouting and recovery after spinal injury. J Neurosci. 2004a;24:10511–10520. doi: 10.1523/JNEUROSCI.2828-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kim JE, Budel S, Hampton TG, Strittmatter SM. Transgenic inhibition of Nogo-66 receptor function allows axonal sprouting and improved locomotion after spinal injury. Mol Cell Neurosci. 2005;29:26–39. doi: 10.1016/j.mcn.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Walus L, Rabacchi SA, Jirik A, Chang E, Schauer J, Zheng BH, Benedetti NJ, Liu BP, Choi E, Worley D, Silvian L, Mo W, Mullen C, Yang W, Strittmatter SM, Sah DW, Pepinsky B, Lee DH. A neutralizing anti-Nogo66 receptor monoclonal antibody reverses inhibition of neurite outgrowth by central nervous system myelin. J Biol Chem. 2004b;279:43780–43788. doi: 10.1074/jbc.M401803200. [DOI] [PubMed] [Google Scholar]
- Liebscher T, Schnell L, Schnell D, Scholl J, Schneider R, Gullo M, Fouad K, Mir A, Rausch M, Kindler D, Hamers FP, Schwab ME. Nogo-A antibody improves regeneration and locomotion of spinal cord-injured rats. Ann Neurol. 2005;58:706–719. doi: 10.1002/ana.20627. [DOI] [PubMed] [Google Scholar]
- Liu K, Tedeschi A, Park KK, He Z. Neuronal intrinsic mechanisms of axon regeneration. Annu Rev Neurosci. 2011;34:131–152. doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- McGee AW, Strittmatter SM. The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends Neurosci. 2003;26:193–198. doi: 10.1016/S0166-2236(03)00062-6. [DOI] [PubMed] [Google Scholar]
- McGee AW, Yang Y, Fischer QS, Daw NW, Strittmatter SM. Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science. 2005;309:2222–2226. doi: 10.1126/science.1114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell L, Schwab ME. Axonal regeneration in the rat spinal cord produced by an antibody against myelin-associated neurite growth inhibitors. Nature. 1990;343:269–272. doi: 10.1038/343269a0. [DOI] [PubMed] [Google Scholar]
- Schwab ME, Strittmatter SM. Nogo limits neural plasticity and recovery from injury. Curr Opin Neurobiol. 2014;27:53–60. doi: 10.1016/j.conb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tenney AP, Busch SA, Horn KP, Cuascut FX, Liu K, He Z, Silver J, Flanagan JG. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel CS, Fink KL, Strittmatter SM, Cafferty WB. Plasticity of intact rubral projections mediates spontaneous recovery of function after corticospinal tract injury. J Neurosci. 2015;35:1443–1457. doi: 10.1523/JNEUROSCI.3713-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonen M, Pedersen V, Weinmann O, Schnell L, Buss A, Ledermann B, Christ F, Sansig G, van der Putten H, Schwab ME. Systemic deletion of the myelin-associated outgrowth inhibitor Nogo-A improves regenerative and plastic responses after spinal cord injury. Neuron. 2003;38:201–211. doi: 10.1016/S0896-6273(03)00226-5. [DOI] [PubMed] [Google Scholar]
- Steward O, Zheng B, Tessier-Lavigne M. False resurrections: distinguishing regenerated from spared axons in the injured central nervous system. J Comp Neurol. 2003;459:1–8. doi: 10.1002/cne.10593. [DOI] [PubMed] [Google Scholar]
- Steward O, Zheng B, Banos K, Yee KM. Response to: Kim et al., “axon regeneration in young adult mice lacking Nogo-A/B.”. Neuron. 2007;54:191–195. doi: 10.1016/j.neuron.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Steward O, Sharp K, Yee KM, Hofstadter M. A re-assessment of the effects of a Nogo-66 receptor antagonist on regenerative growth of axons and locomotor recovery after spinal cord injury in mice. Exp Neurol. 2008a;209:446–468. doi: 10.1016/j.expneurol.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Zheng B, Tessier-Lavigne M, Hofstadter M, Sharp K, Yee KM. Regenerative growth of corticospinal tract axons via the ventral column after spinal cord injury in mice. J Neurosci. 2008b;28:6836–6847. doi: 10.1523/JNEUROSCI.5372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Popovich PG, Dietrich WD, Kleitman N. Replication and reproducibility in spinal cord injury research. Exp Neurol. 2012;233:597–605. doi: 10.1016/j.expneurol.2011.06.017. [DOI] [PubMed] [Google Scholar]
- Tuszynski MH, Steward O. Concepts and methods for the study of axonal regeneration in the CNS. Neuron. 2012;74:777–791. doi: 10.1016/j.neuron.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vié MP, Evrard C, Osty J, Breton-Gilet A, Blanchet P, Pomérance M, Rouget P, Francon J, Blondeau JP. Purification, molecular cloning, and functional expression of the human nicodinamide-adenine dinucleotide phosphate-regulated thyroid hormone-binding protein. Mol Endocrinol. 1997;11:1728–1736. doi: 10.1210/mend.11.11.9915. [DOI] [PubMed] [Google Scholar]
- Wang X, Baughman KW, Basso DM, Strittmatter SM. Delayed Nogo receptor therapy improves recovery from spinal cord contusion. Ann Neurol. 2006;60:540–549. doi: 10.1002/ana.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Duffy P, McGee AW, Hasan O, Gould G, Tu N, Harel NY, Huang Y, Carson RE, Weinzimmer D, Ropchan J, Benowitz LI, Cafferty WB, Strittmatter SM. Recovery from chronic spinal cord contusion after Nogo receptor intervention. Ann Neurol. 2011;70:805–821. doi: 10.1002/ana.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Reynolds A, Kirry A, Nienhaus C, Blackmore MG. Overexpression of Sox11 promotes corticospinal tract regeneration after spinal injury while interfering with functional recovery. J Neurosci. 2015;35:3139–3145. doi: 10.1523/JNEUROSCI.2832-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenberg R, Steward O. Nonspecific labeling limits the utility of Cre-Lox bred CST-YFP mice for studies of corticospinal tract regeneration. J Comp Neurol. 2015 doi: 10.1002/cne.23809. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf CJ. No Nogo: now where to go? Neuron. 2003;38:153–156. doi: 10.1016/S0896-6273(03)00233-2. [DOI] [PubMed] [Google Scholar]
- Yasvoina MV, Genç B, Jara JH, Sheets PL, Quinlan KA, Milosevic A, Shepherd GM, Heckman CJ, Özdinler PH. eGFP expression under UCHL1 promoter genetically labels corticospinal motor neurons and a subpopulation of degeneration-resistant spinal motor neurons in an ALS mouse model. J Neurosci. 2013;33:7890–7904. doi: 10.1523/JNEUROSCI.2787-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Atwal J, Ho C, Case L, He XL, Garcia KC, Steward O, Tessier-Lavigne M. Genetic deletion of the Nogo receptor does not reduce neurite inhibition in vitro or promote corticospinal tract regeneration in vivo. Proc Natl Acad Sci U S A. 2005;102:1205–1210. doi: 10.1073/pnas.0409026102. [DOI] [PMC free article] [PubMed] [Google Scholar]