Abstract
Complex Robertsonian rearrangements, with shared arms in different fusions, are expected to prevent gene flow between hybrids through missegregation during meiosis. Here, we estimate gene flow between recently diverged and chromosomally diverse rock-wallabies (Petrogale) to test for this form of chromosomal speciation. Contrary to expectations, we observe relatively high admixture among species with complex fusions. Our results reinforce the need to consider alternative roles of chromosome change, together with genic divergence, in driving speciation.
Keywords: chromosome, speciation, monobrachial homology, gene flow
1. Introduction
Chromosome change has long been regarded as a driver of speciation [1,2]. This can happen in two ways: (I) underdominance—where hybrid individuals have reduced fertility resulting from missegregation during meiosis; and (II) recombination suppression—where genes associated with local adaptation within rearranged regions are linked as a result of reduced recombination within rearranged segments [3]; differences can then accumulate through time and cause reproductive isolation between chromosomally different populations (e.g. [4–6]). The former mechanism is commonly associated with Robertsonian changes and the latter most strongly with inversions.
Robertsonian rearrangements cause little disruption of meiosis when single fusions occur, fixing in populations under weak genetic drift or through meiotic drive [7–9]. However, hybrids between populations differing by multiple fusions with one or more arms in common (monobrachial homology) typically have severely reduced fertility owing to missegregation of complex multivalent chains (model I) [10–12]. Under this scenario, reproductive isolation occurs quickly leading to reduced gene flow between populations [7]. However, the effect of chromosomal rearrangements on gene flow is debated [4,6,13] and in systems with extensive monobrachial homology (e.g. Rattus, Mus, Sorex), the relationship of genic and karyotypic divergence is mixed [8,10,14,15].
Petrogale (rock-wallabies) have been regarded as a classic example of chromosomal speciation through meiotic breakdown (model I) [5,16,17]. The restriction of Petrogale populations to isolated rock outcrops with concomitant low dispersal and small population size has been assumed to facilitate the fixation of novel underdominant rearrangements via genetic drift [18]. Therefore, the use of Petrogale extends the scope of current model systems (e.g. rodents, shrews, Drosophila [4–6]), in which various models of chromosomal speciation can be explored and new theory developed—e.g. to incorporate drift into currently deterministic models. The six parapatric Petrogale species on the northeast coast of Australia show a range of autosomal chromosomal rearrangements, including Robertsonian fusions, inversions and centric shifts (figure 1 and table 1) [20]. The recent and rapid radiation of these taxa (0.44–1.58 Ma) [21] provides an opportunity to test for reduced gene flow in the presence of complex chromosomal changes (i.e. model I). Evidence from hybrids [16,22] indicated that genic as well as chromosomal differences may be involved in reproductive isolation (models I and II) and preliminary analyses using low-resolution methods also suggested introgression between species [23–25].
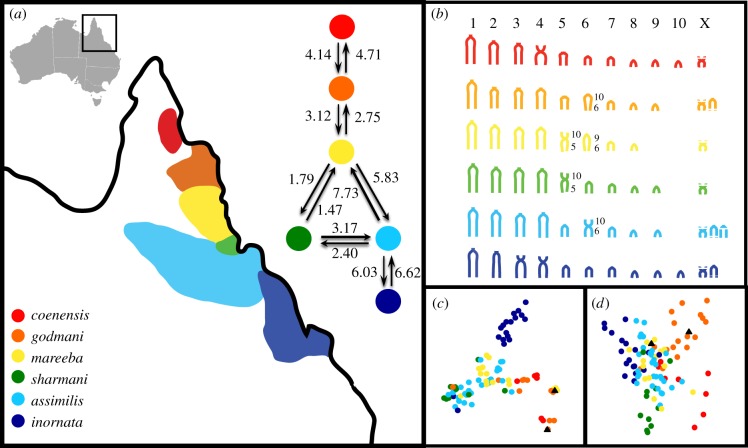
Figure 1.
(a) Distribution of six Petrogale species in northeastern Australia (colours are species-specific for 1a–d; triangles represent natural godmani × mareeba hybrids) and relative mutation-scaled migration rates between adjacent species based on MIGRATE-N [19]. (b) Schematic of species karyotypes highlighting the diagnostic Robertsonian fusions (for more detail on rearrangements refer to electronic supplementary material, table S6). (c) MDS of mtDNA based on genetic distances (percentage of variation: PC1 = 9.23%, PC2 = 7.56%). (d) PCA of microsatellite data based on species genetic distance matrix of species in (a) (percentage of variation: PC1 = 21.29%, PC2 = 18.50%). PC1 is on the X-axis, PC2 is on the Y-axis.
Table 1.
Summary of chromosome differences and known fertility for hybrids among the six northeast Queensland Petrogale. i, inversion; —, fusion; a, shift in centromere to acrocentric; FGD, fixed allozyme differences. Predicted levels of introgression are based on model I (see text).
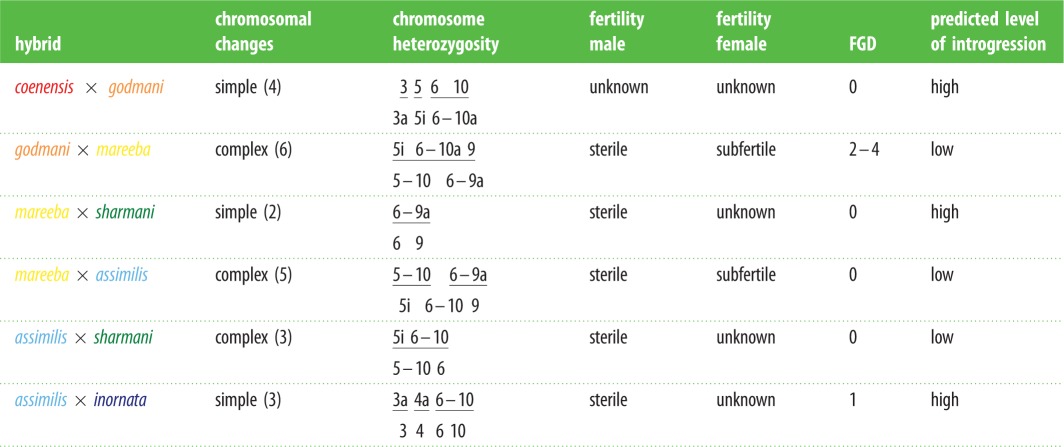 |
Here, we apply fine-scale genetic markers (mitochondrial DNA—mtDNA sequences and microsatellite genotypes) to (i) investigate the concordance between genetic and karyotypic structure across the chromosomally diverse eastern Petrogale; and (ii) test for reduced gene flow between parapatric taxa with complex monobrachial homology, compared to those with simple fusions (table 1) as expected under model I and generally assumed for this system.
2. Material and methods
One hundred and four samples were used in mtDNA and 99 in microsatellite analyses from across the distributions of six parapatric northeastern Australian Petrogale species: P. coenensis (n = 8); P. godmani (n = 17); P. mareeba (n = 16); P. sharmani (n = 11); P. assimilis (n = 31) and P. inornata (n = 18) (figure 1a; electronic supplementary material, figure S1 and table S1). In addition, two naturally occurring F1 chromosomal hybrids (godmani × mareeba) were included and a P. penicillata as an outgroup for phylogenetic analysis. Individuals were identified to species via karyotyping [16,24]. Methods for mtDNA sequencing and genotyping of 17 microsatellite loci are presented in the electronic supplementary material. Two of the 14 loci mapped to available Macropus eugenii data are on rearranged chromosomes in Petrogale (see the electronic supplementary material).
Multidimensional scaling (MDS) of sequence divergence among mtDNA haplotypes was conducted using the package ape [26] in R [27] based on Euclidean genetic distances to visualize genetic differentiation among species. In addition, a principal coordinate analysis (PCA) of microsatellite loci was analysed using a standardized covariance method in GenAlEx 6 [28]. The extent of recent admixture at microsatellite loci was also assessed using Structure (v. 2.1) ([29]; see the electronic supplementary material for details). For PCA and Structure, comparisons were conducted (i) for all species, and (ii) for comparisons between adjacent species, to assess admixture rates among geographically proximate taxa. Genic differentiation between species was also assessed using both ΦST (mtDNA) and FST (microsatellites) calculated in Arlequin (v. 3.11) [30].
Using microsatellite genotypes, we estimated relative mutation-scaled migration rates between adjacent species using coalescence in MIGRATE-N (see the electronic supplementary material). To evaluate whether high similarity among species could be the result of recent separation rather than migration, we used IMa [31] to contrast population-splitting models with and without migration, focusing on the P. assimilis, P. mareeba, P. sharmani (AMS) system (see the electronic supplementary material).
3. Results
Overall, there is no strong mtDNA structuring besides P. inornata (no reciprocal monophyly or separation in MDS—figure 1c; electronic supplementary material, figure S2). Similarly, the microsatellite loci reveal no clear genetic separation among species (PCA—figure 1d) and Structure results suggest only two genetic clusters, largely separating the northern taxa (P. coenensis and P. godmani) from the remainder (electronic supplementary material, figure S3, and tables S2 and S3).
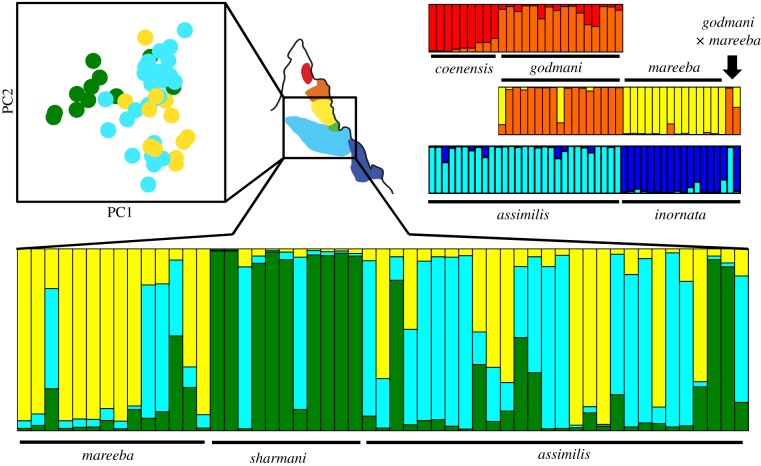
Contrary to expectations, there is no general pattern of stronger suppression of gene flow between taxa with complex versus simple Robertsonian fusions (figures 1a and 2; electronic supplementary material, figures S2 and S3). This is especially evident across the three-way contact between AMS. Admixture and gene flow are highest between P. assimilis and P. mareeba, which differ by complex fusions, whereas P. sharmani clustered separately from P. mareeba despite only a simple chromosomal difference (figures 1a and 2; electronic supplementary material, table S4). IMa nested model comparisons reject the hypothesis that genetic similarity between AMS species is owing to ancestral polymorphism alone (electronic supplementary material, table S5). At other contacts, there is evidence for low admixture, with (P. godmani/P. mareeba) or without (P. assimilis/P. inornata) monobrachial homology of fusions (figure 2; electronic supplementary material, S3). In general, parapatric species pairs have the lowest genetic differentiation (FST, ΦST; electronic supplementary material, table S3) and to some extent greater gene flow (figure 1a; electronic supplementary material, table S4), despite some evidence of separation based on pairwise PCA comparisons between geographically adjacent species (electronic supplementary material, figure S3).
Figure 2.
Genetic clustering of adjacent species of Petrogale (figure 1a) based on Structure [28] and PCA. Highlighted is the three-way comparison of Petrogale assimilis (A), P. mareeba (M) and P. sharmani (S) who share complex (A/M; A/S) and simple (S/M) rearrangements—table 1 (percentage of variation: PC1 = 21.74%, PC2 = 18.43%).
4. Discussion
The empirical results for Petrogale do not support a strong role of monobrachial homology in chromosomal speciation (model I) as has long been assumed. Specifically, we did not observe consistently reduced admixture in the presence of complex (monobrachial) versus simple Robertsonian fusions. This is concordant with other systems (e.g. Sorex, Mus) where complex rearrangements have differential roles as gene flow barriers [8,32]. The (sub)-fertility of some female hybrid Petrogale may allow for backcrossing [33] and thus introgression between species, as has been noted in the well-studied Mus system [8].
In principle, high genetic similarity (and thus high rates of gene flow inferred using MIGRATE-N) can also result from recent separation, as could occur if the fusions resulting in monobrachial homology established quickly [14]. Retained ancestral polymorphism could explain some of the admixture seen, particularly for mtDNA. However, consistent rejection of models with zero migration using IMa (electronic supplementary material, table S5) highlights that recent separation alone cannot entirely explain our results.
The fact that our predictions associated with model I were not supported suggests that other mechanisms are acting independently or synergistically with the chromosomal rearrangements to drive speciation in this system (model II). The increasing scope and sophistication of theory relating chromosome change to gene flow suppression and speciation point to the need for incorporating population genomic data to test for suppression of gene flow and/or adaptive divergence specific to rearranged regions of chromosomes (model II). Such analyses for model systems with high-quality genomes have sometimes supported these predictions (e.g. [34,35]). However, there is a need to extend this approach to systems with diverse life histories to test generality, with an initial step to demonstrate that strong reproductive isolation owing to underdominance (model I) alone does not hold. We now expect that future comparative population genomic analysis of Petrogale would therefore illuminate how different effects of chromosome change and genic divergence interact to drive speciation, and so extend the generality of such studies.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Steve Barker, Robert Close, Gerry Maynes and Geoff Sharman for sample collection, and Janine Deakin and Jason Bragg for discussions.
Data accessibility
DNA sequences: GenBank accessions JQ62304, KT726229–KT726317. Microsatellite genotypes: electronic supplementary material.
Authors' contributions
S.P. participated in design of the study, carried out the molecular laboratory work, data analysis and wrote the paper. C.M. helped interpret results and with writing the manuscript. M.D.B.E. conceived the study, participated in interpretation of the results and helped draft the manuscript. All authors gave final approval for publication and agree to be held accountable for all aspects of the work.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by the Chadwick Biodiversity Fellowship and the Australian Museum Research Institute. S.P. and C.M. are further supported by an ARC Laureate award to C.M.
References
- 1.White MJD. 1978. Modes of speciation. San Francisco, CA: W. H. Freeman. [Google Scholar]
- 2.Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 3.Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434. ( 10.1534/genetics.105.047985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358. ( 10.1016/S0169-5347(01)02187-5) [DOI] [PubMed] [Google Scholar]
- 5.Brown JD, O'Neill RJ. 2010. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu. Rev. Genomics Hum. Genet. 11, 291–316. ( 10.1146/annurev-genom-082509-141554) [DOI] [PubMed] [Google Scholar]
- 6.Faria R, Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669. ( 10.1016/j.tree.2010.07.008) [DOI] [PubMed] [Google Scholar]
- 7.Sites JW, Moritz C. 1987. Chromosomal evolution and speciation revisited. Syst. Zool. 36, 153–174. ( 10.2307/2413266) [DOI] [Google Scholar]
- 8.Garagna S, Page J, Fernandez-Donoso R, Zuccotti M, Searle JB. 2014. The Robertsonian phenomenon in the house mouse: mutation, meiosis and speciation. Chromosoma 123, 529–544. ( 10.1007/s00412-014-0477-6) [DOI] [PubMed] [Google Scholar]
- 9.Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24, 2295–2300. ( 10.1016/j.cub.2014.08.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gropp A, Winking H, Redi CA. 1982. Consequences of Robertsonian heterozygosity: segregational impairment of fertility versus male-limited sterility. In Genetic control of gamete production and function (eds Crosignani PG, Rubin BL, Faccaro M), pp. 115–134. London, UK: Academic Press. [Google Scholar]
- 11.Baverstock PR, Gelder M, Jahnke A. 1983. Chromosome evolution in Australian Rattus—G-banding and hybrid meiosis. Genetica 60, 93–103. ( 10.1007/BF00127495) [DOI] [Google Scholar]
- 12.Baker RJ, Bickham JW. 1986. Speciation by monobrachial centric fusions. Proc. Natl Acad. Sci. USA 83, 8245–8248. ( 10.1073/pnas.83.21.8245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarro A, Barton NH. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57, 447–459. ( 10.1111/j.0014-3820.2003.tb01537.x) [DOI] [PubMed] [Google Scholar]
- 14.Britton-Davidian J, Catalan J, Belkhir K. 2002. Chromosomal and allozyme analysis of a hybrid zone between parapatric Robertsonian races of the house mouse: a case of monobrachial homology. Cytogenet. Genome Res. 96, 75–84. ( 10.1159/000063040) [DOI] [PubMed] [Google Scholar]
- 15.Horn A, et al. 2012. Chromosomal rearrangements do not seem to affect the gene flow in hybrid zones between karyotypic races of the common shrew (Sorex araneus). Evolution 66, 882–889. ( 10.1111/j.1558-5646.2011.01478.x) [DOI] [PubMed] [Google Scholar]
- 16.Sharman GB, Close RL, Maynes GM. 1990. Chromosomal evolution, phylogeny and speciation of rock wallabies (Petrogale: Macropodidae). Aust. J. Zool. 37, 351–363. ( 10.1071/ZO9890351) [DOI] [Google Scholar]
- 17.King M. 1993. Species evolution: the role of chromosome change. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 18.Eldridge MDB, Close RL. 1993. Radiation of chromosome shuffles. Curr. Opin. Genet. Dev. 3, 915–922. ( 10.1016/0959-437X(93)90014-G) [DOI] [PubMed] [Google Scholar]
- 19.Beerli P. 2009. How to use migrate or why are Markov chain Monte Carlo programs difficult to use? In Population genetics for animal conservation (eds Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi C), pp. 42–79. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 20.Eldridge MDB, Close RL. 1997. Chromosomes and evolution in rock-wallabies, Petrogale (Marsupialia: Macropodidae). Aust. Mammal. 19, 123–136. [Google Scholar]
- 21.Potter S, Cooper SJ, Metcalfe CJ, Taggart DA, Eldridge MDB. 2012. Phylogenetic relationships of rock-wallabies Petrogale (Marsupialia: Macropodidae) and their biogeographic history within Australia. Mol. Phylogenet. Evol. 62, 640–652. ( 10.1016/j.ympev.2011.11.005) [DOI] [PubMed] [Google Scholar]
- 22.Close RL, Bell JN. 1997. Fertile hybrids in two genera of wallabies: Petrogale and Thylogale. J. Hered. 88, 393–397. ( 10.1093/oxfordjournals.jhered.a023124) [DOI] [PubMed] [Google Scholar]
- 23.Briscoe DA, Calaby JM, Close RL, Maynes GM, Murtagh CE, Sharman GB. 1982. Isolation, introgression and genetic variation in rock wallabies. In Species at risk: research in Australia (eds Groves RH, Ryde WDL), pp. 73–87. Canberra, Australia: Australian Academy of Science. [Google Scholar]
- 24.Bee CA, Close RL. 1993. Mitochondrial DNA analysis of introgression between adjacent taxa of rock-wallabies, Petrogale species (Marsupialia: Macropodidae). Genet. Res. 61, 21–37. ( 10.1017/S0016672300031074) [DOI] [Google Scholar]
- 25.Briscoe DA. 1991. Allozyme data of Petrogale species. Recorded in MDB Eldridge (1991), ‘Chromosomal rearrangements and speciation in rock wallabies’. PhD thesis, Macquarie University, Sydney, Australia.
- 26.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 27.R Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 28.Peakall R, Smouse PE. 2006. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. ( 10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 31.Hey J, Nielsen R. 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl Acad. Sci. USA 104, 2785–2790. ( 10.1073/pnas.0611164104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matveevsky SN, Pavlova SV, Acaeva MM, Kolomiets OL. 2012. Synaptonemal complex analysis of interracial hybrids between the Moscow and Neroosa chromosomal races of the common shrew Sorex araneus showing regular formation of a complex meiotic configuration (ring-of-four). Comp. Cytogen. 6, 301–314. ( 10.3897/compcytogen.v6i3.3701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Close RL, Bell JN, Dollin AE, Harding HR. 1996. Spermatogenesis and synaptonemal complexes of hybrid Petrogale (Marsupialia). J. Hered. 87, 96–107. ( 10.1093/oxfordjournals.jhered.a022982) [DOI] [PubMed] [Google Scholar]
- 34.Giménez MD, White TA, Hauffe HC, Panithanarak T, Searle JB. 2013. Understanding the basis of diminished gene flow between hybridizing chromosome races of the house mouse. Evolution 67, 1446–1462. ( 10.1111/evo.12054) [DOI] [PubMed] [Google Scholar]
- 35.Lohse K, Clarke M, Ritchie MG, Etges WJ. 2015. Genome-wide tests for introgression between cactophilic Drosophila implicate a role of inversions during speciation. Evolution 69, 1178–1190. ( 10.1111/evo.12650) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accessions JQ62304, KT726229–KT726317. Microsatellite genotypes: electronic supplementary material.