Abstract
The endogenous peptides AtPep1-8 in Arabidopsis mature from the conserved C-terminal portions of their precursor proteins PROPEP1-8, respectively. The two homologous leucine-rich repeat-receptor kinases (LRR-RKs) PEPR1 and PEPR2 act as receptors of AtPeps. AtPep binding leads to stable association of PEPR1,2 with the shared receptor LRR-RK BAK1, eliciting immune responses similar to those induced by pathogens. Here we report a crystal structure of the extracellular LRR domain of PEPR1 (PEPR1LRR) in complex with AtPep1. The structure reveals that AtPep1 adopts a fully extended conformation and binds to the inner surface of the superhelical PEPR1LRR. Biochemical assays showed that AtPep1 is capable of inducing PEPR1LRR-BAK1LRR heterodimerization. The conserved C-terminal portion of AtPep1 dominates AtPep1 binding to PEPR1LRR, with the last amino acid of AtPep1 Asn23 forming extensive interactions with PEPR1LRR. Deletion of the last residue of AtPep1 significantly compromised AtPep1 interaction with PEPR1LRR. Together, our data reveal a conserved structural mechanism of AtPep1 recognition by PEPR1, providing significant insight into prediction of recognition of other peptides by their cognate LRR-RKs.
Keywords: PEPR1, LRR, AtPeps, BAK1, repeat-receptor kinase, crystal structure
Introduction
Plant genomes encode a large number of membrane-localized receptor kinases (RKs) that play critical roles in diverse biological processes1. RKs mediate plant immunity by acting as pattern recognition receptors (PRRs) to recognize pathogen-derived signature components (pathogen-associated molecular patterns, PAMPs) or host-derived danger signals (damage-associated molecular patterns, DAMPs)2,3,4. Well-documented PRRs include RKs FLS2, EFR and CERK1 from Arabidopsis, which recognize the conserved N-terminal portion of bacterial flagellin (flg22)5, elongation factor Tu (EF-Tu)6, and fungal cell wall component chitin7,8, respectively. Recognition of ligands by the ectodomains of their respective receptors activates PAMP-triggered immunity (PTI) that confers resistance to a multitude of pathogens. Ligand binding subsequently leads to the heterodimerization of FLS2 and likely EFR with the shared receptor BAK19, another RK. BAK1 belongs to the SERK family and generally acts to form ligand-induced heteromers with other RKs for subsequent signaling10,11,12. However, BAK1 is not involved in chitin-induced CERK1 signaling in Arabidopsis; rather, chitin induces CERK1 homodimerization for its activation13.
AtPep1, a 23-amino acid endogenous peptide, was initially identified as a DAMP in Arabidopsis14. A later study using alanine-scanning analysis showed that an AtPep1-derived peptide with the N-terminal deletion of eight amino acids, AtPep1(9-23), is equally active as the wild-type peptide in inducing PTI-like responses15. AtPep1 and its homologs AtPep2-8 mature from the conserved C-terminal portion of their respective precursors PROPEP1-8, respectively14,16. AtPeps are functionally similar to systemin, an 18-residue peptide identified in tomato, which has a critical role in defense signaling17. While displaying different expression patterns and localizations, all the eight AtPeps have a similar function of inducing plant immunity16. PEPR1 and its homolog PEPR2 are RKs with extracellular leucine-rich repeat (LRR) motifs (also found in FLS2 and EFR) and function as receptors of AtPeps18,19,20, although in vitro reconstitution of these complexes is not available yet. PEPR1 and PEPR2 appear to have different preferences for AtPeps. For example, AtPep1-6 have a similar activity of inducing PEPR1-mediated plant immune responses, whereas PEPR2 is preferentially responsive to AtPep1-220. Ca2+ has been shown to be important for the AtPep/PEPR signaling21,22. Similar to FLS2 and EFR, PEPR1 stably associates with BAK1 in response to treatment with AtPeps23,24. A number of studies suggest that PEPR-mediated immune responses serve to amplify PTI signaling25,26,27,28 via the JA/ET (jasmonic acid-ethylene) and SA (salicylic acid) pathways25,28. A more recent study suggests that PEPR1,2-mediated signaling has a critical role in coupling local and systemic immunity29.
In the present study, we solved a crystal structure of the LRR domain of PEPR1 (PEPR1LRR) in complex with AtPep1. The structure reveals the molecular mechanism underlying AtPep1 recognition by PEPR1. Combined with biochemical data, our structure supports the idea that AtPep1-induced PEPR1 heterodimerization with the shared receptor BAK1 is important for its activation. These mechanisms are largely conserved in flg22-induced FLS2 activation, providing information for predicting recognition of other signaling peptides by their cognate LRR-RKs.
Results
Reconstitution of the PEPR1LRR-AtPep1 and PEPR1LRR-AtPep1-BAK1LRR complexes in vitro
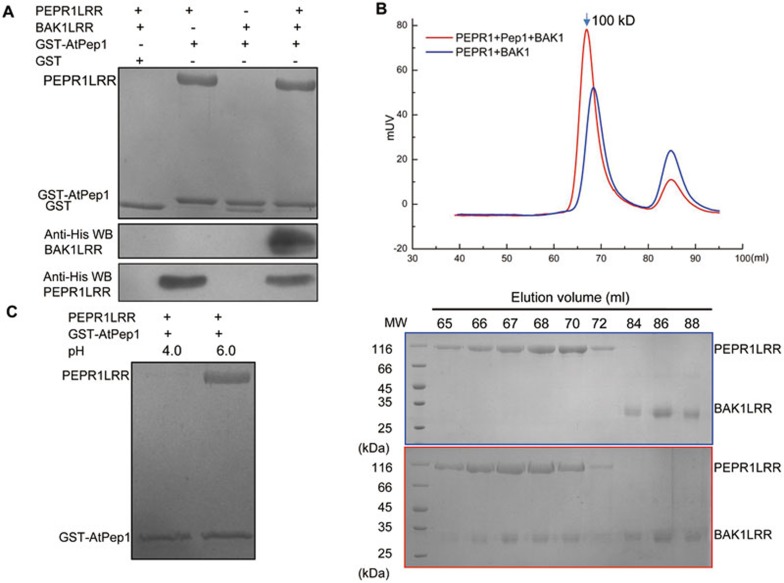
Previous studies showed that AtPep1 binding induced PEPR1-BAK1 heteromerization23,24. However, in vitro reconstitution of an AtPep1-induced complex using purified proteins has not yet been reported. To understand the molecular mechanism underlying AtPep1 recognition by PEPR1, we first set up an in vitro pull-down assay using purified proteins of GST-fused AtPep1 (residues 1-23), PEPR1LRR (residues 1-767) and BAK1LRR (residues 1-220) from Arabidopsis. Consistent with previous reports, GST-AtPep1 but not GST strongly interacted with PEPR1LRR (Figure 1A). By contrast, GST-AtPep1 and BAK1LRR displayed no detectable interaction under the same conditions. The BAK1LRR protein was pulled down by GST-AtPep1 in the presence of PEPR1LRR (Figure 1A), suggesting that formation of the complex containing GST-AtPep1, PEPR1LRR and BAK1LRR is a sequential process. To further confirm the pull-down data, we characterized AtPep1-induced interaction between PEPR1LRR and BAK1LRR using gel filtration. Consistently, the PEPR1LRR and BAK1LRR proteins co-migrated in the assay only in the presence of chemically synthesized AtPep1 (residues 1-23; Figure 1B), suggesting that the interaction between the two LRR proteins was induced by the peptide. The PEPR1LRR-BAK1LRR complex was eluted at the position corresponding to a molecular weight of ∼100 kD in gel filtration (Figure 1B, bottom panel), equivalent to a monomer of the complex. This result demonstrates that AtPep1 binding induced no oligomerization of PEPR1LRR or the PEPR1LRR-BAK1LRR complex. Similar results have been observed in flg22-induced formation of the FLS2-BAK1 complex30 or brassinosteroid (BR)-induced BRI1-BAK1/SERK1 complexes31,32, the latter of which are involved in plant growth and development33. We previously reported that lowering pH in vitro promotes BR-induced formation of the BRI1-BAK1 complex31. We therefore tested whether pH has a role in AtPep1-induced PEPR1LRR-BAK1LRR interaction. Unexpectedly, the interaction between PEPR1LRR and AtPep1 was nearly abolished at pH 4.0 in the pull-down assay (Figure 1C), though they strongly interacted with each other at a higher pH (6.0).
Figure 1.
PEPR1 recognition of AtPep1 induces ectodomain heterodimerization of PEPR1 and BAK1. (A) In vitro reconstitution of AtPep1-induced PEPR1LRR-BAK1LRR heterodimerization using purified proteins. His-tagged PEPR1LRR and BAK1LRR proteins were expressed in insect cells and purified to homogeneity. The GST-AtPep1 was expressed in E. Coli. An equal amount of the purified GST- AtPep1 protein was loaded onto 30 μl of GS4B-resin and then incubated with an excessive amount of His-tagged PEPR1LRR and BAK1LRR on ice for 20 min. After extensive washing, proteins bound to the GS4B resin were detected by Coomassie blue staining or western blot. WB: western blot. GST in line 3 likely resulted from proteolytic removal of AtPep1. (B) AtPep1 induces PEPR1LRR-BAK1LRR heterodimerization in solution. Top panel: superposition of the gel filtration chromatograms of PEPR1LRR and BAK1LRR in the absence (blue) or presence (red) of AtPep1. The vertical and horizontal axes represent ultraviolet absorbance (λ = 280 nm) and elution volume (ml), respectively. The molecular weight for the PEPR1LRR-AtPep1-BAK1LRR complex is indicated. Bottom panels: Coomassie blue staining of the peak fractions shown on the top following SDS-PAGE. (C) AtPep1-PEPR1LRR interaction is pH-dependent. The assay was performed as described in A.
Overall structure of the PEPR1LRR-AtPep1 complex
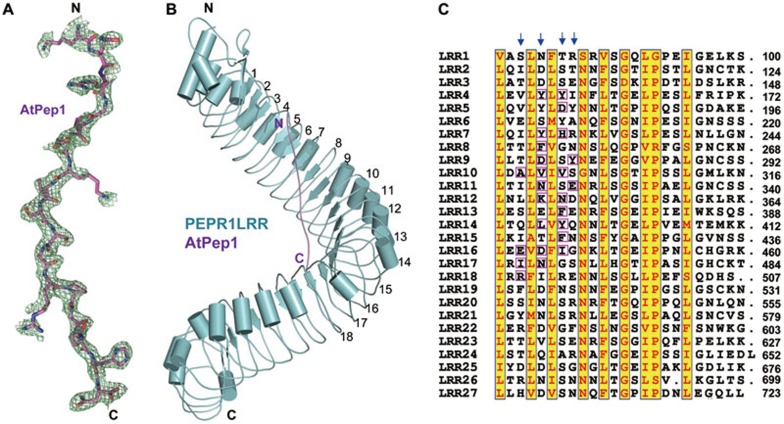
We then solved the crystal structure of chemically synthesized AtPep1 (residues 1-23) in complex with PEPR1LRR (residues 1-767) at 2.6 Å resolution (Supplementary information, Table S1). The structure was determined by molecular replacement using FLS2LRR (PDB code: 4MN8) as the initial search model. After refinement of the initial PEPR1LRR model, the electron density (Figure 2A) was sufficient to build the model of AtPep1 (residues 7-23). The N-terminal six residues of the bound AtPep1 were not well defined presumably because they were disordered in solution. In the crystal, each asymmetric unit contains two copies of the PEPR1LRR-AtPep1 complex, but the interface between the two complexes buries only 336 Å2 in total, which is unlikely to mediate bona fide protein-protein interactions. This structural observation further supports the biochemical data showing that AtPep1 binding induced no oligomerization of PEPR1LRR (Figure 1B). PEPR1LRR that contains 27 canonical LRRs assumes a superhelical structure, reminiscent of other plant LRR structures such as FLS2 and BRI130,34,35 that also carry the plant-specific sequence GxIP (x stands for any amino acid). Nearly in parallel with the central axis of the PEPR1LRR superhelix, the bound AtPep1 adopts a fully extended conformation and interacts with the inner side of the PEPR1LRR helical structure, running across 15 LRRs of PEPR1LRR, from LRR4 to LRR18 (Figure 2B). Many amino acids are highly conserved among the LRRs of PEPR1, but AtPep1 selectively makes contacts with the variable residues on the inner surface of PEPR1LRR (Figure 2C), indicating that these variable residues are the structural determinants for ligand specificity. At the primary sequence level, the AtPep1-interacting amino acids are from the 3rd, 5th, 7th and 8th positions of each LRR motif (Figure 2C). Similar observations were also made in the binding of flg22 to FLS2LRR30.
Figure 2.
AtPep1 binds to the inner surface of PEPR1LRR superhelical structure. (A) Electron density around AtPep1 in the structure of the PEPR1LRR-AtPep1 complex. Omit electron density (2Fo-Fc) around AtPep1 (residues 7-23) contoured at 1.20. “N” and “C” represent the N- and C-terminus, respectively. Color codes are indicated. (B) Overall structure of the PEPR1LRR-AtPep1 complex shown in cartoon. The positions of some LRRs are indicated. (C) Sequence alignment of LRRs of PEPR1LRR. Numbers indicate the positions of the last amino acid of each LRR. The conserved residues are shown with yellow background. Amino acids involved in interaction with AtPep1 are highlighted with magenta frames. Arrows indicate the 3rd, 5th, 7th and 8th positions of each LRR.
Recognition of AtPep1 by PEPR1
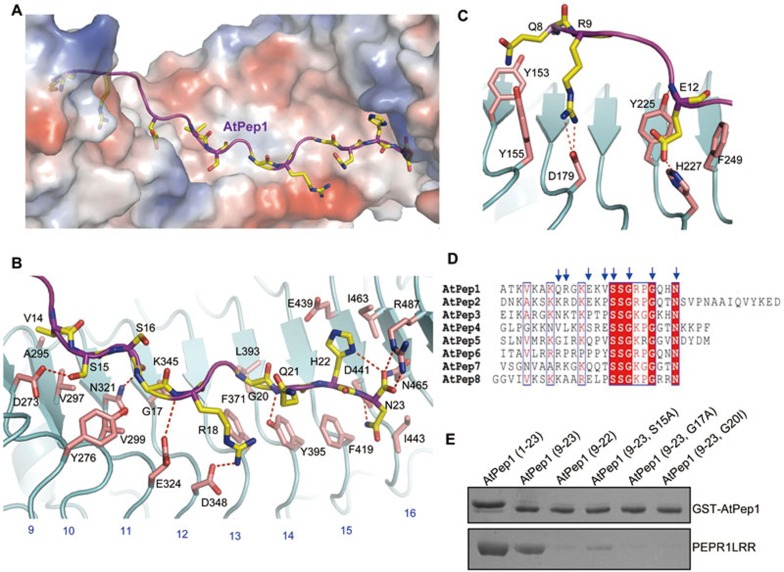
In the structure of the PEPR1LRR-AtPep1 complex, the fully extended AtPep1 closely matches the surface topology of PEPR1LRR and binds to an elongated inner surface groove interspersed with several cavities (Figure 3A). The C-terminal ten residues of AtPep1 (14-23; Figure 3B) form more concentrated interactions with PEPR1LRR than the N-terminal seven residues (7-13; Figure 3C). The conserved amino acids of AtPep1 dominate its interaction with PEPR1LRR (Figure 3D). The last amino acid of AtPep1, Asn23 (AtPep1Asn23) is highly conserved among AtPeps and makes extensive contacts with PEPR1LRR (Figure 3B). The free carboxyl group of AtPep1Asn23 contributes to AtPep1 interaction with PEPR1LRR by forming a pair of salt bridges with Arg487 and a hydrogen bond with Asn465 of PEPR1. PEPR1 Asp441 immediately underneath the peptide makes a bifurcated hydrogen bond with the side chain of AtPep1Asn23 and its amide nitrogen, whereas the aliphatic portion of AtPep1Asn23 makes hydrophobic packing against PEPR1 Ile443. Supporting the structural observations, deletion of AtPep1Asn23 significantly compromised AtPep1-induced immune responses in Arabidopsis cells15 and nearly abolished AtPep1-PEPR1LRR interaction in our pull-down assay (Figure 3E). AtPep1Gly17 and AtPep1Gly20 are highly conserved among the AtPep family peptides (Figure 3D). The Cα atoms of these two amino acids confront the inner side of PEPR1LRR (Figure 3B). Limited by space, mutation of AtPep1Gly17 to any other amino acids is predicted to generate steric clash with its neighboring residues and thus compromise AtPep1 interaction with PEPR1LRR. Indeed, the mutant peptide AtPep1G17A was reported to have a greatly reduced activity in inducing alkalinzation of suspension cells15, and displayed no detectable interaction with PEPR1LRR (Figure 3E). Compared to AtPep1Gly17, the region around AtPep1Gly20 is more spacious (Figure 3B). Consistently, the mutant peptide AtPep1G20A had a comparable activity to the wild-type peptide in inducing immune responses of Arabidopsis cells15. However, mutation of AtPep1Gly20 to the bulkier residue isoleucine resulted in loss of AtPep1 interaction with PEPR1LRR (Figure 3E). AtPep1Ser15 forms a hydrogen bond with PEPR1 Asp273 and makes hydrophobic contact with PEPR1 Val297, whereas AtPep1Ser16, though highly conserved among AtPeps (Figure 3D), is completely solvent exposed (Figure 3B). The modestly conserved AtPep1Arg18 (Figure 3D) contributes to AtPep1-PEPR1LRR interaction by salt-bonding with PEPR1 Asp348 and making π−π packing against PEPR1 Phe371 (Figure 3B).
Figure 3.
Structural basis for recognition of AtPep1 by PEPR1LRR. (A) AtPep1 binds to a surface groove at the inner side of the PEPR1LRR solenoid. PEPR1LRR is shown in electrostatic surface and AtPep1 in cartoon. White, blue and red indicate neutral, positive and negative surfaces, respectively. The side chains of some amino acids from AtPep1 are shown (yellow and stick). (B) Interaction of the C-terminal portion (residues 14-23) of AtPep1 with PEPR1LRR. The side chains of PEPR1LRR and AtPep1 are shown in pink and yellow, respectively. Red dashed lines indicate hydrogen or salt bonds. Numbers in blue indicate the positions of LRRs. (C) Interaction of the N-terminal portion (residues 7-13) of AtPep1 with PEPR1LRR. (D) Sequence alignment of AtPeps from Arabidopsis. The conserved residues are shown with red background. Amino acids with arrows on the top are involved in interaction with PEPR1LRR. (E) Mutagenesis analysis of AtPep1 interaction with PEPR1LRR. The assay was performed as described in Figure 1A.
The non-conserved amino acids of AtPep1 (7-13) further strengthen AtPep1 binding to PEPR1LRR via polar and van der waals interactions (Figure 3C). While much less dense as compared to AtPep1-PEPR1LRR contacts mediated by the C-terminal portion of AtPep1, these non-conserved interactions are likely also important for AtPep1-induced signaling as AtPep1(14-23) is nearly inactive in inducing cell immune responses when tested at lower concentrations15. However, a higher concentration (25 nM) of the peptide, but not AtPep1(15-23), displayed immunogenic activity, though lower than that of the wild-type peptide15. These results suggest that AtPep1(14-15) linking the conserved C-terminal and non-conserved N-terminal regions of AtPep1 may function as a hot spot for AtPep1-PEPR1LRR interaction. Probably for this reason, the mutant peptide AtPep1S15A was reported to have a reduced activity in inducing plant cell immune responses15 and showed a weak interaction with PEPR1LRR (Figure 3E), despite that Ser15 does not form extensive interactions with PEPR1LRR (Figure 3B).
Modeling study of the PEPR1LRR-AtPep1-BAK1LRR complex
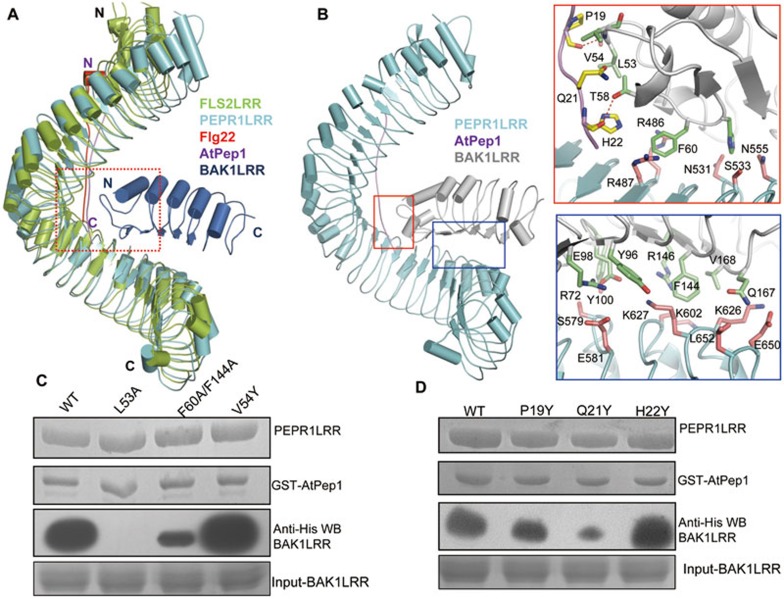
Structural comparison showed that PEPR1LRR and FLS2LRR share a conserved helical structure (Figure 4A), with 22 of their C-terminal LRRs being well aligned. Importantly, AtPep1 and flg22 bind to a similar position at the inner side of their respective receptors with the C-termini of the two peptides similarly oriented (Figure 4A). The FLS2LRR-bound BAK1LRR in the aligned structures contacts the C-terminal portion of AtPep1, suggesting that AtPep1 may employ a similar mechanism as flg22 to induce a stable PEPR1-BAK1 association. The other lateral side of the C-terminal PEPR1LRR, opposite to the BAK1-interacting side observed in FLS2LRR, seems unlikely to be involved in interactions with BAK1LRR, because several glycosylation sites (Asn302, Asn420 and Asn560, not shown) were identified around this region in the structure. The biological significance of these glycosylation sites remains uncertain. We therefore used the FLS2LRR-flg22-BAK1LRR complex as a template to model a structure of the PEPR1LRR-AtPep1-BAK1LRR complex (Figure 4B, left panel). BAK1Leu53 that stabilizes the curved loop (residues 52-56) and is important for flg22-induced stable FLS2-BAK1 association30 also interacts with the C-terminal portion of AtPep1 (Figure 4B). The carbonyl oxygen of AtPep1Pro19 forms a hydrogen bond with the amide nitrogen of BAK1Val54, whereas the side chains of AtPep1Pro19, Gln21, His22 make hydrophobic and van der waals contacts with their neighboring residues from the curved loop of BAK1 (Figure 4B, right top). Similar to the FLS2LRR-flg22-BAK1LRR complex, PEPR1LRR and BAK1LRR also form direct interactions mainly through van der waals packing (Figure 4B, right bottom). To verify the structural observations, we investigated the interaction of the mutant BAK1 L53A with PEPR1LRR using pull-down assays. As with the FLS2LRR-BAK1LRR complex, the mutation BAK1 L53A resulted in no detectable PEPR1LRR-BAK1LRR interaction in the presence of AtPep1 (Figure 4C). BAK1 Phe60 and Phe144 are centers of the amino acids mediating the direct BAK1LRR-PEPR1LRR interaction (Figure 4B, right panels). In support of this structural observation, a mutant BAK1LRR protein with Phe60 and Phe144 substituted with alanine showed a decreased activity in interacting with PEPR1LRR induced by AtPep1 (Figure 4C). Mutation of AtPep1Gln21 that inserts into a cavity around the curved loop of BAK1 to the bulkier residue tyrosine compromised the activity of AtPep1 in inducing PEPR1LRR-BAK1LRR interaction, though the mutant peptide displayed a similar level of interaction with PEPR1LRR compared to the wild-type peptide (Figure 4D). By contrast, mutations of AtPep1Pro19 and AtPep1His22 that appear to have more free space around them (Figure 4B) generated no detectable effect on AtPep1-induced PEPR1LRR-BAK1LRR interaction (Figure 4D), further supporting the modeled structure.
Figure 4.
Modeled structure of the PEPR1LRR-AtPep1-BAK1LRR complex. (A) Structural comparison of PEPR1LRR-AtPep1 with FLS2LRR-flg22-BAK1LRR (PDB code: 4MN8). The C-terminal 20 LRRs of PEPR1LRR were used to aligned with those of FLS2LRR. Color codes are indicated. (B) AtPep1 mediates PEPR1LRR-BAK1LRR heterodimerization. Left panel: overall modeled structure of the PEPR1LRR-AtPep1-BAK1LRR complex shown in cartoon. Right top: interaction of the C-terminal portion of AtPep1 with BAK1LRR. The side chains of PEPR1LRR, AtPep1 and BAK1LRR are shown in pink, yellow and green, respectively. Right bottom: PEPR1LRR and BAK1LRR make direct contacts. (C) Mutations in BAK1 affect AtPep1-induced PEPR1LRR-BAK1LRR interaction. The assay was performed as described in Figure 1A. (D) The mutation AtPep1Q21Y compromises AtPep1-induced PEPR1LRR-BAK1LRR interaction but has little effect on recognition of the peptide by PEPR1LRR. The assay was performed as described in Figure 1A.
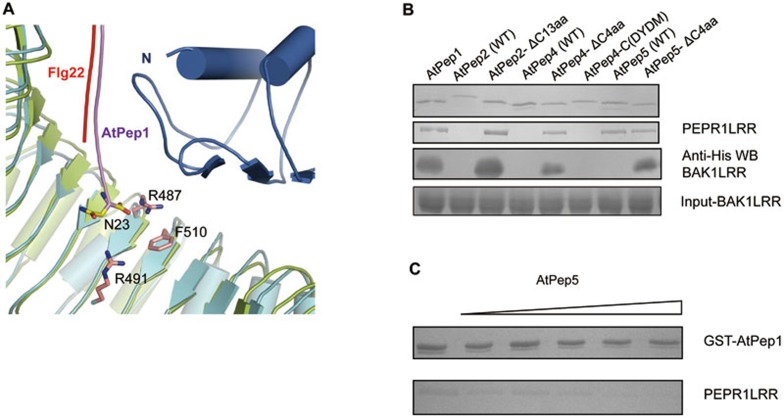
Role of the C-terminal extensions from AtPep2,4,5 in inducing PEPR1-BAK1 heterodimerization
Our structural and biochemical analyses suggest that AtPep1 and flg22 utilize a conserved mechanism to induce PEPR1-BAK1 and FLS2-BAK1 heterodimerization, respectively. However, these two peptides differ from each other in that when binding to their respective receptors, the last residue of flg22 is disordered30, whereas AtPep1Asn23 is well defined and involved in extensive interactions with PEPR1LRR (Figure 3B). Thus, the amino acids extended at the C-terminal portion of flg22 would be flexible. By contrast, because of the rigidity of AtPep1Asn23, in longer peptides harboring additional amino acid residues beyond Asn23, amino acids following Asn23 would be brought into a position between PEPR1LRR and BAK1LRR (Figure 5A). Additionally, peptide-bond formation of the carboxyl group of AtPep1Asn23 with its next residue will weaken the interaction of the residue with PEPR1 Arg487 (Figure 3B). Therefore, we hypothesized that the additional amino acids beyond Asn23 in AtPep2,4,5 (Figure 3D) may interfere with PEPR1LRR-BAK1LRR heterodimerization. In support of this hypothesis, while GST-AtPep1,3,6,7,8 induced PEPR1LRR-BAK1LRR interaction (Supplementary information, Figure S1), GST-AtPep2,4,5 failed to do so under the same conditions (Figure 5B). We hypothesized that deletion of the C-terminal extensions would restore the ability of these three peptides to induce PEPR1LRR-BAK1LRR heterodimerization. Indeed, the truncated forms of these three peptides were active in inducing PEPR1LRR-BAK1LRR interaction (Figure 5B), indicating that the C-terminal extensions were responsible for the inability of these three peptides to induce assembly of the PEPR1LRR-BAK1LRR complex. Similar results were obtained when chemically synthesized peptides were used to assay the PEPR1LRR-BAK1LRR interaction (Supplementary information, Figure S2). Interestingly, AtPep5, but not AtPep2,4, was capable of interacting with PEPR1LRR for reasons unknown (Figure 5B). Swapping of the last four residues between AtPep4 and AtPep5 did not restore the PEPR1-binding activity of AtPep4 (Figure 5B). This result suggests that AtPep5 could compete with other AtPeps for binding to PEPR1LRR. To test this hypothesis, we examined whether AtPep5 interferes with AtPep1 for interaction with PEPR1LRR. Indeed, the AtPep1-PEPR1LRR interaction was progressively reduced with increasing amounts of AtPep5 (Figure 5C).
Figure 5.
The C-terminal extensions in AtpPep2,4,5 interfere with PEPR1LRR-BAK1LRR heterodimerization. (A) Extensions at the C-terminal portion of AtPep1 may interfere with PEPR1LRR-BAK1LRR interaction. A close up view of structural comparison between PEPR1LRR-AtPep1 and FLS2LRR-flg22-BAK1LRR around the C-terminal region of AtPep1. The last amino acid AtPep1Asn23 is shown. (B) AtPep2,4,5 with their C-terminal extensions removed induced PEPR1LRR-BAK1LRR interaction. The assay was performed as described in Figure 1A. AtPep4-C(DYDM) represents a mutant AtPep4 with susbstitution of the C-terminal 4 residues of AtPep4 with those of AtPep5. (C) AtPep5 that interacts with PEPR1LRR but fails to induce PEPR1LRR-BAK1LRR interaction inhibits AtPep1-PEPR1LRR interaction. AtPep1 was first mixed with increasing amount of chemically synthesized AtPep5 and then incubated with the PEPR1LRR protein. The mixture was loaded onto GS4B resin. After extensive washing, proteins bound to the GS4B resin were detected by Coomassie blue staining.
Discussion
In the present study, our biochemical assays demonstrate direct recognition of AtPep1 by PEPR1LRR and AtPep1-induced PEPR1LRR-BAK1LRR heterodimerization (Figure 1). AtPep1 binding to PEPR1LRR is stringently pH-dependent in vitro, with pH 4.0 nearly abolishing their interaction. This is in sharp contrast to BR-induced BRI1-BAK1 heterodimerization, which is promoted at pH 4.0 in vitro31. The pH-dependent PEPR1LRR recognition of AtPep1 appears consistent with the observation that binding of AtPep1 to its receptors is accompanied by an increase in extracellular pH in cultured cells15. The exact mechanism underlying the pH-dependent PEPR1LRR-AtPep1 interaction remains unclear. However, protonation of some key residues such as PEPR1 Arg487 that forms salt bonds with the free carboxy group of AtPep1 (Figure 3B) might contribute to this process. A number of studies suggest that AtPeps function as an amplifier of PTI25,26,27,28, one hallmark of which is an increase in medium pH36,37. Although further in vivo studies are required to investigate whether and how PTI is associated with the pH-dependent PEPR1LRR recognition of AtPep1, this observation indicates that changes in pH during PTI can affect the biochemical activities of signaling molecules involved.
PEPR1 recognition of the conserved C-terminal amino acids of AtPep1 dictates their specific interaction (Figure 3), explaining why PEPR1 is able to recognize other members of AtPeps. However, the variable N-terminal segment is also important for AtPep1-induced signaling as its deletion substantially compromised immune responses induced by AtPep1 in plant cells15. These results suggest that the non-conserved N- and conserved C-terminal regions act cooperatively in signaling initiation. The AtPep1-interacting residues in PEPR1 are highly conserved in PEPR2 (Supplementary information, Figure S3). However, LRR4 in PEPR1 that interacts with the N-terminal portion of AtPep1 (Figure 2C) is missing in PEPR2. These results offer an explanation for the observation that PEPR1 displayed a slightly higher affinity with AtPep1 than PEPR220. Compared to PEPR1, PEPR2 is much less responsive to AtPep3-6 in inducing medium alkalinization20. Consistent with this observation, treatment of the PEPR1-deficient plants with AtPep1-2 but not with AtPep3-8 resulted in significant phosphorylation of MPK3/616. The non-conserved N-terminal sides of AtPeps might contribute to the preferential recognition of AtPep1,2 by PEPR2 over the other AtPeps. For example, AtPep1Glu12 conserved in AtPep2 but not in AtPep3-7 forms hydrogen bonds with PEPR1 Tyr225 and His227 (Figure 3C), two amino acids that are conserved in PEPR2. Factors (such as protein expression levels in plants) other than binding affinity could also have a role in AtPep-induced responses. This has been suggested in the recognition of AtPep1 by PEPR1 and PEPR220. While PEPR1 only has a slightly higher AtPep1-binding activity than PEPR2, PEPR1 is much more efficient than PEPR2 in suppressing Pst DC3000 growth in Arabidopsis. Additionally, as PEPR1,2-mediated signaling involves the participation of BAK1 or other SERK members, differences in assembly of the PEPR1/2-BAK1 (or other SERK members) complexes would also be important for different perception of AtPeps by PEPR1 and PEPR2. Further quantitative studies are needed to characterize interaction of AtPeps with PEPR1,2 and AtPep-induced assembly of the PEPR1/2-BAK1 complexes. It is noteworthy to point out that, while only AtPep1,2 induced medium alkalinization in PEPR2-expressing tobacco cells20, AtPep1,2,3 were equally active in eliciting membrane depolarization of pepr1 mutants of Arabidopsis19.
Our structural and biochemical data suggest that the C-terminal extensions in AtPep2,4,5 interfere with the assembly of the PEPR1-BAK1 complex. Interestingly, AtPep5 was able to compete with AtPep1 for interaction with PEPR1LRR (Figure 5C). These results raise the provocative possibility that AtPep5 might act to negatively regulate the functions of other AtPeps. However, all eight AtPeps are active in inducing PTI-like responses16. Therefore, a better explanation to reconcile these findings would be that in vivo AtPep2,4,5 need proteolytic processing at their C-terminal sides for maturation. Maturation of plant signaling peptides through proteolytic cleavage of both N- and C-terminal sides has been demonstrated in the CLE family of peptides38. Currently, little is known about the dynamics of PROPEP maturation, but it is possible that this might be a rapid process. Thus, AtPep2,4,5 may not manifest differences from the remaining AtPeps in immunogenic activity under the conditions tested16. Future time-course and quantitative examination of the immunogenic activity induced by AtPeps will be required to test the roles of the C-terminal extensions of AtPep2,4,5 in plant immunity.
The structure of PEPR1LRR resembles those of FLS2LRR and BRI1LRR, further supporting the idea that a helical structure may be shared by the plant LRR proteins harboring the specific sequence GxIP. While completely sequence-unrelated, AtPep1 and flg22 contact a conserved site at the inner surface of the helical structures when interacting with PEPR1 and FLS2, respectively. This is strikingly different from ligand recognition by TLRs in which the lateral sides of the solenoid TLR structures are generally utilized for interaction with their cognate ligands39. Peptide ligand recognition as observed in the structures of flg22-FLS2 and AtPep1-PEPR1 complexes involves a few conserved residues of an LRR. This information is useful for predicting binding of other peptide ligands to their cognate LRR-RKs. In contrast to the ligands, the shared receptor BAK1LRR interacts with one lateral side of FLS2LRR30 and BRI1LRR31. Our modeling study and biochemical assays showed that BAK1LRR likely binds to a similar position of PEPR1LRR. Although the exact mechanism underlying the recognition of AtPep1-bound PEPR1LRR by BAK1LRR remains unclear, our present structural information suggests that assembly of the PEPR1-BAK1 signaling complex involves no higher order of oligomerization, further supporting the notion that dimerization is important for plant RK activation4,40.
Materials and Methods
Protein expression and purification
The extracellular LRR domain of Arabidopsis PEPR1 (residues 1-767, PEPR1LRR) with a C-terminal 6× His tag was cloned into the pFastBac™ 1 vector (Invitrogen). The protein was expressed using the Bac-to-Bac baculovirus expression system (Invitrogen) in High Five insect cells. One litre of cells (1.8 × 106 cells/ml cultured in the medium from Expression Systems) was infected with 20 ml of baculovirus. Forty-eight hours after the infection, the medium was harvested for purification of the secreted proteins using Ni-NTA column (Novagen) followed by gel filtration chromatography (Hiload 200, GE Healthcare). Methods previously described were used to express and purify the extracellular LRR domain of BAK1 from Arabidopsis (residues 1-220, BAK1LRR). For crystallization, the purified AtPEPR1LRR protein was concentrated to about 7.0 mg/ml.
Crystallization, data collection, structure determination and refinement
Crystallization experiments were performed with hanging-drop vapor-diffusion methods by mixing equal volumes of protein and reservoir solution at 18 °C. A mixture of PEPR1LRR (∼7.0 mg/ml) and chemically synthesized AtPep1 peptide (residues 1-23, China Scilight Biotechnology; 20 mg/ml) with a molar ratio of 1:5 was used for crystallization. Initial crystals of AtPEPR1-ECD were obtained in buffer containing 0.2 M Trimethylamine N-oxide dehydrate, 0.1 M Tris pH 8.5, 20% w/v Polyethylene Glycol Monomethyl ether 2000. However, the crystals diffracted X-ray poorly. To improve the diffraction ability of the crystals, the PEPR1LRR protein was digested with endoglycosidase F1 and F3 at 18 °C overnight and further cleaned using gel filtration. Similar conditions produced crystals of the de-glycosylated protein, which well diffracted X-ray. Diffraction data were collected on the BL17U1 beam-line of the Shanghai Synchrotron Research Facility (SSRF). The datasets were processed using HKL200041. The structure was determined by the molecular replacement method with PHASER42 and refined with PHENIX43. The structure of FLS2LRR (PDB code: 4MN8) was used as the initial searching model. After refinement of the initial model, AtPep1 (residues 7-23) was built into the electron density in COOT44. The structure of PEPR1LRR-AtPep1 was finally refined to a resolution of 2.59 Å with Rfactor 23.7% and Rfree 28.8%, respectively. All the figures representing structures were prepared with PyMOL.
Gel filtration assay
PEPR1LRR and BAK1LRR proteins purified as described above were subjected to gel filtration analysis (Hiload 200, GE Healthcare) in the absence or presence of chemically synthesized AtPep1 peptide from Arabidopsis. Approximately equal moles of AtPEPR1LRR, AtBAK1LRR and AtPep1 peptide were incubated at 4 °C for 30 min before the gel filtration analysis. Buffer containing 10 mM Bis-Tris pH 6.0, 100 mM NaCl was used for the assay. The assays were performed with a flow rate of 0.8 ml/min and an injection volume of 40 ml at 4 °C. The proteins were subjected to SDS-PAGE and visualized by Coomassie blue staining.
In vitro pull-down assay and western blot
The PEPR1LRR and BAK1LRR proteins were purified as described above. GST-AtPep1 and other AtPeps were expressed in E. Coli. and purified using Glutathione Sepharose 4B beads (GS4B, GE Healthcare). The purified GST-tagged AtPep1 (from Arabidopsis) protein was used to pull down His-tagged PEPR1LRR (residues 1-767) and His-tagged BAK1LRR (residues 1-220) proteins. About 50 μg of GST-taggedAtPep1 was first loaded onto 50 μl GS4B beads. After extensively washed with washing buffer (10 mM Bis-Tris pH6.0, 100 mM NaCl), the beads were incubated with the purified His-tagged PEPR1LRR and/or BAK1LRR proteins on ice for 20 min, followed by washing with 1 ml of washing buffer for three times. The final beads were detected by Coomassie blue staining or western blot. Primary and secondary antibodies used for western blot were purchased from Beijing CoWin Bioscience Corporation (China). The assays were repeated for three times.
The GST–tagged AtPep5 protein was expressed in E.coli and purified as described above. The GST tag was then removed using prescission protease. The protein generated was further purified through gel filtration chromatography (Superdex 200, GE Healthcare). Approximately 50 μg of the His-tagged PEPR1LRR protein was added to mixtures containing an equal amount of GST tagged-AtPep1 protein and varying concentration of AtPep5 peptide. The mixtures were then individually loaded onto GS4B beads. After incubation on ice for 20 min, the beads were washed with 1 ml buffer containing 10 mM Bis-Tris pH 6.0, 100 mM NaCl for three times. The final beads were detected by Coomassie blue staining.
Structural modeling of the PEPR1LRR-AtPep1-BAK1LRR complex
The complex formed by PEPR1LRR, BAK1LRR and AtPep1 was predicted by the docking method HoDock45, which incorporates an initial rigid docking and a refined semi-flexible docking. The structure of the FLS2LRR-flg22-BAK1LRR complex (PDB code: 4MN8) was used as restraints for conformational searching and model selecting. Totally 11 000 complex structures were generated and scored to pick up the final correct complex structure model. Molecular dynamics simulation package Gromacs 4.5246 with OPLS force field was used for the minimization to relax and equilibrate the structure in solution. Then the minimized structure in the last frame fitting best with stereo-chemical restraints was selected as the built model.
Acknowledgments
We thank Yu F and He J at Shanghai Synchrotron Radiation Facility (SSRF). The research was funded by the National Natural Science Foundation of China (31420103906 and 31130063), the Chinese Ministry of Science and Technology (2014CB910101) and the National Outstanding Young Scholar Science Foundation of China (31025008 to JC).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
AtPep1,3,5,6,7,8 interact with PEPR1LRR.
Chemically synthesized AtPep4,5 with their C-terminal extensions deleted but not the wild type ones induce PEPR1LRR-BAK1LRR heterodimerization
Sequence alignment of the extracellular LRR domains of PEPR1 and PEPR2 from Arabidopsis.
Data Collection Statistics
SI Materials and Methods
References
- 1Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004; 16:1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 2009; 60:379–406. [DOI] [PubMed] [Google Scholar]
- 3Ryan CA, Huffaker A, Yamaguchi Y. New insights into innate immunity in Arabidopsis. Cell Microbiol 2007; 9:1902–1908. [DOI] [PubMed] [Google Scholar]
- 4Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell 2014; 54:263–272. [DOI] [PubMed] [Google Scholar]
- 5Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 2000; 5:1003–1011. [DOI] [PubMed] [Google Scholar]
- 6Zipfel C, Kunze G, Chinchilla D, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006; 125:749–760. [DOI] [PubMed] [Google Scholar]
- 7Miya A, Albert P, Shinya T, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 2007; 104:19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Wan J, Zhang XC, Neece D, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008; 20:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Chinchilla D, Zipfel C, Robatzek S, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007; 448:497–500. [DOI] [PubMed] [Google Scholar]
- 10Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. One for all: the receptor-associated kinase BAK1. Trends Plant Sci 2009; 14:535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002; 110:213–222. [DOI] [PubMed] [Google Scholar]
- 12Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002; 110:203–212. [DOI] [PubMed] [Google Scholar]
- 13Liu T, Liu Z, Song C, et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012; 336:1160–1164. [DOI] [PubMed] [Google Scholar]
- 14Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 2006; 103:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Pearce G, Yamaguchi Y, Munske G, Ryan CA. Structure-activity studies of AtPep1, a plant peptide signal involved in the innate immune response. Peptides 2008; 29:2083–2089. [DOI] [PubMed] [Google Scholar]
- 16Bartels S, Lori M, Mbengue M, et al. The family of Peps and their precursors in Arabidopsis: differential expression and localization but similar induction of pattern-triggered immune responses. J Exp Bot 2013; 64:5309–5321. [DOI] [PubMed] [Google Scholar]
- 17Ryan CA, Pearce G. Systemins: a functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci USA 2003; 100 Suppl 2:14577–14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 2006; 103:10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Krol E, Mentzel T, Chinchilla D, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 2010; 285:13471–13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 2010; 22:508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Qi Z, Verma R, Gehring C, et al. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc Natl Acad Sci USA 2010; 107:21193–21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Ma Y, Walker RK, Zhao Y, Berkowitz GA. Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc Natl Acad Sci USA 2012; 109:19852–19857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Postel S, Kufner I, Beuter C, et al. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol 2010; 89:169–174. [DOI] [PubMed] [Google Scholar]
- 24Schulze B, Mentzel T, Jehle AK, et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 2010; 285:9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Huffaker A, Ryan CA. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 2007; 104:10732–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Liu Z, Wu Y, Yang F, et al. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 2013; 110:6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Tintor N, Ross A, Kanehara K, et al. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci USA 2013; 110:6211–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Flury P, Klauser D, Schulze B, Boller T, Bartels S. The anticipation of danger: microbe-associated molecular pattern perception enhances AtPep-triggered oxidative burst. Plant Physiol 2013; 161:2023–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Ross A, Yamada K, Hiruma K, et al. The Arabidopsis PEPR pathway couples local and systemic plant immunity. EMBO J 2014; 33:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Sun Y, Li L, Macho AP, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013; 342:624–628. [DOI] [PubMed] [Google Scholar]
- 31Sun Y, Han Z, Tang J, et al. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 2013; 23:1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 2013; 341:889–892. [DOI] [PubMed] [Google Scholar]
- 33Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 2012; 46:701–724. [DOI] [PubMed] [Google Scholar]
- 34Hothorn M, Belkhadir Y, Dreux M, et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011; 474:467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35She J, Han Z, Kim TW, et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004; 16:3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 1999; 18:265–276. [DOI] [PubMed] [Google Scholar]
- 38Kondo T, Sawa S, Kinoshita A, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 2006; 313:845–848. [DOI] [PubMed] [Google Scholar]
- 39Song DH, Lee JO. Sensing of microbial molecular patterns by toll-like receptors. Immunol Rev 2012; 250:216–229. [DOI] [PubMed] [Google Scholar]
- 40Han Z, Sun Y, Chai J. Structural insight into the activation of plant receptor kinases. Curr Opin Plant Biol 2014; 20C:55–63. [DOI] [PubMed] [Google Scholar]
- 41Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 42McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 44Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 45Gong X, Wang P, Yang F, et al. Protein-protein docking with binding site patch prediction and network-based terms enhanced combinatorial scoring. Proteins 2010; 78:3150–3155. [DOI] [PubMed] [Google Scholar]
- 46Pronk S, Pall S, Schulz R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013; 29:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AtPep1,3,5,6,7,8 interact with PEPR1LRR.
Chemically synthesized AtPep4,5 with their C-terminal extensions deleted but not the wild type ones induce PEPR1LRR-BAK1LRR heterodimerization
Sequence alignment of the extracellular LRR domains of PEPR1 and PEPR2 from Arabidopsis.
Data Collection Statistics
SI Materials and Methods