The first US case of Middle East respiratory syndrome coronavirus was confirmed in May 2014 in a 65-year-old physician who worked in Saudi Arabia and presented to an Indiana hospital on illness day 11. He had bilateral pneumonia and recovered fully.
Keywords: Middle East respiratory syndrome coronavirus, viral pneumonia
Abstract
Background. The Middle East respiratory syndrome coronavirus (MERS-CoV) was discovered September 2012 in the Kingdom of Saudi Arabia (KSA). The first US case of MERS-CoV was confirmed on 2 May 2014.
Methods. We summarize the clinical symptoms and signs, laboratory and radiologic findings, and MERS-CoV–specific tests.
Results. The patient is a 65-year-old physician who worked in a hospital in KSA where MERS-CoV patients were treated. His illness onset included malaise, myalgias, and low-grade fever. He flew to the United States on day of illness (DOI) 7. His first respiratory symptom, a dry cough, developed on DOI 10. On DOI 11, he presented to an Indiana hospital as dyspneic, hypoxic, and with a right lower lobe infiltrate on chest radiography. On DOI 12, his serum tested positive by real-time reverse transcription polymerase chain reaction (rRT-PCR) for MERS-CoV and showed high MERS-CoV antibody titers, whereas his nasopharyngeal swab was rRT-PCR negative. Expectorated sputum was rRT-PCR positive the following day, with a high viral load (5.31 × 106 copies/mL). He was treated with antibiotics, intravenous immunoglobulin, and oxygen by nasal cannula. He was discharged on DOI 22. The genome sequence was similar (>99%) to other known MERS-CoV sequences, clustering with those from KSA from June to July 2013.
Conclusions. This patient had a prolonged nonspecific prodromal illness before developing respiratory symptoms. Both sera and sputum were rRT-PCR positive when nasopharyngeal specimens were negative. US clinicians must be vigilant for MERS-CoV in patients with febrile and/or respiratory illness with recent travel to the Arabian Peninsula, especially among healthcare workers.
The Middle East respiratory syndrome coronavirus (MERS-CoV) was first reported in September 2012 in a Saudi Arabian patient with pneumonia [1]. As of 12 May 2014, MERS-CoV had been confirmed by the World Health Organization (WHO) in 536 patients; all cases were related to residence, recent travel, or contact with a recent traveler from the Arabian Peninsula [2]. Initial reports of clinical course among MERS-CoV case patients from Saudi Arabia revealed high case fatality proportions [3, 4], but subsequent increases in testing of symptomatic and asymptomatic persons as part of contact investigations has revealed that approximately one-fifth to one-quarter of cases are mildly symptomatic or asymptomatic [5, 6]. As of April 2014, travel-associated cases have been detected in 8 countries outside the Arabian Peninsula [7]. We report here on the clinical course and laboratory findings from the first case of MERS-CoV in the United States.
METHODS
Clinical History and Physical Findings
The patient's clinical history and possible exposures to MERS-CoV were elicited by direct interview with the patient and family members. His physical, laboratory, and radiologic findings were extracted from his medical record.
MERS-CoV Laboratory Testing and Genome Sequencing
Specimens were drawn at the hospital and sent on ice to the Indiana Department of Health (IDH) laboratory or the Centers for Disease Control and Prevention (CDC). Initial testing by real-time reverse transcription polymerase chain reaction (rRT-PCR) was performed at the IDH laboratory with confirmatory sequencing performed at the CDC. The CDC rRT-PCR screening assay consists of 2 signatures that target a region upstream of the MERS-CoV envelope protein gene (upE) and the nucleocapsid gene (N2) [8]. A positive test result with either or both assays is then confirmed with a third rRT-PCR assay also targeting the nucleocapsid gene (N3). Serology was done using a recombinant MERS-CoV nucleocapsid protein–based enzyme-linked immunosorbent assay (ELISA) developed by the CDC. This ELISA was developed using a modified version of the HKU5.2 N ELISA as described [9]. In brief, sera were considered positive when the optical density (OD) values were at or above the 0.36 cutoff value (mean absorbance at 405 nm of sera from US blood donors plus 3 standard deviations). The overall specificity of the assay was determined after screening 555 serum samples from donors in the United States and the Middle East and persons with other non-MERS respiratory infections (eg, human coronavirus [HCoV] OC43, HCoV-229E, severe acute respiratory syndrome [SARS] coronavirus, HCoV-NL63, rhinovirus, human metapneumovirus, H1N1). The assay specificity was 98.1% (544/555). Serum from HKU1 human sera was not available for evaluation; however, HKU1 mouse hyperimmune serum did not cross-react with the MERS-CoV N protein. At a screening dilution of 1:400, sera with OD values at or near the cutoff were titrated with serial 2- to 4-fold dilutions (1:100–1:6400). The assay sensitivity was determined by screening a limited number of serum samples from individuals with confirmed MERS-CoV infection (sera provided by Public Health England, Robert Koch Institute, and the Jordan Ministry of Health). Confirmatory testing for MERS-CoV–specific antibodies was done on all positive ELISAs by MERS-CoV immunofluorescence and microneutralization assays [10]. Both confirmatory assays were evaluated using similar panels of sera as described above with similar specificities. Serologic testing was done at the CDC.
The full genome sequence was determined from RNA obtained directly from sputum collected on day of illness (DOI) 13 by generating tiling amplicons across the virus genome followed by Sanger sequencing. The sequence was deposited into the GenBank database as Indiana/USA-1_Saudi Arabia_2014 (accession number KJ813439).
RESULTS
Clinical History
The patient is a 65-year-old male physician living in the Kingdom of Saudi Arabia (KSA) who reported onset of fatigue and mild myalgia on 18 April 2014 (DOI 1), which curtailed his daily exercise program. He reported a low-grade fever of <38.0°C but no respiratory symptoms. On DOI 1, the patient began acetaminophen and naproxen for myalgias and also began a 5-day course of ciprofloxacin due to a history of prostatitis with no improvement. He went to the emergency department in KSA on DOI 2, where he had a normal complete blood count (CBC) and normal chest radiograph by report. He was not tested for MERS-CoV.
The patient flew from KSA to London on 24 April (DOI 7) and then from London to Chicago, arriving the same day. He took a bus to his family's home in Indiana. Upon arrival, the patient's sister noticed that he appeared fatigued, which she attributed to jet lag. On DOI 8, the patient recorded an oral temperature of 38.6°C, and family suggested that he start oseltamivir for possible influenza. A nonproductive cough developed on DOI 10, and on DOI 11 he developed visible dyspnea and tachypnea. He used an albuterol meter–dosed inhaler on both days without improvement, which prompted his sister to drive him to the emergency department of a local hospital.
Medical History
The patient has hypertension and coronary artery disease, for which he had 2 stents placed 5 and 17 years ago. He also has benign prostatic hypertrophy and had prostatitis 2 years ago. His medications include valsartan, atenolol, atorvastatin, and clopidogrel. He does not smoke or drink alcohol.
Exposure History
The patient works full-time at a large hospital in Riyadh. He attends patients in both the outpatient and inpatient setting, including the emergency department. He does not recall directly treating any patients with known MERS-CoV infection from 1 to 23 April, but was aware of MERS-CoV–positive patients in the hospital during the month of April. He entered the rooms of several intubated patients as part of his work, but did not have extensive direct contact with these patients. He recalls examining a few patients while they were undergoing nebulizer treatments, while wearing a surgical mask. He denies being present during intubations or respiratory suctioning. His last day of work was 23 April, the day before he traveled.
In Saudi Arabia, he lives with 3 family members and a household employee, none of whom were ill in the 2 weeks before his symptom onset. The patient denied contact with known MERS-CoV patients outside of work. He also denied physical contact with or consumption of camels or camel products.
Physical Exam and Clinical Course
On presentation to the emergency department of an Indiana hospital on 28 April (DOI 11), the patient had an oral temperature of 37.1°C, blood pressure 158/93 mm Hg, heart rate 83 beats/minute, respiratory rate 20 breaths/minute, and oxygen saturation (O2Sat) of 90% on room air. Pulmonary exam revealed right lower quadrant rhonchi with decreased breath sounds. Cardiovascular, abdominal, skin, neurological and musculoskeletal exams were unremarkable.
His admission laboratory data were remarkable for lymphopenia (total lymphocyte count of 0.81 × 109/L), mildly elevated liver function tests, slight hyponatremia, and mildly elevated inflammatory markers (Table 1). Other laboratory results were within normal limits. His chest radiography on admission showed right lower lobe infiltrates (Figure 1).
Table 1.
Admission Laboratory Results for a Patient With Middle East Respiratory Syndrome Coronavirus, 28 April 2014
| Test | Result | Normal Range |
|---|---|---|
| Complete blood count | ||
| White blood cell count | 7.01 × 109/L | 4.50–11 × 109/L |
| Neutrophil %, count | 80%, 5.62 × 109/L | 55%–72%, 2.48–7.92 × 109/L |
| Lymphocyte %, count | 11.6%, 0.81 × 109/L | 20%–40%, 1.5–4.0 × 109/L |
| Immature granulocyte %, count | 0.7%, 0.05 × 109/L | 0.0%–0.4%, 0.0–0.04 × 109/L |
| Hemoglobin/hematocrit | 13.2%/38.6% | 13.5%–16.5/41%–50% |
| Platelets, μL | 224 000 | 100 000–450 000 |
| Electrolytes | ||
| Sodium, mEq/L | 132 | 135–145 |
| Potassium, mEq/L | 4.2 | 3.4–5.1 |
| Chlorine, mEq/L | 96 | 96–106 |
| CO2, mEq/L | 22 | 20–29 |
| BUN, mg/dL | 15 | 7–21 |
| Creatinine, mg/dL | 0.75 | 0.50–1.20 |
| Glucose, mg/dL | 167 | <140 |
| Liver function tests | ||
| ALT (SGPT), U/L | 80 | 0–41 |
| AST (SGOT), U/L | 95 | 0–37 |
| Alkaline phosphatase, U/L | 270 | 35–116 |
| Direct bilirubin, mg/dL | 0.6 | 0.0–0.3 |
| Total bilirubin, mg/dL | 1.0 | 0.0–1.2 |
| Inflammatory markers | ||
| Erythrocyte sedimentation rate, mm/h | 44 | 0–15 |
| C-reactive protein, mg/dL | 10 | 0.0–0.5 |
| Procalcitonin, mg/dL | 0.54 | 0.0–0.5 |
Bolded values are outside of normal range.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CO2, carbon dioxide; SGOT, serum glutamic oxaloacetic transaminase; SGPT, serum glutamate-pyruvate transaminase.
Figure 1.
Chest radiographs of the patient with Middle East respiratory syndrome coronavirus on admission, Indiana, 28 April 2014.
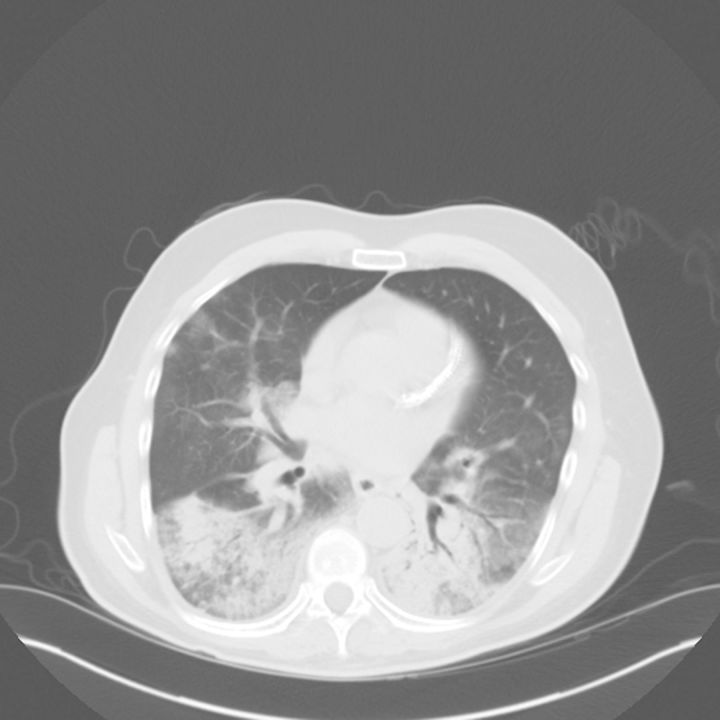
The patient was initially placed on 2 L/minute of oxygen (O2) with nasal cannula, given antibiotics (vancomycin, piperacillin/tazobactam) for hospital-acquired pneumonia, and admitted to the medical floor. The patient had a maximum temperature of 38.6°C on DOI 12, when his O2 requirement increased to 6 L/minute. Computed tomography of the chest from DOI 12 showed bilateral infiltrates predominantly in the lower lobes (Figure 2).
Figure 2.
Chest computed tomography of the patient with Middle East respiratory syndrome coronavirus, Indiana, 29 April 2014.
On DOI 13, levofloxacin was added for coverage of atypical pneumonia pathogens given his continued fevers, but was replaced with ceftriaxone on DOI 14 when his Legionella and Mycoplasma pneumoniae tests returned negative. On DOI 14, the patient was afebrile and had a decreasing O2 requirement (5 L/minute) while maintaining an O2Sat of 95%. He also received 2 doses of 100 mg/kg of intravenous immunoglobulin (IVIG) on DOI 14 and 15. On DOI 16, the patient was thought to have volume overload; a chest radiograph showed worsening bilateral infiltrates, and he had an increasing O2 requirement to 10 L/minute. The same day, furosemide was started with brisk diuresis via a Foley catheter, the oxygen was weaned rapidly to 6 L/minute, and oral antibiotics (linezolid and levofloxacin) were started. On DOI 18, the patient no longer required oxygen (O2Sat of 96%–97% on room air), and had an improving chest radiograph on DOI 21.
His total lymphocyte count remained low throughout the hospital stay with a nadir of 0.69 × 109/L and a discharge value of 1.44 × 109/L. All other elements of the CBC and electrolytes, including renal function, remained within normal limits on subsequent testing. He had several other microbiologic tests apart from MERS-CoV testing, including a negative blood culture from DOI 11, negative sputum culture from DOI 12, negative multiplex PCR for common respiratory pathogens (Biofire Diagnostics, Utah) from 29 April, and negative urine antigen tests for pneumococcus, M. pneumoniae, and Legionella pneumophila from DOI 12.
The patient was discharged home in stable condition on DOI 22.
MERS-CoV Testing and Genome Sequencing
The patient was initially positive by rRT-PCR for MERS-CoV in the serum at the first collection on 29 April (DOI 12), although the nasopharyngeal (NP) sample was negative on that date. The serum antibody titer was 1:3200. The patient had 3 additional positive samples: sputum (DOI 13), oropharyngeal swab (DOI 14), and plasma (DOI 15) (Table 2) [8]. The viral load in sputum on DOI 13 was 5.31 × 106 copies/mL. On DOI 16, viral load decreased to 1.26 × 105 copies/mL in the sputum sample, and antibody titers were >1:6400. Antibody titers remained high until the last day of collection (DOI 31). Stool and urine tested negative for MERS-CoV. To date, attempts to culture the virus from sputum (DOI 13) sample have been unsuccessful.
Table 2.
Middle East Respiratory Syndrome Coronavirus Real-Time Reverse Transcription Polymerase Chain Reaction Test Results of Index Patient Specimen Types
| Date of Collection | NP | NP/OP | Sputum | Stool | Urine | Serum |
|---|---|---|---|---|---|---|
| April 29 (DOI 12) | Neg | … | … | … | … | Pos (36.0) |
| April 30 (DOI 13) | … | … | Pos (26.5)a | Neg | … | … |
| May 1 (DOI 14) | … | Pos (31.2)b | … | … | … | … |
| May 2 (DOI 15) | … | … | … | … | … | Pos (38.7) |
| May 3 (DOI 16) | Neg | Neg | Pos (31.8)c | … | Neg | Neg |
| May 4 (DOI 17) | … | … | … | Neg | … | Neg |
| May 5 (DOI 18) | Neg | Neg | Neg | Neg | Neg | Neg |
| May 7 (DOI 19) | Neg | … | Neg | Neg | Neg | … |
Cycle threshold (CT) values for nucleocapsid protein gene signature are shown in parentheses.
Abbreviations: DOI, day of illness; Neg, negative; NP, nasopharyngeal; OP, oropharyngeal; Pos, positive.
a Envelope protein gene target CT 28.1.
b Only OP swab taken on this day.
c Envelope protein gene target CT 33.7.
The genome sequence (30 123 nt) was similar (>99%) to other known MERS-CoV sequences and clustered most closely with human-derived MERS-CoV strains obtained in Riyadh and Hafr-Al-Batin from summer 2013 (Figure 3 and Supplementary Figure 1).
Figure 3.
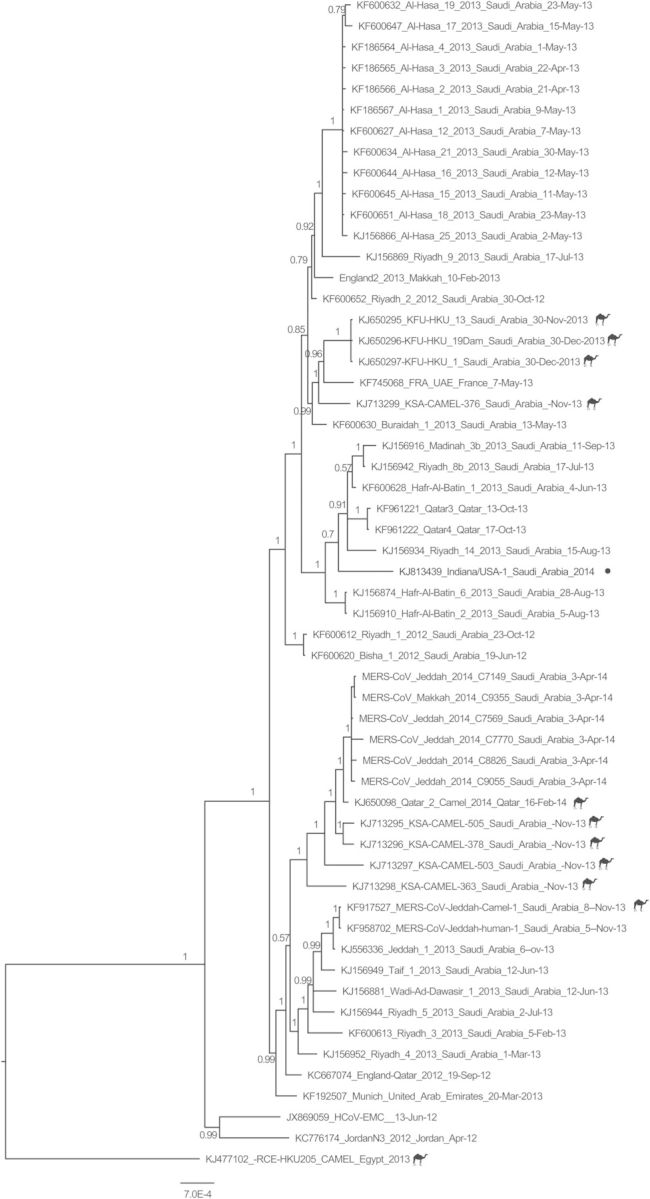
Phylogenetic analysis of the Indiana/USA-1_Saudi Arabia_2014 strain of Middle East respiratory syndrome coronavirus (MERS-CoV). The sequence alignment was generated using 56 nearly complete genome sequences published at (i) GenBank, (ii) the Health Protection Agency (available at: http://www.hpa.org.uk/webw/HPAweb&HPAwebStandard/HPAweb_C/1317136246479), and (iii) the Institut Für Virologie (available at: http://www.virology-bonn.de/index.php?id=46). The phylogenetic reconstructions were performed using MrBayes v3.2 under a general time-reversible (GTR) model of nucleotide substitution with 4 categories of γ-distributed rate heterogeneity and a proportion of invariant sites (GTR+4 + I). The Indiana/USA-1_Saudi Arabia_2014 strain is highlighted in red. The camel MERS-CoV sequences are labeled with a camel icon. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The Bayesian posterior probabilities (>0.5) are shown at nodes.
Infection Control Procedures
The patient spent 2.5 hours in the emergency department, the entire time in a private triage room. He was admitted to the general medical floor into a private room without airborne or contact precautions for approximately 20 hours. He was placed on airborne precautions on DOI 12 (hospital day 2) but remained in a private room that was not negative pressure relative to the hallway. On DOI 13, contact precautions were added and the patient was moved to a negative-pressure room; these precautions were maintained throughout the remainder of his hospital stay.
DISCUSSION
We report on the first case of MERS-CoV in the United States. There are several aspects of this patient's clinical presentation and course that provide insight into MERS-CoV disease. First, he had a relatively prolonged period of systemic symptoms of malaise, myalgia, and low-grade fever, which lasted 10 days, before he developed his first respiratory symptoms. Although in a series of critically ill MERS-CoV patients, the median time from illness onset to admission was 1 day, a protracted, nonrespiratory illness of >1 week has been described in a patient who eventually had significant bilateral lung infiltrates, as did this patient [3, 11, 12]. Although the patient in Indiana eventually developed a dry cough, which has occurred in approximately half of MERS-CoV patients, the cough was never prominent despite extensive lung parenchymal involvement [4].
Second, the patient's clinical presentation and radiologic and laboratory findings had no distinguishing features of a MERS-CoV infection and could easily have been diagnosed as having other, more common viral or bacterial pneumonia etiologies. Whereas this patient had the 3 most prevalent symptoms observed in the largest series of 47 MERS-CoV patients from Saudi Arabia [4]—fever (98%), cough (83%), and shortness of breath (72%)—he lacked upper respiratory tract (eg, sore throat, rhinorrhea) and gastrointestinal tract symptoms (eg, diarrhea, vomiting), which have been seen in approximately one-quarter of MERS-CoV patients. The patient's laboratory data were unremarkable except for a mild elevation in liver enzymes, which has been seen in up to 15% of patients [4], lymphopenia, which has been seen in 34%–86% of patients [4, 9], and mild elevation in inflammatory markers. The key feature to the diagnosis of MERS-CoV in this patient was the history of practice in and recent travel from KSA [13]. At the time of this case, approximately 88% of the MERS-CoV cases worldwide had occurred in KSA, and infection among healthcare workers is well documented [6, 9, 11]. The number of confirmed MERS-CoV cases identified in KSA increased substantially from March to early May 2014 (318 cases confirmed by WHO from 1 March to 12 May). Because MERS-CoV cases are prevalent in the Arabian Peninsula, US clinicians need to be ever more vigilant for MERS-CoV among patients with compatible clinical presentation (eg, fever and severe respiratory illness), travel history (ie, illness onset within 14 days after travel to the Arabian Peninsula or nearby countries), and close contact with a confirmed or probable case [7]. (Although this patient did not recall close contact, his work in a healthcare setting in KSA suggested the potential for unrecognized close contact.) Ten days after this patient's confirmed diagnosis, a second imported case of MERS-CoV was detected in Florida [7] in another healthcare worker who also had worked in KSA. No other US cases have occurred to date.
Once the Indiana patient was suspected of having MERS-CoV infection, the diagnosis was made within 24 hours. This virus was first detected in the patient's serum, when his NP swab was negative [14, 15]. NP swabs, because of their ease of collection, have been the most common sample used in making a diagnosis of MERS-CoV [4]. Nonetheless, there is evidence from some MERS-CoV cases that even when high load of virus is detected in lower respiratory tract specimens, upper respiratory tract specimens can be weakly positive or negative [15, 16]. In this patient, the initial NP sample was negative at a time when the sputum was positive by rRT-PCR at MERS-CoV concentrations that were as high as those from tracheobronchial secretions in other patients who were more critically ill [16]. Moreover, the sputum remained positive for up to 2 days longer than the pharyngeal samples. The serum was also positive by rRT-PCR on 2 occasions during a 4-day period when the nasopharyngeal sample was negative, suggesting a prolonged viremia. Blood and serum samples have been found to be rRT-PCR positive in other patients with MERS-CoV, and these might be more sensitive diagnostic specimens in MERS-CoV patients than in SARS patients [9, 15–18]. In contrast to SARS, MERS-CoV has been rarely detected in stool, and when detected was present in relatively low concentration [15, 16, 19]. For diagnostic testing of persons under investigation for MERS-CoV, the CDC states that lower respiratory specimens, which can include induced or expectorated sputa, are preferred; however, collecting NP/oropharyngeal (OP) specimens, as well as stool and serum, are also recommended as soon as possible after symptom onset [20]. If symptom onset was ≥14 days ago, a single serum specimen for serologic testing in addition to a lower respiratory specimen and an NP/OP specimen is recommended, although the kinetics of the antibody response to MERS-CoV infection needs further clarification [20].
Despite having bilateral pulmonary infiltrates, the patient required supplemental oxygen only by nasal cannula and was able to be weaned to room air 8 days after admission. There is no evidence yet for any effective treatment for MERS-CoV infection [21]. In the case series of 47 Saudi MERS-CoV patients, no bacterial coinfection was diagnosed [4]. Although bacterial pneumonia superinfection commonly seen following influenza virus infection has not been demonstrated with novel coronaviruses, WHO has a permissive recommendation for antibiotics based on clinical judgment in patients with novel coronavirus infections [21, 22]. IVIG has not been evaluated in MERS-CoV patients, but is unlikely to have been effective in this patient's recovery given his nonsevere clinical status and the expected absence of MERS-CoV antibodies in pooled sera in the United States. Any immunomodulatory effect of IVIG in this patient is unknown. Although IVIG was administered to patients with SARS-CoV infection during 2003, the effectiveness of IVIG treatment for SARS patients is unknown due to confounders, variable severity of illness when treatment was initiated, and the uncontrolled study design [23]. Due to its potential risks and unknown benefit, IVIG is not recommended to treat MERS-CoV patients. Whereas interferon alfa-2b and ribavirin combined therapy has shown some therapeutic potential in cell culture and animal experiments [24, 25], this treatment has not been shown to be effective in MERS-CoV patients. However, these agents have only been implemented late in the course of illness. Steroids are also not recommended for the treatment of MERS-CoV [21].
The most closely genetically related strain to this patient's is from Riyadh in July 2013, which is where the Indiana patient lived and worked in a hospital. Of interest, the next most closely related virus is the June 2013 index case in a community cluster in Hafr Al-Batin [26], a town in northeast KSA near the Kuwaiti border, approximately 500 km from Riyadh (Supplementary Figure 1). Few sequences from 2014 are present in the GenBank database, particularly from Riyadh.
The ongoing threat of the spread of MERS-CoV into the United States requires the vigilance of astute clinicians and public health departments to detect MERS-CoV–infected patients and respond rapidly to prevent spread in healthcare facilities and the community [13]. People who are traveling to provide healthcare services in the Arabian Peninsula should be familiar with recommendations for infection control of confirmed or suspected MERS cases.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We acknowledge the following staff members of Community Hospital, Munster, Indiana, for their contribution to the care of this patient: Fadi Layous, Kinan Attassi, Marlene Madrigal, and Infection Prevention Coordinator Ronda Mackey. At Indiana State Department of Health (IDH), many staff contributed to the diagnosis and contact tracing for this patient, including Donna Allen, Steve Allen, Jennifer Brown, Stephanie Dalenberg, Brian Pope, and Michelle Sandoval, Lieutenant Commander, US Public Health Service. We thank Richard Dunville, David Kuhar, Tim Uyeki, and Satish Pillai of the Centers for Disease Control and Prevention (CDC) for their assistance with manuscript preparation. We acknowledge the work of Krista Queen, Yan Li, and Clint Paden on genome sequencing, and Hayat Caidi, Congrong Miao, Jennifer Harcourt, Azaibi Tamin, and Suvang Trivedi for serologic testing.
Disclaimer. The findings and conclusions are those of the authors and do not necessarily represent the views of the CDC.
Financial support. This work was supported by Community Hospital, Munster, Indiana; the IDH; and the CDC.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Coronavirus infections. Available at: http://www.who.int/csr/disease/coronavirus_infections/en/ . Accessed 12 May 2014.
- 3.Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160:389–97. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 4.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752–61. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Updated information on the epidemiology of Middle East respiratory syndrome coronavirus (MERS-CoV) infection and guidance for the public, clinicians, and public health authorities, 2012–2013. MMWR Morb Mortal Wkly Rep. 2013;62:793–6. [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Global alert and response. Middle East respiratory syndrome coronavirus (MERS-CoV) summary and literature update—as of 9 May 2014. Available at: http://www.who.int/csr/disease/coronavirus_infections/MERS_CoV_Update_09_May_2014.pdf?ua=1 . Accessed 15 July 2014.
- 7.Centers for Disease Control and Prevention. First confirmed Middle East respiratory syndrome coronavirus (MERS-CoV) cases in the United States and updated information on the epidemiology of MERS-CoV infection and guidance for the public, clinicians, and public health authorities, May 2014. 2014;19:431–6. MMWR Morb Mortal Wkly Rep. [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X, Whitaker B, Sakthivel SK, et al. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Abdallat MM, Payne D, Alqasrawi S, et al. Hospital-associated outbreak of Middle East respiratory syndrome coronavirus: a serologic, epidemiologic, and clinical description. Clin Infect Dis. 2014;59:1225–33. doi: 10.1093/cid/ciu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sui J, Li W, Murakami A, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101:2536–41. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assiri A, McGeer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–16. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368:2487–94. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Interim guidance for health professionals. Available at: http://www.cdc.gov/coronavirus/mers/interim-guidance.html . Accessed 12 May 2014.
- 14.World Health Organization. Laboratory testing for Middle East respiratory syndrome coronavirus. Interim recommendations—September 2013. Available at: http://www.who.int/csr/disease/coronavirus_infections/MERS_Lab_recos_16_Sept_2013.pdf . Accessed 12 May 2014.
- 15.Guery B, Poissy J, el Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–72. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–51. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Z, Zhuang H, Zhao C, Dong Q, Peng G, Dwyer DE. Using patient-collected clinical samples and sera to detect and quantify the severe acute respiratory syndrome coronavirus (SARS-CoV) Virol J. 2007;4:32. doi: 10.1186/1743-422X-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang P, Louie M, Richardson SE, et al. Interpretation of diagnostic laboratory tests for severe acute respiratory syndrome: the Toronto experience. CMAJ. 2004;170:47–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng PK, Wong DA, Tong LK, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. MERS-CoV case definitions. Available at: http://www.cdc.gov/coronavirus/mers/case-def.html . Accessed 13 June 2014.
- 21.World Health Organization. Clinical management of severe acute respiratory infections when novel coronavirus is suspected: what to do and what not to do. Available at: http://www.who.int/csr/disease/coronavirus_infections/InterimGuidance_ClinicalManagement_NovelCoronavirus_11Feb13.pdf . Accessed 12 May 2014.
- 22.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19:571–82. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falzarano D, de Wit E, Rasmussen AL, et al. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–7. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Memish ZA, Cotten M, Watson SJ, et al. Community case clusters of Middle East respiratory syndrome coronavirus in Hafr Al-Batin, Kingdom of Saudi Arabia: a descriptive genomic study. Int J Infect Dis. 2014;23:63–8. doi: 10.1016/j.ijid.2014.03.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.