Abstract
Chromatin, the structure formed by the wrapping of approximately 146 base pairs of DNA around an octamer of histones, has a profound impact on numerous DNA-based processes. Chromatin modifications and chromatin remodellers have recently been implicated in important aspects of the DNA damage response including facilitating the initial sensing of the damage as well as subsequent recruitment of repair factors. Radiation is an effective cancer therapy for a large number of tumours, and there is considerable interest in finding approaches that might further increase the efficacy of radiotherapy. The use of radiation leads to the generation of DNA damage and, therefore, agents that can affect the sensing and repair of DNA damage may have an impact on overall radiation efficacy. The chromatin modifications as well as chromatin modifiers that have been associated with the DNA damage response will be summarized in this review. An emphasis will be placed on those processes that can be pharmacologically manipulated with currently available inhibitors. The rationale for the use of these inhibitors in combination with radiation will also be described.
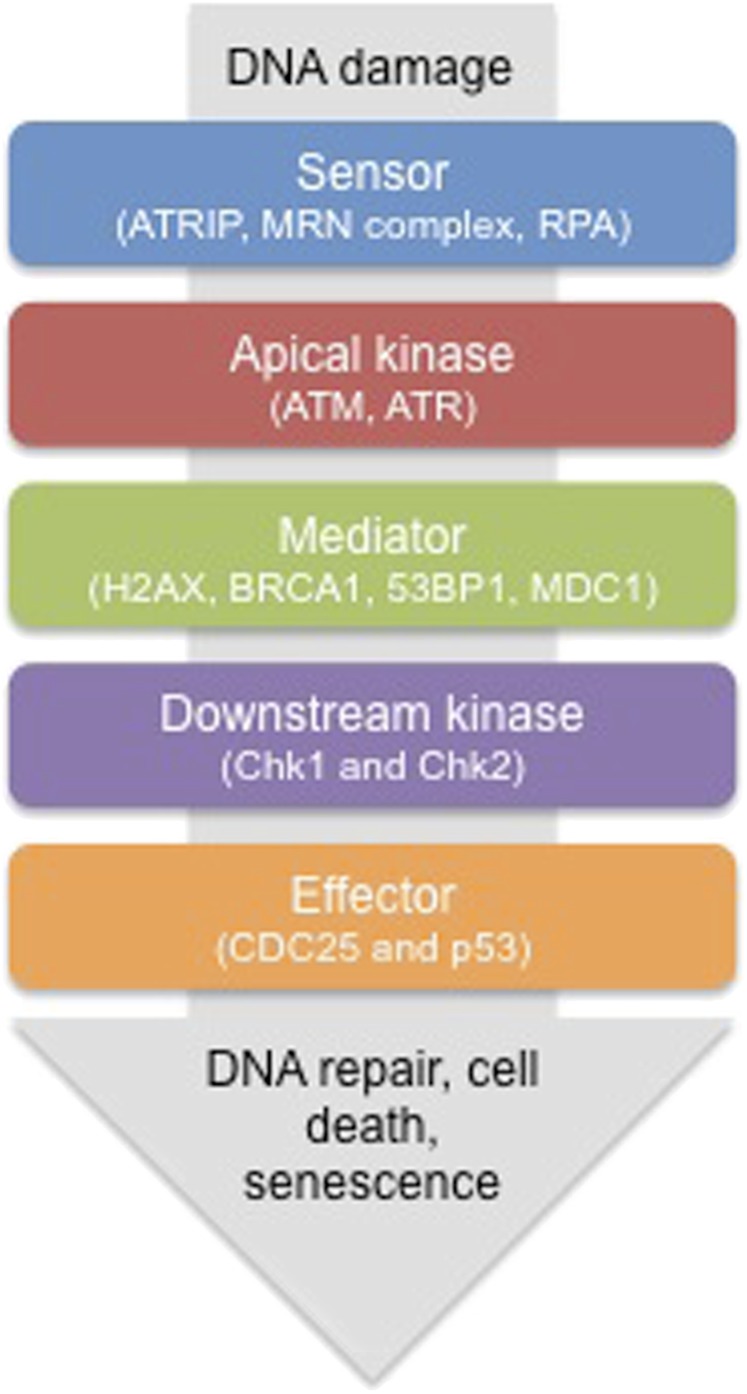
Radiotherapy is an effective therapy for around 50% of patients.1 Radiation leads to the direct damage of DNA as well as the formation of radicals, which result in the generation of lesions such as single-stand break (SSB), double-strand break (DSB) and base changes. These DNA lesions result in the activation of the DNA damage response (DDR), which is a complex signalling cascade.2 This response is induced in an attempt to halt the cell cycle in order to repair the damage or induce apoptosis or senescence if the damage is too severe (shown schematically in Figure 1 and reviewed in Sulli et al3). The DDR is initiated by signalling via apical kinases including ATR, ATM and DNA-PK.4 Targeting the DDR has been proposed as a method of enhancing the effects of radiation. Inhibitors of critical components to the DDR such as CHK1, ATR or ATM have shown promise in a number of cancer cell types in vitro as well as in vivo.5–9
Figure 1.
Schematic representation of the DNA damage response (DDR). A simplified representation of the DDR is shown to illustrate the signalling cascade that is triggered in response to DNA damage. Examples of the molecular components are given. As a result of the activation of the DDR, cells can attempt to repair the DNA damage. If the damage is unrepairable, the cell can be directed to apoptosis or senescence to ensure mutations are not transmitted to daughter cells.
DNA damage does not occur in isolation but rather within the context of the surrounding chromatin. Chromatin refers to the structure formed by the wrapping of approximately 146 base pairs of DNA around an octamer of histones consisting of H2A, H2B, H3 and H4. It is now becoming apparent that chromatin changes can have a profound impact on the DDR.3,10,11 This can be by physically protecting the DNA from damage, affecting the spread of DDR signalling or by regulating the mechanism of response to DNA damage within specific chromatin environments.3 Evidence suggests, for example, that following radiation, DNA that is lacking histones or other chromatin components sustains a greater degree of damage than does DNA associated with high chromatin compaction.12,13
In this review, chromatin changes that have been linked to changes in DDR signalling and radiation sensitivity will be described. Emphasis will be placed on those chromatin modifications that can be targeted through the use of inhibitors and how the use of such compounds could be combined with radiation to improve radiotherapy outcomes.
CHROMATIN MODULATION
The term “epigenetics” was originally coined by Conrad Waddington to describe changes in the cellular phenotype that, while heritable, were observed to be independent of alterations to the DNA sequence. Today, modifications involving DNA, histones, histone variants as well as chromatin remodellers are known to be both dynamic and highly regulated.14–17 Post-translational modification of histones has been widely characterized with histone acetylation and methylation typically receiving most attention. The N-acetylation of histones (especially on lysine residues), catalysed by histone acetyltransferases (HATs), has been characterized and is involved in transcriptional regulation, chromatin structure control and DNA repair. The addition of an acetyl group neutralizes the positive charge of lysine and is thought to weaken the electrostatic interaction present between charged DNA and histones. Therefore, histone acetylation is associated with more open chromatin structures and active transcription. Furthermore, lysine acetylation allows for binding of proteins with bromodomains and plant homeodomain fingers capable of recognizing this covalent modification.18 Not surprisingly, the removal or “erasing” of these marks by histone deacetylases (HDACs) leads to a condensation of chromatin and correlates with transcriptional repression/silencing.19
Histone methylation can occur on basic amino acid residues such as lysine and arginine. Methylation is carried out by histone methyltransferases (HMTs) and is dependent on the donation of a methyl group from S-adenosyl methionine (SAM).20 Lysine residues may be mono-, di- or tri-methylated by the lysine methyltransferases (KMTs), a group of enzymes that were originally categorized by the presence of either a suppressor of variation 3–9, enhancer of zeste trithorax (SET) or DOT1-like (DOT1L) domain.20,21 Protein arginine methyltransferases are responsible for the methylation of arginine residues and are grouped into two types dependent on the symmetry of their methyl group addition.20 Histone methylation does not alter the charge of the histone tail (unlike acetylation), but instead modifies the hydrophobicity and basicity of histones and therefore the affinity for other proteins, such as transcription factors.22 Histone methylation is highly contextual in its biological effects and may result in either activation or repression of transcription dependant on the location of the modification. For example, methylation of lysine residues has been correlated with activation of transcription (H3K4/36/79) as well as transcriptional silencing (H3K9, H4K20).20 The complex code of histone methylation, in addition to the role of these enzymes within the context of multiprotein complexes, makes these enzymes more difficult to target pharmacologically. As a direct result, HMT inhibitors (HMTi) have not yet reached the clinic, although there is evidence for their roles in cancer, therefore ensuring continued investigation of their therapeutic potential.23
As their name implies histone demethylases (HDMs) are responsible for histone demethylation. To date this group of enzymes consist of lysine-specific demethylases (KDM) and a single arginine-specific demethylase (RDM). The KDM include two families: those that contain the amine oxidase-like (AOL) domain and those with the Jumonji domain (JmjC).23 Only two members of the AOL domain family are known, KDMIA (LSDI) and KDMIB (LSD2).24 These enzymes specifically catalyse the removal of mono- and di-methylation from H3K4, but change specificity when in complex with different accessory proteins.25 The Jumonji C (JmjC) family currently has approximately 30 members, that are divided into 7 groups. They have been found to demethylate mono-, di- and tri-methylated lysines.20
CHROMATIN AND THE DNA DAMAGE RESPONSE
Any damage to DNA occurs within a particular chromatin context. In the case of a DSB for instance, initial sensing of the damage and activation of upstream kinases can be influenced and facilitated by chromatin modifiers. The interaction of the chromatin context with the DDR is extremely complex and only a subset of events (and references) are shown in Table 1 (for a more comprehensive recent review of the topic, see Sulli et al3). Upstream/apical kinases such as ATM, ATR and DNA-PK can mediate chromatin changes themselves, perhaps the most widely characterized of these being H2AX phosphorylation (γH2AX).38,39 Mediator proteins acting downstream of these apical kinases help create a positive-feedback loop to sustain the DDR signalling. Some of these mediators such as MDC1 recruit E3 ubiquitin ligases such as UBC13-RNF8 to allow H2AX ubiquitylation, an important event for the recruitment of factors that facilitate repair such as 53BP1 or BRCA1.40–43 HATs such as TIP60 can also relax chromatin at DSBs by mediating acetylation on H4 and H2A.44–46 TIP60-dependent acetylation of H2AX on Lys 5 is required for ubiquitylation of H2AX.29 Furthermore, HDAC1 and HDAC2 localize to sites of damage leading to a reduction in acetylation on H3K56. Functionally, cells where HDAC1 and HDAC2 have been depleted display marked defects in non-homologous end joining (NHEJ) and may also have defective homologous recombination-mediated repair.35
Table 1.
An overview of selected chromatin modifiers in DNA damage response (DDR) signalling and repair
| Name | Type | Specificity | Role in repair/DDR | Reference |
|---|---|---|---|---|
| KDM2A/FBXL11/JHDM1A | JmjC HDM | H3K36me1 H3K4me3 |
Enhances NHEJ-mediated repair | Fnu et al26 |
| KDM4B/JMJD2B/JHDM3B | JmjC HDM | H3K9me2/3 | Increased KDM4B leads to less IR-induced H2AX and increased cell survival | Young et al27 |
| DOT1L | HMT | H3K79me2 | 53BP1 recruitment | Huyen et al28 |
| TIP60 | HAT | Various including ATM, p53 and H2AK5ac | Lysine 5 acetylation of H2AX is necessary for ubiquitylation of H2AX ATM acetylation facilitates ATM autophosphorylation and activation |
Ikura et al,29 Sun et al,30 Kaidi and Jackson31 |
| CHD3 | ATP-ase chromatin remodeller | Various | Mediates heterochromatin formation preventing DSB expansion | Goodarzi et al32 |
| CHD4 | ATP-ase chromatin remodeller | Various | Promotes DSB repair possibly specifically during S/G2 phase | Chou et al,33 Larsen et al34 |
| HDAC | HDAC | Various | Facilitates HR and NHEJ Facilitates ATM signalling |
Miller et al35 |
| SWI/SNF | ATP-ase chromatin remodeller | Various | Facilitates DSB repair | van Attikum et al36 |
| CBP | HAT | H3K56ac | Checkpoint and chromatin assembly recovery | Vempati et al37 |
DSB, double strand break; HAT, histone acetyltransferase; HDAC, histone deacetylase; HDM, histone demethylase; HMT, histone methyltransferase; HR, homologous recombination; IR, ionizing radiation; JmjC, Jumonji domain; NHEJ, non-homologous end joining.
While H2A histone modifications are important for co-ordinating an appropriate DDR, histone methylation has also been shown to play an important role. H3K9me3 appears important for TIP60-dependent ATM acetylation, an event that facilitates ATM activation.30,47 Furthermore, DOT1L histone H3K79 methyltransferase-dependent dimethylation may facilitate 53BP1 recruitment following DNA damage through its tandem tudor domain.28 DOT1L depletion may result in reductions in H3K79me2 and 53BP1 foci formation. It is interesting to note, however, that H3K79me2 levels are not altered following radiation.28 More recently, it has become apparent that 53BP1 appears to have a greater affinity for H4K20me1 than for H3K79me2 and silencing of the SET domain-containing KMT 8 (SETD8/PRSET7), the methyltransferase catalysing H4K20me1, impairs 53BP1 recruitment. In addition, multiple myeloma SET domain-containing protein (MMSET/WHSC1), the methyltransferase responsible for H4K20 di- and trimethylation, is also required for 53BP1 localization. Both MMSET and PRSET7 are recruited to sites of DNA damage and consequently levels of H4K20me1/2/3 are induced around a DNA break site.48 Recruitment of PRSET7 has recently been shown to be dependent on Ku70 and appears important for appropriate NHEJ.49 Recruitment of PRSET7 also facilitates dimethylation at H4K20 by the Suv4-20 methyltransferase an event that is again required for 53BP1 nucleation.49 The importance of H4K20 methylation in 53BP1 recruitment has been demonstrated further by the finding that the tandem tudor domain of the HDM JMJD2A can recognize H4K20me2, thereby preventing 53BBP1 recruitment in the absence of damage. RNF8 and RNF168 target this demethylase for ubiquitylation and proteasomal degradation following ionizing radiation (IR) to allow 53BP1 foci formation.50 Recently, H4K16 acetylation has been suggested to impair 53BP1 binding to H4K20me2, with histone deacetylation occurring in response to DNA damage facilitating the formation of 53BP1 foci.51
Several other HDMs have also recently been demonstrated to play a role in DNA damage signalling. H3K4me3 levels have been shown to be reduced at the sites of DSBs.52 Interestingly, H3K4me3 demethylase LSD1 is recruited to DSBs, a process facilitated by interaction between LSD1 and RNF168. shRNA-mediated knockdown of LSD1 results in reduced H2A/H2AX ubiquitylation and reduced 53BP1 and BRCA1 recruitment following DNA damage. In addition, LSD1 knockdown confers mild sensitivity to IR.52 Notably, the H3K9me2/3 demethylase KDM4B is also recruited to DSB sites, and KDM4B overexpression results in reduced γH2AX and increased cell survival following IR.27
Similarly, hampering of DDR signalling has been shown to occur following the overexpression of bromodomain and extra terminal family member BRD4, specifically, BRD4 isoform B.53 This isoform has recently been implicated in maintaining chromatin compaction around DSBs by recruiting the condensing II chromatin remodelling complex to acetylated histones, and thereby preventing efficient γH2AX signalling following IR. Inhibition of BRD4 using JQ1 (a BRD4 inhibitor) results in decreased sensitivity to IR presumably through rapid checkpoint recovery following the increased chromatin relaxation achieved by decreased BRD4 isoform B activity. It is important to note, however, that inhibiting chromatin-modifying factors such as BRD4 might have differential effects depending on the cell type.53
INHIBITING CHROMATIN CHANGES AND RADIOSENSITIVITY
Of the chromatin modifications, DNA methylation, in addition to histone methylation and acetylation, is generally the most widely characterized and has therefore been the major focus of novel therapeutic strategies in this area. Despite current questions and lingering gaps in knowledge, inhibitors of the enzymes involved in these modifications have and are being developed, and some show promise for cancer therapy.54
DNA METHYLTRANSFERASE INHIBITORS
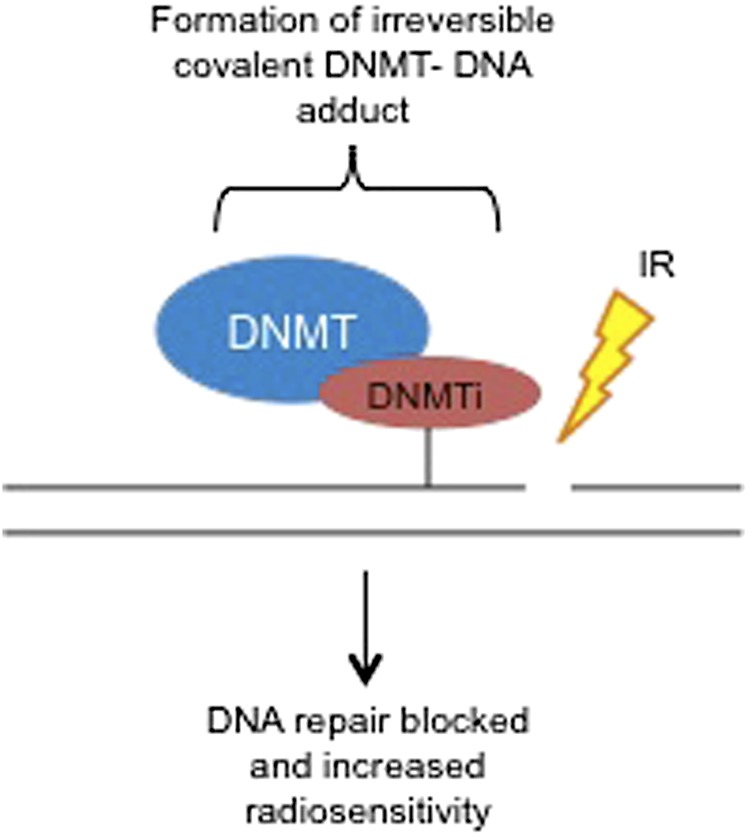
Some DNA methyltransferase inhibitors (DNMTi) have demonstrated clinical efficacy in the treatment of certain haematological malignancies.55 Their use as single agents, however, has raised some concerns owing to mild efficacy and the profile of associated side effects. The use of lower doses of some of these inhibitors in combination with chemotherapy or radiotherapy has been proposed as a potentially useful means of using such inhibitors.56 A number of mechanisms have been suggested as a rationale for the efficacy of these agents in combination with radiation (Table 2).56 For example, the DNMTi that are nucleoside analogues, such as 5-Aza-cytidine (azacitidine), 5-aza-2'-deoxycytidine (decitabine) and zebularine, can create an irreversible DNA adduct (where the DNMT is bound to DNA and is therefore inhibited).60,61 When this occurs in close proximity to radiation-induced damage, such as a SSB, the break is more difficult to repair (Figure 2).62 Repair of radiation-induced DNA damage may also be reduced following treatment with nucleoside analogues, since these can hinder the progress of DNA synthesis.63 Furthermore, it has been proposed that DNMTi could synchronize cells in the more radiosensitive phases of the cell cycle such as G2/M. Moreover, since DNA methylation can lead to gene silencing, the use of a DNMTi can result in the reactivation of certain genes. Induction of apoptosis has been proposed to accompany DNMTi treatment potentially owing to the re-expression of specific genes such as RASSF1 or DAPK1 or reactivation of mediators of apoptosis.56 Together, these effects suggest that some DNMTi could be used in a beneficial manner together with radiation.
Table 2.
Reported radiosensitizing effects of selected DNA methyltransferase inhibitors
| Inhibitor | Mechanism of radiosensitization | Cell lines tested | Reference |
|---|---|---|---|
| Decitabine | Prolonged γH2AX expression | A549 and U373MG | Kim et al57 |
| Azacitidine | Demethylation of p16 and MLH1 promoters observed in radiosensitive colorectal cell lines DNA damage response signalling and apoptosis induced in MM cells |
HCT116, SW480, L174T, Co115 No radiosensitizing effect seen in A549 or U373MG MM.1S, MM.1R |
Hofstetter et al,58 Kiziltepe et al59 |
| Zebularine | Prolonged γH2AX expression | A549 and U373MG | Kim et al57 |
MM, multiple myeloma.
Figure 2.
The proposed mechanism of radiosensitization by DNA methyltransferase inhibitors (DNMTi). A schematic representation of the mechanism of action of DNMTi. The presence of a DNMTi following the generation of DNA damage by ionizing radiation (IR) results in hindered DNA repair and increased radiosensitivity.
HISTONE ACETYLTRANSFERASE INHIBITORS
Loss of TIP60 inhibits the effective repair of DNA damage and sensitizes cells to radiation, suggesting that TIP60 inhibitors could be therapeutically effective in combination with radiation.30 Several “global” HAT inhibitors have been demonstrated to possess the ability to radiosensitize cancer cells, including anacardic acid and curcumin. Despite permeability restrictions in vitro, anacardic acid has been found to block the TIP60-dependent activation of the ATM and DNA-PKcs protein kinases by DNA damage in vivo, therefore sensitizing human tumour cells to the cytotoxic effects of ionizing radiation.64
HISTONE DEACETYLASE INHIBITORS
There has been considerable effort in investigating the benefits of HDAC inhibition in the context of cancer treatment. Two of the zinc-dependent HDAC inhibitors (HDACi) (vorinostat and romidespin) have been approved for cutaneous T-cell lymphoma in the USA.65 The biological response to HDACi includes the upregulation of p21 in a p53-independent fashion, which causes cell cycle arrest in the G1 phase.66 The HDACi butyrate and trichostatin A (TSA) have been shown to stabilize p21 mRNA, repress cyclins A and D, and activate p16 and p27, causing cell cycle arrest.67–69 Furthermore, certain HDACi increase the expression of proapoptotic genes, such as TRAIL, DR5, Bax, Apaf-1, Bmf, Bim and TP2 and/or downregulate antiapoptotic genes (such as BCL2, MCL1 and XIAP).67,70 Functionally, treatment with several HDACi also leads to prolonged IR-induced γH2AX foci.71,72 This has been suggested to result, at least in part, owing to the impaired DDR signalling and repair following HDAC inhibition. Indeed, treatment with vorinostat [suberoyl + anilide + hydroxamic acid (SAHA)] together with IR leads to a reduction in levels of RAD50 following IR in melanoma cells or DNA-PK in prostate cells,73 again suggesting compromised DNA repair. Down regulation of ATM gene expression and signalling has also been demonstrated in response to IR following HDAC inhibition.74,75
As radiosensitizing agents, HDACi have shown promise in vitro in a variety of cell lines as well as in xenograft studies76 (Table 3). Vorinostat has been demonstrated to enhance radiation-induced apoptosis at low micromolar concentrations in DU145 prostate cancer cells.76 Vorinostat has also demonstrated effects as a radiosensitizer in melanoma, non-small-cell lung cancer, prostate, glioma, osteosarcoma, rhabdomyosarcoma and glioma cancers and cell lines.73,79–82,88 The HDAC inhibitor etinostat has shown radiosensitizing effects towards prostate cancer in vitro, and this effect was the greatest when the drug was present before and after radiation.87 When erythroleukemic K562 cells were treated with valproic acid 24-h pre-radiation, the cells exhibited enhanced radiosensitivity, leading to increased radiation-induced cell death and apoptosis.89 The HDACi valproic acid radiosensitizes colorectal cancer cell lines,77 while TSA has been found to radiosensitize cancers such as melanoma and squamous carcinoma.85 The HDACi H6CAHA has demonstrated radiosensitizing effects against prostate cancer cell lines,75 and the compound PC1-24781 radiosensitizes lung adenocarcinoma, large-cell lung and colon cancer cell lines.85 The HDACi Scriptaid (Karus Therapeutics, Pbingdon, UK) radiosensitizes squamous carcinoma cancer cell lines.86
Table 3.
Reported radiosensitizing effects of selected histone deacetylase inhibitors
| Inhibitor | Mechanism of radiosensitization | Cells lines tested | Reference |
|---|---|---|---|
| Valproic acid | Radiosensitization dependent on p53 (colorectal cell lines) Inhibition of DNA double-strand break repair and prolonged γH2AX foci |
LS174T, HCT116, K562, SF539 and U251 | Chen et al,77 Camphausen et al78 |
| Vorinostat | Appears to inhibit non-homologous end joining following radiation Increases γH2AX foci formation and downregulates Ku70, Ku80, Ku86, DNA-PKcs, Rad51, EGFR, AKT in a variety of cell lines |
A375, MeWo, A549, DU145, U373vIII, KHOS-24OS, SAOS2 (OS), A-204, RD (RMS), MDA-MB-231-BR, NB1691, SY5Y, Tet21 (MYCN overexpressed), Tet21 (MYCN repressed) | Munshi et al,73 Baschnagel et al,79 Karagiannis and El-Osta,80 Blattmann et al,81 Mueller et al,82 Karagiannis and El-Osta83 |
| Trichostatin A (TSA) | Downregulates Ku700, Ku80 and DNA-PKcs Prolongs γH2AX Enhances G2/M cell cycle arrest and apoptosis (via mitochondrial pathway) |
A549, H1650, A375, MeWo, SQ-20B, SCC-35 | Munshi et al,72 Zhang et al84 |
| H6CAHA | Downregulates ATM, BRCA1 and BRCA2 Prolongs γH2AX |
PC3, DU145, LNCaP | Konsoula et al75 |
| PCI-24781 | Downregulates Rad51 | HCT116, NCI-H460, A549 | Adimoolam et al85 |
| Scriptaid | Downregulates Ku80 (but not Ku70) Prolongs γH2AX |
SQ-20B | Kuribayashi et al86 |
| Etinostat | Prolongs γH2AX | DU145, U251 | Camphausen et al87 |
HISTONE METHYLTRANSFERASE INHIBITORS
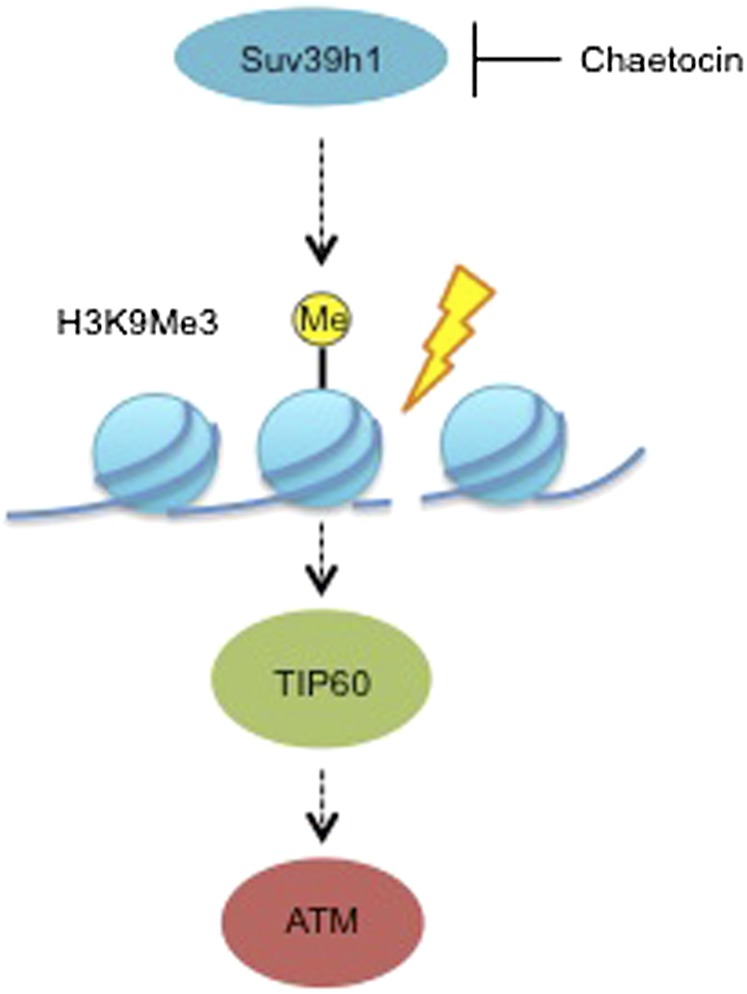
The complex code of histone methylation, in addition to the role of these enzymes within the context of multiprotein complexes, makes these enzymes challenging to target therapeutically.23 Despite current difficulties in targeting the HMTs, our knowledge of their function is rapidly increasing.47,90 As described, several histone methylation marks as well as some of the methyltransferases are emerging as facilitators of the DDR. Decreases in the levels of histone methylation on specific residues might therefore impair DDR signalling and lead to sensitization.47 Since loss of ATM is well known to lead to exquisite radiosensitization, it is tempting to speculate that attenuating ATM signalling through targeting chromatin modifications essential for ATM activation may lead to similar effects, that is, increased radiosensitivity. Inhibition of H3K9me3, the chromatin mark associated with ATM activation, by loss of two important methyltransferases, Suv39h1 and Suv39h2, could therefore lead to radiosensitization. Chaetocin, a fungal metabolite that binds to the SAM binding site, is a currently available Suv39h1 and G9a inhibitor (Figure 3).91 G9a inhibitors such as BIX-01294, UNC0321, UNC0638 or UNC0646 are also available, but only limited pre-clinical testing has been carried out with these to date.23,92 As more and more specific inhibitors of the HMTs are developed in the future, it will be interesting to test them in combination with radiation.
Figure 3.
The proposed mechanism of radiosensitization by the histone methyltransferase chaetocin. Regions of H3K9me3 are important for TIP60-dependent ATM acetylation, an event that facilitates ATM activation following DNA damage. Chaetocin inhibits H3K9me3 methyltransferases including Suv39h1 and may thereby reduce H3K9me3 levels, TIP60-dependent ATM acetylation and the subsequent ATM activation. Failure to activate ATM appropriately may lead to increased sensitivity to DNA damage induced by ionizing radiation. Me, methylation.
The mixed lineage leukaemia 1 (MLL) methyltransferase is associated with chromosomal translocations resulting in the expression of chimeric fusion proteins and deregulated recruitment of DOT1L. The DOT1L inhibitor, EPZ004777 (a SAM analogue) has been showed to be effective in cell lines and xenografts with MLL translocations.93 Since DOT1L is known to be important for 53BP1 recruitment and would be predicted to impact NHEJ-mediated repair, the use of a DOTL1 inhibitor could be envisioned to also be of therapeutic benefit in combination with radiation.28
However, it is clear that more investigation is needed regarding the development of HMTi that is both highly selective and well tolerated before it will be possible to assess the full therapeutic potential of such treatments and their ability to act as radiosensitizers. Structural diversity of SAM binding sites amongst all methyltransferases suggests selectivity could be achieved.28,94,95
HISTONE DEMETHYLASE INHIBITORS
A number of HDM inhibitors (HDMi) have been developed in recent years. The involvement of both LSD1 as well as some of the members of the JmjC family in DDR signalling suggests that targeting these may also impact the DDR and therefore, once again, could lead to radiosensitization if used in combination with IR. Currently available HDMi can be classified according to their ability to inhibit the AOL domain-containing family or the JmjC demethylases. Current JmjC inhibitors compete with the cofactor (2-oxoglutarate) and bind iron in the active site.95 Interestingly, HDACi have also displayed HDM inhibitory properties, and this overlap is thought to be owing to the shared enzymatic characteristics of co-ordinating metal ions. For example, SAHA has displayed moderate inhibition of JMJD2E but is much less effective as a JmjC inhibitor than as a HDACi.20
CONCLUSION
As described above, a variety of chromatin remodellers and chromatin-remodelling processes have been shown to affect DNA damage sensing, signalling and repair. It is tempting to speculate that combining some of the inhibitors that have been developed to target these processes in combination with DNA damaging agents such as IR could be therapeutically beneficial. Indeed some of these compounds, such as HDACi, have already been shown to radiosensitize different tumour types at least in pre-clinical models. Before these approaches can be taken any further, the safety profiles of compounds will need to be investigated. For instance, the impaired DDR signalling and repair expected to result from the combination of some of these inhibitors with radiotherapy also raises the question of whether such combinations would also result in increased normal tissue toxicity. Nevertheless, it is important to bear in mind that in contrast to tumour cells, normal cells would not have compromised DNA repair pathways and may therefore be less affected by these combinations.96,97 Furthermore, differences in expression of chromatin remodelers between tumour and normal cells may also increase the tumour sensitivity towards therapeutic strategies targeting chromatin.67,68 Establishing a comprehensive safety profile will, however, be particularly important if inhibitors of chromatin modifiers that have also been implicated in tumour suppression are to be used in combination with radiotherapy (such as TIP60). The timing of administration of these inhibitors in relation to irradiation protocols as well as the choice of tumour type will also be critical in devising the most effective radiosensitizing strategies. It will be important to investigate whether the use of these compounds for short treatment courses, perhaps limited to just before or for the duration of radiation therapy, may alleviate some of the safety concerns. It is clear, however, that an area of novel and exciting possibilities is likely to emerge from the study of future therapeutic strategies involving the chromatin modulation together with radiotherapy.
FUNDING
This work was supported by a Cancer Research UK grant awarded to EMH. MMO is supported by the MRC with an additional MRC Centenary award (awarded to MMO).
Acknowledgments
ACKNOWLEDGMENTS
We would like to thank all members of the Hammond laboratory for insightful comments and discussion and apologize to those authors whom we have not been able to cite here.
REFERENCES
- 1.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer 2005; 104: 1129–37. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461: 1071–8. doi: 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulli G, Di Micco R, d'Adda di Fagagna F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 2012; 12: 709–20. doi: 10.1038/nrc3344 [DOI] [PubMed] [Google Scholar]
- 4.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013; 14: 197–210. [PubMed] [Google Scholar]
- 5.Pires IM, Olcina MM, Anbalagan S, Pollard JR, Reaper PM, Charlton PA, et al. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br J Cancer 2012; 107: 291–9. doi: 10.1038/bjc.2012.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis 2012; 3: e441. doi: 10.1038/cddis.2012.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batey MA, Zhao Y, Kyle S, Richardson C, Slade A, Martin NM, et al. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol Cancer Ther 2013; 12: 959–67. doi: 10.1158/1535-7163.MCT-12-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst GR, McLaughlin M, Kyula JN, Neijenhuis S, Khan A, Good J, et al. Targeted radiosensitization by the Chk1 inhibitor SAR-020106. Int J Radiat Oncol Biol Phys 2013; 85: 1110–18. doi: 10.1016/j.ijrobp.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 9.Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther 2012; 13: 1072–81. doi: 10.4161/cbt.21093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 2012; 46: 722–34. doi: 10.1016/j.molcel.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 11.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 2008; 31: 167–77. doi: 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- 12.Elia MC, Bradley MO. Influence of chromatin structure on the induction of DNA double strand breaks by ionizing radiation. Cancer Res 1992; 52: 1580–6. [PubMed] [Google Scholar]
- 13.Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput Biol 2007; 3: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 2011; 11: 726–34. doi: 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 2011; 25: 2436–52. doi: 10.1101/gad.179184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell 2007; 128: 693–705. [DOI] [PubMed] [Google Scholar]
- 17.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011; 146: 1016–28. doi: 10.1016/j.cell.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 2007; 14: 1025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner JM, Hackanson B, Lübbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics 2010; 1: 117–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bojang P, Jr, Ramos KS. The promise and failures of epigenetic therapies for cancer treatment. Cancer Treat Rev 2014; 40: 153–69. doi: 10.1016/j.ctrv.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell 2007; 131: 633–6. [DOI] [PubMed] [Google Scholar]
- 22.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol 2001; 13: 263–73. [DOI] [PubMed] [Google Scholar]
- 23.Tian X, Zhang S, Liu HM, Zhang YB, Blair CA, Mercola D, et al. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets 2013; 13: 558–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, et al. Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci U S A 2006; 103: 13956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trewick SC, McLaughlin PJ, Allshire RC. Methylation: lost in hydroxylation? EMBO Rep 2005; 6: 315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A 2011; 108: 540–5. doi: 10.1073/pnas.1013571108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young LC, McDonald DW, Hendzel MJ. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J Biol Chem 2013; 288: 21376–88. doi: 10.1074/jbc.M113.491514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 2004; 432: 406–11. [DOI] [PubMed] [Google Scholar]
- 29.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol 2007; 27: 7028–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 2005; 102: 13182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature 2013; 498: 70–4. doi: 10.1038/nature12201 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 2011; 18: 831–9. doi: 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- 33.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A 2010; 107: 18475–80. doi: 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 2010; 190: 731–40. doi: 10.1083/jcb.200912135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010; 17: 1144–51. doi: 10.1038/nsmb.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 2007; 26: 4113–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem 2010; 285: 28553–64. doi: 10.1074/jbc.M110.149393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cellule 2003; 114: 359–70. [DOI] [PubMed] [Google Scholar]
- 39.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 2003; 5: 675–9. [DOI] [PubMed] [Google Scholar]
- 40.Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 2003; 421: 952–6. [DOI] [PubMed] [Google Scholar]
- 41.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003; 421: 961–6. [DOI] [PubMed] [Google Scholar]
- 42.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 2004; 23: 2674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cellule 2005; 123: 1213–26. [DOI] [PubMed] [Google Scholar]
- 44.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 2004; 16: 979–90. [DOI] [PubMed] [Google Scholar]
- 45.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 2004; 119: 767–75. [DOI] [PubMed] [Google Scholar]
- 46.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004; 119: 777–88. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 2009; 11: 1376–82. doi: 10.1038/ncb1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 2014; 15: 7–18. doi: 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- 49.Tuzon CT, Spektor T, Kong X, Congdon LM, Wu S, Schotta G, et al. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep 2014; 8: 430–8. doi: 10.1016/j.celrep.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 2012; 31: 1865–78. doi: 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol 2013; 5: 157–65. doi: 10.1093/jmcb/mjs066 [DOI] [PubMed] [Google Scholar]
- 52.Mosammaparast N, Kim H, Laurent B, Zhao Y, Lim HJ, Majid MC, et al. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J Cell Biol 2013; 203: 457–70. doi: 10.1083/jcb.201302092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 2013; 498: 246–50. doi: 10.1038/nature12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: an update. Mol Oncol 2012; 6: 657–82. doi: 10.1016/j.molonc.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song SH, Han SW, Bang YJ. Epigenetic-based therapies in cancer: progress to date. Drugs 2011; 71: 2391–403. doi: 10.2165/11596690-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 56.Gravina GL, Festuccia C, Marampon F, Popov VM, Pestell RG, Zani BM, et al. Biological rationale for the use of DNA methyltransferase inhibitors as new strategy for modulation of tumor response to chemotherapy and radiation. Mol Cancer 2010; 9: 305. doi: 10.1186/1476-4598-9-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HJ, Kim JH, Chie EK, Young PD, Kim IA, Kim IH. DNMT (DNA methyltransferase) inhibitors radiosensitize human cancer cells by suppressing DNA repair activity. Radiat Oncol 2012; 7: 39. doi: 10.1186/1748-717X-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hofstetter B, Niemierko A, Forrer C, Benhattar J, Albertini V, Pruschy M, et al. Impact of genomic methylation on radiation sensitivity of colorectal carcinoma. Int J Radiat Oncol Biol Phys 2010; 76: 1512–19. doi: 10.1016/j.ijrobp.2009.10.037 [DOI] [PubMed] [Google Scholar]
- 59.Kiziltepe T, Hideshima T, Catley L, Raje N, Yasui H, Shiraishi N, et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol Cancer Therapeutics 2007; 6: 1718–27. [DOI] [PubMed] [Google Scholar]
- 60.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A 1984; 81: 6993–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2'-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A 1994; 91: 11797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferguson AT, Vertino PM, Spitzner JR, Baylin SB, Muller MT, Davidson NE. Role of estrogen receptor gene demethylation and DNA methyltransferase. DNA adduct formation in 5-aza-2'deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem 1997; 272: 32260–6. [DOI] [PubMed] [Google Scholar]
- 63.Smits KM, Melotte V, Niessen HE, Dubois L, Oberije C, Troost EG, et al. Epigenetics in radiotherapy: where are we heading? Radiother Oncol 2014; 111: 168–77. doi: 10.1016/j.radonc.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 64.Sun Y, Jiang X, Chen S, Price BD. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett 2006; 580: 4353–6. [DOI] [PubMed] [Google Scholar]
- 65.Fatkins DG, Zheng W. Substituting N(epsilon)-thioacetyl-lysine for N(epsilon)-acetyl-lysine in peptide substrates as a general approach to inhibiting human NAD(+)-dependent protein deacetylases. Int J Mol Sci 2008; 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandor V, Senderowicz A, Mertins S, Sackett D, Sausville E, Blagosklonny MV, et al. P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer 2000; 83: 817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006; 5: 769–84. [DOI] [PubMed] [Google Scholar]
- 68.Espino PS, Drobic B, Dunn KL, Davie JR. Histone modifications as a platform for cancer therapy. J Cell Biochem 2005; 94: 1088–102. [DOI] [PubMed] [Google Scholar]
- 69.Wharton W, Savell J, Cress WD, Seto E, Pledger WJ. Inhibition of mitogenesis in Balb/c-3T3 cells by Trichostatin A. Multiple alterations in the induction and activation of cyclin-cyclin-dependent kinase complexes. J Biol Chem 2000; 275: 33981–7. [DOI] [PubMed] [Google Scholar]
- 70.Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP, et al. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis 2013; 4: e519. doi: 10.1038/cddis.2013.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang F, Zhang T, Teng ZH, Zhang R, Wang JB, Mei QB. Sensitization to gamma-irradiation-induced cell cycle arrest and apoptosis by the histone deacetylase inhibitor trichostatin A in non-small cell lung cancer (NSCLC) cells. Cancer Biol Ther 2009; 8: 823–31. [DOI] [PubMed] [Google Scholar]
- 72.Munshi A, Kurland JF, Nishikawa T, Tanaka T, Hobbs ML, Tucker SL, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res 2005; 11: 4912–22. [DOI] [PubMed] [Google Scholar]
- 73.Munshi A, Tanaka T, Hobbs ML, Tucker SL, Richon VM, Meyn RE. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol Cancer Ther 2006; 5: 1967–74. [DOI] [PubMed] [Google Scholar]
- 74.Thurn KT, Thomas S, Raha P, Qureshi I, Munster PN. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Mol Cancer Ther 2013; 12: 2078–87. doi: 10.1158/1535-7163.MCT-12-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konsoula Z, Cao H, Velena A, Jung M. Adamantanyl-histone deacetylase inhibitor H6CAHA exhibits favorable pharmacokinetics and augments prostate cancer radiation sensitivity. Int J Radiat Oncol Biol Phys 2011; 79: 1541–8. doi: 10.1016/j.ijrobp.2010.11.057 [DOI] [PubMed] [Google Scholar]
- 76.Sharma NL, Groselj B, Hamdy FC, Kiltie AE. The emerging role of histone deacetylase (HDAC) inhibitors in urological cancers. BJU Int 2013; 111: 537–42. doi: 10.1111/j.1464-410X.2012.11647.x [DOI] [PubMed] [Google Scholar]
- 77.Chen X, Wong P, Radany E, Wong JY. HDAC inhibitor, valproic acid, induces p53-dependent radiosensitization of colon cancer cells. Cancer Biother Radiopharm 2009; 24: 689–99. doi: 10.1089/cbr.2009.0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Camphausen K, Cerna D, Scott T, Sproull M, Burgan WE, Cerra MA, et al. Enhancement of in vitro and in vivo tumor cell radiosensitivity by valproic acid. Int J Cancer 2005; 114: 380–6. [DOI] [PubMed] [Google Scholar]
- 79.Baschnagel A, Russo A, Burgan WE, Carter D, Beam K, Palmieri D, et al. Vorinostat enhances the radiosensitivity of a breast cancer brain metastatic cell line grown in vitro and as intracranial xenografts. Mol Cancer Ther 2009; 8: 1589–95. doi: 10.1158/1535-7163.MCT-09-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karagiannis TC, El-Osta A. Modulation of cellular radiation responses by histone deacetylase inhibitors. Oncogene 2006; 25: 3885–93. [DOI] [PubMed] [Google Scholar]
- 81.Blattmann C, Oertel S, Ehemann V, Thiemann M, Huber PE, Bischof M, et al. Enhancement of radiation response in osteosarcoma and rhabdomyosarcoma cell lines by histone deacetylase inhibition. Int J Radiat Oncol Biol Phys 2010; 78: 237–45. doi: 10.1016/j.ijrobp.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 82.Mueller S, Yang X, Sottero TL, Gragg A, Prasad G, Polley MY, et al. Cooperation of the HDAC inhibitor vorinostat and radiation in metastatic neuroblastoma: efficacy and underlying mechanisms. Cancer Lett 2011; 306: 223–9. doi: 10.1016/j.canlet.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karagiannis TC, El-Osta A. Clinical potential of histone deacetylase inhibitors as stand alone therapeutics and in combination with other chemotherapeutics or radiotherapy for cancer. Epigenetics 2006; 1: 121–6. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Jung M, Dritschilo A, Jung M. Enhancement of radiation sensitivity of human squamous carcinoma cells by histone deacetylase inhibitors. Radiat Res 2004; 161: 667–74. [DOI] [PubMed] [Google Scholar]
- 85.Adimoolam S, Sirisawad M, Chen J, Thiemann P, Ford JM, Buggy JJ. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc Natl Acad Sci U S A 2007; 104: 19482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuribayashi T, Ohara M, Sora S, Kubota N. Scriptaid, a novel histone deacetylase inhibitor, enhances the response of human tumor cells to radiation. Int J Mol Med 2010; 25: 25–9. [PubMed] [Google Scholar]
- 87.Camphausen K, Burgan W, Cerra M, Oswald KA, Trepel JB, Lee MJ, et al. Enhanced radiation-induced cell killing and prolongation of gammaH2AX foci expression by the histone deacetylase inhibitor MS-275. Cancer Res 2004; 64: 316–21. [DOI] [PubMed] [Google Scholar]
- 88.Karagiannis TC, El-Osta A. The paradox of histone deacetylase inhibitor-mediated modulation of cellular responses to radiation. Cell Cycle 2006; 5: 288–95. [DOI] [PubMed] [Google Scholar]
- 89.Karagiannis TC, Kn H, El-Osta A. The epigenetic modifier, valproic acid, enhances radiation sensitivity. Epigenetics 2006; 1: 131–7. [DOI] [PubMed] [Google Scholar]
- 90.Olcina MM, Foskolou IP, Anbalagan S, Senra JM, Pires IM, Jiang Y, et al. Replication stress and chromatin context link ATM activation to a role in DNA replication. Mol Cell 2013; 52: 758–66. doi: 10.1016/j.molcel.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cherblanc FL, Chapman KL, Brown R, Fuchter MJ. Chaetocin is a nonspecific inhibitor of histone lysine methyltransferases. Nat Chem Biol 2013; 9: 136–7. doi: 10.1038/nchembio.1187 [DOI] [PubMed] [Google Scholar]
- 92.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol 2011; 7: 566–74. doi: 10.1038/nchembio.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20: 53–65. doi: 10.1016/j.ccr.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dowden J, Hong W, Parry RV, Pike RA, Ward SG. Toward the development of potent and selective bisubstrate inhibitors of protein arginine methyltransferases. Bioorg Med Chem Lett 2010; 20: 2103–5. doi: 10.1016/j.bmcl.2010.02.069 [DOI] [PubMed] [Google Scholar]
- 95.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 2012; 11: 384–400. doi: 10.1038/nrd3674 [DOI] [PubMed] [Google Scholar]
- 96.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–74. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 97.Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood 2011; 117: 6074–82. doi: 10.1182/blood-2011-01-313734 [DOI] [PubMed] [Google Scholar]