Abstract
Arterial spin labeling (ASL) MRI provides an accurate and reliable measure of cerebral blood flow (CBF). A rapidly growing number of CBF measures are being collected both in clinical and research settings around the world, resulting in a large volume of data across a wide spectrum of study populations and health conditions. Here, we describe a central CBF data repository with integrated processing workflows, referred to as the Cerebral Blood Flow Biomedical Informatics Research Network (CBFBIRN). The CBFBIRN provides an integrated framework for the analysis and comparison of CBF measures across studies and sites. In this work, we introduce the main capabilities of the CBFBIRN (data storage, processing and sharing), describe what types of data are available, explain how users can contribute to the data repository and access existing data from it, and discuss our long term plans for the CBFBIRN.
Introduction
Healthy brain function is critically dependent on well-regulated cerebral blood flow (CBF) for the delivery of oxygen and glucose. Regional alterations in CBF have been observed in a wide range of health conditions, including acute and chronic cerebrovascular disease (e.g. stroke, transient ischemic attacks), Alzheimer’s disease, mild cognitive impairment, epilepsy, HIV-related cognitive impairment, multiple sclerosis, depression, schizophrenia, post-traumatic stress disorder, traumatic brain injury, obsessive-compulsive disorder, and vascular dementia (Brown et al., 2007; Taber et al., 2005; Wintermark et al., 2005). Over the past decade, arterial spin labeling (ASL) magnetic resonance imaging (MRI) has emerged as a robust and non-invasive method for acquiring regional CBF maps with a relatively short scan duration (5 to 10 minutes). Because of its non-invasive nature and ease of use, a growing number of research and clinical sites are now using ASL and are collectively generating a rich dataset of CBF measures, spanning a broad range of health conditions and populations. As an initial step towards harnessing the potential of the available data, we have developed a CBF data repository with integrated processing workflows (referred to as the Cerebral Blood Flow Biomedical Informatics Research Network (CBFBIRN)) to integrate CBF datasets from a wide range of ongoing and completed studies. By providing a unified framework for the analysis and comparison of CBF measures across studies, the CBFBIRN has the potential to significantly advance efforts to more fully characterize this key physiological quantity in both health and disease.
A main hindrance to the faster adoption of ASL has been a lack of standardized data acquisition and processing methods. Pooling of ASL data across different sites into a central repository facilitates the careful evaluation of different acquisition and processing methods, leading to 1) improved understanding of how these differences affect the accuracy and reliability of quantified CBF measures; 2) support for standard ASL protocols and a common CBF processing method that minimizes inter-site differences and promotes faster adoption of ASL by the neuroimaging and clinical communities.
In this paper, we introduce the main capabilities of the CBFBIRN, describe what types of data are available, and explain how users can contribute to the data repository and access existing data from it.
Methods
Overview of the CBFBIRN Functionality
The CBFBIRN database architecture is based on the Human Imaging Database (HID) framework (Ozyurt et al., 2010), originally developed for the Functional Biomedical Research Network (FBIRN) to support federated clinical and imaging data management across multiple institutions. For the CBFBIRN, the HID architecture was adapted to accommodate additional requirements. The CBFBIRN data repository is a centralized system where the raw and derived data are maintained in one centralized location at the University of California San Diego (UCSD) Center for Functional MRI (CFMRI) with a fully mirrored backup server located at the UCSD Supercomputer Center, simplifying system resource maintenance and data curation. In addition to being a data repository, the CBFBIRN incorporates two data process workflows that provide: 1) support for the conversion of the ASL raw image data into calibrated CBF maps; 2) infrastructure for basic statistical tests (i.e. group analysis with ANOVA) using system-generated CBF maps and subject metadata (clinical assessments and subject demographics). A detailed description of the CBFBIRN system data model and architecture is described in another publication (Shin et al., 2013a).
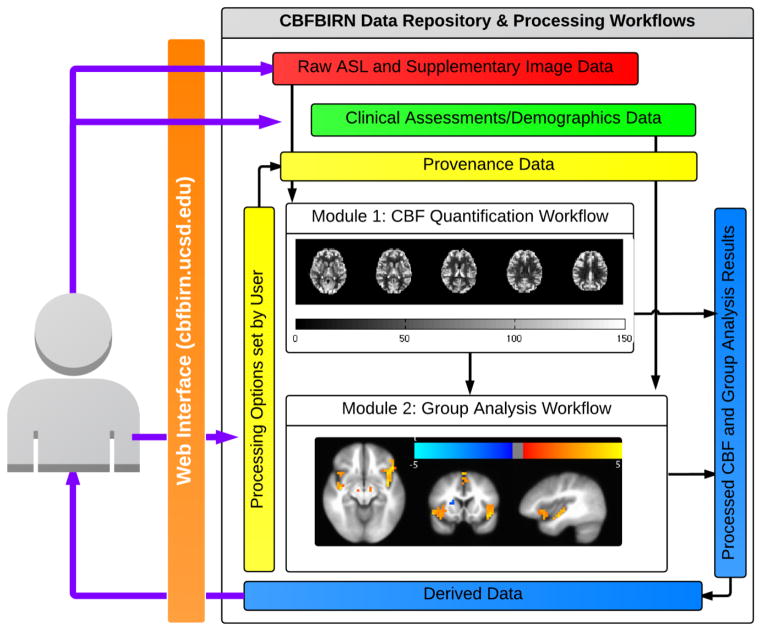
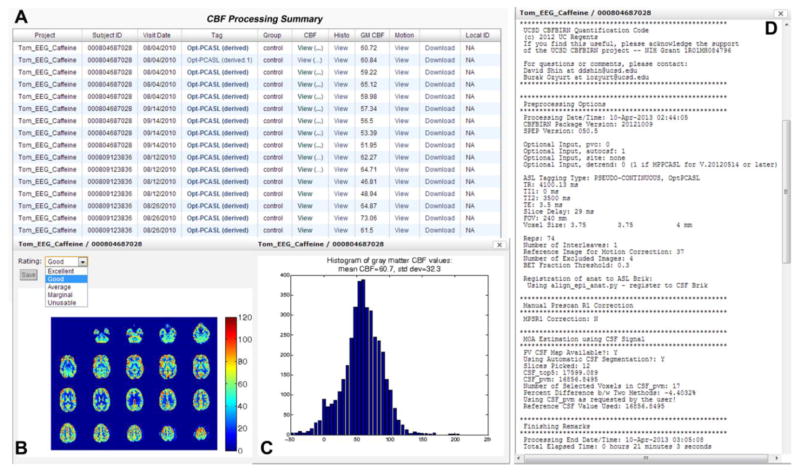
A general overview of the CBFBIRN functionality is presented in Fig. 1. All aspects of user interaction with the system are achieved via a web browser using a secure Internet connection (URL: https://cbfbirn.ucsd.edu). The raw ASL and supplemental image data, e.g. anatomical and field maps (red box) are uploaded to the data repository directly through the web browser either as a zip or tar file using the Representational State Transfer (REST) protocol (Fielding and Taylor, 2000). The user then initiates a job on the CBF processing workflow (Module 1) to generate a calibrated CBF map. The set of specific processing options selected by the user is captured as provenance data (yellow box). Figure 2 (Shin et al., 2013a) shows a data summary page that allows the user to review a list of processed jobs.
Figure 1.
A schematic overview of CBFBIRN functionality.
Figure 2.
CBF Processing Summary Page that presents a table (A) containing the complete list of successfully processed jobs. For each job, the table provides additional details such as Subject ID, scan date, experimental condition, type of ASL used, and whole-brain mean gray matter CBF value. A CBF map (B), a histogram of gray matter CBF values (C), and detailed processing logs (D) are shown from a representative job, all of which are accessible directly from the table.
The CBFBIRN currently supports the integration and analysis of baseline ASL data acquired with either pulsed ASL (e.g. FAIR) or pseudocontinuous ASL (PCASL) methods (Dai et al., 2008; Kim and Tsekos, 1997). For FAIR ASL, data acquired with 2D acquisitions (single-shot 2D spiral or EPI) on GE and Siemens MRI systems are currently supported. For PCASL data, data acquired with either 2D or 3D acquisition schemes (single-shot 2D spiral or 3D stack of spirals) on GE MRI systems are currently supported. Given the flexible architecture of the CBFBIRN, modifications to support baseline ASL data from other vendor platforms or software versions are rather straightforward. The capability to handle additional ASL data acquisition schemes, such as multiple inversion time ASL data, is currently under development.
The CBFBIRN provides two standard calibration methods (as user selectable options) for estimating the equilibrium magnetization of arterial blood (M0A). The M0A map is necessary for conversion of the perfusion signal into physiological units (ml/100g-min). The first method calculates M0A using the ventricular CSF signal from a separately acquired proton density (PD) image (Chalela et al., 2000) while the second method creates a voxel-wise M0A map using λ, the partition coefficient, in combination with the reference PD image (Alsop and Detre, 1996; Detre et al., 1992). Using the CBFBIRN, it is also possible to process the same data twice using both methods and compare the CBF values.
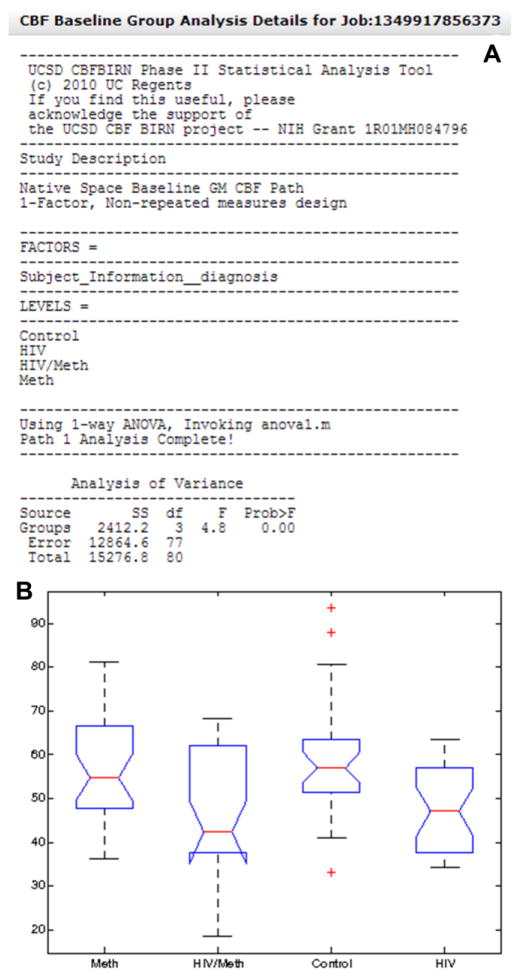
The user can also add clinical data as a CSV file and associate them to existing subjects in the repository (green box in Figure 1). The calibrated CBF maps, clinical data, and existing provenance data are combined together when a job is initiated in the group analysis workflow (Module 2), which generates a statistics summary and associated plots. See Figure 3 for an example of a system generated output (Shin et al., 2013a). Three types of group analysis are currently supported, i.e. 1) mean gray matter CBF analysis; 2) regional CBF analysis using user-specified masks; 3) voxelwise CBF analysis in standard Talairach space.
Figure 3.
A. The system generated group analysis report from an example study looking at the effect of HIV and methamphetamine use on whole brain gray matter CBF. Also shown in the report is a box plot (B) summarizing the range and mean of gray matter CBF values across factor levels.
Calibrated CBF maps and results of a group analysis are stored as derived data in the repository (blue box in Figure 1) and can be downloaded to a local computer at any time by the user. The CBFBIRN provides extensive querying capabilities based on clinical assessments, demographics data, image metadata, and provenance data (e.g. processing options used and quality of CBF maps), allowing the user quick access to the desired data elements.
The source code underlying the CBFBIRN data repository and workflows management system is available on the Neuroimaging Informatics Tools and Resource Clearinghouse (NITRC) site (http://www.nitrc.org/projects/cbfdap). We also have registered the CBFBIRN as a resource on NITRC (http://www.nitrc.org/projects/cbfbirn).
Since its initial release in April 2011, the CBFBIRN data repository has been growing steadily. We have successfully integrated legacy data (e.g. ASL data from completed FBIRN studies). Currently, the CBFBIRN is actively used by several ongoing projects, primarily for generation of calibrated CBF maps and for data storage.
CBFBIRN Data Repository Data Sets
To date, the CBFBIRN data repository hosts more than 2,000 data sets from 34 different projects. Table 1 shows a summary of data based on the types of studies available. For each type of study, the number of datasets, participating institutes, type of ASL data acquired, and MRI systems used are included. The repository hosts CBF measures across a wide range of health conditions and populations.
Table 1.
Summary of currently existing data in the CBFBIRN repository.
| Type of Study | Datasets | Institutes | ASL Type | MRI Vendor |
|---|---|---|---|---|
| Schizophrenia FBIRN Phase 3 | 278 | Duke, UCSF, UCI, UCSD, MRN, UMN, UIowa | FAIR | Siemens, GE |
| Eating Disorder | 195 | UCSD | FAIR | GE |
| Twin Study | 167 | UCSD | PCASL | GE |
| Adolescent Depression | 151 | UCSF | PCASL | GE |
| Bipolar Disorder | 140 | UCSD | PCASL | GE |
| Sleep Disorder | 138 | UCSD | FAIR | GE |
| TBI | 138 | UCSD | FAIR/PCASL | GE |
| Methamphetamine/AIDS | 101 | UCSD | FAIR | GE |
| Multisite Reproducibility FBIRN WCTS | 94 | UCI, UCSD | FAIR | Siemens, GE |
| Alzheimer’s | 92 | UCSD | FAIR/PCASL | GE |
| Depression | 81 | UCSD | FAIR/PCASL | GE |
| Caffeine | 80 | UCSD | PCASL | GE |
| Aphasia | 74 | SDSU | FAIR | GE |
| MCI | 66 | UCSD | PCASL | GE |
| Multisite Reproducibility FBIRN ECTS | 66 | Duke, Yale, BWH, MGH | FAIR | Siemens, GE |
| Hypoxia | 52 | UCSD | PCASL | GE |
| ADHD/Depression | 45 | UCSD | FAIR | GE |
| Aging | 45 | UCSD | FAIR/PCASL | GE |
| Dementia | 24 | UCSD | FAIR/PCASL | GE |
| Stroke | 12 | Chang Gung University | FAIR/PCASL | GE |
| Total | 2039 |
Table 2 shows a summary of data types and corresponding data formats currently supported by the CBFBIRN. Note that we may add support for other data formats to accommodate new users who are using a different acquisition protocol.
Table 2.
Summary of Data Types and Data Format.
| Data Types | Data Format | Input/output |
|---|---|---|
| Raw ASL | AFNI/Pfile (GE) DICOM (Siemens) |
Input (required) |
| Supplementary Image Data (Anatomical and/or Field map) | DICOM | Input (optional) |
| Image Metadata | AFNI header (GE) DICOM header (Siemens) |
Input (required) |
| Subject Metadata | Gender and Age are user specified during data upload | Input (required) |
| Clinical Data | CSV file | Input (optional) |
| Derived Data | Motion corrected raw ASL (AFNI) Quantified CBF map (AFNI and PNG) Process log (TXT) |
Output (default) |
| Registered Anatomical Map (AFNI) Tissue Probability Maps (AFNI) Mean Gray Matter CBF Measure (TXT) Histogram of Gray Matter CBF (PNG) |
Output (if anatomical images were uploaded) | |
| Statistical Analysis report (TXT) Statistical Graphs/Plots (PNG). Tabulated Data used in group analysis as input data (CSV) |
Output (if group analysis was performed) |
Quality Control of the CBFBIRN Data Sets
For multisite studies, most of the sites (UCSD, UCI, Duke, Yale, MWH, MGH, UCSF, UIowa, MRN, UMN) that have contributed data to the CBFBIRN use both the FBIRN QA and ASL protocols that were designed to minimize inter-site differences for fMRI and ASL studies (Glover et al., 2012; Liu et al., 2008). A description of the ASL protocol used for the multisite FBIRN studies is provided in (Liu et al, 2008) with further description of the specific protocol used on Siemens MRI systems available at http://www.nmr.mgh.harvard.edu/~jjchen/ASL.html.
Single site studies that have contributed to the CBFBIRN have typically used protocols developed at UCSD and described at http://fmri.ucsd.edu/Howto/3T/asl.html.
Data curation is performed at the time of data upload (including confirmation of data de-identification, verification of complete image file uploads, and data readability). The verification of these steps at the upload level helps maintain the overall data quality (both raw and derived), prevents unintended accumulation of corrupted files, and minimizes processing errors on the workflows.
The CBFBIRN allows users to assign quality ratings to all system generated CBF maps. The ratings are stored in the database as provenance data. This rating information can be used as an exclusion criterion prior to running group analysis (a feature currently available in the CBFBIRN) or for generating a QA report (e.g. across participating sites, different imaging systems, types of ASL used, etc.).
CBFBIRN Access and Data Download
Request for a user ID to the CBFBIRN data repository can be made via the project web page (https://cbfbirn.ucsd.edu/site/showcontact.action). After a review process that validates the requestor’s affiliation to a research institution, access is granted with provision of a user ID and password. Access to specific projects is determined by the data contributors. At the highest level, the user data are organized by projects and each project has an administrator (usually a PI) who specifies the details of data access. In this way the CBFBIRN supports both private and shared data (Shin et al., 2013a).
When the PI grants access to another user or to the public, only the derived data associated with the project are shared via the CBFBIRN web interface. The derived data includes the calibrated CBF map for each subject as well as motion corrected ASL time series data and segmented gray matter, white matter, and cerebrospinal fluid maps (if anatomical data were uploaded as supplementary image data). These data are provided to allow the recipient to carry out additional post processing off-line. If the data owner initiated a job on the group analysis workflow, additional derived data from this workflow also become available for download in the form of CSV and PNG image files (Fig. 1 and Fig. 3).
Note that through the CBFBIRN web interface, users can only download the derived data (Table 2). This minimizes the download time particularly when a project contains a large number of subjects. However, the raw anonymized data are stored permanently in the data repository (in addition to an offsite backup) and can be accessed by the PI upon request.
For data download, compressed archive files are generated on demand and streamed to the end user’s web browser on a per subject basis. Since downloads occur at a subject level, a failed download due to interrupted connections and/or system shutdowns can be reinitiated by the user. Project-level batch download capability is currently not available via the web interface but we do support batch transfer of data to a user-designated server using the Secure Shell (SSH) Protocol upon request. The downloaded datasets currently do not come with citable DOI’s/URI’s.
Currently, most of the datasets hosted in the data repository are associated with ongoing NIH-funded projects, and most of the sharing occurs through PI-specified private data access arrangements. If other registered users are interested in accessing datasets owned by a PI, we refer them directly to the PI, who can then choose to provide read and/or write (ability to contribute new data) privileges to them upon request to the CBFBIRN administrators. Currently, all datasets require explicit communication with the PI. However, as studies are completed, we expect a growing number of the datasets will become available for public access, at which point, a formal agreement enumerating terms and conditions for data usage will be put in place.
In the future, as the size of datasets and users grow, we plan on supporting large-scale data distribution, and an up-to-date reporting mechanism containing information regarding withdrawn, revised, and added data to the repository.
Contributing New Data to the CBFBRIN Data Repository
Anyone with a user account on the CBFBIRN can contribute new data to the CBFBIRN. However, the data repository currently only accepts data acquired either using the ASL protocols distributed directly via the CBFBIRN (https://cbfbirn.ucsd.edu/cbfbirnweb/welcome.action) or the FBIRN FAIR protocol used by the participating FBIRN sites (see Table 1 for the list of sites). The CBFBIRN checks for data integrity and data format compliance during upload and will alert the user if the data are identified as unrecognizable. This mechanism prevents accumulation of unusable data in the database and also ensures that the subsequent CBF processing is carried out without errors associated with data compatibility issues.
Long Term Plans for the CBFBIRN Data Repository
In the future, we plan to integrate more clinical ASL data, beginning with data acquired within the UCSD Health System. We are also actively adding new processing options, evaluating existing ones using the data already available in the repository, and making improvements to the existing workflows.
Discussion and Conclusions
With the public release of the source code for the CBFBIRN system framework via NITRC, neuroinformatics researchers can not only replicate the system we have implemented, but can also adapt and extend it for many applications where a web-based database with data management and processing capabilities is desirable. While we have used the system to promote storing, processing and sharing of ASL data, the system can be extended to handle other types of scientific data. Given the growing trend for data sharing, the system infrastructure presented here may be a useful resource for other scientific research disciplines.
Aside from data sharing, the CBFBIRN promotes standardization of data acquisition and processing, which is particularly timely for the ASL community. Recently, the ISMRM Perfusion Study Group published the first white paper (Alsop et al., 2014) with recommended data acquisition and processing guidelines for clinical applications, citing that the “overabundance of choices is an impediment to the acceptance of ASL by the clinical community, complicating the implementation of ASL in standard care, comparisons between sites and the establishment of meaningful clinical trials.” The CBFBIRN data sharing and processing capabilities dovetail well with this concerted effort toward standardization. The combined efforts of the CBFBIRN and the Perfusion Study Group can help accelerate the rate of adoption of ASL by researchers and clinicians.
Current users have reported that the CBFBIRN greatly reduces the time it takes from data acquisition to the reporting of study findings. Since its introduction to the research community in 2011, several investigators have published studies that have used the CBFBIRN as the primary tool for data storage and processing (Ho et al., 2013; Ho et al., 2012; Shin et al., 2013b; Wierenga et al., 2012).
We have seen a moderate adoption of the CBFBIRN by the research community since its introduction in 2011, particularly with the availability of ASL protocols (for GE MRI systems) provided via the CBFBIRN. However, adoption of the CBFBIRN by clinicians has been slow. Based on our experience, the primary concerns for data sharing revolve around maintaining the privacy of subject and patient information as well as misgivings about sharing valuable data in a public data repository prior to publication of findings. Note that we anonymize subject information during data upload and never share data publicly without the explicit permission and consent of the data contributor. Ultimately, the viewpoint of the scientific community with regards to data sharing must be fundamentally changed (Poldrack and Gorgolewski, 2014). The success of this cultural shift will be heavily determined by the efforts of federal funding agencies such as the NIH and research consortia like the ISMRM Perfusion Study Group (http://www.ismrm.org/chapter-study-groups).
In conclusion, the CBFBIRN is an open-access online platform that supports data storage, processing, and sharing of ASL data. While it is designed to support the needs of the ASL community for CBF processing, group analysis, and data sharing, the system architecture can be extended to include additional types of data, such as resting-state fMRI data. The full potential of the CBFBIRN will be realized by the active participation of the neuroimaging and clinical communities in the process of contributing and sharing data.
Highlights.
We present a CBF data repository for storing, processing, and sharing ASL data.
CBFBIRN provides a framework for analysis and comparison of CBF measures across studies/sites.
The CBFBIRN supports data acquired with pulsed ASL and pseudocontinous ASL methods.
Standard CBF quantification and group analysis options are available.
CBFBIRN hosts more than 2,000 data sets from 34 different projects and is growing.
Acknowledgments
This work was supported by the National Institutes of Health (grant number: R01MH084796). The authors would like to thank the FBIRN collaborators (U24-PR021992) for sharing the ASL and clinical assessment data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alsop DC, Detre JA. Reduced transit-time sensitivity in noninvasive magnetic resonance imaging of human cerebral blood flow. J Cereb Blood Flow Metab. 1996;16:1236–1249. doi: 10.1097/00004647-199611000-00019. [DOI] [PubMed] [Google Scholar]
- Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez-Garcia L, Lu H, Macintosh BJ, Parkes LM, Smits M, van Osch MJ, Wang DJ, Wong EC, Zaharchuk G. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2014 doi: 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GG, Clark C, Liu TT. Measurement of cerebral perfusion with arterial spin labeling: Part 2. Applications. J Int Neuropsychol Soc. 2007;13:526–538. doi: 10.1017/S1355617707070634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalela JA, Alsop DC, Gonzalez-Atavales JB, Maldjian JA, Kasner SE, Detre JA. Magnetic resonance perfusion imaging in acute ischemic stroke using continuous arterial spin labeling. Stroke. 2000;31:680–687. doi: 10.1161/01.str.31.3.680. [DOI] [PubMed] [Google Scholar]
- Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med. 2008;60:1488–1497. doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. 1992;23:37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- Fielding RT, Taylor RN. Proceedings of the 22nd international conference on Software engineering. ACM; Limerick, Ireland: 2000. Principled design of the modern Web architecture; pp. 407–416. [Google Scholar]
- Glover GH, Mueller BA, Turner JA, van Erp TG, Liu TT, Greve DN, Voyvodic JT, Rasmussen J, Brown GG, Keator DB, Calhoun VD, Lee HJ, Ford JM, Mathalon DH, Diaz M, O’Leary DS, Gadde S, Preda A, Lim KO, Wible CG, Stern HS, Belger A, McCarthy G, Ozyurt B, Potkin SG. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J Magn Reson Imaging. 2012;36:39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TC, Wu J, Shin DD, Liu TT, Tapert SF, Yang G, Connolly CG, Frank GKW, Max JE, Wolkowitz O, Eisendrath S, Hoeft F, Banerjee D, Hood K, Hendrin RL, Paulus MP, Simmons AN, Yang TT. Altered Cerebral Perfusion in Executive, Affective, and Motor Networks During Adolescent Depression. Journal of the American Academy of Child and Adolescent Psychiatry. 2013 doi: 10.1016/j.jaac.2013.07.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TC, Wu J, Shin DD, Yang G, Chan M, Hoang N, Simmons AN, Yang TT. Amygdala Hypoperfusion in Depressed Adolescents: An Optimized Pseudo-Continuous Arterial Spin Labeling Study. Proceedings of the American Academy of Child & Adolescent Psychiatry 59th Annual Meeting; San Francisco, CA, USA. 2012. Program #3.11. [Google Scholar]
- Kim SG, Tsekos NV. Perfusion imaging by a flow-sensitive alternating inversion recovery (FAIR) technique: application to functional brain imaging. Magn Reson Med. 1997;37:425–435. doi: 10.1002/mrm.1910370321. [DOI] [PubMed] [Google Scholar]
- Liu TT, Wierenga CE, Mueller BA, Wang JJ, Glover GH, Voyvodic JT, Greve D, Turner JA. Reliability and Reproducibility of Arterial Spin Labeling Perfusion Measures with a Multi-Center Study. Proceedings of the International Society for Magnetic Resonance in Medicine 16th Annual Meeting; Toronto, Ontario, Canada. 2008. Program #3338. [Google Scholar]
- Ozyurt IB, Keator DB, Wei D, Fennema-Notestine C, Pease KR, Bockholt J, Grethe JS. Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics. 2010;8:231–249. doi: 10.1007/s12021-010-9078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Gorgolewski KJ. Making big data open: data sharing in neuroimaging. Nat Neurosci. 2014;17:1510–1517. doi: 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- Shin DD, Ozyurt IB, Liu TT. The Cerebral Blood Flow Biomedical Informatics Research Network (CBFBIRN) database and analysis pipeline for arterial spin labeling MRI data. Front Neuroinform. 2013a;7:21. doi: 10.3389/fninf.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DD, Rasmussen JM, Ozyurt IBJB, Van Erp TG, Vaidya J, Mathalon DH, Mueller BA, Voyvodic JT, Greve DN, Ford JM, Glover GH, Brown GG, Potkin SG, Liu TT. CBF Differences between Healthy and Schizophrenic Brains – FBIRN Phase 3 Multisite Study at 3T using CBFBIRN Database and Analysis Pipelines. Proceedings of the International Society for Magnetic Resonance in Medicine 21st Annual Meeting; Salt Lake City, Utah, USA. 2013b. Program #736, 129. [Google Scholar]
- Taber KH, Black KJ, Hurley RA. Blood flow imaging of the brain: 50 years experience. J Neuropsychiatry Clin Neurosci. 2005;17:441–446. doi: 10.1176/jnp.17.4.441. [DOI] [PubMed] [Google Scholar]
- Wierenga CE, Dev SI, Shin DD, Clark LR, Bangen KJ, Jak AJ, Rissman RA, Liu TT, Salmon DP, Bondi MW. Effect of mild cognitive impairment and APOE genotype on resting cerebral blood flow and its association with cognition. J Cereb Blood Flow Metab. 2012;32:1589–1599. doi: 10.1038/jcbfm.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermark M, Sesay M, Barbier E, Borbely K, Dillon WP, Eastwood JD, Glenn TC, Grandin CB, Pedraza S, Soustiel JF, Nariai T, Zaharchuk G, Caille JM, Dousset V, Yonas H. Comparative overview of brain perfusion imaging techniques. J Neuroradiol. 2005;32:294–314. doi: 10.1016/s0150-9861(05)83159-1. [DOI] [PubMed] [Google Scholar]