Abstract
There has been an increasing effort to correlate electrophysiology data with imaging in patients with refractory epilepsy over recent years. IEEG.org provides a free-access, rapidly growing archive of imaging data combined with electrophysiology data and patient metadata. It currently contains over 1200 human and animal datasets, with multiple data modalities associated with each dataset (neuroimaging, EEG, EKG, de-identified clinical and experimental data, etc.). The platform is developed around the concept that scientific data sharing requires a flexible platform that allows sharing of data from multiple file-formats. IEEG.org provides high and low-level access to the data in addition to providing an environment in which domain experts can find, visualize, and analyze data in an intuitive manner. Here, we present a summary of the current infrastructure of the platform, available datasets and goals for the near future.
1. INTRODUCTION
Intracranial Electroencephalography (iEEG) and Magnetic Resonance Imaging (MRI) are equally important neuroimaging modalities for studying neural activity and structure [1], [2]. Accurate dynamics of neural activities and interactions can be studied across large number of electrodes and with high temporal resolution using iEEG [3]–[6]. Precise localization of this neuronal activity along with structural descriptions of neuronal pathways can be determined using MRI with different contrasts [7]. In addition, complementary features of neuronal activity can be studied using fMRI [8]–[10]. The explosion of technology in these modalities has spurred interest in their use in studying neuroscience and advanced translational research in neurology, particularly epilepsy.
Joint analysis of these multimodal datasets allows neuroscientists to study functional and structural relationships and has great potential to lead to vital discoveries in epilepsy otherwise not captured when studying modalities independently [11]. Additional data-modalities further impact the overall richness of a dataset, such as patient clinical history, genetics, electrocardiogram (EKG), etc. Integrating, analyzing, and sharing these complex datasets pose unique challenges to the data science community. Individual institutions have access to small data sets. Significant advancements in this field of study will be made using large data sets shared across multiple institutions. Unique challenges of sharing human biomedical data arise such as data format interoperability, de-identification of protected health information (PHI), and adherence to mandated government regulations.
The ability to share clinical metadata along with high-resolution data, such as iEEG and neuroimaging across multiple sites, often geographically sparsely distributed, requires novel infrastructure with a focus on data integration. A simple approach such as allowing collaborators to download the data to their local site for analysis is often not feasible, especially with the terabytes (TB) of data that comprise these datasets.
Over the past four years, our team of neuroscience and computer science experts has established a cloud-based resource for data sharing and collaboration, http://IEEG.org [12]. This platform provides data sharing and analysis capabilities to the neuroscience community, particularly in the epilepsy domain. Multiple neuroscience research centers are making their data available through the platform for collaborations where data access is controlled, and access to data is controlled by the data contributors.
Currently, the IEEG portal contains high quality iEEG and multimodal imaging from over 1200 subjects including 576 animal models (dog, mouse, rat, sheep, primate) and 733 patients with epilepsy. As this time of writing this article, there are 487 public datasets, 667 registered users, and 162 publicly accessible clinical datasets. These data were collected across multiple institutions throughout the world. Users originate from all 5 continents and represent institutions like UCLA in Los Angeles, CA and University Hospital Motol in Prague, Czech Republic. Table 1 summarizes the data available on the IEEG-Portal.
Table 1.
The number of patients by Imaging Modality per Institution (from the IEEG Portal dataset)
| Number of Patients by Imaging Modality per Institution | Mayo Clinic | Hospital of the University of Pennsylvania |
|---|---|---|
| Structural Modalities | ||
| 3T MRI - T1W | 23 | 23 |
|
|
||
| T2W | 2 | 19 |
|
|
||
| FLAIR | 10 | 19 |
|
|
||
| DWI/DTI | 8 | |
|
|
||
| ECoG Pre-Implant Imaging (MRI/CT) | 23 | 14 |
|
|
||
| ECoG Post-Implant Imaging (MRI/CT) | 23 | 21 |
|
|
||
| 6 Months Post-Implant Imaging (MRI/CT) | 13 | |
|
|
||
| 7T MRI | 20 | |
|
|
||
| HARDI Imaging | 20 | |
| Metabolic/Functional Modalities | ||
| SPECT (Ictal and Interictal) | 20 | |
|
|
||
| 18(F)-FDG PET | 1 | |
|
|
||
| BOLD | 20 | |
|
|
||
| ASL | 20 | |
|
|
||
| MRS | 20 | |
Abbreviations for Imaging: Positron Emission Tomography (18(F)-FDG PET), T1-Weighted MRI (T1W), T2-Weighted MRI (T2W), Fluid Attentuated Inversion Recovery sequence (FLAIR MRI), Diffusion Weighted Imaging (DWI)/Diffusion Tensor Imaging (DTI), X-Ray Computed Tomography (CT), Single-Photon Emission Computed Tomography (SPECT), Blood oxygenation level dependent (BOLD), Arterial Spin Labeling (ASL), High angular resolution diffusion-weighted imaging (HARDI), 1(H) Magnetic Resonance Spectroscopy (MRS). All image sequences available as 3D NIfTI files.
Each human dataset can contain up to 100 electrodes and are recorded for 1–4 weeks continuously using sample rates as high as 32 kHz. Standard clinical epilepsy protocol images are also provided, including T1-weighted (T1W) isotropic axial, T2-weighted (T2W) coronal, FLAIR coronal and diffusion weighted image sequences. In addition, de-identified clinical metadata, such as patient medical and family history, medication history, Epilepsy Monitoring Unit reports and scalp EEG findings are available for a subset of the patients on the portal. There are currently 46 patients with intractable epilepsy who have at least intracranial EEG, pre-operative T1W/T2W/DWI MRI, ECoG post-implant MRI or CT as well as a full clinical report. Half of these patients are from the Hospital of the University of Pennsylvania and the other half are from the Mayo Clinic. Table 2 shows four example patients along with a sampling of the data available on the portal for these patients. All patients who have clinical reports available have lesional findings on histopathology and neuroimaging reported in the respective sections in the reports. These include lesions such as malformations of cortical development (focal cortical dysplasia or schizencephaly), vascular malformations (AVMs or cavernomas), and low-grade glial tumors. A significant minority of the patients currently on the portal has these lesional findings present on either histopathology or imaging.
Table 2.
Sample Patient Profiles from the IEEG Portal.
| Patient ID | I002_P002 | Mayo Clinic Study 005 | I002_P005 | I002_P006 |
|---|---|---|---|---|
| Institution | Hospital of the University of Pennsylvania | Mayo Clinic | Hospital of the University of Pennsylvania | Hospital of the University of Pennsylvania |
| Epilepsy Type | Localization-related; Left frontal onset, left cortical dysplasia | Bi-Temporal Onset | Localization- related; Left temporal onset (secondary to hemorrhagic HSV encephalitis) | Localization-related; Symptomatic meningitis |
| Engel Outcome | 1 | 1 | 4 | 1 |
| Seizure Type | Simple partial and generalized tonic- clonic of left temporal onset | Focal Complex partial-endpoint of seizure- not evolving to secondary generalized | Complex partial seizures of left temporal onset and status epilepticus | Complex partial seizures of right temporal onset with secondarily generalized tonic-clonic seizure |
| # of Seizures | 1 | 0 | 5 | |
| Age during First Seizure | 4 | 21 | 30 | 11 |
| Age at Admission | 20 | 26 | 35 | 32 |
| Sample Past Anti-Epileptic Medications | carbamazepine, topiramate, levetiracetam, clobazam, valproic acid | - | phenytoin, valproic acid, levetiracetam | carbamazepine, zonisamide |
| Imaging available | T1 MPRAGE, T2 FLAIR, Post-Implant T1 MPRAGE and FLAIR, Post-Implant CT, 6 months post- op T1, T2, FLAIR, DWI | MRI T1 pre- implant, SPECT ictal/interictal, MRI T1 post implant, CT post implant | Pre-Implant T1 MPRAGE and T2, Post-Implant T1 MPRAGE and FLAIR, Post-Implant Head CT | T1 MPRAGE, T2 FLAIR, T2 Susceptibility-Weighted, DTI, Pre-Implant T1 MPRAGE and T2 FLAIR, Post-Implant T1 MPRAGE and FLAIR, Post-Implant CT, 6 months post-op T1, T2, FLAIR |
Four patients and their profiles on the IEEG Portal. Dates of hospital admission for epilepsy monitoring have been standardized to start on Jan-01-2000. All other dates have been adjusted accordingly. Note that some data is not available and marked by -. All patients who have lesional epilepsy will have lesion findings noted under MRI report summaries in the respective patients’ clinical reports. All lesional patients will have appropriate pathology and imaging findings reported, whether lesions are malformations of cortical development, vascular, or low-grade glial tumors.
Analyzing large scale EEG and neuroimaging data requires substantial computational resources. Leveraging cloud resources provides a scalable solution to benchmark experiments, share gold standard datasets, and advance towards more integrative collaborative research in the neuroscience community.
IEEG.org, like other databases such as the Human Connectome Project [13], the European EEG database (http://epilepsy-database.eu) [14], [15] and LONI IDA [16] are critical to the standardization of neuroimaging data analyses, avoiding bias, and allowing for significant research advances [15]. Benchmarking experiments (i.e. testing algorithms on novel data) requires a central body to curate “gold standard” training data and withhold testing data. IEEG.org has been developed to allow users to share data and use its resources to validate and benchmark new algorithms.
2. Architecture of IEEG.org
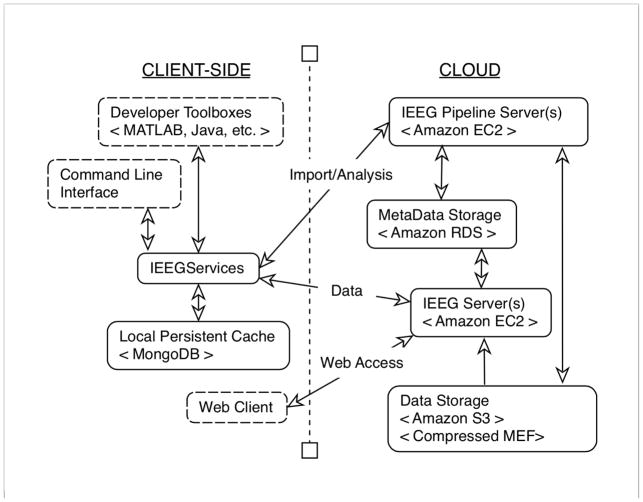
The IEEG-Portal is developed using the Google Web Toolkit in Java, and JavaScript. It is hosted on Amazon’s EC2 service and the data is stored on the Amazon S3 service with reduced redundancy. Figure 1 shows an abstract schematic of the various components of the IEEG.org infrastructure. All meta-information associated with the datasets as well as user-information and portal-state information is hosted using on Amazon RDS. Separate pipeline servers handle data import, wrangling and cloud-based data analysis. The IEEG.org platform has been developed with a continuous focus on scalability and security. Users have full control over who can see their data and track their usage. A variety of tools are available to upload, access, edit, and annotate data by the user in both a web-environment and through multiple API’s. MongoDB (http://www.mongodb.org) can be used as a persistent cache to increase efficiency and reduce traffic/cost from the cloud. Accessing data through MATLAB is easy and fast; the toolbox fetches 470,000 values within 10 milliseconds utilizing the local cache. Fetching the same amount directly from the portal takes about 25 milliseconds (depending on bandwidth). Efficiency of data transfer increases when larger blocks of data are requested through the MATLAB toolbox. Images, which are currently saved as zip files, are quickly downloaded over FTP.
Figure 1.
IEEG Portal Setup
Schematic of the cloud infrastructure supporting IEEG.org platform. While a local persistent cache is available for clients to work with parts of data locally, majority of the data computation occurs on the cloud, including storage and pipelining.
The platform provides mechanisms to automatically import and wrangle datasets from various data-formats. The import pipeline process converts the data to a standardized format, associate meta-information with the datasets, and makes them available through the platform. Files that are not recognized as imaging or time series data are made available as direct downloads. All EEG and imaging are then de-identified again by stripping all header information in data files. In addition, clinical metadata and reports are re-written by removing any identifiable information, including physician names and recorded seizure times.
Investigators interested in using the portal can request a user account by visiting http://www.IEEG.org. All users of the portal have to agree to a Data Usage agreement. Data providers can set permissions for each dataset specifying which users can find, view, and edit their datasets, or they can make the datasets globally visible to the portal user community with a click of a button. This makes the platform ideally suited for inter-institutional collaborations where data is selectively shared with a research team.
Recognizing the various user-groups of the platform (data scientist, clinicians, educators) and levels of expertise in these groups, we provide an architecture with data and metadata services that may be invoked from a Web browser or from a remote MATLAB or Java “toolbox”. In addition, to facilitate the wide range of scenarios regarding data sharing, the platform provides flexible authorization and access control capabilities. The platform will support OAuth authorization in the near future, allowing users to login using their LinkedIn, Google, or Facebook account (by March 2015).
2.1 Web Console and Viewer
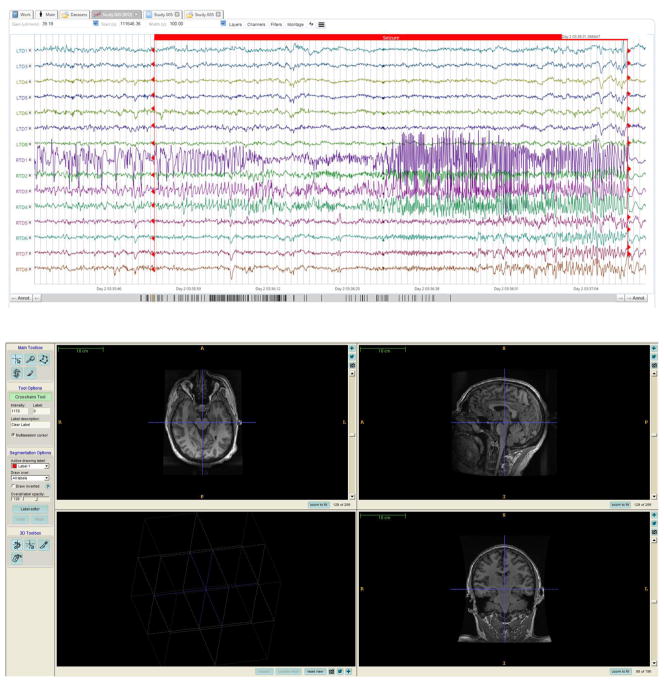
The web-console provides the ability to view time series and imaging data (Figure 2). Users can download additional files that are associated with these datasets, such as clinical metadata, zipped imaging data, other metadata. Users can also track usage of their datasets by other users of the platform and engage in group-discussions within “Project Groups”. The web-interface also allows users to upload data and edit information for each dataset they own.
Figure 2.
Annotated figure of a seizure (from EEG.org)
A screenshot of the IEEG.org portal’s web viewer. The annotated window (top) shows a recorded seizure from Patient ‘Mayo Clinic Study 005’ (Table 2). On the bottom is an example of imaging dataset: T1-weighted MPRAGE image for this patient. In this example, the patient is viewed using the radiological orientation (LAS).
The EEG Viewer runs in all major web browser and provides comprehensive functionality with a small local computational footprint. It provides the ability to search for a particular dataset, or for particular events within datasets to allow the user to quickly select only segments of data that are of interest to them. This feature is essential in the large data sets available on IEEG.org. For instance, this search feature can be used to filter the dataset to a particular imaging sequence or animal dataset.
The documentation for interfacing with the portal is available both on http://www.IEEG.org as well as supplementary material S1. The documentation describes the functionality of the IEEG-Portal and its exposed API for Java and Matlab. It contains multiple example tutorials and a thorough explanation of the terminology used on the IEEG-Portal.
2.2 APIs for 3rd party software and Other Tools
The IEEG.org platform provides a set of APIs to interact, and manage data on the platform. This includes methods to upload datasets and annotations, download datasets and stream data to a local host. Although most API methods are targeting time series at this point, we intent to provide similar programmable access to all imaging data. A MATLAB toolbox is available that allows users to easily interface with the platform from their MATLAB session. Returned content can be locally cached using MongoDB to minimize redundant traffic from the cloud platform.
3. Imaging Datasets
Most currently available images are the standard epilepsy imaging sequences used at clinical institutions. These images are saved in NIFTI images and are not altered in any other way from the original DICOM images. Figure 2 shows an example MPRAGE MRI image sequence for patient ‘I002_P002_D001’, viewed in ITK-SNAP [17]. All identifiable header information is stripped from the images. The names of the images identify the image contrast and imaging modality. All images are set in the original coordinate space of the scanner. The origin of each image is similarly dependent on the origin set during scan time. All images can be reoriented into the appropriate coordinate space and displayed in the radiological orientation (LAS) using ITK-SNAP. Table 3 summarizes some of the different modalities of imaging for some of the patients on the portal.
Table 3.
Imaging Modalities and the associated contrast/sequence.
| Modality | Contrast/Sequence | Notes |
|---|---|---|
| 3T MRI | Localizer | |
|
|
||
| 3T MRI | T1 (MPRAGE) | Standard 1mm isotropic |
|
|
||
| 3T MRI | T2 | T2-weighted |
|
|
||
| 3T MRI | FLAIR | |
|
|
||
| 3T MRI | ASL | Pseudo-Continuous Arterial Spin Labeling |
|
|
||
| 3T MRI | DWI/DTI | Diffusion Weighted Imaging |
|
|
||
| 3T MRI | Susceptibility-Weighted | |
|
|
||
| 3T MRI | Contrast Enhanced (Gadolinium) | |
|
|
||
| 7T MRI | T1 (MPRAGE) | 7T images taken at research scanners |
|
|
||
| CT | Pre-Implant Stealth | CT Scan taken right before electrode grid implantations |
|
|
||
| CT | Post-Implant Stealth | |
|
|
||
| PET | (18)F-FDG | |
Notes indicate details on the imaging sequence used.
There are a number of imaging modalities and contrasts available for the datasets, depending on the clinical epilepsy imaging protocol used at the institution. For instance, the Mayo Clinic (Table 1) regularly obtains interictal and ictal SPECT for SISCOM imaging. Most institutions obtain Head CTs both pre-implant and post-implant but not all institutions acquire MRI images post-electrode implantation.
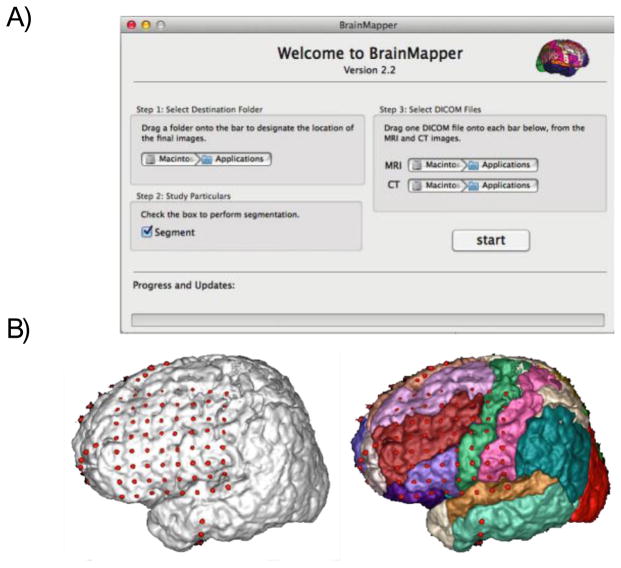
In addition to these iEEG tools, the web console also contains a BrainMapper application, which is an easy to use “drag and drop” application that automatically co-registers CT and MRI electrode implantation images in order to provide an accurate and reliable 3D reconstruction of the electrodes on the surface of a patient’s brain [18]. Figure 3a shows a snapshot of the BrainMapper application. Within the application, the co-registered images can be viewed in 3-dimensional space. Figure 3b shows the final image in the application, which uses rigid and non-rigid registration along with prior-based probabilistic segmentation in order to minimize errors caused by parenchymal shift during the implantation procedure as well as “unbury” surface electrodes that appear buried in cortical tissue.
Figure 3.
BrainMapper Application
IEEG.org provides a desktop tool to coregister MRI and CT to visualize subdural electrode locations on a 3D parcellated brain. This tool has been quantitatively validated and is available for free download [18]. (A) Screenshot of BrainMapper application, which shows an intuitive user interface along with a progress bar (B) Sample results that can be visualized in BrainMapper. Cortical parcellations allow users to visualize location of each electrode in an implant.
The application uses the atlas of Non-Rigid Image Registration Evaluation Project (NIREP http://www.nirep.org) from the University of Iowa, which is based on 16 normal adult T1-weighted brain scans and contains 32 cortical gray matter labels. Skull and other non-brain tissues in the subject MR images are stripped by applying the BET [19]. The patient MR image is non-rigidly registered to the NIREP atlas using cross-correlation as the similarity metric. The high level of accuracy of our co-registration method was quantitatively confirmed [18]. The images are all in the patient’s original T1 image coordinate frame.
In the future, there will be multiple expanded tools for images for all patients on the portal. Specifically, we are in the process of implementing an online DICOM and 3D surface render viewer using WebGL API. This tool will allow users to view imaging datasets on the portal directly in their web browser rather than simply downloading the dataset. The pipeline for the BrainMapper will be included in this viewer, allowing for users to view the co-registered electrodes on the surface of brains in all patients who have Pre-Implant and Post-Implant MRI and CT imaging. In addition to these electrodes, important computational iEEG and imaging biomarkers, such as seizure onset zones, marked structural dysplasias and BOLD activated regions will be displayed on cortical surface renderings as density heat maps. This tool can display other user-generated computed surface maps, allowing for improved integration of EEG and multimodal imaging.
4. Patient Consent and De-Identification
All patients from the Hospital of the University of Pennsylvania and the Mayo Clinic are consented post-surgery in allowing their clinical metadata, ECoG recordings and neuroimaging to be uploaded onto the portal. Since the University of Pennsylvania and the Mayo Clinic are founding members of the portal, our institutional IRBs allow us to upload patient data from consenting patients. Different institutions that have contributed and would like to contribute in the future can consent their patients through permission of their institutional IRB.
Other institutions that would like to contribute should make sure they first get consent from the patient and has an approved IRB to included these patients. In addition, these institutions should de-identify all data before uploading on the portal. IEEG.org provides a tool that can de-identify DICOM images before they are uploaded to the platform. In the supplementary materials, we have included a copy of the user agreement (Figure S2) as well as a copy of the agreement that an owner of a dataset agrees to when contributing data through the portal (Figure S3) to the supplementary information of this article. Finally, a basic diagram of the flow of data from submission to loading onto the portal is shown in Figure S4.
5. Long term goals
Table 4 shows a timetable of the expected changes we hope to implement in the coming year. The platform has been developed from the beginning with sustainability in mind, and currently these resources are provided to the scientific community free of charge. Additional funding strategies may be implemented in the near future to ensure its continuation. For example, having a free tier of data-access, plus additional ‘pay-as-you-go’ services where costs are passed onto the user depending on the amount of data that is downloaded/analyzed.
Table 4.
Timetable for upcoming changes and improvements to the IEEG.com portal
| Expected Date | Updates and Improvements | Notes |
|---|---|---|
| Fall 2015 | Interactive Web-based DICOM, and NIFTI viewer, linking between EEG and Imaging data. | Allows viewing of BrainMapper results (electrode grid location), results from image analysis tools (registration/segmentation) |
|
|
||
| Late 2015 | Image analysis on the cloud | Running various image analysis pipelines over imported imaging data. |
|
| ||
| Search textual and numeric content of documents, metadata | ||
|
| ||
| LinkedIn, Facebook, and Mendeley | ||
|
|
||
| Within 2 years | Sophisticated Search Capability, Interlink with external databases, Social Networking, scaling of service. | Databases include LONI IDA, GenBank, Pubmed, Google Scholar |
|
|
||
The IEEG.org platform initially focused primarily on large iEEG datasets, but now handles many other data formats including imaging. Over the next several years, imaging will become an increasingly larger part of the platform and, specifically, integration between iEEG and imaging datasets. A particular effort will be to interlink with other, external databases that already exist such as GenBank (genetics) and LONI (imaging). Providing a ‘single sign-on’ could be an initial step towards providing a truly integrated user-experience.
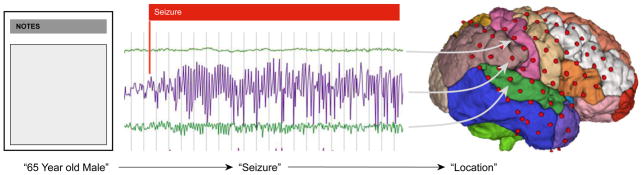
As the platform becomes more diversified, through inclusion of new data-modalities and support for additional data-formats, it becomes more important to provide useful methods to search for particular information within these multi-modal datasets. For example, one can be interested in all imaging data from people with frontal lobe epilepsy that have a minimum of 3 number of seizures in the associated iEEG. This requires matching of keywords in text (Patient Information), expert annotations (‘Seizures’), and return specific imaging data (Figure 4). We are developing sophisticated search capabilities, which will facilitate search over the textual and numeric content of documents, metadata, and data associated with a data set – and queries linking these resources [20].
Figure 4.
Querying information on the Portal
Data graph relating patient notes, electrophysiology and imaging. Key-word search capabilities allow users to query across data modalities such as documentation, annotations in iEEG and Electro-placement information in 3D models of MRI Imaging.
Management of data on the IEEG.org platform will rely predominantly on the users of the platform and specifically the owners of the data hosted on the platform. We envision that there will be a subset of datasets on the platform that will be used more often by the community and, similar to Wikipedia articles, be meticulously maintained over time. Datasets that are less used might be archived to cheaper storage (i.e. Amazon Glacier Storage) but remain accessible.
As mentioned prior, we plan to provide integration with several professional platforms, such as LinkedIn, Facebook, and Mendeley to emphasize the social aspects of data sharing. This will allow users to look up, and contact owners of particular datasets and find publications that are associated with particular datasets. The entire platform is developed as an open-source project. The modular framework of the platform fully supports the development of additional viewers, analysis, or conversion tools.
A short-term goal is to provide an interactive Web-based DICOM, and NIFTI viewer which users can use to view and annotate imaging data. We expect to have this capability before the end of 2015. Novel search capabilities, will also be available to search for imaging datasets that meet specific search criteria (e.g. “has 7T MRI” or “contain Radiologist-identified lesions”).
We believe that over time, some users will want to customize the platform to their own needs. Therefore, the entire platform is provided as an open-source project that users can deploy on their own machines. Source-code is available at: https://code.google.com/p/braintrust/. The portal continues to grow thanks to contributions from many epilepsy researchers across the world. Though initially geared towards epilepsy research, the database is capable of handling any large dataset. For instance, neuroscience research datasets including combined EEG and fMRI can be easily uploaded on the portal. High fidelity long-term recordings from animals are already uploaded for use in seizure prediction and detection analysis. The platform is already capable of handling scalp EEG data, which will allow storage of gold standard scalp EEG datasets. These gold standard EEG datasets can validate inverse problem methods that users can run using the available APIs. Such methods already exist for iEEG [21], [22], which can be further developed and validated on the patients currently on the portal.
5. Conclusion
We introduce the platform IEEG.org (www.IEEG.org) and highlight its added value for sharing and analyzing large complex biosignal datasets, including imaging. The development of the platform is focused on providing a truly scalable solution for connecting large, complex scientific data with domain experts, and to provide intuitive access for various user communities (i.e. educational, data-scientists, clinicians). IEEG.org leverages the unique novel features that today’s ‘cloud-environments’ support to provide a scalable, cost-efficient solution to sharing and analyzing datasets.
Taking advantage of the power and utility of data assembled in a collaborative way, this method of data sharing allows investigators to test specific algorithms (such as seizure predication algorithms in the field of epilepsy) on a wide range of patients and disease patterns while maintaining balance in the dataset. In addition, novel biomarkers can be discovered efficiently by quickly accessing large collections of data, such as the EEG and neuroimaging (MRI/CT/PET/SPECT) currently available on the portal. A large investment by the NIH allowed for a design of a fast, secure and easy-to-use portal. We expect that the platform will continue to grow in both size and functionality over the upcoming years, and extend to fields outside of epilepsy research.
Supplementary Material
Highlights.
We introduce an online epilepsy data platform capable of handling large datasets.
There are more than 1200 animal and human datasets (both public and private).
IEEG.org provides toolboxes for analysis of EEG and neuroimaging data.
IEEG.org is a versatile platform for neuroscience data sharing.
Acknowledgments
The authors would like to acknowledge Dr. Gregory Worrell (Mayo Clinic), Dr. Ben Brinkmann (Mayo Clinic), Dr. Zachary Ives (UPenn) and Dr. Brian Litt (UPenn) for their support in the development of the IEEG.org portal. The International Epilepsy Electrophysiology Portal is funded by the NIH 5-U24-NS-063930-05 and NIH 1K01ES025436-01. Additional grant funding provided by P20 NS12006 (NIH NINDS).
Abbreviations
- iEEG
Intracranial Electroencephalogram
- EEG
Scalp Electroencephalogram
- EKG
Electrocardiogram
- API
Application Program Interface
- T1W
T1-Weighted MRI
- T2W
T2-Weighted MRI
- FLAIR
Fluid-Attenuated Inversion Recovery MRI
- BOLD
Blood Oxygen Level Dependent imaging
- ASL
Arterial Spin Labeling
- HARDI
High angular resolution diffusion imaging
- MRS
Magnetic Resonance Spectroscopy
- PET
Positron Emission Tomography
- SPECT
Single-photon Emission Computed Tomography
- MP-RAGE
Magnetization-Prepared Rapid Gradient-Echo MRI
- DICOM
Digital Imaging and Communications in Medicine
- NIfTI
Neuroimaging Informatics Technology Initiative Data Format
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kathryn A. Davis, Email: Kathryn.Davis@uphs.upenn.edu.
Joost B. Wagenaar, Email: joostw@seas.upenn.edu.
References
- 1.Liu Z, Ding L, He B. Integration of EEG/MEG with MRI and fMRI. IEEE Eng Med Biol Mag. Jan;25(4):46–53. doi: 10.1109/memb.2006.1657787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dale AM, Halgren E. Spatiotemporal mapping of brain activity by integration of multiple imaging modalities. Curr Opin Neurobiol. 2001 Apr;11(2):202–8. doi: 10.1016/s0959-4388(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 3.Dale AM, Liu AK, Fischl BR, Buckner RL, Belliveau JW, Lewine JD, Halgren E. Dynamic statistical parametric mapping: combining fMRI and MEG for high-resolution imaging of cortical activity. Neuron. 2000 Apr;26(1):55–67. doi: 10.1016/s0896-6273(00)81138-1. [DOI] [PubMed] [Google Scholar]
- 4.Hämäläinen M, Hari R, Ilmoniemi RJ, Knuutila J, Lounasmaa OV. Magnetoencephalography—theory, instrumentation, and applications to noninvasive studies of the working human brain. Rev Mod Phys. 1993 Apr;65(2):413–497. [Google Scholar]
- 5.Cohen D, Palti Y, Cuffin BN, Schmid SJ. Magnetic fields produced by steady currents in the body. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1447–51. doi: 10.1073/pnas.77.3.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson SJ, Kaufman L, Brénner D. Latency of the neuromagnetic response of the human visual cortex. Vision Res. 1978 Jan;18(1):107–10. doi: 10.1016/0042-6989(78)90084-6. [DOI] [PubMed] [Google Scholar]
- 7.Kwong KK, Chesler DA, Weisskoff RM, Donahue KM, Davis TL, Ostergaard L, Campbell TA, Rosen BR. MR perfusion studies with T1-weighted echo planar imaging. Magn Reson Med. 1995 Dec;34(6):878–87. doi: 10.1002/mrm.1910340613. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci U S A. 1992 Jul;89(13):5951–5. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong KK, Belliveau JW, Chesler DA, Goldberg IE, Weisskoff RM, Poncelet BP, Kennedy DN, Hoppel BE, Cohen MS, Turner R. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belliveau JW, Kennedy DN, McKinstry RC, Buchbinder BR, Weisskoff RM, Cohen MS, Vevea JM, Brady TJ, Rosen BR. Functional mapping of the human visual cortex by magnetic resonance imaging. Science. 1991 Nov;254(5032):716–9. doi: 10.1126/science.1948051. [DOI] [PubMed] [Google Scholar]
- 11.So EL. Integration of EEG, MRI, and SPECT in Localizing the Seizure Focus for Epilepsy Surgery. Epilepsia. 2000 Mar;41(s3):S48–S54. doi: 10.1111/j.1528-1157.2000.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 12.Wagenaar JB, Brinkmann BH, Ives Z, Worrell GA, Litt B. A multimodal platform for cloud-based collaborative research. International IEEE/EMBS Conference on Neural Engineering, NER; 2013. pp. 1386–1389. [Google Scholar]
- 13.Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E. The Human Connectome Project: a data acquisition perspective. Neuroimage. 2012 Oct;62(4):2222–31. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klatt J, Feldwisch-Drentrup H, Ihle M, Navarro V, Neufang M, Teixeira C, Adam C, Valderrama M, Alvarado-Rojas C, Witon A, Le Van Quyen M, Sales F, Dourado A, Timmer J, Schulze-Bonhage A, Schelter B. The EPILEPSIAE database: an extensive electroencephalography database of epilepsy patients. Epilepsia. 2012 Sep;53(9):1669–76. doi: 10.1111/j.1528-1167.2012.03564.x. [DOI] [PubMed] [Google Scholar]
- 15.Ihle M, Feldwisch-Drentrup H, Teixeira CA, Witon A, Schelter B, Timmer J, Schulze-Bonhage A. EPILEPSIAE - a European epilepsy database. Comput Methods Programs Biomed. 2012 Jun;106(3):127–38. doi: 10.1016/j.cmpb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Dinov I, Lozev K, Petrosyan P, Liu Z, Eggert P, Pierce J, Zamanyan A, Chakrapani S, Van Horn J, Parker DS, Magsipoc R, Leung K, Gutman B, Woods R, Toga A. Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PLoS One. 2010 Jan;5(9):e13070. doi: 10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006 Jul;31(3):1116–28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Azarion AA, Wu J, Davis KA, Pearce A, Krish VT, Wagenaar J, Chen W, Zheng Y, Wang H, Lucas TH, Litt B, Gee JC. An open-source automated platform for three-dimensional visualization of subdural electrodes using CT-MRI coregistration. Epilepsia. 2014 Dec;55(12):2028–37. doi: 10.1111/epi.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002 Nov;17(3):143–55. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ives Zachary G, Yan Zhepeng, Zheng Nan, Litt Brian, Wagenaar Joost B. Looking at Everything in Context. CIDR. 2015 [Google Scholar]
- 21.Caune V, Ranta R, Le Cam S, Hofmanis J, Maillard L, Koessler L, Louis-Dorr V. Evaluating dipolar source localization feasibility from intracerebral SEEG recordings. Neuroimage. 2014 Sep;98:118–133. doi: 10.1016/j.neuroimage.2014.04.058. [DOI] [PubMed] [Google Scholar]
- 22.Ramantani G, Dümpelmann M, Koessler L, Brandt A, Cosandier-Rimélé D, Zentner J, Schulze-Bonhage A, Maillard LG. Simultaneous subdural and scalp EEG correlates of frontal lobe epileptic sources. Epilepsia. 2014 Feb;55(2):278–88. doi: 10.1111/epi.12512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.