ABSTRACT
The sialyl-T antigen sialylα2-3Galβ1-3GalNAc is a common O-glycan structure in human glycoproteins and is synthesized by sialyltransferase ST3Gal1. The enterohemorrhagic Escherichia coli serotype O104 has the rare ability to synthesize a sialyl-T antigen mimic. We showed here that the wbwA gene of the E. coli O104 antigen synthesis gene cluster encodes an α2,3-sialyltransferase WbwA that transfers sialic acid from CMP-sialic acid to Galβ1-3GalNAcα-diphosphate-lipid acceptor. Nuclear magnetic resonance (NMR) analysis of purified WbwA enzyme reaction product indicated that the sialyl-T antigen sialylα2-3Galβ1-3GalNAcα-diphosphate-lipid was synthesized. We showed that the conserved His-Pro (HP) motif and Glu/Asp residues of two EDG motifs in WbwA are important for the activity. The characterization studies showed that WbwA from E. coli O104 is a monofunctional α2,3-sialyltransferase and is distinct from human ST3Gal1 as well as all other known sialyltransferases due to its unique acceptor specificity. This work contributes to knowledge of the biosynthesis of bacterial virulence factors.
IMPORTANCE This is the first characterization of a sialyltransferase involved in the synthesis of an O antigen in E. coli. The enzyme contributes to the mimicry of human sialyl-T antigen and has unique substrate specificity but very little sequence identity to other sialyltransferases. Thus, the bacterial sialyltransferase is related to the human counterpart only by the similarity of biochemical activity.
INTRODUCTION
Sialic acids are ubiquitous in nature and contribute to the acidity and hydration of a glycoprotein, metal ion binding, and epitope exposure. Sialylation affects the adhesive properties of cells and has been implicated in the functions of cell surface receptors (1). Sialic acids are also found in O antigens of bacterial lipopolysaccharides (LPS) or lipooligosaccharides, for example, in Neisseria, Campylobacter, Pasteurella, or Photobacterium spp. (2–5). In Escherichia coli, O antigens containing sialic acid linkages are rare and have only been found in serotypes O24, O55, O104, O145, and O171 (ECODAB). Specific proteins in the mammalian nervous system as well as a number of bacterial capsules carry polysialic acids that regulate adhesive properties of the cell. Masking their own antigens with human-like structures derived from glycolipids and glycoproteins may help bacteria to escape the host immune system. For example, Campylobacter jejuni has lipooligosaccharides resembling human gangliosides (2). However, cross-reactivity in infected humans can lead to autoimmune disease affecting the nervous system, for example the Guillain-Barré syndrome (3). O-acetylation of sialic acids controls the exposure of sialyl epitopes (1).
The enterohemorrhagic, Shiga toxin-producing E. coli serotype O104 is the causative agent of outbreaks of bloody diarrhea and hemolytic-uremic syndrome with significant mortality (6, 7). The E. coli O104 antigen has the unusual repeating unit structure (d-Galα1-4Neu5,7,9Ac3α2-3-d-Galβ1-3-d-GalNAcβ1)n (ECODAB) (8), which mimics the human sialyl-T antigen (sialylα2-3Galβ1-3GalNAc-R) found in large amounts on cancer cell and tumor glycoproteins. However, the sialic acid residue in the mature E. coli O104 antigen is 7- and 9-O-acetylated, and the sialyl-T antigen is therefore masked. Although O antigens have a number of biological functions in bacterial adhesion, colonization, and interactions with host defenses (4), the specific biological role of the mimicry of the sialyl-T antigen structure in E. coli O104 is not clear. The enzymes synthesizing the sialyl-T antigen in bacteria could be of interest in the synthesis of tumor-selective sialyl-T and T antigens.
Sialyltransferases (SiaTs) are ubiquitous in higher eukaryotes and in certain bacteria (9). The bacterial SiaTs share conserved Asp(Glu)-Asp(Glu)-Gly and His-Pro (HP) motifs which may contain the amino acids involved in substrate binding and catalysis (10). These bacterial SiaTs are diverse, inverting glycosyltransferases (GTs) classified in either the CAZy GT42 family with a GT-A fold or GT52 or GT80 family with a predicted GT-B or unknown fold. Polysialyltransferases are in the GT38 and GT4 families, possibly having a GT-B fold. Bacterial SiaTs act on terminal Gal or sialic acid residues and can be monofunctional (11). Certain SiaTs can be promiscuous with respect to their acceptor substrates or have multiple activities: e.g., α2,3-, α2,6-, or α2,8-SiaT activities, or significant levels of CMP-sialic acid hydrolase, trans-sialidase, and neuraminidase activities (12).
All of the known eukaryotic SiaTs are members of the large GT29 family of inverting GTs, having predicted variants of the GT-A fold, that transfer sialic acid to oligosaccharides. Four or more peptide sequences have been identified as sialyl motifs in eukaryotic SiaTs: a large L motif, small S motif, very small VS motif, and motif III (13). The catalytic acid/base residues in α2,3- and α2,6-sialyltransferases have been shown to include His residues. The major human SiaT that synthesizes the sialyl-T antigen from the T antigen, Galβ1-3GalNAc, is the α2,3-sialyltransferase ST3Gal1. The activity and expression of ST3Gal1 are increased in cancer cells and tissues, contributing to high levels of sialyl-T antigens of cancer glycoproteins and mucins, which appears to be related to cancer cell survival and tumorigenesis (14). The crystal structure of the soluble domain of porcine ST3Gal1 with CMP and Galβ1-3GalNAc suggested that His319 is the catalytic base in motif III (15), proposed to interact with carbon-2 of the sialic acid moiety of CMP-sialic acid. The enzyme was shown to assume a GT-A fold. However, the detailed substrate specificity and metal ion dependence of purified human ST3Gal1 have not yet been determined.
The O104 antigen synthesis gene cluster contains genes encoding GTs, those for O-antigen processing enzymes, and those involved in nucleotide sugar biosynthesis (16): galF→wckD→nnaB→nnaC→nnaA→wbwA→wzy→wbwB→wzx→wbwC→gnd.
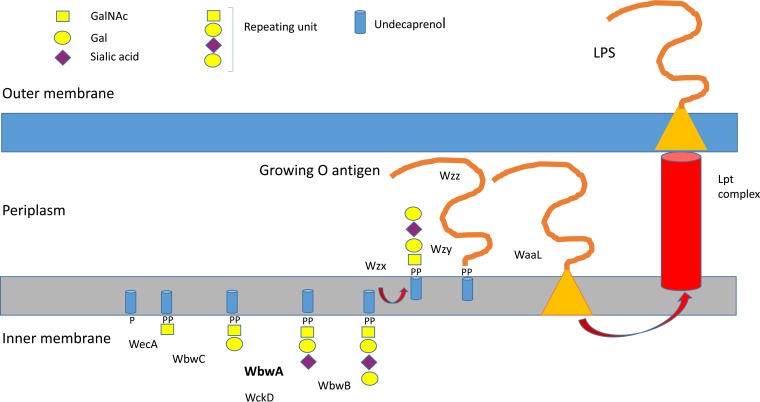
The GTs include previously characterized β1,3-Gal-transferase WbwC, putative α2,3-sialyltransferase WbwA, and putative α1,4-Gal-transferase WbwB. The nnaA, nnaB, and nnaC genes are expected to be involved in the synthesis of CMP-Neu5Ac donor substrate. GalF may also be involved in nucleotide sugar synthesis, while gnd encodes a putative 6-phosphogluconate dehydrogenase. The acetyltransferase responsible for the acetylation of sialic acid appears to be encoded by the wckD gene. The presence of wzx and wzy genes that encode putative translocase and polymerase, respectively, suggests that the O104 antigen synthesis follows the Wzy polymerase-dependent biosynthesis pathway (4, 16). In this pathway, the natural acceptor substrate upon which the O antigen is assembled in the cytoplasmic compartment is membrane-bound undecaprenol-phosphate (Fig. 1). The repeating units are initiated in the cytoplasm by the addition of sugar phosphate to undecaprenol-phosphate to form sugar-diphosphate-undecaprenol. This natural acceptor substrate is then extended by GTs that transfer individual sugar residues to form the O-antigen repeating unit. These units are translocated across the membrane by Wzx to the periplasm, where they are polymerized by Wzy before being ligated to the outer core structure linked to lipid A. The completed LPS molecule is then transported by the Lpt complex to the outer membrane, where the O antigen is exposed to the environment. While the LPS is essential for maintenance of membrane structure and bacterial growth, O antigens are targets for protective immunity by the host and are important virulence factors.
FIG 1.
Proposed O-antigen repeating unit synthesis pathway of E. coli serotype O104 involving sialyltransferase WbwA. In the proposed O104 antigen pathway, WecA may transfer GalNAc-phosphate from UDP-GalNAc to undecaprenol-phosphate (P-Und). Alternatively, GlcNAc-phosphate is transferred and 4-epimerase forms GalNAcα-PP-Und, the substrate for WbwC. WbwC then adds a Gal residue in β1-3 linkage to GalNAc-PP-Und. WbwA transfers sialic acid from CMP-sialic acid to the Gal residue of Galβ1-3GalNAc-PP-Und. Further conversion is catalyzed by yet uncharacterized α4-Gal-transferase WbwB and putative sialate O-acetyltransferase WckD to synthesize the O104 antigen repeating unit d-Galα1-4Neu5,7,9Ac3α2-3-d-Galβ1-3-d-GalNAc. Flippase Wzx transfers the oligosaccharide to the periplasm, where it is polymerized by polymerase Wzy to form the O antigen. Chain-length terminator Wzz may help to restrict the number of repeating units assembled by polymerase. The O antigen is then ligated to the outer core structure of the growing lipopolysaccharide by ligase WaaL. The Lpt complex extrudes LPS to the outer membrane.
In this article, we identify the biochemical function of the SiaT WbwAO104 encoded by the wbwA gene of the O-antigen synthesis gene cluster of the highly pathogenic E. coli strain O104. We show that the enzyme is a monofunctional α2,3-SiaT that transfers sialic acid from CMP-sialic acid to Galβ1-3GalNAcα-diphosphate-phenylundecyl (Galβ1-3GalNAc-PP-PhU) to synthesize the sialyl-T antigen. WbwAO104 has a unique acceptor specificity for a diphosphate aglycone group not seen in any other SiaT.
MATERIALS AND METHODS
Materials.
Reagents and materials were obtained from Sigma-Aldrich unless otherwise stated. Radioactive nucleotide sugars were from American Radiolabeled Chemicals. GalNAc and GlcNAc derivatives and inhibitors were synthesized as described previously (17–21). Glycopeptides and Galβ1-3GalNAcα-Bn derivatives were synthesized and kindly donated by Hans Paulsen, University Hamburg, Germany. The WbwA substrate Galβ1-3GalNAc-PP-PhU was synthesized using WbwC as described previously (20). Briefly, bacterial lysates containing Gal-transferase WbwC were obtained by sonication in 50 mM sucrose. Lysates were incubated with 1 mM UDP-Gal and 0.1 mM GalNAc-PP-PhU, 0.125 M MES (morpholineethanesulfonic) acid buffer, pH 7, and 5 mM MnCl2 for 1 h. Product was prepurified by C18 Sep-Pak and purified by high-performance liquid chromatography (HPLC) using a C18 column and quantified by HPLC in comparison to GalNAc-PP-PhU at known concentrations.
Expression, purification, and assays of ST3Gal1.
The ST3Gal1 ectodomain, including a portion of the stem and the entire catalytic domain, was expressed as an N-terminally 8× histidine-tagged product in the baculovirus-insect cell system (22). The supernatant from infected cells was harvested at 48 h postinfection, and the secreted product was isolated by nickel-affinity chromatography using HisPur Ni-nitrilotriacetic acid (NTA) resin (Thermo-Fisher Scientific). After elution with imidazole, the purified enzyme was buffer exchanged into 100 mM Tris-HCl (pH 7)–0.125% Triton X-100 on a Sephadex G25 column (PD10; GE Healthcare Bio-Sciences Corp.). SDS-PAGE and Western blots were carried out as described previously (20). The ST3Gal1 assays were carried out in at least duplicate determinations and contained in a total volume of 40 μl 1 μl enzyme (40 ng), 20 nmol Galβ1-3GalNAcα-Bn, 0.1 M Tris, pH 7, 0.8 mM CMP-[3H]sialic acid (2,400 cpm/nmol), and 5 mM MnCl2. The mixtures were incubated for 1 h at 37°C. Water was added (700 μl), and mixtures were passed through a C18 Sep-Pak column. CMP-sialic acid and sialic acid were eluted with 4 ml water, and product was eluted with 3 ml MeOH and quantified by scintillation counting as described previously (23). The inhibition was tested under the standard assay conditions, having inhibitor with 10% MeOH in the assay mixture and 10% MeOH in control assays. ST3Gal1 product using Galβ1-3GalNAc-Bn acceptor and nonradioactive CMP-sialic acid was prepared as described above, but using nonradioactive CMP-sialic acid, and subjected to analysis by hydrophobic and anion-exchange chromatography and mass spectrometry (electrospray ionization [ESI] with an Orbitrap Velos Pro in positive-ion mode). To determine the optimal pH for the enzyme, Tris buffer was used at pH range 7 to 9 and MES buffer at pH 5 to 7, with overlap of both buffers at pH 7.
Synthesis of the wbwA gene and expression of the WbwA protein.
Plasmid pRSETA, having an ampicillin resistance gene and containing the wbwA gene, was obtained from GeneArts (Life Technologies). The WbwA protein had 6 His residues at the amino terminus. Plasmid was transformed into BL21 bacteria, which were grown overnight for 16 h on a plate containing LB agar with 100 μg/ml ampicillin. A single colony was picked and grown in 5 ml LB broth with ampicillin. Bacteria from a 6-ml overnight culture were grown in 250 ml of LB containing 100 μg/ml of ampicillin at 37°C with 200 rpm shaking until the optical density at 600 nm was 0.8. The expression of His-tagged protein was induced with 1 mM isopropylthiogalactoside for 4 h at 37°C at 200-rpm shaking and was confirmed by SDS-PAGE and Western blotting as described before (20). Bacterial cells were obtained by centrifugation at 3,500 × g for 10 min. The pellet was resuspended in phosphate-buffered saline (PBS)–20% glycerol and frozen in aliquots at −20°C. WbwA was solubilized and purified by affinity chromatography using Ni-NTA resin (20).
Bacterial sialyltransferase assays.
WbwA-containing bacteria were stored at −20°C in 20% glycerol in PBS, thawed and isolated by centrifugation, and then disrupted in either 50 mM sucrose or PBS by sonication for 20 s (3 times) with 2-min intervals on ice (23). WbwA was assayed in a total volume of 40 μl containing 10 μl homogenate (3 to 4 mg protein), 5 mM MnCl2, 0.125 M Tris-HCl, pH 7, 0.35 to 1 mM CMP-[3H]Neu5Ac (1,450 cpm/nmol), and 0.1 mM Galβ1-3GalNAcα-PP-PhU (prepared with β1,3-Gal-transferase WbwCO104 and GalNAc-PP-PhU acceptor) (20). The incubation time was 30 min at 37°C. Controls lacked the acceptor substrate. The mixture was applied to C18 Sep-Pak columns in water; the reaction product was eluted with MeOH and quantified by scintillation counting as described before (23). Assays were carried out in duplicate determinations with <10% difference between assays. To determine the apparent Km and Vmax values for the acceptor, 0.8 mM CMP-sialic acid was used in the assays, and for the donor substrate, 0.1 mM acceptor substrate was used. Kinetic parameters were determined using the program GraphPad Prism. The inhibition of WbwA was tested under the standard assay conditions, as described above for ST3Gal1.
Production of large-scale WbwA enzyme product.
A large-scale production of WbwA product was carried out in a one-pot, two-step reaction using WbwC to synthesize Galβ1-3GalNAcα-PP-PhU from GalNAcα-PP-PhU and WbwA to synthesize sialylα2-3Galβ1-3GalNAcα-PP-PhU. The reaction tube contained 7 ml of WbwC homogenate in 50 mM sucrose, 7 ml of WbwA homogenate in 50 mM sucrose, 7 μmol GalNAcα-PP-PhU, 20 μmol CMP-sialic acid, 28 μmol UDP-Gal, 0.14 μmol MnCl2, and 3.5 μmol MES buffer, pH 7, in a total volume of 28 ml. After incubation for 1 h at 37°C, 30 ml water was added, the mixture was centrifuged at 12,000 × g, and supernatants were subjected to purification on 60 Sep-Pak C18 columns. Hydrophilic compounds were eluted from each column with 4 ml water. Fractions (3 ml) eluted with MeOH were collected, and MeOH was removed by rotor evaporation followed by lyophilization. Three milliliters of water was added, and insoluble residue was removed by filtration through 0.45-μm-pore syringe filters. Aliquots were injected into an HPLC chromatograph using an analytical C18 column and 9/91 acetonitrile-water as the mobile phase at 1 ml/min. Product fractions were pooled, and concentrated by lyophilization. Radioactive WbwA reaction product was prepared in parallel, but in a 70× smaller reaction volume and using radioactive CMP-[3H]sialic acid as the donor substrate and 8 Sep-Pak C18 columns. Purified WbwA product was analyzed for its molecular mass by electrospray ionization mass spectrometry (ESI-MS) in the negative-ion mode. To determine the linkage of sialic acid synthesized by WbwA, product was analyzed by nuclear magnetic resonance (NMR). Product was dissolved and concentrated 3 times in D2O and analyzed as described before by 600-MHz NMR using a Bruker spectrometer (20).
RESULTS
Sequence comparisons.
WbwAO104 (see Table S1 in the supplemental material) has been classified in the CAZy data bank as an inverting glycosyltransferase within family GT52 with a yet unknown fold. This GT52 family contains a number of characterized SiaTs with low sequence identities (between 12.4 to 14.6%) to WbwA (3, 5). All of these SiaTs have HP and E(D)E(D)G motifs, which are conserved in bacterial SiaTs. WbwAO104 also has low (13.6%) sequence identity to human ST3Gal1.
In Gram-negative bacteria, the GTs involved in O-antigen synthesis appear to be associated with the inner membrane. Thus, hydrophobicity plots indicate that WbwAO104 has short hydrophobic regions but does not have a significant transmembrane domain. As expected, no mammalian sialyl motifs were detected in WbwAO104, but an HP motif (His236-Pro237) is present near the N terminus (see Fig. S1 in the supplemental material) (24, 25). An EDG motif (Glu206-Asp207-Gly208) typical of bacterial SiaT is located more centrally in WbwA. An additional EDG motif (Glu111-Asp112-Gly113) is found N terminal to this motif. The two EDG motifs may potentially include the catalytic base.
Analyses by PHYRE2 modeling (26) generated a model based on the template c2yk5A with 98.9% confidence. The model covered 87% of the WbwA sequence (amino acid residues 6 to 283). The PHYRE 2 model predicted that the structure of WbwA forms a GT-B fold, similar to that of Neisseria lipooligosaccharide-specific bifunctional α2,3/α2,6-SiaT (NST2) in complex with CMP, in spite of low (17%) sequence identity (27). The HP and EDG motifs of WbwA are within the peptide domains of similarity.
Development of sialyltransferase assays for WbwAO104.
The molecular mass of the His6-tagged WbwAO104 was calculated to be 38.02 kDa. SDS-PAGE and Western blots before and after affinity chromatography on Ni-NTA showed that the enzyme indeed was expressed with an approximate molecular mass of 38 kDa (see Fig. S2 in the supplemental material).
The sialic acid residue of the O104 repeating unit is triacetylated (8), and it was not clear if the sialic acid in the donor substrate had to be triacetylated as well. Known eukaryotic and bacterial SiaTs utilize oligosaccharides as acceptor substrates and do not require a diphosphate group in the acceptor. Thus, it seemed feasible that the acceptor substrates would be Galβ1-3GalNAcα derivatives that served well as substrates for human ST3Gal1. WbwAO104 is the third enzyme to act in the O104 repeating unit pathway. Previously studied GTs that act as the third enzyme acted on mono- or oligosaccharide acceptors and did not require the diphosphate-lipid group (28, 29). To establish an assay for SiaT activity of WbwAO104, we first used CMP-[3H]sialic acid as the donor substrate and Galβ1-3GalNAcα-Bn as the acceptor. To isolate the enzyme product after the incubation, the assay mixture was separated using C18 Sep-Pak columns. These assay conditions were appropriate for human ST3Gal1. However, no SiaT activity of WbwA was observed with 0.5 to 1 mM Galβ1-3GalNAcα-Bn or -p-nitrophenyl, Galβ1-3(6-deoxy) GalNAcα-Bn, glycopeptide AHGVT-(Galβ1-3GalNAcα)SAPDTRPAPGSTAPNA, or other glycopeptides as acceptors in the presence or absence of 5 mM MnCl2 or MgCl2 in the assay mixture. It was therefore possible that WbwAO104 required an acceptor having the diphosphate-lipid structure expected in the natural acceptor substrate Galβ1-3GalNAcα-PO3-PO3-undecaprenol. To produce the natural WbwA acceptor analog Galβ1-3GalNAc-PP-PhU, we used Gal-transferase WbwCO104, with UDP-Gal as the donor and GalNAc-PP-PhU as the acceptor (20), followed by extensive purification. In contrast to all other Galβ1-3GalNAc derivatives tested, Galβ1-3GalNAc-PP-PhU at 0.1 mM concentration in the assay was very active as an acceptor for SiaT WbwA. The WbwC substrate GalNAc-PP-PhU and its analog GlcNAc-PP-PhU did not serve as substrates for WbwA at 0.1 to 1 mM concentrations, showing that the addition of sialic acid was dependent on the presence of the Gal residue in Galβ1-3GalNAc-PP-PhU.
Characterization of WbwAO104 activity.
The SiaT activity in the bacterial lysate was linear up to 1 h of incubation time, and the activity was stable for several days at 4°C and for several weeks at −20°C. The apparent Km value for the acceptor substrate Galβ1-3GalNAc-PP-PhU was 0.02 mM, with an apparent Vmax of 50 nmol/h/mg (see Fig. S3A in the supplemental material) for the activity in bacterial lysates. After passing the lysate through the Ni-NTA affinity column, SDS-PAGE showed a major band at about 38 kDa, and a single band was observed by Western blots (see Fig. S2 in the supplemental material). Although the enzyme did not appear to be degraded after the purification procedure, it lost activity, and only 10% of the enzyme appeared to be soluble and adhering to the Ni-NTA column. It is possible that the enzyme in the lysate aggregated and precipitated or the protein structure was unstable and lacked components of the membrane necessary for its activity. Since the purified enzyme showed relatively low activity, and only a small amount of purified, soluble enzyme was isolated, we used the bacterial homogenate as the enzyme source for WbwA characterization. The apparent Km value for CMP-sialic acid was 0.013 mM, with an apparent Vmax of 90 nmol/h/mg (see Fig. S3B). UDP-Gal, UDP-GalNAc, UDP-GlcNAc, GDP-Fuc, and GDP-Man could not substitute for CMP-sialic acid and were ineffective in transferring a sugar residue to Galβ1-3GalNAc-PP-PhU. This indicates that WbwA has a strict specificity for its donor substrate, CMP-sialic acid.
To determine the requirement of WbwA for divalent metal ions, assays were carried out without metal ion addition, with 5 mM EDTA, or with the addition of various metal ions at a 5 mM concentration. Clearly, the activity was not metal ion dependent, but the activities varied, with MnCl2 giving the highest activity with 169% of product yield and cobalt acetate giving the lowest activity (77%) compared to assays without metal ion addition (see Fig. S4 in the supplemental material). It is therefore possible that the presence of metal ions affected protein structure and activity or, alternatively, that metal ions differentially affected the structure of the negatively charged donor and/or acceptor substrates. We therefore added MnCl2 in subsequent assays. The optimal pH of the buffer used in assays was 7 (see Fig. S5 in the supplemental material). The CMP-sialic acid hydrolase activity of WbwA was determined at pH 5 and 7 with CMP-[3H]sialic acid. CMP-[3H]sialic acid was separated from [3H]sialic acid by ascending paper chromatography in Leloir solvent (1 M NH4-acetate [pH 7.5]–95% EtOH at 3:7). The amount of sialic acid released from CMP-[3H]sialic acid was at similar levels in controls and in assays containing WbwA, showing that WbwA is not an active CMP-sialic acid hydrolase. Neuraminidase activity of WbwA was tested in assays using radioactive WbwA and ST3Gal1 products, [3H]sialylα2-3Galβ1-3GalNAc-PP-PhU and [3H]sialylα2-3Galβ1-3GalNAcα-Bn, respectively, as potential neuraminidase substrates at pH 5 and 7. The released [3H]sialic acid and uncleaved radioactive SiaT products were separated by C18 Sep-Pak columns. No significant release of [3H]sialic acid above background (lacking WbwA) was observed.
Bis-imidazolium chloride or mesylate salts (21) have two positively charged imidazolium groups linked by an aliphatic chain. For both enzymes, WbwAO104 and humal ST3Gal1 (hST3Gal1), very little or no inhibition was seen, with chloride salts having an aliphatic linker chain of 4 to 14 carbons, at five times the acceptor concentration. However, compounds with at least 15 carbons in the linker chains inhibited WbwA, and those with at least 16 carbons inhibited ST3Gal1. The inhibitory activity of compounds increased with the number of carbons in the aliphatic linker chains and was highest with compounds having 20 or 22 carbons (see Table S2 in the supplemental material). The mesylate salt having 22 carbons in the aliphatic linker chain had 50% inhibitory concentrations (IC50s) of 0.2 mM for WbwA and 0.04 mM for ST3Gal1.
Analysis of WbwAO104 reaction product.
To produce several micromoles of WbwAO104 reaction product for structural analyses, WbwC product Galβ1-3GalNAc-PP-PhU was used as an acceptor and CMP-sialic acid as a donor substrate. We observed that in a coupled one-pot synthesis of WbwC and WbwA products, using both enzymes simultaneously with the initial acceptor GalNAc-PP-PhU, UDP-Gal, and radioactive CMP-[3H]sialic acid, about 4 times more WbwA product was obtained, compared to the standard WbwA reaction. This highly efficient synthesis was a strong indication that the combination of the two enzymes had a stimulatory effect on the conversion of GalNAc-PP-PhU to the first reaction product, Galβ1-3GalNAc-PP-PhU, and subsequently to the second reaction product, [3H]sialyl-Galβ1-3GalNAc-PP-PhU.
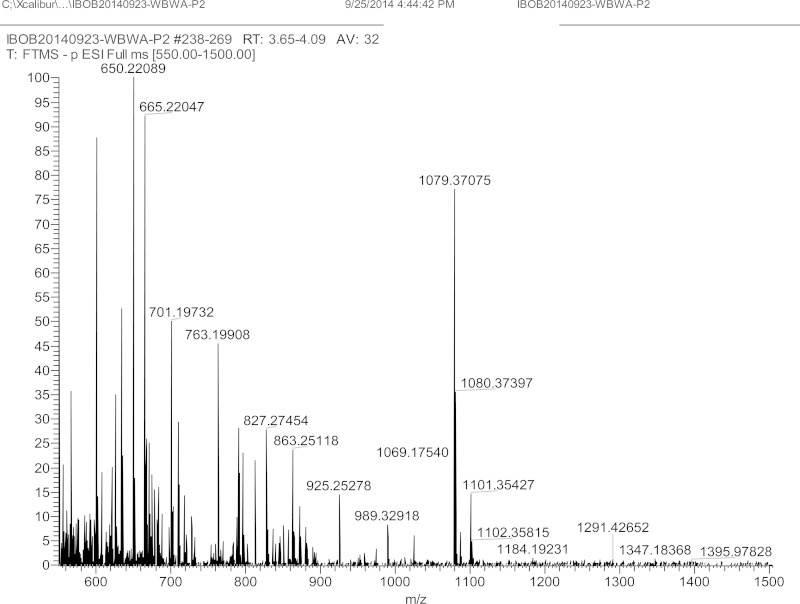
In the nonradioactive large-scale production, a total of 3.6 μmol of WbwA reaction product was obtained in 1 h from 7 μmol of GalNAc-PP-PhU (51% conversion of initial acceptor to WbwA product). The WbwA product was isolated by C18 Sep-Pak followed by HPLC. With 9/91 acetonitrile-water as the mobile phase, the sialylated product eluted in a broad peak at 45 min. Electrospray ionization-mass spectrometry (ESI-MS [negative-ion mode]) of the reaction product sialyl-Gal-GalNAc-PP-PhU showed m/z of 1,079.4 for [M-H]− and 1,101.4 for [M-H + Na]−, indicating that WbwA had added one residue of sialic acid to Gal-GalNAc-PP-PhU (Fig. 2).
FIG 2.
Mass spectrum for WbwA reaction product sialyl-Galβ1-3GalNAcα-PP-PhU. The purified WbwAO104 reaction product was analyzed by ESI-MS in the negative-ion mode. The ion m/z 1,079.4 indicates the presence of enzyme product sialyl-Gal-GalNAc-PP-PhU [M-H]−. The monosodium salt is seen as m/z 1,101.4 [M-H + Na]−.
NMR analysis of the WbwAO104 reaction product.
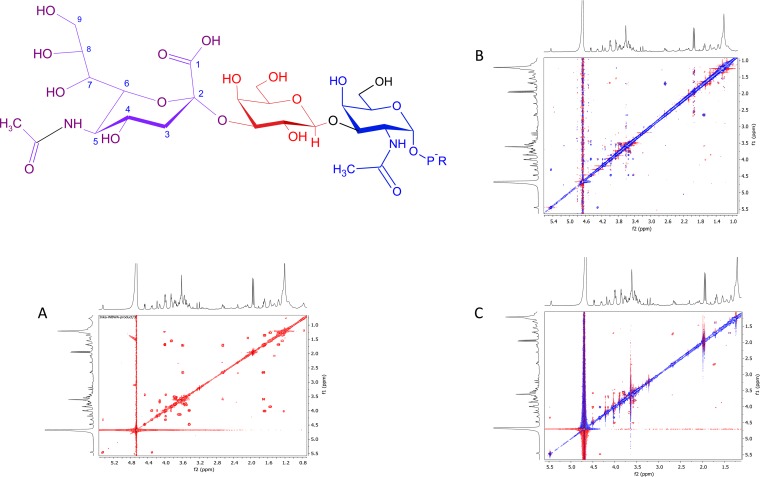
To determine the anomeric linkage of the newly added sialic acid residue, the WbwAO104 reaction product was dissolved in D2O and subjected to one-dimensional (1D) and 2D nuclear magnetic resonance (NMR) spectral analyses (Table 1 and Fig. 3A to C; see Fig. S6A and B in the supplemental material). The proton and carbon shifts were obtained from correlation spectroscopy (COSY), total correlated spectroscopy (TOCSY), and heteronuclear single quantum coherence (HSQC) spectroscopy. Although the spectra indicated the presence of a single compound, not all parameters could be assigned due to crowding and overlapping signals. In nuclear Overhauser effect spectroscopy (NOESY) and rotating frame Overhauser effect spectroscopy (ROESY) spectra, cross peaks were found between the axial H-3 of sialic acid and the equatorial H-3 and between the axial H-3 of sialic acid and the axial H-3 of Gal (Fig. 3B and C). In addition, the product spectrum, in comparison to that of the substrate (20), showed that the chemical shift of Gal H-3 had changed most significantly, compared to the other Gal signals, from 3.55 ppm to 3.98 ppm. The 13C chemical shift of Gal C-3 also showed a large change from 72.1 ppm in the substrate to 75.4 ppm in the product. These results strongly suggest that the WbwAO104 reaction product is sialylα2-3Galβ1-3GalNAcα-PP-PhU and that WbwA is an α2,3-SiaT acting on the Gal residue of Galβ1-3GalNAc-PP-PhU.
TABLE 1.
Six-hundred-megahertz NMR parameters of WbwAO104 reaction product sialylα2-3Galβ1-3GalNAcα-PO3-PO3-phenylundecyla
| Residue | NMR result (ppm) |
|
|---|---|---|
| 1H | 13C | |
| GalNAc-1 | 5.45 (J1,2 = 3.3, 6.7 Hz) | 94.7 |
| GalNAc-2 | 4.31 (J = 11.1 Hz) | 48.4 |
| GalNAc-3 | 3.98 | 77.1 |
| GalNAc-4 | 4.18 | 68.2 |
| GalNAc-5 | 4.13 | 71.6 |
| GalNAc-6 | ∼3.66 | 60.5 |
| N-Acetyl | 1.9 | |
| Gal-1 | 4.47 (J1,2 = 7.8 Hz) | 104.2 |
| Gal-2 | 3.44 | 68.7 |
| Gal-3 | 3.98 | 75.4 |
| Gal-4 | 3.86 | 66.8 |
| Sialyl-3ax | 1.70 (J = 12.5 Hz) | |
| Sialyl-3eq | 2.67 (J = 12.5, 4.5 Hz) | |
| Sialyl-4 | 3.58 | 66.1 |
| Sialyl-5 | 3.75 | 51.4 |
| Sialyl-6 | 3.54? | |
| N-Acetyl | 1.9 | |
| P-O-CH2 | 3.85 | |
The chemical shifts of the reaction product were adjusted to match GalNAc H-1 of the acceptor substrate Galβ1-3GalNAcα-PO3-PO3-phenylundecyl (20).
FIG 3.
(A) Six-hundred-megahertz COSY NMR spectrum of WbwAO104 product sialylα2-3Galβ1-3GalNAcα-PO3-PO3-(CH2)11-O-phenyl. WbwA product (structure shown) was isolated by HPLC as described in Materials and Methods, dissolved in D2O, and analyzed by 1D and 2D experiments. The axial H-3 of sialic acid appears to be close in space to the axial H-3 of Gal. (B) Six-hundred-megahertz NOESY NMR spectrum of WbwAO104 product sialylα2-3Galβ1-3GalNAcα-PO3-PO3-(CH2)11-O-phenyl. The axial H-3 of sialic acid shows nuclear Overhauser effects (NOEs) to the equatorial H-3 of sialic acid and to the H-3 of Gal. (C) Six-hundred-megahertz ROESY NMR spectrum of WbwAO104 product sialylα2-3Galβ1-3GalNAcα-PO3-PO3-(CH2)11-O-phenyl. As in the NOESY spectrum, the axial H-3 of sialic acid shows NOEs to the equatorial H-3 of sialic acid and to the H-3 of Gal.
Role of the conserved HP and EDG motifs in WbwAO104 mutational analyses.
Six enzymes with mutations of highly conserved residues with a focus on the HP and the two EDG motifs were prepared and compared to the wild-type WbwAO104 enzyme. All of the mutants and wild-type enzyme were well expressed and showed one band on Western blots using anti-His tag antibody (see Fig. S2 in the supplemental material). The P237A and the H236A mutants were found to be inactive, suggesting that His and Pro of the HP motif may be critical residues, as is the case for NST SiaT (27). In WbwAO104, there are two conserved D(E)D(E)G motifs (see Fig. S1 in the supplemental material) (24, 25). Point mutations of the acidic residues showed drastic decreases in activity, with E111A and E112A having undetectable activity and E206A and D207A having <12% activity (Fig. 4). This suggests that both EDG motifs in WbwAO104 have an important function, either in SiaT activity or in the proper folding of the protein.
FIG 4.
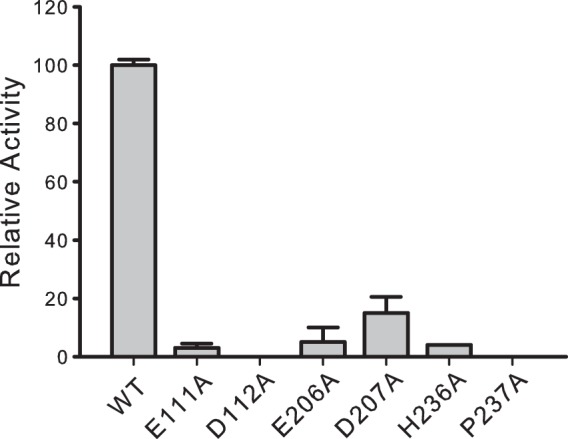
Relative activities of site-specific WbwA mutants of the HP and EDG motifs. Mutants of WbwAO104 were produced by replacing the His and Pro residues of the HP motif and Asp and Glu residues of the EDG motifs with Ala, as described in Materials and Methods. All mutants were then assayed as described in Materials and Methods in duplicate determinations. The error bars indicate the variation between duplicates. Western blots confirmed that all mutants were expressed (see Fig. S2 in the supplemental material). The E206A and D207A mutants had residual activities, but the H236A and P237A mutants from the HP motif, as well as the E111A and D112A mutants from the first EDG motif were inactive. Relative activities are shown.
Properties of human ST3Gal1.
Human ST3Gal1 has a similar activity to WbwA in synthesizing the sialyl-T antigen with a low amino acid sequence identity. However, the catalytic sites of these enzymes may have similar properties. Human ST3Gal1 has not been carefully characterized before. To compare the properties of the human enzyme to those of WbwA, human ST3Gal1 was expressed and purified. Purified His-tagged ST3Gal1 showed a single band at 37 kDa by SDS-PAGE. The activity of ST3Gal1 was stable for several days at 4°C and for several weeks at −20°C. No metal ions were required for activity, and the enzyme was fully active in the presence of EDTA. The buffer pH optimum for ST3Gal1 activity was between 6 and 6.5.
An evaluation of the acceptor substrate specificity of ST3Gal1 showed that the enzyme is specific for the Galβ1-3GalNAc structure of O-glycan core 1 (Galβ1-3GalNAc) and core 2 (GlcNAcβ1-6 [Galβ1-3] GalNAc). The exception was Galβ1-3GalNAc-PP-PhU, the acceptor substrate for WbwA, which has the core 1 structure but was not a substrate for ST3Gal1 (Table 2), probably due to the negative charge of the diphosphate group. All other compounds having different aglycone groups, including benzyl, p-nitrophenyl, allyl, or peptides were ST3Gal1 acceptors to various degrees. Using glycopeptide acceptors, Galβ1-3GalNAc in both α- and β-linkages to Thr and in α-linkage to Ser were similarly active, indicating that the anomeric configuration of the GalNAc-residues or the O-glycosylated amino acid was not important for ST3Gal1 activity. However, glycopeptides having the core 1 structure linked to Ser or Thr were generally not as effective as Galβ1-3GalNAc-Bn, suggesting that ST3Gal1 preferred the hydrophobic character of the aglycone group in the acceptor. The apparent Km value for the standard acceptor Galβ1-3GalNAcα-Bn was 0.05 mM, with an apparent Vmax value of 0.9 μmol/min/mg, while the glycopeptide AHGVT-(Galβ3GalNAcα)SAPDTRPAPGSTAPTA had a higher apparent Km value of 0.2 mM, with an apparent Vmax value of 3 μmol/min/mg. For the acceptor Galβ1-3(4-deoxy-)GalNAcα-Bn, the apparent Km value was 0.09 mM, with an apparent Vmax value of 1 μmol/min/mg, whereas the acceptor Galβ1-3(6-deoxy-)GalNAcα-Bn had an apparent Km value of 0.07 mM and an apparent Vmax value of 0.97 μmol/min/mg, which was similar to the values obtained for the standard acceptor Galβ1-3GalNAcα-Bn. Replacement of the Gal moiety of the acceptor with GlcNAc or GalNAc or changing the Gal linkage to β1-6 drastically reduced activity. However, Galβ1-6GalNAcα-Bn served as an acceptor substrate for porcine ST3Gal1 (15). This is probably due to the fact that the human and porcine enzymes are different proteins, having about 15% identity difference in their amino acid sequences and possibly having different properties of their acceptor binding sites. The 4-hydroxyl of the GalNAc residue was also important since Galβ1-3GlcNAcα-Bn and Galβ1-3GlcNAcβ-Bn had undetectable activity. Since Galβ1-3(6-deoxy-)GalNAcα-Bn and core 2-containing glycopeptides were good substrates, the 6-hydroxyl of GalNAc was apparently not recognized by the enzyme and could even be substituted for with a large GlcNAc residue. These studies show significant differences between the acceptor specificities of WbwA and ST3Gal1.
TABLE 2.
ST3Gal1 has a broad acceptor substrate specificity
| Compound (0.5 mM)a | Relative activity (%)b |
|---|---|
| Galβ3GalNAcα-Bn | 100 |
| Galβ3GalNAcα-pnp | 99–100 |
| Galβ3GalNAcα-O-allyl | 10 |
| Galβ3GalNAcα-PO3-PO3-(CH2)11-O-phenyl | <1 |
| Galβ3(4-deoxy-)GalNAcα-Bn | 106 |
| Galβ3(6-deoxy-)GalNAcα-Bn | 88 |
| GalNAcβ3GalNAcα-Bn | <7 |
| Galβ6GalNAcα-Bn | <1 |
| GlcNAcβ3GalNAcα-pnp | <1 |
| Galβ3Galβ-allyl | <3 |
| Galα3Galβ-O-allyl | <1 |
| Galβ3GlcNAcβ-Bn | <3 |
| Galβ4GlcNAcβ-Bn | <4 |
| Galβ3GlcNAcα-Bn | <2 |
| Galβ3GlcNAcβ3Gal-β3GalNAcα-Bn | <1 |
| AHGVT-(Galβ3GalNAcα)SAPESRPAPGSTAPNA | 64 |
| AHGVT-(Galβ3GalNAcα)SAPDTRPAPGSTAPTA | 48 |
| AHGVT-(Galβ3GalNAcα)SAPETRPAPGSTAPTA | 71 |
| AHGVT-(Galβ3GalNAcα)SAPDTRPAPGSTAPNA | 91 |
| AHGVT-(Galβ3GalNAcα)SAPDTRPAPGSTAP-(Galβ3GalNAcα)TA | 92 |
| TTTVTP-(Galβ3GalNAcα)TPTG | 20 |
| TT-(Galβ3GalNAcα)TVTPTPTG | 16 |
| Ac-P-(GalNAcα)T-(Galβ3GalNAcα)T-(GalNAcα)TPIST-NH2 | 45 |
| TT-(GlcNAcβ6[Galβ3]GalNAcα)TVTPTPTG | 46 |
| T-(GlcNAcβ6[Galβ3]GalNAcα)TTVTPTPTG | 55 |
| Ac-G-(Galβ3GalNAcβ)TTTPIST-NH2 | 65 |
| Ac-PTT-(Galβ3GalNAcβ)TPIST-NH2 | 40 |
| Ac-PT-(Galβ3GalNAcβ)TTPIST-NH2 | 33 |
| Ac-GT-(Galβ3GalNAcβ)TTPIST-NH2 | 46 |
Sialyltransferase assays were carried out as described in Materials and Methods. Enzyme products were estimated by their radioactivity levels after elution from the Sep-Pak column. Galβ1-3GalNAcα-PO3-PO3-(CH2)11-O-phenyl was synthesized using WbwC (20). pnp, p-nitrophenyl; Ac-, acetyl; -NH2, amide at the C terminus; boldface S or T, glycosylated amino acid.
One hundred percent activity for ST3Gal1 was 0.9 μmol/min/mg.
DISCUSSION
This article reports the first biochemical study of WbwAO104, a SiaT encoded by the O-antigen synthesis gene cluster of the pathogenic E. coli serotype O104. This work contributes to the detailed understanding of the biosynthesis of an important virulence factor in an intestinal pathogen. We have clearly shown that the WbwA enzyme transfers one sialic acid residue from CMP-sialic acid to the terminal Gal residue of Galβ1-3GalNAcα-PP-PhU, forming a sialylα2-3 linkage. The sialylα2-3Galβ1-3GalNAcα structure of the reaction product is identical to the human sialyl-T antigen and is consistent with the structure of the O104 antigen (8). The diphosphate group can easily be removed with pyrophosphatases and phosphatases, thus creating a reducing end of the sialyl-T antigen with a potential for further derivatization. For example, tumor-selective epitopes could be created with the sialyl-T antigen linked to a specific mucin peptide, which could increase its immunogenicity (30). Therefore, recombinant WbwAO104 could be used to synthesize stereo- and linkage-specific bacterial structures as well as the human sialyl-T antigen for functional studies or development of a tumor-selective vaccine targeting the sialyl-T epitope. Given the abundance of sialyl-T on cancer glycoproteins, the benefit of immunogenicity toward sialyl-T might outweigh potential cross-reactions toward normal glycoproteins.
In the repeating unit synthesized on PP-Und, the GalNAc residue at the nonreducing end is present in an α-linkage (8). However, in the polymerized O antigen, GalNAc is found in β linkage. It is likely that polymerase Wzy links GalNAc of the oligosaccharide to the Galα residue at the nonreducing end of another unit with inversion of configuration. This suggests that Wzy from E. coli O104 is an inverting oligosaccharyltransferase.
WbwCO104 is the second enzyme in the pathway of O104 antigen synthesis and requires the diphosphate in the acceptor (GalNAc–PP-PhU). Thus far, the second enzymes in the repeating unit assembly sequence have all been shown to have an absolute requirement for at least one phosphate group in the acceptor (20, 23, 31–34). WbwAO104 is the first example of the third enzyme in the pathway and of a eukaryotic or bacterial SiaT that has a requirement for diphosphate in the acceptor. Future mutational and structural studies may identify the amino acids responsible for the unique acceptor specificity of WbwA.
Similar to the sialic acid residue in the O antigen of E. coli serotype O171, the sialic acid moiety of the O104 antigen is 7- and 9-O-acetylated. The gene responsible for O-acetylation in E. coli O104 is likely to be wckD, which is found in the O104 antigen synthesis gene cluster (16) and encodes a putative sialate O-acetyltransferase. O-acetyltransferases, including WckDO104, are thought to transfer O-acetyl groups from acetyl coenzyme A to CMP-sialic acid. Acetylated CMP-sialic acid would then be the donor substrate for WbwA to create a 7,9-di-O-acetyl-sialic acid-containing oligosaccharide. Since CMP-sialic acid served as a donor substrate for WbwA, the enzyme may have a relaxed specificity toward the structure of sialic acid in the donor substrate and utilizes both CMP-sialic acid and CMP-7,9-di-O-acetyl-sialic acid. Our preliminary data showed that the next GT in the pathway, α4-Gal-transferase WbwB, acts on sialyl-oligosaccharides. Thus, WbwB may also have a relaxed specificity toward sialyl- and acetylated sialyl-oligosaccharides. However, the detailed substrate specificities of these enzymes and biosynthetic pathways remain to be examined.
In SiaTs, His residues appear to complex the nucleotide sugar phosphate groups instead of divalent metal ions that are used by many other GTs having a DXD motif (35). Indeed, WbwAO104 has a critically important His236 residue within the HP sequence that is conserved in bacterial SiaTs (10). The His236 residue could be a general acid (27) or could be involved in binding the donor substrate CMP-sialic acid. The Pro237 residue, due to its unique properties in affecting protein structure, may serve to ensure the function of the His236 residue. Glu111, Glu112, and Asp206 residues in two of the conserved EDG motifs possibly play a role in substrate binding and catalysis or by ensuring the proper protein conformation. Future protein structure and modeling studies could confirm the roles of acidic residues in WbwAO104.
In spite of the similarities, the analysis of amino acid sequences suggests that human ST3Gal1 and WbwAO104 do not have a common evolutionary origin. The overall protein structure and the biochemical roles of critically important His, Asp, Glu, and other residues that are responsible for the unique acceptor specificity to WbwA remain to be determined. This may confirm the hypothesis that bacterial enzymes and their mammalian counterparts can have similar but characteristic geometries and functions of amino acids in the substrate binding and catalytic sites in spite of different overall sequences and folds.
ST3Gal1 has an α2,3-SiaT activity that is similar to that of WbwA, and both enzymes synthesize the common sialyl-T antigen, are monofunctional, and do not require divalent metal ions for activity. Both enzymes are inverting GTs (albeit of different GT families and having different overall folds), bind to CMP-sialic acid donor substrate and Galβ1-3GalNAc acceptor substrates (17, 36), and are effectively inhibited by bis-imidazolium salts having at least 16 carbons in the aliphatic linker chain between positively charged groups. The mechanism by which these inhibitors selectively block GT activities is under investigation and may involve binding to anionic and hydrophobic pockets of the enzyme proteins. This knowledge may be used in the development of antibacterial strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hans Paulsen, University of Hamburg, Germany, for providing glycopeptides and sugar derivatives and John Schutzbach for critical advice. The NMR analyses were carried out by F. Sauriol, Department of Chemistry, Queen's University.
This work was supported by an operating grant from the Canadian Institutes of Health Research (to I.B. and W.A.S.) and a Discovery grant from the Natural Science and Engineering Research Council of Canada (to I.B.). The isolation of baculovirus vectors and production and purification of human ST3Gal1 were supported by the Glycorepository Project (NIH P41GM103390 and P41RR005351).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00521-15.
REFERENCES
- 1.Schauer R. 2009. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol 19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houliston RS, Vinogradov E, Dzieciatkowska M, Li J, St Michael F, Karwaski MF, Brochu D, Jarrell HC, Parker CT, Yuki N, Mandrell RE, Gilbert M. 2011. Lipooligosaccharide of Campylobacter jejuni: similarity with multiple types of mammalian glycans beyond gangliosides. J Biol Chem 286:12361–12370. doi: 10.1074/jbc.M110.181750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuki N, Odaka M. 2005. Ganglioside mimicry as a cause of Guillain-Barré syndrome. Curr Opin Neurol 18:557–561. doi: 10.1097/01.wco.0000174604.42272.2d. [DOI] [PubMed] [Google Scholar]
- 4.Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 5.Brockhausen I. 2014. Crossroads between bacterial and mammalian glycosyltransferases. Front Immunol 492:1–21. doi: 10.3389/fimmu.2014.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J, Cui Y, Zhao X, Rohde H, Liang T, Wolters M, Li D, Belmar Campos C, Christner M, Song Y, Yang R. 2011. Identification of the Shiga toxin-producing Escherichia coli O104:H4 strain responsible for a food poisoning outbreak in Germany by PCR. J Clin Microbiol 49:3439–3440. doi: 10.1128/JCM.01312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubino S, Cappuccinelli P, Kelvin DJ. 2011. Escherichia coli (STEC) serotype O104 outbreak causing haemolytic syndrome (HUS) in Germany and France. J Infect Dev Ctries 5:437–440. doi: 10.3855/jidc.2172. [DOI] [PubMed] [Google Scholar]
- 8.Gamian A, Romanowska E, Ulrich J, Defaye J. 1992. The structure of the sialic acid-containing Escherichia coli O104 O-specific polysaccharide and its linkage to the core region in lipopolysaccharide. Carbohydr Res 236:195–208. doi: 10.1016/0008-6215(92)85016-S. [DOI] [PubMed] [Google Scholar]
- 9.Harduin-Lepers A, Mollicone R, Delannoy P, Oriol R. 2005. The animal sialyltransferases and sialyltransferase-related genes: a phylogenetic approach. Glycobiology 15:805–817. doi: 10.1093/glycob/cwi063. [DOI] [PubMed] [Google Scholar]
- 10.Freiberger F, Claus H, Gunzel A, Oltmann-Norden I, Vionnet J, Muhlenhoff M, Vogel U, Vann WF, Gerardy-Schahn R, Stummeyer K. 2007. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol Microbiol 65:1258–1275. doi: 10.1111/j.1365-2958.2007.05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schur MJ, Lameignere E, Strynadka NC, Wakarchuk WW. 2012. Characterization of α2,3- and α2,6-sialyltransferases from Helicobacter acinonychis. Glycobiology 22:997–1006. doi: 10.1093/glycob/cws071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmölzer K, Ribitsch D, Czabany T, Luley-Goedl C, Kokot D, Lyskowski A, Zitzenbacher S, Schwab H, Nidetzky B. 2013. Characterization of a multifunctional α2,3-sialyltransferase from Pasteurella dagmatis. Glycobiology 23:1293–1304. doi: 10.1093/glycob/cwt066. [DOI] [PubMed] [Google Scholar]
- 13.Patel RY, Balaji PV. 2006. Identification of linkage-specific sequence motifs in sialyltransferases. Glycobiology 16:108–116. doi: 10.1093/glycob/cwj046. [DOI] [PubMed] [Google Scholar]
- 14.Brockhausen I, Gao Y. 2012. Structural glycobiology: applications in cancer research, p 177–213. CRC Press, Taylor & Francis Group, Abingdon, Oxford, United Kingdom. [Google Scholar]
- 15.Rakić B, Rao FV, Freimann K, Wakarchuk W, Strynadka NCJ, Withers SG. 2013. Structure-based mutagenic analysis of mechanism and substrate specificity in mammalian glycosyltransferases: porcine ST3Gal-I. Glycobiology 23:536–545. doi: 10.1093/glycob/cwt001. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Briggs CE, Rothemund D, Fratamico P, Luchansky JB, Reeves PR. 2001. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 270:231–236. doi: 10.1016/S0378-1119(01)00471-1. [DOI] [PubMed] [Google Scholar]
- 17.Kuhns W, Rutz V, Paulsen H, Matta KL, Baker MA, Barner M, Granovsky M, Brockhausen I. 1993. Processing O-glycan core 1, Galbeta1–3GalNAc alpha-R. Specificities of core 2, UDP-GlcNAc:Galbeta1–3GalNAc-R(GlcNAc to GalNAc) beta 6-N-acetylglucosaminyltransferase and CMPsialic acid:Galbeta1–3GalNAc-R alpha 3-sialyltransferase. Glycoconj J 10:381–394. [DOI] [PubMed] [Google Scholar]
- 18.Montoya-Peleaz PJ, Riley JG, Szarek WA, Valvano MA, Schutzbach JS, Brockhausen I. 2005. Identification of a UDP-Gal:GlcNAc-R galactosyltransferase activity in Escherichia coli VW187. Bioorg Med Chem Lett 15:1205–1211. doi: 10.1016/j.bmcl.2004.11.077. [DOI] [PubMed] [Google Scholar]
- 19.Chandrasekaran EV, Xue J, Xia J, Chawda R, Piskorz C, Locke RD, Neelamegham S, Matta KL. 2005. Analysis of the specificity of sialyltransferases toward mucin core 2, globo, and related structures. Identification of the sialylation sequence and the effects of sulfate, fucose, methyl, and fluoro substituents of the carbohydrate chain in the biosynthesis of selectin and siglec ligands, and novel sialylation by cloned alpha2,3(O)sialyltransferase. Biochemistry 44:15619–15635. doi: 10.1021/bi050246m. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Czuchry D, Liu B, Vinnikova AN, Gao Y, Vlahakis JZ, Szarek WA, Wang L, Feng L, Brockhausen I. 2014. Characterization of two UDP-Gal:GalNAc-diphosphate-lipid β1,3-galactosyltransferases WbwC from Escherichia coli serotypes O104 and O5. J Bacteriol 196:3122–3133. doi: 10.1128/JB.01698-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Vlahakis JZ, Szarek WA, Brockhausen I. 2013. Selective inhibition of glycosyltransferases by bivalent imidazolium salts. Bioorg Med Chem 21:1305–1311. doi: 10.1016/j.bmc.2012.12.034. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis DL. 2014. Recombinant protein expression in baculovirus-infected insect cells. Methods Enzymol 536:149–163. doi: 10.1016/B978-0-12-420070-8.00013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Liu B, Strum S, Schutzbach JS, Druzhinina TN, Utkina NS, Torgov VI, Danilov LL, Veselovsky VV, Vlahakis JZ, Szarek WA, Wang L, Brockhausen I. 2012. Biochemical characterization of WbdN, a beta1,3-glucosyltransferase involved in O-antigen synthesis in enterohemorrhagic Escherichia coli O157. Glycobiology 22:1092–1102. doi: 10.1093/glycob/cws081. [DOI] [PubMed] [Google Scholar]
- 24.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Systems Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas KB, Nicholas HB Jr, Deerfield DW II. 1997. GeneDoc: analysis and visualization of genetic variation, EMBNEW. NEWS 4:14. [Google Scholar]
- 26.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 27.Lin LYC, Rakic B, Chiu CPC, Lameignere E, Wakarchuk WW, Withers SG, Strynadka NCJ. 2011. Structure and mechanism of the lipooligosaccharide sialyltransferase from Neisseria meningitidis. J Biol Chem 286:37237–37248. doi: 10.1074/jbc.M111.249920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi W, Shen J, Zhou G, Li J, Wang PG. 2008. Bacterial homologue of human blood group A transferase. J Am Chem Soc 130:14420–14421. doi: 10.1021/ja805844y. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Shen J, Liu X, Shao J, Yi W, Chow CS, Wang PG. 2008. Identification of a new alpha1,2-fucosyltransferase involved in O-antigen biosynthesis of Escherichia coli O86:B7 and formation of H-type 3 blood group antigen. Biochemistry 47:11590–11597. doi: 10.1021/bi801067s. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser A, Gaidzik N, Westerlind U, Kowalczyk D, Hobel A, Schmitt E, Kunz H. 2009. A synthetic vaccine consisting of a tumor-associated sialyl-T(N)-MUC1 tandem-repeat glycopeptide and tetanus toxoid: induction of a strong and highly selective immune response. Angew Chem Int Ed Engl 48:7551–7555. doi: 10.1002/anie.200902564. [DOI] [PubMed] [Google Scholar]
- 31.Riley JG, Menggad M, Montoya-Peleaz PJ, Szarek WA, Marolda CL, Valvano MA, Schutzbach JS, Brockhausen I. 2005. The wbbD gene of E. coli strain VW187 (O7:K1) encodes a UDP-Gal:GlcNAc alpha-pyrophosphate-R beta1,3-galactosyltransferase involved in the biosynthesis of O7-specific lipopolysaccharide. Glycobiology 15:605–613. doi: 10.1093/glycob/cwi038. [DOI] [PubMed] [Google Scholar]
- 32.Brockhausen I, Liu B, Hu B, Lau K, Szarek WA, Wang L, Feng L. 2008. Characterization of two beta-1,3-glucosyltransferases from Escherichia coli serotypes O56 and O152. J Bacteriol 190:4922–4932. doi: 10.1128/JB.00160-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C, Liu B, Hu B, Han Y, Feng L, Allingham JS, Szarek WA, Wang L, Brockhausen I. 2011. Biochemical characterization of UDP-Gal:GlcNAc-pyrophosphate-lipid β-1,4-galactosyltransferase WfeD, a new enzyme from Shigella boydii type 14 that catalyzes the second step in O-antigen repeating-unit synthesis. J Bacteriol 193:449–459. doi: 10.1128/JB.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Hao Y, Lam JS, Vlahakis JZ, Szarek WA, Vinnikova A, Veselovsky V, Brockhausen I. 2015. Biosynthesis of the common polysaccharide antigen of Pseudomonas aeruginosa PAO1: characterization and role of GDP-d-rhamnose:GlcNAc/GalNAc-diphosphate-lipid alpha1,3-d-rhamnosyltransferase WbpZ. J Bacteriol 197:2012–2019. doi: 10.1128/JB.02590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breton C, Fournel-Gigleux S, Palcic MM. 2012. Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol 22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Rearick JI, Sadler JE, Paulson JC, Hill RL. 1979. Enzymatic characterization of beta D-galactoside alpha2 leads to 3 sialyltransferase from porcine submaxillary gland. J Biol Chem 254:4444–4451. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.