Abstract
Although tuberculosis (TB) is a reemerging disease that affects people in developing countries and immunocompromised populations in developed countries, the current diagnostic methods are far from optimal. Metabolomics is increasingly being used for studies on infectious diseases. We performed metabolome profiling of plasma samples to identify potential biomarkers for diagnosing TB. We compared the plasma metabolome profiles of TB patients (n = 46) with those of community-acquired pneumonia (CAP) patients (n = 30) and controls without active infection (n = 30) using ultrahigh-performance liquid chromatography–electrospray ionization-quadrupole time of flight mass spectrometry (UHPLC-ESI-QTOFMS). Using multivariate and univariate analyses, four metabolites, 12R-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid [12(R)-HETE], ceramide (d18:1/16:0), cholesterol sulfate, and 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol, were identified and found to have significantly higher levels in TB patients than those in CAP patients and controls. In a comparison of TB patients and controls, the four metabolites demonstrated area under the receiver operating characteristic curve (AUC) values of 0.914, 0.912, 0.905, and 0.856, sensitivities of 84.8%, 84.8%, 87.0%, and 89.1%, specificities of 90.0%, 86.7%, 86.7%, and 80.0%, and fold changes of 4.19, 26.15, 6.09, and 1.83, respectively. In a comparison of TB and CAP patients, the four metabolites demonstrated AUC values of 0.793, 0.717, 0.802, and 0.894, sensitivities of 89.1%, 71.7%, 80.4%, and 84.8%, specificities of 63.3%, 66.7%, 70.0%, and 83.3%, and fold changes of 4.69, 3.82, 3.75, and 2.16, respectively. 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol combined with 12(R)-HETE or cholesterol sulfate offered ≥70% sensitivity and ≥90% specificity for differentiating TB patients from controls or CAP patients. These novel plasma biomarkers, especially 12(R)-HETE and 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol, alone or in combination, are potentially useful for rapid and noninvasive diagnosis of TB. The present findings may offer insights into the pathogenesis and host response in TB.
INTRODUCTION
Tuberculosis (TB) is a disease caused by the bacterium Mycobacterium tuberculosis. Although it is a well-known disease that has been around for much of human history, there are still millions of new TB cases occurring per year worldwide, and TB remains a leading cause of deaths worldwide, especially in developing countries. Since the 1980s, TB has reemerged as a result of the AIDS epidemic and increasing use of immunosuppressants. In recent years, a higher incidence of extrapulmonary disease in immunocompromised hosts and the emergence of multidrug-resistant strains have further complicated diagnosis and treatment (1–3). Despite the medical importance of TB, diagnosis is still associated with many unresolved problems. The traditional gold standard methods are smear and culture for acid-fast bacilli from clinical specimens. Although culture offers higher sensitivity and specificity than those of smears, it is not useful for culture-negative cases, especially in early, disseminated, or extrapulmonary disease (4, 5). Moreover, it often takes 2 to 6 weeks before culture is positive and even longer for definitive species identification. While newer diagnostic modalities, such as adenosine deaminase levels in pleural fluid, lipoarabinomannan levels in urine, PCR, and Xpert MTB/RIF assays, have been developed (6–12), there are still limitations in terms of their sensitivity and/or specificity.
Metabolomics is an emerging platform for studies of infectious diseases or pathogens (13–19). For TB, the technique has been applied on cultured isolates for differentiation from other Mycobacterium species and studies on the biology and virulence of tuberculosis (14, 15, 20–22). For example, lipidomics studies have revealed novel metabolites potentially associated with growth and virulence of M. tuberculosis (23, 24). We also recently identified extracellular metabolites specific to M. tuberculosis (25), supporting the potential of metabolomics in exploring novel biomarkers to better understand its biology and pathogenesis. On the other hand, metabolomics applied on direct patient samples may reveal specific diagnostic markers or be used to monitor treatment response (26–29). It has been shown that volatile organic compounds (VOCs) in the urine of TB patients can be distinguished from those of healthy subjects (26). A study using nuclear magnetic resonance spectroscopy-based metabolomics on serum samples from TB patients demonstrated discrimination between patients and healthy controls (27). In another metabolomics study, several abundant metabolites were identified, including two mycobacterium-derived cell wall glycolipids, in EDTA-plasma from TB patients that were not identified in household contacts (28). However, patients with other bacterial infections were not included as controls in these studies, and thus, the potential of such a metabolic profile for use in the diagnosis of TB remains to be determined.
To identify potential biomarkers for noninvasive diagnosis of TB, we applied state-of-the-art technology for metabolomics profiling of plasma samples from TB patients, using ultrahigh-performance liquid chromatography–electrospray ionization-quadrupole time of flight mass spectrometry (UHPLC-ESI-QTOFMS). Multi- and univariate statistical analyses were used to identify metabolites with significantly higher levels in plasma from TB patients than in plasma from patients with community-acquired pneumonia (CAP) or controls without active infection. The diagnostic performance of the identified biomarkers was assessed using whole-metabolome receiver operating characteristic curve (ROC) analysis.
MATERIALS AND METHODS
Patient and control samples.
Clinical samples were collected from hospitalized adult patients (≥18 years of age) at Queen Mary Hospital, Hong Kong. A total of 46 plasma samples from 37 patients with newly diagnosed TB, and 60 plasma samples from 30 patients with CAP and 30 controls without active infection were recruited for UHPLC-QTOFMS analysis. Plasma samples from TB patients were collected before the commencement of antimycobacterial treatment. The plasma samples from CAP patients were collected at admission. This study was approved by the institutional review board of the University of Hong Kong/Hospital Authority of Hong Kong West Cluster (reference no. UW 13-265).
Diagnostic criteria.
A diagnosis of TB was based on compatible clinical features, together with the presence of the following microbiological evidence: (i) positive stain for acid-fast bacilli, (ii) positive culture for M. tuberculosis, and/or (iii) positive PCR for M. tuberculosis from clinical samples. The diagnosis of CAP was based on compatible clinical features and radiological evidence of lung infiltrates, with disease onset occurring within 48 h of hospital admission. The causative agents of CAP included Acinetobacter baumannii (n = 1), Escherichia coli (n = 2), Haemophilus influenzae (n = 2), Klebsiella pneumoniae (n = 1), Moraxella catarrhalis (n = 1), Pseudomonas aeruginosa (n = 1), Streptococcus pneumoniae (n = 4), and influenza B virus (n = 1), while the etiological agent was unknown for the remaining cases. The controls consisted of patients without any clinical evidence of active infection.
Chemicals and reagents.
Liquid chromatography-mass spectrometry (LC-MS)-grade water, methanol, and acetonitrile were purchased from J.T. Baker (Center Valley, PA, USA). HPLC-grade ethanol and acetone were purchased from Merck (Darmstadt, Germany). Formic acid was of American Chemical Society (ACS) reagent grade and obtained from Sigma-Aldrich, (St. Louis, MO, USA). Ceramide (d18:1/16:0) was purchased from Avanti Polar Lipid (Alabaster, AL, USA). 12R-Hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid [12(R)-HETE] and cholesterol sulfate standards were purchased from Cayman Chemical (Ann Arbor, MI, USA).
Sample preparation.
Blood samples were collected in heparin bottles, transferred immediately to the laboratory, and centrifuged at 3,000 rpm and 4°C for 10 min to obtain the plasma fractions. For metabolomics analysis, 100 μl of plasma was thawed at 4°C, and plasma proteins were precipitated with 400 μl of a methanol-ethanol-acetone mixture at a ratio of 1:1:1 (vol/vol/vol). The sample extract was vigorously vortexed for 1 min and centrifuged at 14,000 rpm and 4°C for 10 min. The supernatant was collected for UHPLC-ESI-QTOFMS analysis. All specimens were immediately kept at −80°C until analysis and stored within 1 week. The thawed specimens were analyzed within 48 h in a random manner to prevent the batch effect.
Untargeted metabolomics profiling of patient plasma using UHPLC-ESI-QTOFMS.
The metabolomic profiling of plasma supernatants was performed using Agilent 1290 Infinity UHPLC (Agilent Technologies, Waldbronn, Germany) coupled with the Agilent 6540 UHD accurate-mass QTOF system (Agilent Technologies, Santa Clara, CA, USA) accompanied by the MassHunter Workstation software for QTOF (version B.03.01 for data acquisition; Agilent Technologies, USA). A Waters Acquity UPLC BEH C18 column (2.1 by 100 mm, 1.7 μm) (Waters, Milford, MA, USA) was used for the separation, with an injection volume of 5 μl. The column and autosampler temperatures were maintained at 45°C and 10°C, respectively. The separation was performed at a flow rate of 0.4 ml/min under a gradient program in which mobile phase A was composed of LC-MS-grade water containing 0.1% formic acid (vol/vol), and mobile phase B was composed of acetonitrile. The gradient program was applied as follows: t = 0 min, 5% B; t = 0.5 min, 5% B; t = 7 min, 48% B; t = 20 min, 78% B; t = 27 min, 80% B; t = 31 min, 99.5% B; t = 36.5 min, 99.5% B; and t = 36.51 min, 5% B. The stop time was 40 min. The ESI mass spectra were acquired in both positive and negative ion modes using Agilent Jet Stream ESI source, with capillary voltages of +3,800 V and −3,500 V, respectively. Other source conditions were kept constant in all experiments: the gas temperature was kept constant at 300°C, drying gas (nitrogen) was set at the rate of 7 liters/min, and the pressure of nebulizer gas (nitrogen) was 40 lb/in2. The sheath gas was kept at a flow rate of 10 liters/min at a temperature of 350°C. The voltages of the Fragmentor, Skimmer 1, and Octopole RF peak were 135 V, 50 V, and 500 V, respectively. The mass data were collected between m/z 80 and 1,700 at an acquisition rate of 2 scans/s. Two reference masses at m/z 121.0509 (protonated molecular ion of C5H4N4) and m/z 922.0098 (protonated molecular ion of C18H18O6N3P3F24) for positive mode, and m/z 119.0363 (deprotonated molecular ion of C5H4N4) and m/z 966.0007 (formate adduct of C18H18O6N3P3F24) for negative mode, were used as constant mass corrections during the LC-MS run. Product ion scanning (PIS) experiments were performed with the Acquity UPLC I-class system coupled with the Waters Synapt G2-Si QTOF system (Waters) operating in both positive and negative electrospray ionization modes. The mass data were collected between m/z 50 and 1,000 at an acquisition rate of 3 scans/s. The voltages of the capillary, sampling cone, and source offset were 3 kV, 30 V, and 80 V, respectively. Other source conditions were kept as follows: source temperature, 120°C; desolvation temperature, 380°C; cone gas, 10 liters/h; desolvation gas, 800 liters/h; and nebulizer gas, 6.5 × 105 Pa. Intermittent injections of leucine enkephalin with lock masses of m/z 556.2771 at positive mode and m/z 554.2615 at negative mode at a concentration of 400 pg/μl in 50% acetonitrile at a flow rate of 6 μl/min were used for accurate mass measurements. Tandem mass spectrometry (MS/MS) analysis was performed using ultrahigh-purity argon at a collision energy at 10, 20, or 40 eV to generate the best-quality MS/MS spectra for putative identification and structural elucidation of the significant metabolites.
Data processing and statistical analysis.
Multivariate and univariate analyses were performed to identify molecular features that discriminate TB patients from CAP patients and controls. The multivariate analysis was applied to a total of 106 LC-MS data for plasma samples collected from three groups (46, 30, and 30 samples from newly diagnosed TB patients, CAP patients, and controls without active infection, respectively). The raw LC-MS data were converted into mzXML format using msConvert (ProteoWizard) and subsequently processed using the open-source XCMS package (http://www.bioconductor.org/packages/2.8/bioc/html/xcms.html) operating in R (http://www.r-project.org/), which adopted different peak detection and alignment, as well as data filtering with centWave algorithms. The data were further processed with normalization, scaling, filtering, and statistical analysis using MetaboAnalyst 3.0 (www.metaboanalyst.ca). The data were mean centered, square root scaled, and normalized, such that the sum of squares for each chromatogram equaled on statistical analysis (30). Insignificant features between TB patients and CAP patients or controls were filtered out using both univariate and multivariate analyses. For multivariate analysis, principal component analysis (PCA) was performed for unsupervised analysis, while partial-least-squares discrimination analysis (PLS-DA) was performed for supervised analysis to identify features with discriminative power. A score plot was applied to reduce the dimensionality of the data for grouping of the samples, in which each point in the score plot represented the individual sample, and similar data sets exhibited clustering, while different sets separated farther apart. PLS-DA models were validated based on accuracy, the multiple-correlation coefficient (R2), and cross-validated R2 (Q2) in cross-validation. The significance of the biomarkers was ranked using the variable importance in projection (VIP) score (>1) from the PLS-DA model that was responsible for the separation of TB patients from non-TB groups, i.e., CAP patients and controls. Significant features were further identified by a significance analysis of microarray (SAM), in which the false discovery rate (FDR) was determined by running multiple tests on high-dimensional data that distinguish between TB and non-TB groups, with an FDR of <0.05 considered to be significant (31, 32).
For univariate analysis, the statistical significance of features was determined among TB patients, CAP patients, and controls without active infection using one-way analysis of variance (ANOVA) with Fisher's post hoc test by MetaboAnalyst 3.0. A P value of <0.05 was considered to be statistically significant. Common significant features from PLS-DA, SAM, and one-way ANOVA were included for univariate ROC analysis using Web-based ROCEET (33). The classical ROC curve analysis was performed, and the area under the ROC curve (AUC) was calculated with the predefined model. In addition, the optimal cutoffs for the given metabolite were computed to obtain the sensitivity, specificity, and confidence intervals at different cutoffs for an evaluation of the recognition and prediction abilities with respect to each variable. Protonated ions ([M+H]+) for ceramide (d18:1/16:0) and 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol and deprotonated ions ([M-H]−) for 12(R)-HETE and cholesterol sulfate were used for AUC and abundance calculations, respectively. Significant features with an AUC of >0.85 for a comparison between TB patients and controls without active infection were identified, and ROC curve analysis for the identified metabolites was further performed to compare TB and CAP patients. Box-and-whisker plots were generated, and P values were calculated by the Student t test using the Analyse-it software (Leeds, United Kingdom).
Metabolite identification.
Features with significant differences were selected for PIS experiments. The MS/MS spectra of the potential biomarkers and commercially available reference standards, including ceramide (d18:1/16:0), 12(R)-HETE, and cholesterol sulfate, were processed using the Waters MassLynx version 4.1 software (Waters Corp., Milford, MA). A potential molecular formula based on the accurate mass and isotopic pattern recognitions of parent and fragment ions were generated. All putative identities were confirmed by matching with entries in the METLIN (http://metlin.scripps.edu/), Human Metabolome Database (HMDB) (http://www.hmdb.ca/), MassBank (http://www.massbank.jp/), LipidMaps (http://www.lipidmaps.org/), Mtb Lipid, KEGG (http://www.genome.jp/kegg), MycoMass, and MycoMap databases (http://www.brighamandwomens.org/research/depts/medicine/rheumatology/Labs/Moody/default.aspx) using exact molecular weights, nitrogen rule, or MS/MS fragmentation pattern data and a literature search. Efforts were made to distinguish metabolites from the other isobaric compounds whenever possible by its elution order and degree of difference in fragmentation pattern corresponding to its structural characteristics. The putative identities of the biomarkers were confirmed by comparing their chromatographic retention times (RTs) and MS/MS spectra with those obtained from commercially available standards of ceramide (d18:1/16:0), 12(R)-HETE, and cholesterol sulfate.
RESULTS
Metabolomic profiling of plasma samples and omics-based statistical and bioinformatic analysis.
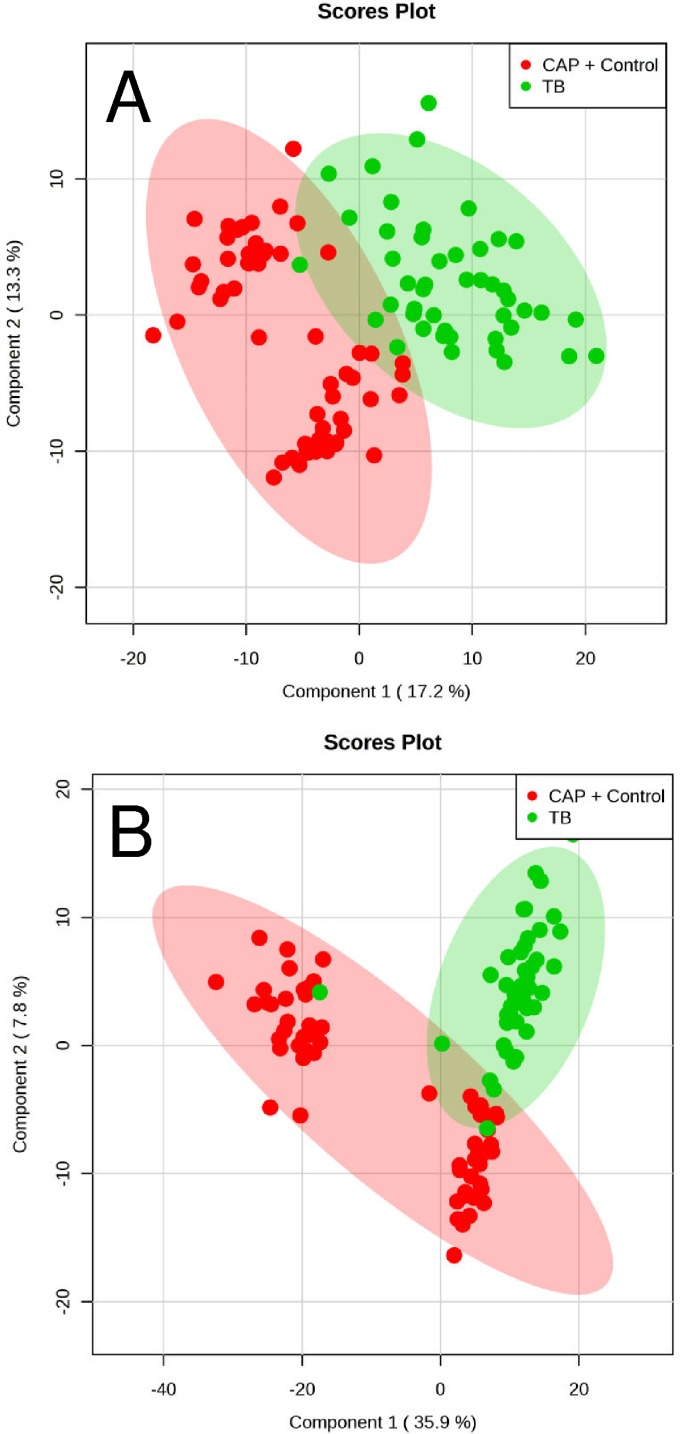
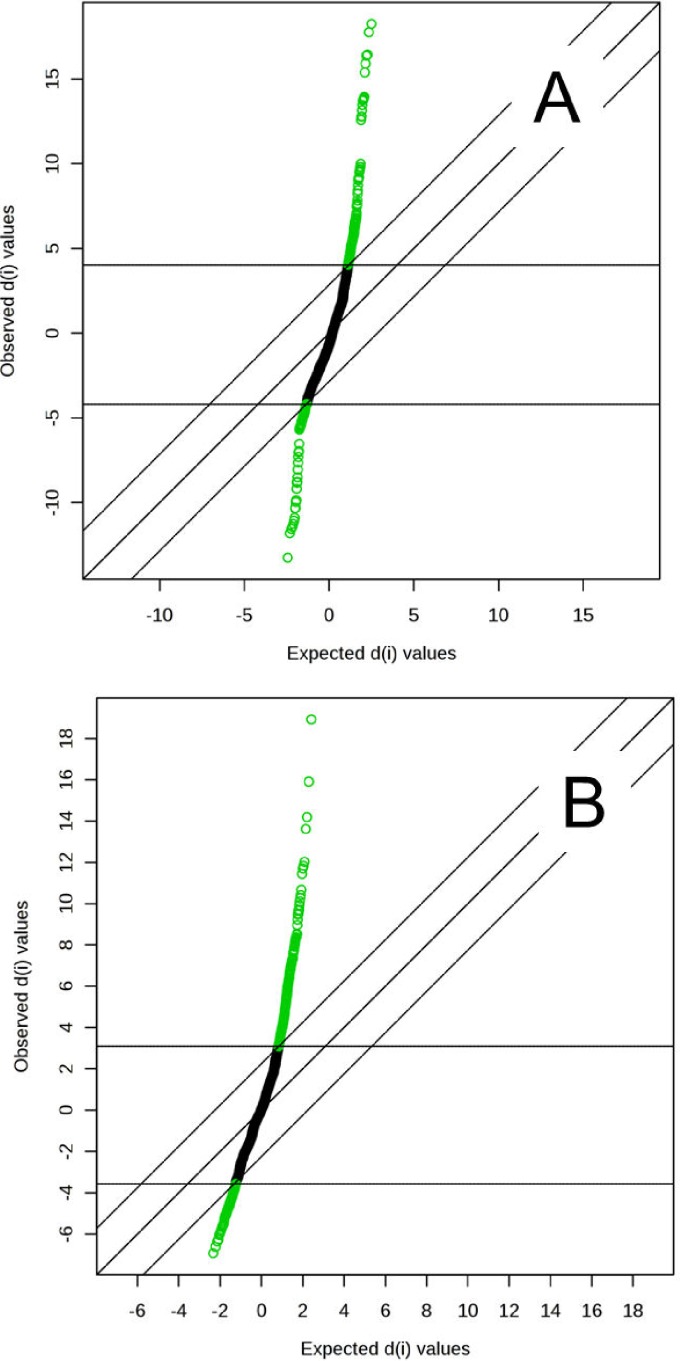
The metabolomes of 106 plasma samples from the three groups (46 samples from newly diagnosed TB patients, 30 samples from CAP patients, and 30 samples from controls without active infection) were characterized and compared. A total of 1,626 and 1,598 molecular features in positive and negative modes, respectively, were obtained and subjected to statistical analysis using MetaboAnalyst 3.0 (see Tables S1 and S2 in the supplemental material). For multivariate analysis, PCA revealed that samples from TB patients and CAP patients clustered separately. However, using this unsupervised method, ambiguous separation was observed between TB patients and controls. Therefore, a supervised method using PLS-DA was performed, which showed that the TB group can be distinguished from the non-TB group (CAP patients and controls) in both positive and negative mode data (Fig. 1A and B), with accuracies of 99% and 97.2%, multiple-correlation coefficients of 92.5% and 88.6%, and cross-validated R2 values of 82.7% and 77.4%, respectively. Significant features for the separation between TB and non-TB groups were ranked, yielding 242 and 243 features with a VIP score of >1 in positive and negative ionization modes, respectively. SAM methods were also used to select the most discriminative features responsible for the separation between TB and non-TB groups, yielding 233 and 324 significant features with an FDR of <0.05 in positive and negative ionization modes, respectively (Fig. 2A and B).
FIG 1.
PLS-DA score plot based on human plasma from 46 TB patients, 30 CAP patients, and 30 controls without active infection. (A) PLS-DA score plot of positive mode data, with accuracy of 99%, multiple correlation coefficient (R2) of 92.5%, and cross-validated R2 (Q2) of 82.7%. (B) PLS-DA score plot of negative mode data, with accuracy of 97.2%, multiple correlation coefficient (R2) of 88.6%, and cross-validated R2 (Q2) of 77.4%.
FIG 2.
Significance analysis of microarray (SAM) plots in positive (A) and negative (B) ionization modes. The false discovery rate (FDR) was determined when running multiple tests on high-dimensional data that distinguish between TB and non-TB groups. An FDR of <0.05 was considered significant. d(i) indicates the SAM score. The middle diagonal line indicates equal observed d(i) and expected d(i) values, and the lines parallel to this line are drawn at delta d(i) distances of 2.85 (A) and 2.25 (B).
Univariate analysis using one-way ANOVA identified 434 and 577 statistically significant molecular features (P < 0.05) in positive and negative mode, respectively. A total of 76 and 84 common significant features in positive and negative ionization modes, respectively, with a VIP score of >1 by PLS-DA, SAM, and one-way ANOVA were manually inspected. Those features not due to interference were included for univariate ROC analysis to select the most discriminant biomarkers. Four metabolites with an AUC of >0.85, two each in positive and negative modes, were identified, which exhibited upregulation in TB patients compared to controls (Table 1).
TABLE 1.
Plasma metabolites with higher levels in TB patients than those in CAP patients and controls without active infection
| Compound | Experimental mass (m/z) | Retention time (min) | Ion | MS/MS fragment masses (m/z) | VIP scorea | Elemental composition | Putative identity |
|---|---|---|---|---|---|---|---|
| 1 | 538.5190 | 33.02 | M+H | 252.2672, 264.2682, 282.2794, 502.4987, 520.5095, 538.5193 | 1.37 | C34H67NO3 | Ceramide (d18:1/16:0) |
| 2 | 429.3738 | 32.22 | M+H | 149.0976, 165.0924, 177.0935, 191.1083, 205.1228, 219.1382, 303.2339, 401.3434, 411.3598, 429.3733 | 4.54 | C29H48O2 | 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol |
| 3 | 319.2276 | 12.73 | M-H | 135.1145, 179.1069, 257.2274, 301.2181, 319.2274 | 1.02 | C20H32O3 | 12(R)-HETE |
| 4 | 465.3039 | 33 | M-H | 79.9580, 96.9594, 465.3059 | 1.29 | C27H46O4S | Cholesterol sulfate |
VIP, variable importance in projection score in PLS-DA analyses. A VIP score of >1 is considered to be statistically significant.
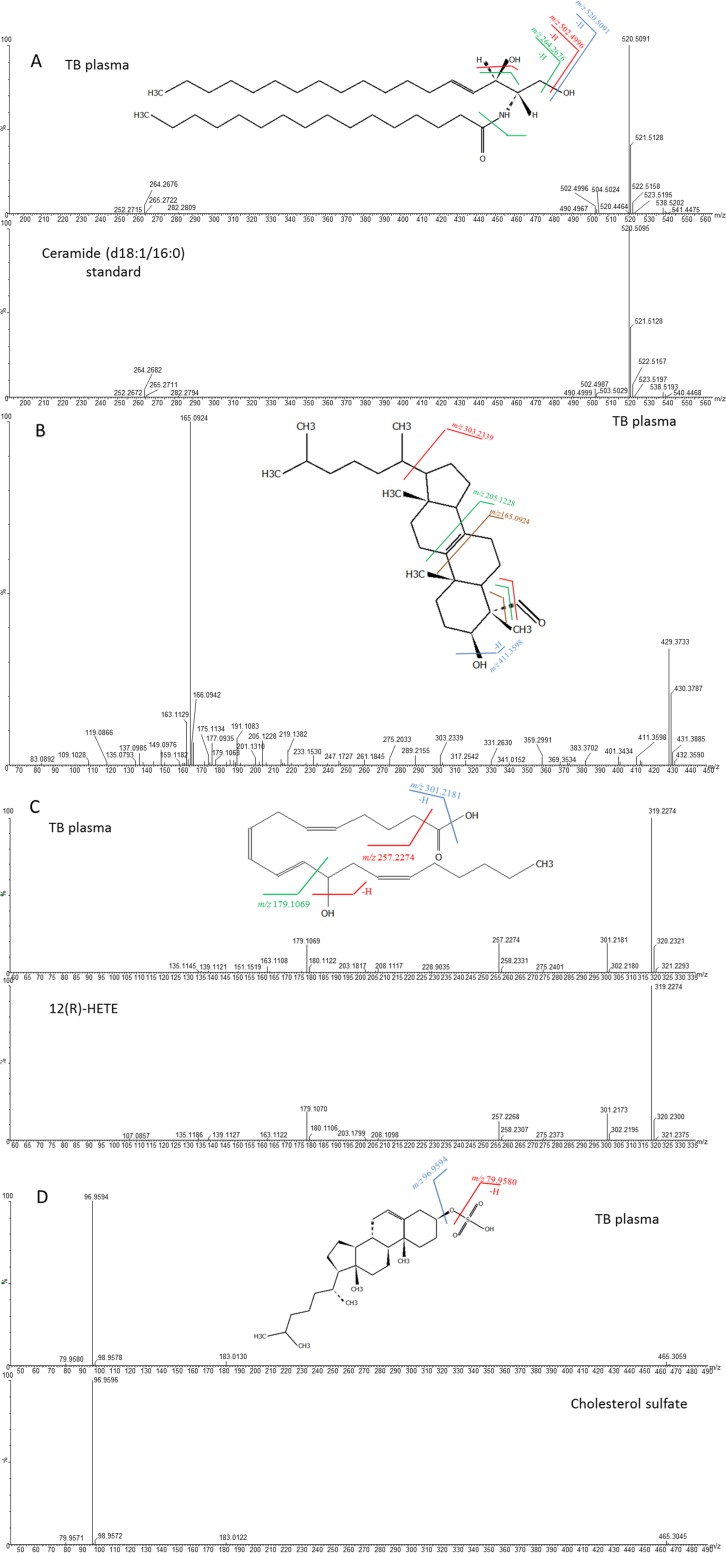
The first metabolite, with m/z 538.5190 and retention time (RT) of 33.02 min in positive mode, was identified as ceramide (d18:1/16:0) by the molecular formula and MS/MS fragmentation pattern corresponding to its structural characteristics in the Human Metabolome Database (HMDB), and this was confirmed by matching the RT and MS/MS spectrum with a commercially available authentic chemical standard of ceramide (d18:1/16:0) (Fig. 3A). The peaks at m/z 520.5091 and 502.4996 corresponded to fragments after the neutral loss of one water molecule and two water molecules from the parent ion, respectively. The peak at m/z 264.2676 referred to the fragment after a combined loss of two water molecules and a hexadecanoyl group.
FIG 3.
MS/MS mass spectra and predicted structures with expected fragmentation profiles of the four biomarkers in TB patient plasma and standards: ceramide (d18:1/16:0) (A), 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol (B), 12(R)-HETE (C), and cholesterol sulfate (D).
The second metabolite, with m/z 429.3733 at an RT of 32.22 min in positive mode, was identified as 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol by the molecular formula and MS/MS fragmentation pattern corresponding to its characteristics in HMDB (Fig. 3B). The peak at m/z 411.3598 was the fragment after the neutral loss of a water molecule at C-3, while the peak at m/z 303.2339 represented the fragment after cleavage of the bond between C-17 and C-20 and the loss of a 4-β-methyl group. The peak at m/z 205.1228 was the fragment resulting from the cleavage of bonds between C-9 and C-11 and between C-8 and C-14, while the peak at m/z 165.0924 was the fragment resulting from the cleavage of bonds between C-9 and C-10 and between C-7 and C-8 and the loss of 4-β-methyl group. However, an authentic chemical standard of 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol was not commercially available for comparison.
The third metabolite, with m/z 319.2274 at an RT of 12.73 min in negative mode, was identified as 12R-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid [12(R)-HETE] [12(R)-HETE] by matching the molecular formula and MS/MS spectra provided in METLIN and LipidMaps, and it was confirmed by matching the RT and MS/MS spectra using a commercially available authentic chemical standard of 12(R)-HETE (Fig. 3C). The peaks at m/z 301.2181 and 257.2274 were fragments after the neutral loss of one water molecule and a combined loss of a water molecule at C-12 and formate group, respectively. The peak at m/z 179.1069 was the fragment resulting from cleavage of the aliphatic group and the neutral loss of a water molecule at C-12.
The fourth metabolite, with m/z 465.3059 at an RT of 33.0 min in negative mode, was identified as cholesterol sulfate by matching the molecular formula in HMDB, and it was confirmed by matching RT and MS/MS spectra using commercially available authentic cholesterol sulfate (Fig. 3D). Two fragments, at m/z 79.9580 and 96.9594, referred to SO3− and HSO4− ions, respectively.
Diagnostic performance of metabolites.
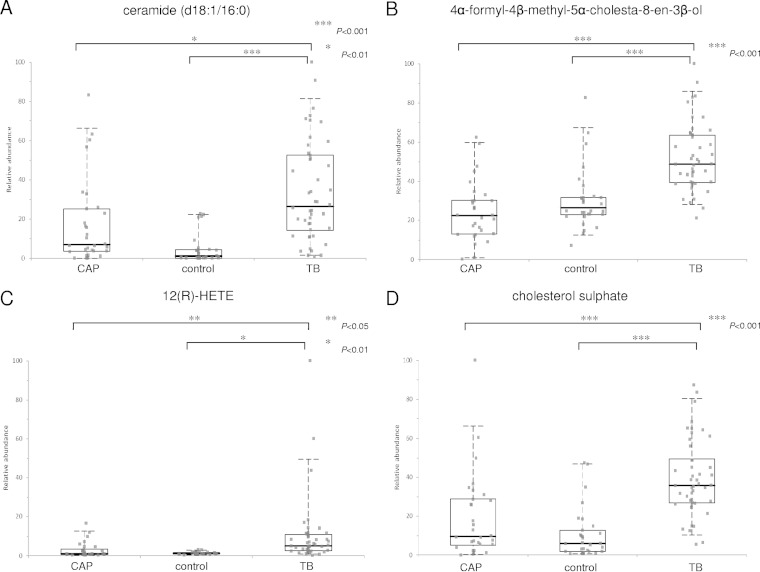
The AUC, sensitivity, and specificity for ROC curves calculated at optimal cutoffs for the four metabolites are summarized in Table 2. Box-and-whisker plots were generated for the three groups (Fig. 4). In a comparison of TB patients and controls, three metabolites, ceramide (d18:1/16:0), 12(R)-HETE, and cholesterol sulfate, demonstrated an AUC of >0.9. In a comparison of TB patients and controls, 12(R)-HETE showed the largest AUC, at 0.914, with 84.8% sensitivity and 90.0% specificity, but in a comparison of TB and CAP patients, it showed a relatively lower AUC, at 0.793, with 89.1% sensitivity and 63.3% specificity. Ceramide (d18:1/16:0) and cholesterol sulfate demonstrated AUC values of 0.912 and 0.905 in a comparison of TB patients and controls, respectively, but they also showed relatively lower AUC values of 0.717 and 0.802 in a comparison of TB and CAP patients, respectively. In contrast, 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol demonstrated better discrimination between TB and CAP patients, with a higher AUC of 0.894, 84.8% sensitivity, and 83.3% specificity compared to that between TB patients and controls.
TABLE 2.
AUC, sensitivity, and specificity for ROC curves calculated at optimal cutoff for ceramide (d18:1/16:0), 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol, 12(R)-HETE, and cholesterol sulfate
| Significant metabolite by comparison group | AUCa | 95% CIb | Sensitivity (%) | Specificity (%) | P valuec | Fold change |
|---|---|---|---|---|---|---|
| TB vs controls | ||||||
| Ceramide (d18:1/16:0) | 0.912 | 0.850–0.974 | 84.8 | 86.7 | 3.90E-08 | 26.15 |
| 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol | 0.856 | 0.758–0.853 | 89.1 | 80.0 | 5.55E-07 | 1.83 |
| 12(R)-HETE | 0.914 | 0.846–0.981 | 84.8 | 90.0 | 5.63E-03 | 4.19 |
| Cholesterol sulfate | 0.905 | 0.829–0.981 | 87.0 | 86.7 | 4.73E-10 | 6.09 |
| TB vs CAP | ||||||
| Ceramide (d18:1/16:0) | 0.717 | 0.595–0.840 | 71.7 | 66.7 | 5.24E-03 | 3.82 |
| 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol | 0.894 | 0.816–0.972 | 84.8 | 83.3 | 9.78E-10 | 2.16 |
| 12(R)-HETE | 0.793 | 0.683–0.904 | 89.1 | 63.3 | 2.32E-02 | 4.69 |
| Cholesterol sulfate | 0.802 | 0.693–0.911 | 80.4 | 70.0 | 6.46E-05 | 3.75 |
AUC, area under the receiving operating characteristic curve.
CI, confidence interval.
All P values were calculated using Student's t test.
FIG 4.
Box-and-whisker plots representing relative abundance of ceramide (d18:1/16:0) (A), 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol (B), 12(R)-HETE (C), and cholesterol sulfate (D) in the plasma from TB patients, CAP patients, and controls without active infections. The relative abundance of each metabolite in the plasma from TB patients was significantly higher than that of the other two groups using Student's t test. The horizontal line represents the median, the bottom and the top of the box represent the 25th and the 75th percentiles, and the whiskers represent the 5th and 95th percentiles.
Using the same optimal cutoffs for each metabolite, we calculated the sensitivities and specificities of different combinations of two or three of the four metabolites for diagnosing TB (Table 3). The specificities of combined metabolites were generally higher than those of individual metabolites, while the sensitivities of combined metabolites were generally lower than those of individual metabolites for differentiating TB patients and controls or CAP patients. Two combinations, 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol plus 12(R)-HETE, and 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol plus cholesterol sulfate, offered ≥70% sensitivity and ≥90% specificity for differentiating TB patients from controls and CAP patients.
TABLE 3.
Sensitivities and specificities of combinations of two or three metabolites for differentiation between TB and controls or CAP
| Metabolites | TB vs controls |
TB vs CAP |
||
|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |
| Ceramide (d18:1/16:0) + 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol | 76.1 | 90.0 | 65.2 | 90.0 |
| Ceramide (d18:1/16:0) + 12(R)-HETE | 69.6 | 100.0 | 60.9 | 76.7 |
| Ceramide (d18:1/16:0) + cholesterol sulfate | 84.8 | 86.7 | 67.4 | 73.3 |
| 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol + 12(R)-HETE | 73.9a | 100.0 | 73.9 | 96.7 |
| 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol + cholesterol sulfate | 78.3 | 90.0 | 71.7 | 96.7 |
| 12(R)-HETE + cholesterol sulfate | 71.7 | 100.0 | 67.4 | 80.0 |
| Ceramide (d18:1/16:0) + 12(R)-HETE + cholesterol sulfate | 69.6 | 100.0 | 56.5 | 80.0 |
| 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol + 12(R)-HETE + cholesterol sulfate | 63.0 | 100.0 | 60.9 | 100.0 |
| Ceramide (d18:1/16:0) + 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol + 12(R)-HETE | 60.1 | 100.0 | 54.4 | 96.7 |
| Ceramide (d18:1/16:0) + 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol + cholesterol sulfate | 76.1 | 90.0 | 63.0 | 96.7 |
Combinations with sensitivity of ≥70% and specificity of ≥90% for both comparisons (TB versus controls and TB versus CAP) are in bold.
DISCUSSION
Using a metabolomics approach, we identified four novel biomarkers with significantly higher levels in plasma from TB patients than those in CAP patients and controls. Of the four metabolites, 12(R)-HETE showed the largest AUC, at 0.914, with 84.8% sensitivity, 90.0% specificity, and a fold change of 4.19 compared to controls. However, compared to CAP patients, the AUC dropped to 0.793, with only 63.3% specificity. The diagnostic performance of ceramide (d18:1/16:0) was also promising, with an AUC of 0.912, 84.8% sensitivity, 86.7% specificity, and a fold change of 26.15 compared to controls. However, the AUC dropped to 0.717, with 71.7% sensitivity, 66.7% specificity, and a fold change of 3.82 compared to CAP patients. Similarly, cholesterol sulfate showed a relatively high AUC of 0.905, 87.0% sensitivity, 86.7% specificity, and a fold change of 6.09 compared to controls. However, the AUC dropped to 0.802, with 80.4% sensitivity, 70% specificity, and a fold change of 3.75 compared to CAP patients. In contrast, 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol showed good diagnostic performance in distinguishing TB patients from both CAP patients and controls, with an AUC of >0.85 and >80% sensitivity/specificity, although the fold changes were only 2.16 and 1.83, respectively. Therefore, 12(R)-HETE, ceramide (d18:1/16:0), and cholesterol sulfate should offer better diagnostic performance than that of 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol in differentiating between TB patients and controls. In particular, the >20-fold change of ceramide (d18:1/16:0) suggests that it may be a promising candidate for use in the diagnosis of TB. However, these biomarkers may also be elevated in CAP patients, although not to levels as high as those in TB patients. On the other hand, 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol may offer better discrimination between TB and CAP patients. When 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol is combined with 12(R)-HETE or cholesterol sulfate, the specificities were ≥90% for differentiating TB patients from both controls and CAP patients, although the sensitivities were <80%. If these biomarkers or their combinations are adopted for the diagnosis of TB, the results should be interpreted and correlated with clinical findings. For example, in patients with clinical features compatible with TB while CAP is considered unlikely, elevated plasma ceramide (d18:1/16:0) levels are highly suggestive of TB. However, in situations in which CAP cannot be excluded, 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol alone or combined with 12(R)-HETE or cholesterol sulfate may be a more specific indicator for empirical treatment while waiting for culture results. Although the present biomarkers exhibit only modest discriminatory power between TB and CAP patients or controls, they may represent promising biomarkers to assist in the diagnosis of TB, especially in disseminated infections for which culture and PCR are often negative. One limitation of these biomarkers is that they do not offer value for drug resistance profiles, unlike traditional culture and genotypic diagnostic methods. Nevertheless, further studies involving larger patient sets should be performed to better assess their diagnostic accuracy and potential prognostic value.
The present results support that ceramides are involved in the host response to pathogens, including TB. Ceramides are produced from the hydrolysis of sphingomyelin or are synthesized from serine and palmitate (34). A ceramide-rich membrane has diverse functions during bacterial infections, including phagocytosis, apoptosis, phagosome trafficking, and macrophage maturation (35–41). However, the roles of different ceramides during infections caused by different pathogens are poorly understood. We showed here that the level of ceramide (d18:1/16:0) in plasma samples from TB and CAP patients was higher than that in controls. Furthermore, the level of ceramide (d18:1/16:0) in TB patients was higher than that in CAP patients, which may reflect the more robust innate immune response in TB. Interestingly, in a recent metabolomic profiling study (42), ceramide (d18:1/16:0) has also been found in the pleural fluid from TB patients at higher levels than those in lung cancer patients. Further studies are required to determine if ceramide (d18:1/16:0) is a promising biomarker for the diagnosis of TB, which may be applied to plasma and pleural fluid samples.
The elevated levels of plasma 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol in TB patients may suggest enhanced cholesterol biosynthesis during TB infections. 4α-Formyl-4β-methyl-5α-cholesta-8-en-3β-ol is an intermediate during cholesterol biosynthesis II. Cholesterol plays key roles in the pathogenesis of TB, such as phagocytosis and macrophage and phagosome maturation (43–45). However, increased plasma 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol levels have not been reported in TB or other infections. It would be interesting to investigate if the intermediate, 4α-formyl-4β-methyl-5α-cholesta-8-en-3β-ol, during cholesterol biosynthesis activation in TB serves a specific function in pathogenesis or the innate immune response.
The elevated levels of plasma 12(R)-HETE may reflect induced arachidonic acid metabolism during TB. 12(R)-HETE is an arachidonic acid metabolite produced by arachidonate 12-lipoxygenase (12-LOX), which is expressed in human macrophages and induced by interleukin 4 (IL-4) (46). Increased IL-4 production by human CD8+ T cells and bronchoalveolar cells has been observed in TB patients (47, 48). The present study represents the first to report increased plasma 12(R)-HETE levels in TB patients, which may be related to IL-4 induction of 12-LOX in macrophages or other cells. In M. tuberculosis-infected mice, high serum levels of lipoxin A4, another arachidonic acid metabolite produced by 5-lipoxygenase (5-LOX), have been observed (49). Moreover, mice deficient in 5-LOX were more resistant to TB and harbored fewer bacteria in the lungs (49). Since arachidonic acid has been shown to enhance mycobacteria killing via activation of NF-κB (50), further studies are warranted to elucidate the role of arachidonic acid metabolism, especially 12(R)-HETE or 12-LOX, in host defense against M. tuberculosis.
Cholesterol sulfate, a component of low-density lipoproteins (51), may represent a less specific biomarker than the other three metabolites for the diagnosis of TB, since plasma cholesterol sulfate may be increased in other pathological conditions, such as cirrhosis of the liver, hypercholesterolemia, and hypothyroidism (52–54). During skin invasion by pathogens, cholesterol sulfate has been shown to facilitate keratinocyte differentiation (55). Our findings of increased plasma cholesterol sulfate levels in TB patients may also suggest a role in the host response to M. tuberculosis. However, given that cholesterol sulfate may be elevated in other disease conditions, it may be a less promising candidate as a diagnostic biomarker for TB.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wing-Keung Ma and members of the Department of Microbiology and Department of Pathology, Queen Mary Hospital, for facilitation and assistance in sample and data collection. We thank Andrea L. Wu for her assistance in data processing.
This work is partly supported by the HKSAR Research Fund for the Control of Infectious Diseases (commissioned study HK-09-01-10) of the Food and Health Bureau; the University Development Fund and Strategic Research Fund, The University of Hong Kong; and donations from Eunice Lam, Michael Tong, and Arthur Lui on emerging infectious disease research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01568-15.
REFERENCES
- 1.Lai CC, Liu WL, Tan CK, Huang YC, Chung KP, Lee MR, Hsueh PR. 2011. Differences in drug resistance profiles of Mycobacterium tuberculosis isolates causing pulmonary and extrapulmonary tuberculosis in a medical centre in Taiwan, 2000–2010. Int J Antimicrob Agents 38:125–129. doi: 10.1016/j.ijantimicag.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Y, Dou X, Zhang W, Liu H, Zhao X, Wang H, Lian L, Yu Q, Zhang J, Li G, Chen C, Wan K. 2013. Genetic diversity of antigens Rv2945c and Rv0309 in Mycobacterium tuberculosis strains may reflect ongoing immune evasion. FEMS Microbiol Lett 347:77–82. doi: 10.1111/1574-6968.12222. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Ding H, Guo FB, Huang J. 2010. Prediction of subcellular location of mycobacterial protein using feature selection techniques. Mol Divers 14:667–671. doi: 10.1007/s11030-009-9205-1. [DOI] [PubMed] [Google Scholar]
- 4.Getahun H, Harrington M, O'Brien R, Nunn P. 2007. Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: informing urgent policy changes. Lancet 369:2042–2049. doi: 10.1016/S0140-6736(07)60284-0. [DOI] [PubMed] [Google Scholar]
- 5.Marais BJ, Pai M. 2007. New approaches and emerging technologies in the diagnosis of childhood tuberculosis. Paediatr Respir Rev 8:124–133. doi: 10.1016/j.prrv.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Cheng VC, Yam WC, Hung IF, Woo PC, Lau SK, Tang BS, Yuen KY. 2004. Clinical evaluation of the polymerase chain reaction for the rapid diagnosis of tuberculosis. J Clin Pathol 57:281–285. doi: 10.1136/jcp.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore DF, Curry JI. 1995. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by Amplicor PCR. J Clin Microbiol 33:2686–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawn SD, Kerkhoff AD, Vogt M, Wood R. 2012. Diagnostic accuracy of a low-cost, urine antigen, point-of-care screening assay for HIV-associated pulmonary tuberculosis before antiretroviral therapy: a descriptive study. Lancet Infect Dis 12:201–209. doi: 10.1016/S1473-3099(11)70251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mdivani N, Li H, Akhalaia M, Gegia M, Goginashvili L, Kernodle DS, Khechinashvili G, Tang YW. 2009. Monitoring therapeutic efficacy by real-time detection of Mycobacterium tuberculosis mRNA in sputum. Clin Chem 55:1694–1700. doi: 10.1373/clinchem.2009.124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohamed KH, Mobasher AA, Yousef AR, Salah A, El-Naggar IZ, Ghoneim AH, Light RW. 2001. BAL neopterin: a novel marker for cell-mediated immunity in patients with pulmonary tuberculosis and lung cancer. Chest 119:776–780. doi: 10.1378/chest.119.3.776. [DOI] [PubMed] [Google Scholar]
- 11.Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol 48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-López EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivier I, Loots DT. 2011. An overview of tuberculosis treatments and diagnostics. What role could metabolomics play? J Cell Tissue Res 11:2655–2671. [Google Scholar]
- 14.Olivier I, Loots DT. 2012. A metabolomics approach to characterise and identify various Mycobacterium species. J Microbiol Methods 88:419–426. doi: 10.1016/j.mimet.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Meissner-Roloff RJ, Koekemoer G, Warren RM, Loots DT. 2012. A metabolomics investigation of a hyper- and hypo-virulent phenotype of Beijing lineage M. tuberculosis. Metabolomics 8:1194–1203. doi: 10.1007/s11306-012-0424-6. [DOI] [Google Scholar]
- 16.Schoeman JC, Loots DT. 2011. Improved disease characterisation and diagnostics using metabolomics: a review. J Cell Tissue Res 11:2673–2683. [Google Scholar]
- 17.Tam EW, Chen JH, Lau EC, Ngan AH, Fung KS, Lee KC, Lam CW, Yuen KY, Lau SK, Woo PC. 2014. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: characterization by internal transcribed spacer, β-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 52:1153–1160. doi: 10.1128/JCM.03258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.To KK, Fung AM, Teng JL, Curreem SO, Lee KC, Yuen KY, Lam CW, Lau SK, Woo PCY. 2013. Characterization of a Tsukamurella pseudo-outbreak by phenotypic tests, 16S rRNA sequencing, pulsed-field gel electrophoresis, and metabolic footprinting. J Clin Microbiol 51:334–338. doi: 10.1128/JCM.02845-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo PC, Lam CW, Tam EW, Leung CK, Wong SS, Lau SK, Yuen KY. 2012. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Neglect Trop Dis 6:e1871. doi: 10.1371/journal.pntd.0001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. 2010. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol 17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Dang NA, Kuijper S, Walters E, Claassens M, van Soolingen D, Vivo-Truyols G, Janssen HG, Kolk AH. 2013. Validation of biomarkers for distinguishing Mycobacterium tuberculosis from non-tuberculous mycobacteria using gas chromatography-mass spectrometry and chemometrics. PLoS One 8:e76263. doi: 10.1371/journal.pone.0076263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, Cheng TY, Annand JW, Kim K, Shamputa IC, McConnell MJ, Debono CA, Behar SM, Minnaard AJ, Murray M, Barry CE III, Matsunaga I, Moody DB. 2011. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol 18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madigan CA, Cheng TY, Layre E, Young DC, McConnell MJ, Debono CA, Murry JP, Wei JR, Barry CE III, Rodriguez GM, Matsunaga I, Rubin EJ, Moody DB. 2012. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 109:1257–1262. doi: 10.1073/pnas.1109958109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layre E, Lee HJ, Young DC, Martinot AJ, Buter J, Minnaard AJ, Annand JW, Fortune SM, Snider BB, Matsunaga I, Rubin EJ, Alber T, Moody DB. 2014. Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc Natl Acad Sci U S A 111:2978–2983. doi: 10.1073/pnas.1315883111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau SKP, Lam CW, Curreem SOT, Lee KC, Lau CCY, Chow WN, Ngan AHY, To KKW, Chan JFW, Hung IFN, Yam WC, Yuen KY, Woo PCY. 2015. Identification of specific metabolites in culture supernatant of Mycobacterium tuberculosis using metabolomics: exploration of potential biomarkers. Emerg Microbes Infect 4:e6. doi: 10.1038/emi.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banday KM, Pasikanti KK, Chan EC, Singla R, Rao KV, Chauhan VS, Nanda RK. 2011. Use of urine volatile organic compounds to discriminate tuberculosis patients from healthy subjects. Anal Chem 83:5526–5534. doi: 10.1021/ac200265g. [DOI] [PubMed] [Google Scholar]
- 27.Zhou A, Ni J, Xu Z, Wang Y, Lu S, Sha W, Karakousis PC, Yao YF. 2013. Application of 1H NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res 12:4642–4649. doi: 10.1021/pr4007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frediani JK, Jones DP, Tukvadze N, Uppal K, Sanikidze E, Kipiani M, Tran VT, Hebbar G, Walker DI, Kempker RR, Kurani SS, Colas RA, Dalli J, Tangpricha V, Serhan CN, Blumberg HM, Ziegler TR. 2014. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS One 9:e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahapatra S, Hess AM, Johnson JL, Eisenach KD, DeGroote MA, Gitta P, Joloba ML, Kaplan G, Walzl G, Boom WH, Belisle JT. 2014. A metabolic biosignature of early response to anti-tuberculosis treatment. BMC Infect Dis 14:53. doi: 10.1186/1471-2334-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basanta M, Jarvis RM, Xu Y, Blackburn G, Tal-Singer R, Woodcock A, Singh D, Goodacre R, Thomas CL, Fowler SJ. 2010. Non-invasive metabolomics analysis of breath using differential mobility spectrometry in patients with chronic obstructive pulmonary disease and healthy smokers. Analyst 135:315–320. doi: 10.1039/b916374c. [DOI] [PubMed] [Google Scholar]
- 31.Zhang A, Sun H, Han Y, Yan G, Wang X. 2013. Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS One 8:e64381. doi: 10.1371/journal.pone.0064381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H, Zhang A, Yan G, Piao C, Li W, Sun C, Wu X, Li X, Chen Y, Wang X. 2013. Metabolomic analysis of key regulatory metabolites in hepatitis C virus-infected tree shrews. Mol Cell Proteomics 12:710–719. doi: 10.1074/mcp.M112.019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Broadhurst DI, Wilson M, Wishart DS. 2013. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics 9:280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tserng KY, Griffin RL. 2004. Ceramide metabolite, not intact ceramide molecule, may be responsible for cellular toxicity. Biochem J 380:715–722. doi: 10.1042/bj20031733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grassmé H, Gulbins E, Brenner B, Ferlinz K, Sandhoff K, Harzer K, Lang F, Meyer TF. 1997. Acidic sphingomyelinase mediates entry of N. gonorrhoeae into nonphagocytic cells. Cell 91:605–615. doi: 10.1016/S0092-8674(00)80448-1. [DOI] [PubMed] [Google Scholar]
- 36.Esen M, Schreiner B, Jendrossek V, Lang F, Fassbender K, Grassmé H, Gulbins E. 2001. Mechanisms of Staphylococcus aureus induced apoptosis of human endothelial cells. Apoptosis 6:431–439. doi: 10.1023/A:1012445925628. [DOI] [PubMed] [Google Scholar]
- 37.Grassmé H, Jendrossek V, Riehle A, von Kürthy G, Berger J, Schwarz H, Weller M, Kolesnick R, Gulbins E. 2003. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat Med 9:322–330. doi: 10.1038/nm823. [DOI] [PubMed] [Google Scholar]
- 38.Cannon CL, Kowalski MP, Stopak KS, Pier GB. 2003. Pseudomonas aeruginosa-induced apoptosis is defective in respiratory epithelial cells expressing mutant cystic fibrosis transmembrane conductance regulator. Am J Respir Cell Mol Biol 29:188–197. doi: 10.1165/rcmb.4898. [DOI] [PubMed] [Google Scholar]
- 39.Anes E, Kühnel MP, Bos E, Moniz-Pereira J, Habermann A, Griffiths G. 2003. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat Cell Biol 5:793–802. doi: 10.1038/ncb1036. [DOI] [PubMed] [Google Scholar]
- 40.Okino N, Ichinose S, Omori A, Imayama S, Nakamura T, Ito M. 1999. Molecular cloning, sequencing, and expression of the gene encoding alkaline ceramidase from Pseudomonas aeruginosa. Cloning of a ceramidase homologue from Mycobacterium tuberculosis. J Biol Chem 274:36616–36622. [DOI] [PubMed] [Google Scholar]
- 41.Morell P, Radin NS. 1970. Specificity in ceramide biosynthesis from long chain bases and various fatty acyl coenzyme A's by brain microsomes. J Biol Chem 245:342–350. [PubMed] [Google Scholar]
- 42.Lam CW, Law CY. 2014. Untargeted mass spectrometry-based metabolomic profiling of pleural effusions: fatty acids as novel cancer biomarkers for malignant pleural effusions. J Proteome Res 13:4040–4046. doi: 10.1021/pr5003774. [DOI] [PubMed] [Google Scholar]
- 43.Vilhardt F, van Deurs B. 2004. The phagocyte NADPH oxidase depends on cholesterol-enriched membrane microdomains for assembly. EMBO J 23:739–748. doi: 10.1038/sj.emboj.7600066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clemens DL, Lee BY, Horwitz MA. 2000. Mycobacterium tuberculosis and Legionella pneumophila phagosomes exhibit arrested maturation despite acquisition of Rab7. Infect Immun 68:5154–5166. doi: 10.1128/IAI.68.9.5154-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huynh KK, Gershenzon E, Grinstein S. 2008. Cholesterol accumulation by macrophages impairs phagosome maturation. J Biol Chem 283:35745–35755. doi: 10.1074/jbc.M806232200. [DOI] [PubMed] [Google Scholar]
- 46.Wuest SJ, Crucet M, Gemperle C, Loretz C, Hersberger M. 2012. Expression and regulation of 12/15-lipoxygenases in human primary macrophages. Atherosclerosis 225:121–127. doi: 10.1016/j.atherosclerosis.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 47.Smith SM, Klein MR, Malin AS, Sillah J, McAdam KP, Dockrell HM. 2002. Decreased IFN-gamma and increased IL-4 production by human CD8(+) T cells in response to Mycobacterium tuberculosis in tuberculosis patients. Tuberculosis (Edinb) 82:7–13. doi: 10.1054/tube.2001.0317. [DOI] [PubMed] [Google Scholar]
- 48.Nolan A, Fajardo E, Huie ML, Condos R, Pooran A, Dawson R, Dheda K, Bateman E, Rom WN, Weiden MD. 2013. Increased production of IL-4 and IL-12p40 from bronchoalveolar lavage cells are biomarkers of Mycobacterium tuberculosis in the sputum. PLoS One 8:e59461. doi: 10.1371/journal.pone.0059461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bafica A, Scanga CA, Serhan C, Machado F, White S, Sher A, Aliberti J. 2005. Host control of Mycobacterium tuberculosis is regulated by 5-lipoxygenase-dependent lipoxin production. J Clin Invest 115:1601–1606. doi: 10.1172/JCI23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutierrez MG, Gonzalez AP, Anes E, Griffiths G. 2009. Role of lipids in killing mycobacteria by macrophages: evidence for NF-kappaB-dependent and -independent killing induced by different lipids. Cell Microbiol 11:406–420. doi: 10.1111/j.1462-5822.2008.01263.x. [DOI] [PubMed] [Google Scholar]
- 51.Strott CA, Higashi Y. 2003. Cholesterol sulfate in human physiology: what's it all about? J Lipid Res 44:1268–1278. doi: 10.1194/jlr.R300005-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Huang YS, Eid K, Davignon J. 1981. Cholesteryl sulfate: measurement with β-sitosteryl sulfate as an internal standard. Can J Biochem 59:602–605. doi: 10.1139/o81-083. [DOI] [PubMed] [Google Scholar]
- 53.Tamasawa N, Tamasawa A, Takebe K. 1993. Higher levels of plasma cholesterol sulfate in patients with liver cirrhosis and hypercholesterolemia. Lipids 28:833–836. doi: 10.1007/BF02536238. [DOI] [PubMed] [Google Scholar]
- 54.van Doormaal JJ, Muskiet FA, Jansen G, Wolthers BG, Sluiter WJ, Doorenbos H. 1986. Increase of plasma and red cell cholesterol sulfate levels in induced hypothyroidism in man. Clin Chim Acta 155:195–200. doi: 10.1016/0009-8981(86)90238-X. [DOI] [PubMed] [Google Scholar]
- 55.Hanley K, Wood L, Ng DC, He SS, Lau P, Moser A, Elias PM, Bikle DD, Williams ML, Feingold KR. 2001. Cholesterol sulfate stimulates involucrin transcription in keratinocytes by increasing Fra-1, Fra-2, and Jun D. J Lipid Res 42:390–398. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.