Abstract
We consider dependent functional data that are correlated because of a longitudinal-based design: each subject is observed at repeated times and at each time a functional observation (curve) is recorded. We propose a novel parsimonious modeling framework for repeatedly observed functional observations that allows to extract low dimensional features. The proposed methodology accounts for the longitudinal design, is designed to study the dynamic behavior of the underlying process, allows prediction of full future trajectory, and is computationally fast. Theoretical properties of this framework are studied and numerical investigations confirm excellent behavior in finite samples. The proposed method is motivated by and applied to a diffusion tensor imaging study of multiple sclerosis.
Keywords: Dependent functional data, Diffusion Tensor Imaging, Functional principal component analysis, Longitudinal design, Multiple Sclerosis
1. Introduction
Longitudinal functional data consist of functional observations (such as profiles or images) observed at several times for each subject of many. Examples of such data include the Baltimore Longitudinal Study of Aging (BLSA), where daily physical activity count profiles are observed for each subject at several consecutive days (Goldsmith et al., 2014; Xiao et al., 2015) and the longitudinal diffusion tensor imaging (DTI) study, where modality profiles along well-identified tracts are observed for each multiple sclerosis (MS) patient at several hospital visits (Greven et al., 2010). As a result of an increasing number of such applications, longitudinal functional data analysis has received much attention recently; see for example Morris et al. (2003); Morris & Carroll (2006); Baladandayuthapani et al. (2008); Di et al. (2009); Greven et al. (2010); Staicu et al. (2010); Li & Guan (2014).
Our motivation is the longitudinal DTI study, where the objective is to investigate the evolution of the MS disease as measured by the dynamics of a common DTI modality profile - fractional anisotropy (FA) - along the corpus callosum (CCA) of the brain. Every MS subject in the study is observed over possibly multiple hospital visits and at each visit the subject's brain is imaged using DTI. In this paper we consider summaries of FA at 93 equally spaced locations along the brain's CCA, which we refer to as CCA-FA profile. The change over time in the CCA-FA profiles is informative of the progression of the MS disease, and thus a model that accounts for all the dependence sources in the data has the potential to be a very useful tool in practice. We propose a modeling framework that captures the process dynamics over time and provides prediction of a full CCA-FA trajectory at a future visit.
Existing literature in longitudinal functional data can be separated into two categories, based on whether or not it accounts for the actual time Tij at which the profile Yij(·) is observed; here i indexes the subjects and j indexes the repeated measures of the subject. Moreover, most methods that incorporate the time Tij focus on modeling the process dynamics (Greven et al., 2010) and only few can do prediction of a future full trajectory. Chen & Müller (2012) considered the latter issue and introduced an interesting perspective, but their method is very computationally expensive and its application in practice is limited as a result. We propose a novel parsimonious modeling framework to study the process dynamics and prediction of future full trajectory in a computationally feasible manner.
In this paper we focus on the case where the sampling design of Tij's is sparse (hence sparse longitudinal design) and the subject profiles are observed at fine grids (hence dense functional design). We propose to model Yij(·) as:
| (1) |
where and are closed compact sets, μ(·, Tij) is an unknown smooth mean response corresponding to Tij, Xi(·, Tij) is a smooth random deviation from the mean at Tij, and εij(·) is a residual process with zero-mean and unknown covariance function to be described later. The bivariate processes Xi(·, ·)'s are independent and identically distributed (iid), the error processes εij's are iid and furthermore are independent of Xi's. For identifiability we require that Xi comprises solely the random deviation that is specific to the subject; the repeated time-specific deviation is included in εij. Here {ϕk(·)}k is an orthogonal basis in and ξik(Tij)'s are the corresponding basis coefficients that have zero-mean, are uncorrelated over i, but correlated over j. We assume that the set of visit times of all subjects, {Tij : i, j}, is dense in . Full model assumptions are given in Section 2.
The class of model (1) is rich and includes many existent models, as we illustrate now. (i) If ξik(Tij) = ζ0,ik + Tijζ1,ik for appropriately defined random terms ζ0,ik and ζ1,ik, model (1) can be represented as in Greven et al. (2010). (ii) If cov(ξik(T), ξik(T′)) = λkρk(|T – T′|; ν) for some unknown variance λk, known correlation function ρk(·; ν) with unknown parameter ν, and n = 1, model (1) resembles to Gromenko et al. (2012) and Gromenko & Kokoszka (2013) for spatially indexed functional data. (iii) If ξik(Tij) = with orthogonal basis functions ψikl(T)'s and the corresponding coefficients ζikl's, then model (1) is similar to Chen & Müller (2012) who used time-varying basis functions ϕk(·|T) instead of our proposed ϕk(·) in model (1) and assumed a white noise residual process εij.
The use of time-invariant orthogonal basis functions is one key difference between the proposed framework and Chen & Müller (2012); another important difference is the flexible error structure that our approach accommodates. The key difference leads to several major advantages of the proposed method. First, by using a time-invariant basis functions, the basis coefficients, ξik(Tij)'s extract the low dimensional features of these massive data. The longitudinal dynamics is emphasized only through the time-varying coefficients ξik(Tij)'s of (1) and, thus, this perspective makes the study of the process dynamics easier to understand. Second, our approach involves at most two dimensional smoothing and as a result is computationally very fast; in contrast, the time-varying basis functions {ϕk(·|T)}k at each T, require three dimensional smoothing which is not only complex but also computationally intensive and slow.
Nevertheless, selecting the time-invariant basis is nontrivial. One option is to use a pre-specified basis; Zhou et al. (2008) considered this approach in modeling paired of sparse functional data. Another option is to use data-driven basis functions, such as eigenbasis of some covariance. The challenge is: what covariance to use ? We take the latter direction and propose to determine {ϕk(·)}k using an appropriate marginal covariance. In this regard, let c((s, T), (s′, T′)) be the covariance function of Xi(s, T) and g(T) be the density of Tij's. Define Σ(s, s′) = for s, s′ ∈ : we show that this bivariate function is a proper covariance function (Horváth & Kokoszka, 2012). Section 2 shows that the proposed basis {ϕk(·)}k has optimal properties with respect to some appropriately defined criterion. From this view point, the model representation (1) is optimal. The idea of using the eigenbasis of the pooled covariance can be related to Jiang & Wang (2010) and Pomann et al. (2013), who considered independent functional data.
The rest of paper is organized as follows. Section 2 introduces the proposed modeling framework. Section 3 describes the estimation methods and implementation. The methods are studied theoretically in Section 4 and then numerically in Section 5. Section 6 discusses the application to the tractography DTI data.
2. Modeling longitudinal functional data
Let [{Tij, Yij(sr) : r = 1, . . . , R} : j = 1, . . . , mi,] be the observed data for the ith subject, where Yij(·) is the jth profile at random time Tij for subject i, and each profile is observed at the fine grid of points {s1, . . . , sR}. For convenience we use the generic index s instead of sr. The number of ‘profiles’ per subject, mi is relatively small to moderate and the set of time points of all subjects, {Tij : for all i, j}, is dense in . Without loss of generality, we set . We model the response Yij(·) using (1), where we assume that εij(s) is the sum of independent components εij(s) = ε1,ij(s) + ε2,ij(s). Here ε1,ij(·) is a random square integrable function which has smooth covariance function Γ(s, s′) = cov{ε1,ij(s),ε1,ij(s′)} and ε2,ij(s) is white noise with covariance cov{ε2,ij(s), ε2,ij(s′)} = σ2 if s = s′ and 0 otherwise.
Let c((s, T), (s′, T′)) = E[Xi(s, T)Xi(s′, T′)] be the covariance function of the process Xi(·,·) and let Σ(s, s′) = ∫ c((s, T), (s′, T))g(T)dT, where g(·) is the sampling density of Tij. In Section 4 we show that Σ(s, s′) is a proper covariance function (Horváth & Kokoszka, 2012); due to its definition we call Σ as the marginal covariance function induced by Xi. The unpublished work Chen et al. (2015) independently considered a similar marginal covariance in a related setting. Denote by Wi(s, Tij) = Xi(s, Tij) + ε1,ij(s); Wi is a bivariate process defined on [0, 1]2 and its induced marginal covariance is Ξ(s, s′) = Σ(s, s′) + Γ(s, s′). Let {ϕk(s), λk}k be the eigencomponents of Ξ(s, s′), where {ϕk(·) : k} forms an orthogonal basis in L2[0, 1] and λ1 ≥ λ2 ≥ . . . ≥ 0. Using arguments similar to the standard functional principal component analysis (FPCA), the eigenbasis functions {ϕk(·) : k = 1, . . . , K} are optimal in the sense that they minimize the following weighted mean square error: MSE(θ1(·), . . . , θK(·)) = , where is the usual inner product in L2[0, 1].
Using the orthogonal basis in L2[0, 1] {ϕk(·)}k, we can represent the square integrable smooth process Wi(·, T) as Wi(s, Tij) = , where ξW,ijk = ∫Wi(s, Tij)ϕk(s)ds = ξik(Tij) + eijk, and ξW,ijk are not necessarily uncorrelated over k. Here ξik(Tij) = ∫Xi(s, Tij)ϕk(s)ds and eijk = ∫ ε1,ij(s)ϕk(s)ds are specified by the definition of Wi; for fixed k these terms are mutually independent due to the independence of the processes Xi and ε1,ij. For each k, one can easily show that, ξik(·) is a smooth zero-mean random process in L2[0, 1] and is iid over i. Furthermore eijk are zero-mean iid random variables over i, j; denote by their finite variance.
One way to model the dependence of the coefficients, ξik(Tij)'s, is by using common techniques in longitudinal data analysis; for example by assuming a parametric covariance structure. As we discussed in Section 1, this leads to models similar to Greven et al. (2010); Gromenko et al. (2012); Gromenko & Kokoszka (2013). We consider this approach in the analysis of the DTI data, Section 6. Another approach is to assume a nonparametric covariance structure and employ a common functional data analysis technique. We detail the latter approach in this section.
For each k ≥ 1 denote by Gk(T, T ′) = cov{,ik(T), ,ik(T′)} the smooth covariance function in [0, 1] × [0, 1]. Mercer's theorem provides the following convenient spectral decomposition , where ηk1 ≥ ηk2 ≥ . . . ≥ 0 and {ψkl(·)}l≥1 is an orthogonal basis in L2[0, 1]. Using the Karhunen-Loève (KL) expansion, we represent ξik(·) as: ξik(Tij) = , where ζikl = ∫ ξik(T)ψkl(T)dT, have zero-mean, variance equal to ηkl, and are uncorrelated over l. By collecting all the components, we represent the model (1) as Yij(s) = μ(s, Tij) + , for . In practice we truncate this expansion. Lek K and L1,...,LK such that Yij(s) is well approximated by the following truncated model based on the leading K and respective basis functions
| (2) |
where . The truncated model (2) gives a parsimonious representation of the longitudinal functional data. It allows to study its dependence through two sets of eigenfunctions: one dependent solely on s and one solely on Tij. This approach involves two main challenges: first, determining consistent estimator of the marginal covariance and second determining consistent estimators of the time-varying coefficients ξik(·).
3. Estimation of model components
We discuss estimation of all model components. The mean estimation is carried out using existing methods (Chen & Müller, 2012; Scheipl et al., 2014); here we briefly describe it for completeness. Our focus and novelty is the estimation of the marginal covariance function and of the eigenfunctions ϕk(·)'s (see Section 3.2), as well as the the estimation of the time-varying basis coefficients ξik(·)'s (see Section 3.3). Prediction of Yi(s, T) is detailed in Section 3.4.
3.1. Step 1: Mean function
As in Scheipl et al. (2014) we estimate the mean function, μ(s, T), using bivariate smoothing via bivariate tensor product splines (Wood, 2006) of the pooled data Yijr = Yij(sr)'s. Consider two univariate B-spline bases, {Bs,1(s), . . . , Bs,ds(s)} and {BT,1(T), . . . , BT,dT(T)}, where ds and dT are their respective dimensions. The mean surface is represented as a linear combination of a tensor product of the two univariate B-spline bases μ(s, T ) = Bs,q1(s)BT,q2(T)βq1q2 = B(s, T)Tβ, where B(s, T) is the known dsdT-dimensional vector of Bs,q1(s)BT,q2(T)'s, and β is the vector of unknown parameters, βq1q2's. The bases dimensions, ds and dT, are set to be sufficiently large to accommodate the complexity of the true mean function, and the roughness of the function is controlled through the size of the curvature in each direction separately, i.e. in direction s, and in T. The penalized criterion to be minimized is ,, where λs and λT are smoothing parameters that control the trade-off between the smoothness of the fit and the goodness of fit. The smoothing parameters can be selected by the restricted maximum likelihood (REML) or generalized cross-validation (GCV). The estimated mean function is = . This method is a very popular smoothing technique of bivariate data.
Other available bivariate smoothers can be used to estimate the mean μ(s, T): for example, kernel-based local linear smoother (Hastie et al., 2009), bivariate penalized spline smoother (Marx & Eilers, 2005) and the sandwich smoother (Xiao et al., 2013). The sandwich smoother (Xiao et al., 2013) is especially useful in the case of very high dimensional data for its appealing computational efficiency, in addition to its estimation accuracy.
3.2. Step 2: Marginal covariance. Data-based orthogonal basis
Once the mean function is estimated, let be the demeaned data. We use the demeaned data to estimate the marginal covariance function induced by Wi(s, Tij), Ξ(s, s′) = Σ(s, s′) + Γ(s, s′). The estimation of Ξ(s, s′) consists of two steps. In the first step, a raw covariance estimator is obtained; the pooled sample covariance is a suitable choice, if all the curves are observed on the same grid of points:
| (3) |
As data Yijr's are observed with white noise, ε2,ij(sr), the ‘diagonal’ elements of the sample covariance, , are inflated by the variance of the noise, σ2. In the second step, the preliminary covariance estimator is smoothed by ignoring the ‘diagonal’ terms; see also Staniswalis & Lee (1998) and Yao et al. (2005) who used similar technique for the case of independent functional data. In our simulation and data application we use the sandwich smoother (Xiao et al., 2015). To ensure the positive semi-definiteness of the estimator the negative eigenvalues are zero-ed. The resulting smoothed covariance function, , is used as an estimator of . In Section 4, we show that is an unbiased and consistent estimator of in two settings: 1) the data are observed fully and without noise, i.e. ∈ij(s) ≡ 0 and 2) the data are observed fully and with measurement error of type ∈1,ij(s), i.e ∈ij(s) ≡ ∈1,ij(s).
Let be the pairs of eigenvalues/eigenfunctions obtained from the spectral decomposition of the estimated covariance function, . The truncation value K is determined based on pre-specified percentage of variance explained (PVE); specifically, K can be chosen as the smallest integer such that is greater than the pre-specified PVE (Di et al. 2009; Staicu et al., 2010).
3.3. Step 3: Covariance of the time-varying coefficients
Let be the projection of the jth repeated demeaned curve of the ith subject onto the direction for k = 1, . . . , K. Since is observed at dense grids of points {sr : r = 1, . . . , R} in [0, 1] for all i and j, is approximated accurately through numerical integration. It is easy to see that the version of that uses μ(s, Tij) in place of and ϕk(s) in place of converges to ξW,ijk with probability one, as R diverges. The time-varying terms are proxy measurements of ξik(Tij); they will be used to study the temporal dependence along the direction ϕk(·), Gk(T, T′) = cov{,ik(T), ,ik(T′)}, and furthermore to obtain prediction for all times T ∈ [0, 1].
Consider now as the ‘observed data’. One viable approach is to assume a parametric structure for Gk(·, ·) such as AR(1) or a random effects model framework; this is typically preferable when mi is very small and the longitudinal design is balanced. We discuss random effects model for estimating the longitudinal covariance in the data application. Here we consider a more flexible approach and estimate the covariance Gk(·,·) nonparametrically, by employing FPCA techniques for sparse functional data (Yao et al., 2005).
Let be the pairs of eigenfunctions and eigenvalues of the covariance Gk; we model the proxy observations as = where ζikl's are random variables with zero mean and variances equal to ηkl, 's are iid with zero-mean and variance equal to and independent of ζikl. Following Yao et al. (2005), we first obtain the raw sample covariance, = . Then the estimated smooth covariance surface, , is obtained by using bivariate smoothing of . Kernel-based local linear smoothing (Yao et al., 2005) or penalized tensor product spline smoothing (Wood, 2006) can be used at this step. The diagonal terms are removed because the noise leads to inflated variance function. Let be the pairs of eigenvalues/eigenfunctions of the estimated covariance surface, . The truncation value, Lk, is determined based on pre-specified PVE; using similar ideas as in Section 3.2. The variance is estimated as the average of the difference between a smooth estimate of the variance based on and ; Yao et al. (2005) discusses an alternative that dismisses the terms at the boundary when estimating the error variance.
Once the eigenbasis functions , eigenvalues ηkl's, and error variance are estimated, the above model framework can be viewed as a mixed effects model and the random components ζikl can be predicted using conditional expectation and a jointly Gaussian assumption for ζijk's and eijk's. In particular, = = , where is the mi-dimensional column vector of the evaluations of at {Tij : j = 1, . . . , mi}, is a mi × mi - matrix with (j, j′)th element equal to , for j = j′ and otherwise, and is the mi - dimensional column vector of 's. The predicted time-varying coefficients corresponding to a generic time T are obtained as . Yao et al. (2005) proved the consistency of the eigenfunctions and predicted trajectories when ξW,ijk's are observed. In Section 4 we extend these results to the case when the proxy 's are used instead and when the profiles Yij(·) are fully observed and the noise is of the type εij(s) = ε1,ij(s); i.e. the data Yij(·) are observed with smooth error.
3.4. Step 4: Trajectories reconstruction
We are now able to predict the full response curve at any time point T ∈ [0, 1] by: , where s ∈ [0, 1]. In Section 4 we show the consistency of .
4. Theoretical properties
Next we discuss the asymptotic properties of the estimators and the predicted trajectories. Our setting - sparse longitudinal design and dense functional design - requires new techniques than the ones commonly used for theoretical investigation of repeated functional data such as Chen & Müller (2012). Since the mean estimation has been studied previously, we assume that the response trajectories, Yij(·)'s, have zero-mean and focus on the estimation of the model covariance. Throughout this section we assume that Yij(·) is observed fully as a function over the domain, . Section 4.1 discusses the main theoretical results when data are observed without error, i.e. εij(s) ≡ 0 for s ∈ [0, 1]. Section 4.2 extends the results to the case when the data are corrupted with smooth error process εij(s)≡ ε1,ij(s). The proofs are detailed in the Supplementary Material; also in the Supplementary Material we include a discussion on how to relax some of the assumptions. Throughout this section we use and to distinguish between the domains.
We assume that the bivariate process Xi(s, T) is a realization of a true random process, X(s, T), with zero-mean and smooth covariance function, c((s, T),(s′, T ′)), which satisfies some regularity conditions:
-
(A1.)
X = {X(s, T) : (s, T) ∈ is a square integrable element of the , i.e. , where and are compact sets.
-
(A2.)
The sampling density g(T) is continuous and supT∈T|g(T)| < ∞.
Under (A1.) and (A2.), the function Σ(s, s′) defined above (i) is symmetric, (ii) is positive definite, and (iii) has eigenvalues λk's with . Thus Σ(·, ·) is a proper covariance function (Horváth & Kokoszka, 2012, p.24).
4.1. Response curves measured without error
Assume εij(s) ≡ 0 and thus Yij(s) = Xi(s, Tij) for s ∈ . The sample covariance of Yij(s) is . The following assumptions regard the moment behavior of X and are commonly used in functional data analysis (Yao et al., 2005; Chen & Müller, 2012); we require them in our study.
-
(A3.)
E[X(s, T)X(s′, T)X(s, T′)X(s′, T′)] < ∞ for arbitrary s, s′ ∈ and T, T′ ∈ .
-
(A4.)
E[[∥X(·, T)∥4] < ∞ for each T ∈ .
Theorem 1
Assume (A1.) - (A3.) hold. Then as n diverges. If in addition (A4.) holds, then
| (4) |
where ∥k(·,·)∥s = is the Hilbert-Schmidt norm of k(·,·).
(A5.) Let and for k ≥ 2, where k is the kth largest eigenvalues of Σ(s, s′). Assume that and λk > 0 for all k (no crossing or ties among eigenvalues).
Using Theorem 4.4 and Lemma 4.3 of Bosq (2000, p.104), the consistency result (4) implies that, if furthermore (A5.) holds, the eigen-elements of are consistent estimators of the corresponding eigen-elements of Σ(s, s′).
Corollary 1
Under the assumptions (A1.)-(A5.), for each k we have , and as n diverges.
Next, we focus on the estimation of the covariance Gk(T, T′), which describes the longitudinal dynamics. We first show the uniform consistency of ; the result follows if supj,s|Yi(s, Tij)| is bounded almost surely. which is ensured if (A6.) holds. Then, we use this result to show that the estimator of Gk(T, T′) based on 's is asymptotically identical to that based on . Consistency results of the remaining model components follow directly from Yao et al. (2005). The Gaussian assumption (A8.) is needed to show the consistency of .
-
(A6.)
E[sups,T|X(s, T)|a] ≤ Ma for a constant, M > 0, and an arbitrary integer, a ≥ 1; This is equivalent to assume that X(s, T) is absolutely bounded almost surely.
-
(A7.)
Let bk1 = (ηk1 − ηk2) and bkl = max[(ηk(l–1) – ηkl), (ηkl – ηk(l+1)))] for l ≥ 2, where ηkl is the lth largest eigenvalues of Gk(T, T′). Assume that 0 < bkl < ∞ and ηkl > 0 for all k and l.
-
(A8.)
ζikl and eijk are jointly Gaussian.
Theorem 2
Under the assumptions (A1.) - (A6.), for each k and as n diverges. In fact a stronger result also holds, namely as n diverges.
Corollary 2
Assume (A1.) - (A8.) hold for each k and l. Then the eigenvalues and eigenfunctions of satisfy , and as n diverges. Uniform convergence of also holds: . Furthermore, as n diverges, we have and , where and is the mi-dimensional column vector of 's.
The consistency results for all model components imply prediction consistency.
Thorem 3
Assume (A1.) - (A8.), for each (s, T) ∈ , Then as n, K and Lk's → ∞.
4.2. Response curves measured with smooth error
Assume next that Yij(s) are observed with smooth error εij(s) ≡ ε1,ij(s) and thus Yij(s) = Xi(s, Tij) + ε1,ij(s) for s ∈ and ε1,i (·) ∈ L2 (). The main difference from Section 4.1 is that the sample covariance of Yij(s) is an estimator of Ξ(s, s′) = Σ(s, s′) + Γ(s, s′), not of (Σs, s′); we denote the sample covariance of Yij(s) by = . Using similar arguments as earlier, we show that is an unbiased estimator of Ξ(s, s′). Moreover similar arguments can be used to show the pointwise consistency as well as the Hilbert-Schmidt norm consistency of . Additional assumptions are required.
-
(A9.)
Assume εij(·) is realization of ε = {ε(s) : s ∈ }, which is square integrable process in L2().
-
(A10.)
-
(A11.)
for a constant, M > 0, and an arbitrary integer, a ≥ 1.
Corollary 3
Under the assumptions (A1.) - (A3.), and (A9.), for each(s, s′) – as n diverges. And under the assumptions (A1.)-(A4.), (A9.)-(A11.), and .
The proofs of these results are detailed in the Supplementary Material. As the smooth error process ε1,ij(s) is correlated only along the functional argument, s, and ε1,ij(s) are iid over i, j, it follows that the theoretical properties of the predictions - of the time-varying coefficients and the response curve - hold without any modification.
The theoretical results are based on the assumptions that data Yij(s)'s are observed fully, without white noise, ε2,ij(s) ≡ 0 for all s, and have mean zero. Some of these assumptions are quite common in theoretical study involving functional data; Cardot et al. (2003, 2004); Chen & Müller (2012). They are discussed in the Supplementary Material.
5. Simulation study
We study our approach in finite samples and compare its performance with Chen & Müller (2012) denoted by CM. We generate Nsim = 1000 samples from model (1) with K = 2, Yij(s) = μ(s, Tij) + ξi1(Tij)ϕ1(s) + ξi2(Tij)ϕ2(s) + εij(s), where μ(s, T) = 1 + 2s + 3T + 4sT, and ϕ1(s) = 1 and ϕ2(s) = . The grid of points for s is the set of 101 equispaced points in [0, 1]. For each i, there are mi profiles associated with visit times, {Tij : j = 1, . . . , mi}; Tij's are randomly sampled from 41 equally spaced points in [0, 1]. ξik(T) are generated from various covariance structures: (a) non-parametric covariance (NP) where ξik(T) = ζik1 ψk1(T) + ζik2 ψk2(T); (b) random effects model (REM) ξik(T) = bik0 + bik1T, and (c) exponential autocorrelation (Exp) . Errors are generated from εij(s) = eij1ϕ1(s) + eij2ϕ2(s) + ε2,ij(s), where eij1, eij2 and ε2,ij(s) are mutually independent with zero- mean and variances equal to and σ2, respectively; the white noise variance, σ2, is set based on the signal to noise ratio (SNR). The details of the models are specified in the Supplementary Material. For each sample of size n we form a training set and a test set. The test set contains 10 profiles and is obtained as follows: randomly select 10 subjects from the sample and collect the subjects’ last profile. The remaining profiles for the 10 subjects and the data corresponding to the rest (n – 10) of the subjects form the training set. Our model is fitted using the training set and the methods of Section 3. The mean function, μ(s, T), is modeled using 50 cubic spline basis functions obtained from the tensor product of ds = 10 basis functions in direction s and dT = 5 in T . The smoothing parameters are selected via REML. The finite truncations K and Lk's are all estimated using the pre-specified level PVE = 0.95.
Estimation accuracy for the model components is evaluated using integrated mean squared errors (IMSE), while prediction performance is assessed through in-sample integrated prediction errors (IN-IPE) and out-of-sample IPE (OUT-IPE). Table 1 shows the results for different covariance models for ξik(T), different number of repeated curve measurements per subject, different SNRs, complex error process, and varying sample sizes. The performance of the proposed estimation (see columns for μ, ϕ1, and ϕ2 of this table) is slightly affected by the covariance structure of ξik(T)'s and mi, but in general is quite robust to the factors we investigated. As expected the estimation accuracy improves with larger sample size; see the 3 × 3 top left block of IMSE results corresponding to n = 100, n = 300, and n = 500. Moreover both the prediction of ξik(T )'s and that of Yij(·) are considered; see columns labeled ξ1, ξ2, IN-IPE and OUT-IPE of Table 1. The underlying covariance structure of ξik(T)'s affects the prediction accuracy. Furthermore increasing the number of repeated curve measurements mi improves the accuracy more than increasing the sample size n. This observation should not be surprising, as with larger number of repeated measurements the estimation of the covariance of the longitudinal process ξik(T)'s improves and as a result it yields superior prediction. We compared our results with another, rather naïve approach: predict a subject's profile by the average of all previously observed profiles for that subject. The naïve approach (see columns IN-IPEnaive and OUT-IPEnaive) is very sensitive to the covariance structure of ξik(T). In all the cases studied the prediction accuracy is inferior to the proposed method.
Table 1.
Estimation and prediction accuracy results based on Nsim = 1000 simulations
|
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| μ | ϕ 1 | ϕ 2 | ξ 1 | ξ 2 | IN-IPE | IN-IPEnaive | OUT-IPE | OUT-IPEnaive | ||
| NP (a) | n = 100 | 0.092 | 0.003 | 0.011 | 0.338 | 0.224 | 0.406 | 7.790 | 0.988 | 11.478 |
| n = 300 | 0.031 | 0.001 | 0.009 | 0.226 | 0.138 | 0.313 | 7.773 | 0.559 | 11.349 | |
| n = 500 | 0.019 | 0.001 | 0.009 | 0.199 | 0.117 | 0.288 | 7.779 | 0.455 | 11.262 | |
| REM (b) | n = 100 | 0.114 | 0.027 | 0.033 | 0.376 | 0.314 | 0.328 | 1.199 | 1.011 | 2.160 |
| n = 300 | 0.040 | 0.008 | 0.013 | 0.216 | 0.162 | 0.265 | 1.197 | 0.675 | 2.160 | |
| n = 500 | 0.024 | 0.005 | 0.010 | 0.181 | 0.133 | 0.247 | 1.197 | 0.571 | 2.150 | |
| Exp (c) | n = 100 | 0.095 | 0.022 | 0.030 | 0.399 | 0.540 | 0.554 | 1.528 | 1.426 | 2.520 |
| n = 300 | 0.031 | 0.007 | 0.015 | 0.289 | 0.412 | 0.508 | 1.531 | 1.143 | 2.498 | |
| n = 500 | 0.019 | 0.004 | 0.013 | 0.266 | 0.383 | 0.494 | 1.530 | 1.074 | 2.492 | |
|
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| μ | ϕ 1 | ϕ 2 | ξ 1 | ξ 2 | IN-IPE | IN-IPEnaive | OUT-IPE | OUT-IPEnaive | ||
| NP (a) | n = 100 | 0.076 | 0.002 | 0.010 | 0.180 | 0.101 | 0.238 | 7.807 | 0.477 | 10.666 |
| n = 300 | 0.026 | < 0. 001 | 0.009 | 0.120 | 0.065 | 0.183 | 7.796 | 0.282 | 10.728 | |
| n = 500 | 0.016 | < 0. 001 | 0.009 | 0.108 | 0.058 | 0.173 | 7.797 | 0.242 | 10.772 | |
| REM (b) | n = 100 | 0.097 | 0.025 | 0.031 | 0.272 | 0.252 | 0.232 | 0.897 | 0.612 | 1.833 |
| n = 300 | 0.034 | 0.008 | 0.013 | 0.156 | 0.132 | 0.201 | 0.896 | 0.462 | 1.841 | |
| n = 500 | 0.020 | 0.005 | 0.010 | 0.135 | 0.110 | 0.194 | 0.897 | 0.440 | 1.836 | |
| Exp (c) | n = 100 | 0.080 | 0.022 | 0.030 | 0.308 | 0.417 | 0.467 | 1.240 | 1.048 | 2.147 |
| n = 300 | 0.026 | 0.006 | 0.015 | 0.233 | 0.309 | 0.444 | 1.245 | 0.938 | 2.155 | |
| n = 500 | 0.016 | 0.004 | 0.012 | 0.221 | 0.285 | 0.438 | 1.246 | 0.886 | 2.129 | |
|
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| μ | ϕ 1 | ϕ 2 | ξ 1 | ξ 2 | IN-IPE | IN-IPEnaive | OUT-IPE | OUT-IPEnaive | ||
| NP (a) | n = 100 | 0.092 | 0.005 | 0.005 | 0.328 | 0.213 | 0.363 | 7.184 | 0.958 | 10.795 |
| n = 300 | 0.031 | 0.001 | 0.002 | 0.213 | 0.124 | 0.268 | 7.170 | 0.506 | 10.662 | |
| n = 500 | 0.019 | 0.001 | 0.001 | 0.187 | 0.103 | 0.242 | 7.178 | 0.402 | 10.585 | |
| REM (b) | n = 100 | 0.114 | 0.037 | 0.037 | 0.404 | 0.355 | 0.293 | 0.594 | 0.958 | 1.478 |
| n = 300 | 0.040 | 0.010 | 0.011 | 0.218 | 0.167 | 0.235 | 0.595 | 0.627 | 1.476 | |
| n = 500 | 0.024 | 0.006 | 0.007 | 0.180 | 0.135 | 0.219 | 0.596 | 0.529 | 1.467 | |
| Exp (c) | n = 100 | 0.095 | 0.033 | 0.033 | 0.420 | 0.573 | 0.513 | 0.922 | 1.419 | 1.838 |
| n = 300 | 0.031 | 0.010 | 0.010 | 0.290 | 0.412 | 0.466 | 0.929 | 1.109 | 1.814 | |
| n = 500 | 0.019 | 0.006 | 0.006 | 0.264 | 0.378 | 0.453 | 0.929 | 1.033 | 1.807 | |
|
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| μ | ϕ 1 | ϕ 2 | ξ 1 | ξ 2 | IN-IPE | IN-IPEnaive | OUT-IPE | OUT-IPEnaive | ||
| NP (a) | n = 100 | 0.076 | 0.003 | 0.003 | 0.174 | 0.095 | 0.205 | 7.462 | 0.441 | 10.300 |
| n = 300 | 0.026 | 0.001 | 0.001 | 0.113 | 0.057 | 0.147 | 7.453 | 0.239 | 10.359 | |
| n = 500 | 0.016 | < 0. 001 | 0.001 | 0.101 | 0.050 | 0.136 | 7.454 | 0.200 | 10.406 | |
| REM (b) | n = 100 | 0.097 | 0.035 | 0.035 | 0.300 | 0.293 | 0.205 | 0.552 | 0.568 | 1.464 |
| n = 300 | 0.034 | 0.010 | 0.010 | 0.160 | 0.140 | 0.178 | 0.552 | 0.426 | 1.473 | |
| n = 500 | 0.020 | 0.006 | 0.007 | 0.136 | 0.114 | 0.172 | 0.554 | 0.405 | 1.470 | |
| Exp (c) | n = 100 | 0.080 | 0.033 | 0.033 | 0.330 | 0.451 | 0.434 | 0.895 | 1.012 | 1.779 |
| n = 300 | 0.027 | 0.009 | 0.010 | 0.236 | 0.313 | 0.410 | 0.902 | 0.901 | 1.785 | |
| n = 500 | 0.016 | 0.005 | 0.006 | 0.221 | 0.284 | 0.403 | 0.902 | 0.851 | 1.763 | |
Table 2 shows the comparison with CM, when the kernel bandwidth is fixed to h = 0.1 for both mean and covariance smoothing. The prediction using CM is more sensitive to the covariance structure of the underlying time-varying coefficients ξik(T) and its accuracy can be improved by up to 50% using our proposed approach. Computation-wise, there is an order of magnitude difference in the computational cost between the methods: when n = 100 CM takes over 16 minutes, while our approach takes about 7 seconds. The overall conclusion is that the proposed approach provides an improved prediction performance over the existing methods in a computationally efficient manner.
Table 2.
Comparison between the proposed method and Chen & Muller (2012) in the presence of correlated errors. Results based on Nsim = 1000 simulations
|
| |||||||
|---|---|---|---|---|---|---|---|
|
Chen & Muller (2012)
|
Proposed method (from Tables 1 and S2) |
||||||
| IN-IPE | OUT-IPE | time (seconds) | IN-IPE | OUT-IPE | time (seconds) | ||
| NP (a) | n = 100 | 0.880 | 2.221 | 983.872 | 0.406 | 0.988 | 7.369 |
| n = 300 | 0.622 | 1.468 | 1659.611 | 0.313 | 0.559 | 15.892 | |
| n = 500 | 0.556 | 1.298 | 2502.462 | 0.288 | 0.455 | 21.418 | |
| REM (b) | n = 100 | 0.424 | 1.359 | 1084.753 | 0.328 | 1.011 | 9.282 |
| n = 300 | 0.289 | 0.729 | 1955.193 | 0.265 | 0.675 | 11.347 | |
| n = 500 | 0.257 | 0.614 | 2947.126 | 0.247 | 0.571 | 22.559 | |
| Exp (c) | n = 100 | 0.634 | 1.642 | 1556.182 | 0.554 | 1.426 | 7.514 |
| n = 300 | 0.549 | 1.251 | 1959.219 | 0.508 | 1.143 | 16.229 | |
| n = 500 | 0.531 | 1.155 | 2865.041 | 0.494 | 1.074 | 17.109 | |
6. DTI application
DTI is a magnetic resonance imaging technique, which provides different measures of water diffusivity along brain white matter tracts; its use is instrumental especially in diseases that affect the brain white matter tissue, such as MS (see Alexander et al. (2007), Basser et al. (1994), Basser et al. (2000), Basser & Pierpaoli (2011)). In this paper we consider the DTI measure called FA along CCA; specifically we consider one-dimensional summaries of FA along CCA (CCA-FA). The DTI study involves 162 MS patients, which are observed at between one and eight hospital visits, with a total of 421 visits and a median of two visits per subject. At each visit, FA profile is recorded at 93 locations along the CCA. The measurements are registered within and between subjects using standard biological landmarks identified by an experienced neuroradiologist (Scheipl et al., 2014).
Our main objective is twofold: (i) to understand the dynamic behavior of the CCA-FA profile in MS patients over time and (ii) to make accurate predictions of the CCA-FA profile of a patient at their next visit. Various aspects of the DTI study have been also considered in Goldsmith et al. (2011), Staicu et al. (2012), Pomann et al. (2013), and Scheipl et al. (2014). Greven et al. (2010) used an earlier version of the DTI study consisting of data from fewer and possibly different patients and obtained through a different registration technique. They studied the dynamic behavior of CCA-FA over time in MS; however, their method cannot provide prediction of the entire CCA-FA profile at the subject's next visit. By being able to predict the full CCA-FA profile at the subject's future visit, our approach has the potential to shed lights on the understanding of the MS progression over time as well as its response to treatment.
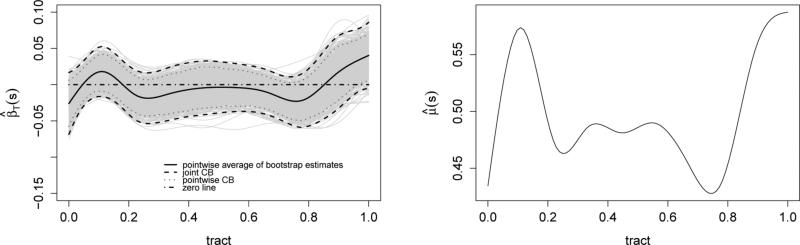
To start with, for each subject we define the hospital visit time Tij by the difference between the reported visit time and the subject's baseline visit time; thus Ti1 = 0 for all subjects i. Then the resulting values are scaled by the maximum value in the study so that Tij ∈ [0, 1] for all i and j. The sampling distribution of the visit times is right-skewed with rather strong skewness; for example there are only few observations Tij's close to 1. The strong skewness of the sampling distribution of Tij's has serious implications on the estimation of the bivariate mean μ(s, T); a completely nonparametric bivariate smoothing would results in unstable and highly variable estimation. This is probably why Greven et al. (2010) first centered the times for each patient i, {Tij : j = 1, . . . , mi}, and then standardized the overall set {Tij : i, j} to have unit variance. However, such subject-specific transformation of Tij's loses interpretability and it is not suited for prediction at unobserved times - which is crucial in our analysis. One way to bypass this issue is to assume a simpler parametric structure along the longitudinal direction, T, for the mean function; based on exploratory analysis we assume linearity in T . Specifically we consider μ(s, Tij) = μ0(s) + βT(s)Tij, where μ0(·) and βT(·) are unknown, smooth functions of s. We estimate μ0(·) and βT(·) using a penalized univariate cubic spline regression with 10 basis functions; the smoothing parameters are estimated using REML. The estimates and are displayed in Figure S1 of the Supplementary Material. Using the bootstrap of subjects - based methods of Park et al. (2015) and B = 1000 bootstrap samples we construct 95% joint confidence bands for ; see Figure 1. The confidence band contains zero for all s, indicating evidence that a mean model μ(s, Tij) = μ0(s) is more appropriate.
Figure 1.
Left panel: 95% pointwise and joint confidence bands of the slope function βT(s) of μ(s, T) using bootstrap; Right: final mean estimate,
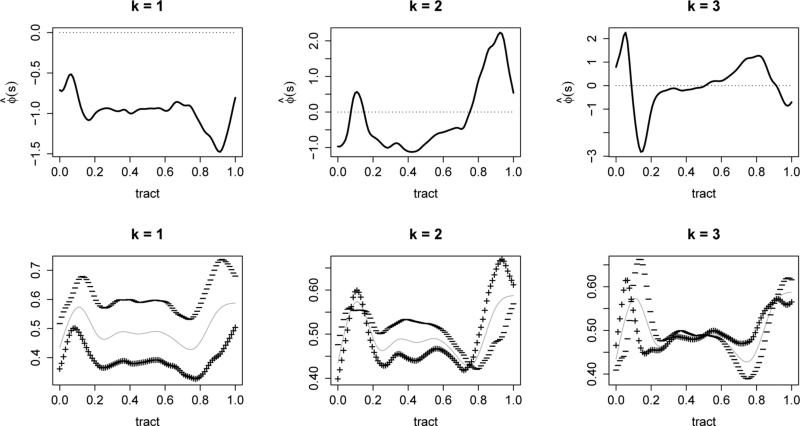
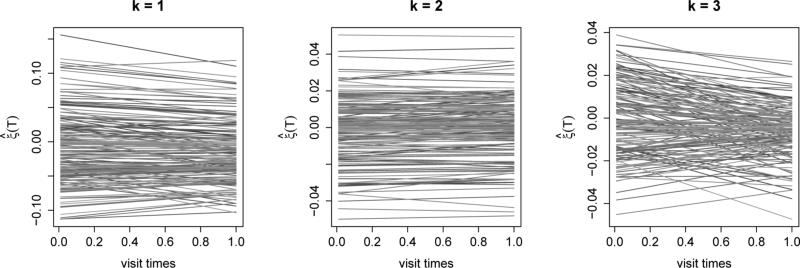
Next we demean the data and estimate the marginal covariance; using a preset level PVE = 0.95 we obtain K = 10 eigenfunctions. Figure 2 shows the leading 3 eigenfunctions that explain in turn 62.69%, 8.37% and 6.77% of the total variance; the rest of the estimated eigenfunctions are given in Figure S3 of the Supplementary Material. Preliminary investigation (not shown here) indicates a simpler model for the longitudinal covariance: a random effects model ξik(Tij) = b0ik + b1ikTij, where var(blik) = for l = 0, 1 and cov(b0ik, b1ik) = œ01k. This resulting model is similar to Greven et al. (2010). The fitted time-varying coefficient functions, , for k = 1, 2 and 3 are shown in Figure 3, and the rest are shown in Figure S4 of the Supplementary Material. The estimated suggest some longitudinal changes, but the signs generally remain constant across time. The results imply that a subject mean profile tends to stay lower than the population mean, if the first eigenfunction corresponding to that individual is positively loaded at baseline, and vise versa. In contrast, , are mostly constant across visit times and imply little changes over time.
Figure 2.
Top: First three eigenfunctions of the estimated marginal covariance; Bottom: estimated mean function (gray line) ± (+ and − signs, respectively)
Figure 3.
Estimated time-varying coefficients for k = 1, 2 and 3 using REM
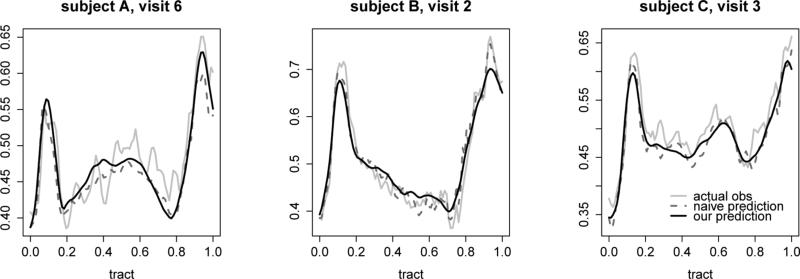
Finally, we assess the goodness-of-fit and prediction accuracy of our final model. For the goodness-of-fit we use the in-sample integrated prediction error (IN-IPE): IN-IPE= , where , and Yij(·)'s are the observed curve data. The square root of the IN-IPE is 2.31 × 10−2 for our model; for comparison Greven et al. (2010) yields 2.66 × 10−2 and Chen & Müller (2012) gives 3.76 × 10−2. For prediction accuracy we use leave-the last-curve-out integrated prediction error (OUT-IPE) calculated for the 106 subjects observed at two hospital visits or more: OUT-IPE = , where is the predicted curve at time Timi for the ith subject using the fitted model based on all the data less the mith curve of the ith subject. Figure 4 shows such predicted curves obtained using our model and the naive model for three randomly selected subjects at their last visit. The square root of OUT-IPE is 3.48 × 10−2 for our model; for comparison Chen & Müller (2012) gives 8.71 × 10−2 and the naïve approach gives 3.52 × 10−2. These results suggest that, in this short term study of MS, there is a small variation of CCA-FA profiles over time.
Figure 4.
Predicted values of FA for the last visits of three randomly selected subjects; actual observations (gray); predictions using our model (black solid) and using the naive approach (black dashed)
Supplementary Material
Acknowledgement
Staicu's research was supported by NSF grant number DMS 1454942 and NIH grant R01 NS085211. We thank Daniel Reich and Peter Calabresi for the DTI tractography data.
Footnotes
Supplementary Material
Detailed proofs of the theoretical results, additional numerical investigations and data analysis results are included in a supplementary material that is available online.
References
- Alexander AL, Lee JE, Lazar M, Field AS. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4(3):316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baladandayuthapani V, Mallick BK, Young Hong M, Lupton JR, Turner ND, Carroll RJ. Bayesian hierarchical spatially correlated functional data analysis with application to colon carcinogenesis. Biometrics. 2008;64(1):64–73. doi: 10.1111/j.1541-0420.2007.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. Mr diffusion tensor spectroscopy and imaging. Biophysical journal. 1994;66(1):259. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using dt-mri data. Magnetic resonance in medicine. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor mri. Journal of magnetic resonance. 2011;213(2):560–570. doi: 10.1016/j.jmr.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Bosq D. Linear processes in function spaces: theory and applications. Vol. 149. Springer; 2000. [Google Scholar]
- Cardot H, Ferraty F, Mas A, Sarda P. Testing hypotheses in the functional linear model. Scandinavian Journal of Statistics. 2003;30(1):241–255. [Google Scholar]
- Cardot H, Goia A, Sarda P. Testing for no effect in functional linear regression models, some computational approaches. Communications in Statistics-Simulation and Computation. 2004;33(1):179–199. [Google Scholar]
- Chen K, Delicado P, Müller HG. Modeling function-valued stochastic processes, with applications to fertility dynamics. 2015 Manuscript submitted. [Google Scholar]
- Chen K, Müller HG. Modeling repeated functional observations. Journal of the American Statistical Association. 2012;107(500):1599–1609. doi:10.1080/01621459.2012.734196. [Google Scholar]
- Di CZ, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel functional principal component analysis. The annals of applied statistics. 2009;3(1):458. doi: 10.1214/08-AOAS206SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith J, Bobb J, Crainiceanu CM, Caffo B, Reich D. Penalized functional regression. Journal of Computational and Graphical Statistics. 2011;20(4) doi: 10.1198/jcgs.2010.10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith J, Zipunnikov V, Schrack J. Generalized multilevel functional-on-scalar regression and principal component analysis. 2014 doi: 10.1111/biom.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greven S, Crainiceanu C, Caffo B, Reich D. Longitudinal functional principal component analysis. Electronic Journal of Statistics. 2010:1022–1054. doi: 10.1214/10-EJS575. doi:10.1214/10-EJS575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromenko O, Kokoszka P. Nonparametric inference in small data sets of spatially indexed curves with application to ionospheric trend determination. Computational Statistics & Data Analysis. 2013;59:82–94. [Google Scholar]
- Gromenko O, Kokoszka P, Zhu L, Sojka J. Estimation and testing for spatially indexed curves with application to ionospheric and magnetic field trends. The Annals of Applied Statistics. 2012;6(2):669–696. doi: 10.1214/11-AOAS524. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J, Hastie T, Friedman J, Tibshirani R. The elements of statistical learning. Vol. 2. Springer; 2009. [Google Scholar]
- Horváth L, Kokoszka P. Inference for functional data with applications. Vol. 200. Springer; 2012. [Google Scholar]
- Jiang CR, Wang JL. Covariate adjusted functional principal components analysis for longitudinal data. The Annals of Statistics. 2010:1194–1226. [Google Scholar]
- Li Y, Guan Y. Functional principal component analysis of spatio-temporal point processes with applications in disease surveillance. Journal of the American Statistical Association. 2014;0(ja) doi: 10.1080/01621459.2014.885434. null, doi:10.1080/01621459.2014. 885434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx BD, Eilers PH. Multidimensional penalized signal regression. Technometrics. 2005;47(1):13–22. [Google Scholar]
- Morris JS, Carroll RJ. Wavelet-based functional mixed models. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2006;68(2):179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Vannucci M, Brown PJ, Carroll RJ. Wavelet-based nonparametric modeling of hierarchical functions in colon carcinogenesis. Journal of the American Statistical Association. 2003;98(463):573–583. [Google Scholar]
- Park SY, Staicu AM, Xiao L, Crainiceanu CM. Simple fixed effects inference for complex functional models. 2015 doi: 10.1093/biostatistics/kxx026. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomann GM, Staicu AM, Ghosh S. Two sample hypothesis testing for functional data. 2013 [Google Scholar]
- Scheipl F, Staicu AM, Greven S. Functional additive mixed models. Journal of Computational and Graphical Statistics. 2014:00–00. doi: 10.1080/10618600.2014.901914. just-accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staicu AM, Crainiceanu CM, Carroll RJ. Fast methods for spatially correlated multilevel functional data. Biostatistics. 2010;11(2):177–194. doi: 10.1093/biostatistics/kxp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staicu AM, Crainiceanu CM, Reich DS, Ruppert D. Modeling functional data with spatially heterogeneous shape characteristics. Biometrics. 2012;68(2):331–343. doi: 10.1111/j.1541-0420.2011.01669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staniswalis JG, Lee JJ. Nonparametric regression analysis of longitudinal data. Journal of the American Statistical Association. 1998;93(444):1403–1418. [Google Scholar]
- Wood SN. Low-rank scale-invariant tensor product smooths for generalized additive mixed models. Biometrics. 2006;62(4):1025–1036. doi: 10.1111/j.1541-0420.2006.00574.x. doi:10.1111/j.1541-0420.2006.00574.x. [DOI] [PubMed] [Google Scholar]
- Xiao L, Huang L, Schrack JA, Ferrucci L, Zipunnikov V, Crainiceanu CM. Quantifying the lifetime circadian rhythm of physical activity: a covariate-dependent functional approach. Biostatistics. 2015;16(2):352–367. doi: 10.1093/biostatistics/kxu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Li Y, Ruppert D. Fast bivariate p-splines: the sandwich smoother. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2013;75(3):577–599. [Google Scholar]
- Xiao L, Ruppert D, Zipunnikov V, Crainiceanu C. Fast Covariance Estimation for High-dimensional Functional Data. Statistics and Computing. 2015 doi: 10.1007/s11222-014-9485-x. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Müller HG, Wang JL. Functional data analysis for sparse longitudinal data. Journal of the American Statistical Association. 2005;100(470):577–590. [Google Scholar]
- Zhou L, Huang JZ, Carroll RJ. Joint modelling of paired sparse functional data using principal components. Biometrika. 2008;95(3):601–619. doi: 10.1093/biomet/asn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.