Abstract
Plasmodium falciparum virulence is linked to its ability to sequester in post-capillary venules in the human host. Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is the main variant surface antigen implicated in this process. Complete loss of parasite adhesion is linked to a large subtelomeric deletion on chromosome 9 in a number of laboratory strains such as D10 and T9-96. Similar to the cytoadherent reference line FCR3, D10 strain expresses PfEMP1 on the surface of parasitized erythrocytes, however without any detectable cytoadhesion. To investigate which of the deleted subtelomeric genes may be implicated in parasite adhesion, we selected 12 genes for D10 complementation studies that are predicted to code for proteins exported to the red blood cell. We identified a novel single copy gene (PF3D7_0936500) restricted to P. falciparum that restores adhesion to CD36, termed here virulence-associated protein 1 (Pfvap1). Protein knockdown and gene knockout experiments confirmed a role of PfVAP1 in the adhesion process in FCR3 parasites. PfVAP1 is co-exported with PfEMP1 into the host cell via vesicle-like structures called Maurer's clefts. This study identifies a novel highly conserved parasite molecule that contributes to parasite virulence possibly by assisting PfEMP1 to establish functional adhesion at the host cell surface.
Introduction
Plasmodium falciparum, a causative agent of human malaria, is an apicomplexan parasite that infects and develops in host erythrocytes. Plasmodium falciparum is the most virulent form of the five Plasmodium species that infect man. It is responsible for over half a million deaths annually, principally in children under the age of 5 (WHO, 2013). Several strategies have been implemented in order to reduce the burden of infection such as transmission control and the development of drugs and vaccines (Eastman and Fidock, 2009; Crompton et al., 2014). However, malaria remains a major public health concern. Determining the factors that contribute to parasite virulence is therefore crucial for designing or improving therapeutic interventions.
Plasmodium falciparum-mediated pathogenesis has been associated with the parasite's ability to mediate binding of its host cell to the vascular endothelium of several organs, notably the brain leading to complications such as cerebral malaria. The mechanisms that lead to pathology are not well elucidated and include the activation of platelets and endothelial cells leading to increased binding of infected erythrocytes (IE) either directly to the vasculature or via platelet bridges (Miller et al., 2013). Infected erythrocyte adhesion to various host cell receptors such as intercellular adhesion molecule-1, CD36, gC1-qR/p32 and chondroitin sulphate A is mediated by the main parasite ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1; Smith et al., 2013), an antigenically variant surface protein, encoded by approximately 60 var genes expressed in a mutually exclusive manner (Scherf et al., 2008). Specific subsets of var genes are associated with IE sequestration to particular tissues in the placenta (Viebig et al., 2005) or the brain in cerebral malaria patients as determined by post-mortem studies (Tembo et al., 2014). Thus, in-depth insight into parasite proteins that contribute to the PfEMP1-mediated adhesion process could enable intervention strategies to reduce cytoadhesion-associated malaria pathogenesis. A systematic gene knockout study of candidate genes coding for proteins trafficked to the IE illustrated that many helper proteins are involved to traffic PfEMP1 from the parasite to the surface of IE (Elsworth et al., 2014).
A different type of cytoadhesion deficient parasites is linked to a large subtelomeric deletion on the right arm of chromosome 9 of cloned lines such as D10 and T9-96 (Day et al., 1993; Chaiyaroj et al., 1994). D10 lacks 25 genes from this region including the cytoadherence-linked asexual gene 9 (clag9), ring exported proteins 1–4 (rex1, rex2, rex3 and rex4), several rifin, two var genes. The export of PfEMP1 in D10 is comparable in parasites with an intact chromosome 9 indicating that the PfEMP1 expressed at the surface of D10 IE lost its virulence feature, namely the adhesion to host receptors (Nacer et al., 2011). This suggests that at least one of the deleted genes of chromosome 9 is important for correct presentation of PfEMP1 at the IE surface. CLAG9 had previously been implicated in cytoadhesion (Trenholme et al., 2000); however, it has been shown that disruption of the clag9 gene does not impair binding to CD36 suggesting that other genes are involved in this process (Nacer et al., 2011). One promising candidate is the resident Maurer's cleft protein REX1. Knockout of rex1 leads to a slight reduction in binding to CD36, and to morphological alterations of the Maurer's cleft similar to those observed in D10 (Dixon et al., 2011; Nacer et al., 2011). Nonetheless, as binding remains partial, this suggests that other genes absent from chromosome 9 in D10 parasites could be implicated in the correct conformation/presentation of PfEMP1 at the IE surface.
In this work, we complemented 12 genes that are bioinformatically predicted to be exported to the IE. We describe the identification of the virulence-associated protein gene (Pfvap1) that restores adhesion of complemented D10 IE to CD36. This gene is unique in P. falciparum and its deletion resulted in a 50% to 60% decrease in binding to CD36. We propose a mechanism by which PfVAP1 interacts with PfEMP1.
Results
Identification of a novel gene implicated in cytoadhesion
We previously showed that non-cytoadherent D10 parasites with a subtelomeric deletion on the right arm of chromosome 9 express PfEMP1 on the IE surface but that the protein has lost its adhesion properties (Nacer et al., 2011). We detected strong surface labelling on IE of D10 and the reference strain for parasite cytoadhesion FCR3 with a pool of hyperimmune human sera (Fig. 1A), known to predominantly recognize PfEMP1 (Chan et al., 2012), thus supporting the notion that the non-binding phenotype of D10 may be due to a conformational defect or incorrect presentation of PfEMP1.
Fig. 1.
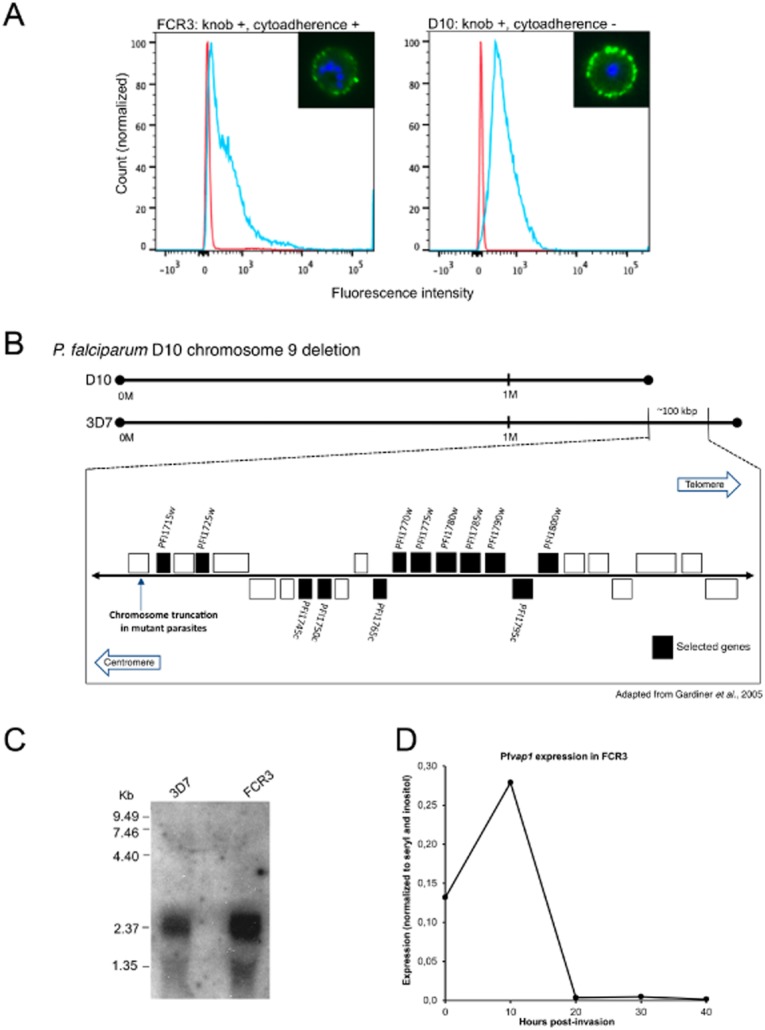
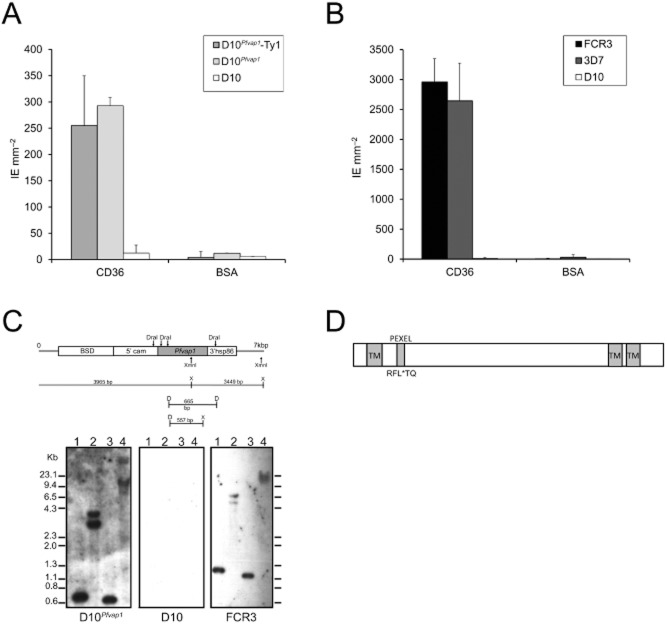
Complementation analysis identified a novel gene that restores adhesion in D10.A. Synchronized FCR3 and D10 parasites (30–35 h) were labelled with a pool of hyperimmune human sera followed by an anti-human IgG Alexa 488 secondary antibody (green). Fluorescence was measured by flow cytometry and imaged by fluorescence microscopy (inset).B. Schematic representation of the chromosome 9 deletion in P. falciparum D10 (D10) compared with P. falciparum 3D7 (3D7). Complementation of D10 using episomal expression of candidate genes (shown in back boxes).C. A transcript of ∼2.3 kb was detected in asynchronous cultures of P. falciparum FCR3 and 3D7.D. qRT-PCR analysis of synchronized FCR3 parasites showed maximal expression in ring stage parasites. Y-axis relative copy number normalized against seryl transferase and inositol. X-axis age of parasites (hours post-invasion).
The criteria chosen to identify genes on chromosome 9 with a potential role in cytoadhesion included the presence of export motifs PNEP, PEXEL/VTS (Marti and Spielmann, 2013) and transcription during the intraerythrocytic cycle (http://www.plasmodb.org). On this basis, 12 genes were chosen to complement D10 (Fig. 1B), none of them have been previously studied. We performed binding assays on purified CD36 receptor coated onto plastic dishes to determine whether episomal expression of a particular candidate could restore adhesion. One out of 12 transfected parasite lines, complemented with PF3D7_0936500 (former ID: PFI1765c), showed a significant restoration of binding to CD36 (20-fold over D10 IE counts). We named this gene Pfvap1. Transcription of Pfvap1 during the intraerythrocytic cycle was confirmed by Northern blot and a 2.3 kb transcript was detected from total RNA of asynchronous cultures of 3D7 and FCR3 (Fig. 1C). Pfvap1 is expressed in ring stages with timing similar to PfEMP1; maximal expression was detected in highly synchronized ring stage cultures by quantitative real-time polymerase chain reaction (qRT-PCR; Fig. 1D) confirming previous reports (Le Roch et al., 2003; Otto et al., 2010). Homology searches failed to identify other homologues for Pfvap1 in P. falciparum and no orthologues were found in other Plasmodium species, suggesting that this single copy gene is specific to P. falciparum. With the exception of trafficking motifs, no other known protein domains could be identified in PfVAP1.
PfVAP1 partially restores binding to CD36
Complementation of D10 parasites with Pfvap1 (D10Pfvap1) resulted in a significant increase of binding to CD36 over background values observed for D10 (Fig. 2A). D10Pfvap1 adhesion values, when compared with previously selected CD36 laboratory lines FCR3 and 3D7 (Fig. 2B), suggest that only partial restoration of adhesion in D10 parasites was achieved. Wild-type D10 parasites do not exist, and thus, the binding capacity of this parasite before chromosome 9 deletion cannot be assessed. We generated a Ty1-tagged Pfvap1 gene and showed that the addition of the tag to the C-terminus did not change the restoration of adhesion levels (Fig. 2A).
Fig. 2.
PfVAP1 partially restores binding to CD36.A. Complementation of D10 with PfVAP1 (episome) leads to a > 20-fold increase in binding.B. Binding of FCR3, 3D7 and D10 to purified CD36. Binding assays performed in duplicate wells spotted with either CD36 or BSA. Error bars (SD) are shown. FCR3 (n = 5), 3D7 (n = 2), D10 (n = 6) and D10Pfvap1 (n = 10) replicates. D10 complemented with PfVAP1 or PfVAP1-Ty1 binds similarly to CD36. Binding assays were performed following four rounds of panning on CD36 purified CD36 or BSA. Error bars represent the standard deviation.C. Diagram showing the position of restriction sites for DraI and XmnI used for digestion of the genomic DNA of complemented D10Pfvap1. Southern blot confirming the complementation of D10 with Pfvap1. Genomic DNAs were digested with DraI (lane 1), XmnI (lane 2), DraI + XmnI (lane 3) or not digested (lane 4). Complemented D10Pfvap1 parasites, D10 and FCR3 are shown. The absence of a reaction on D10 confirmed the specificity of the probe.D. Diagram of PfVAP1 showing the predicted protein structure of the 31.1 kDa protein: the N-terminal transmembrane domain precedes the PEXEL and signal sequence. Two transmembrane domains are predicted in the C-terminus between amino acids 168–182 and 188–206.
Complementation was confirmed by Southern blot of D10Pfvap1 genomic DNA digested with DraI and XmnI hybridized with a Pfvap1 probe where fragments of expected sizes were observed (Fig. 2C). Pfvap1 has a typical exported protein gene organization with a PEXEL motif in the beginning of the second exon (Sargeant et al., 2006), encoding for a protein with a predicted molecular weight of 31.1 kDa with two predicted C-terminal transmembrane domains (Fig. 2D). A single nucleotide polymorphism resulting in a non-synonymous amino acid substitution at position 117 (T for K) has been documented and occurs in 8 out of 10 of the laboratory strains examined (Aurrecoechea et al., 2009).
PfVAP1 is exported to the red blood cell (RBC) membrane
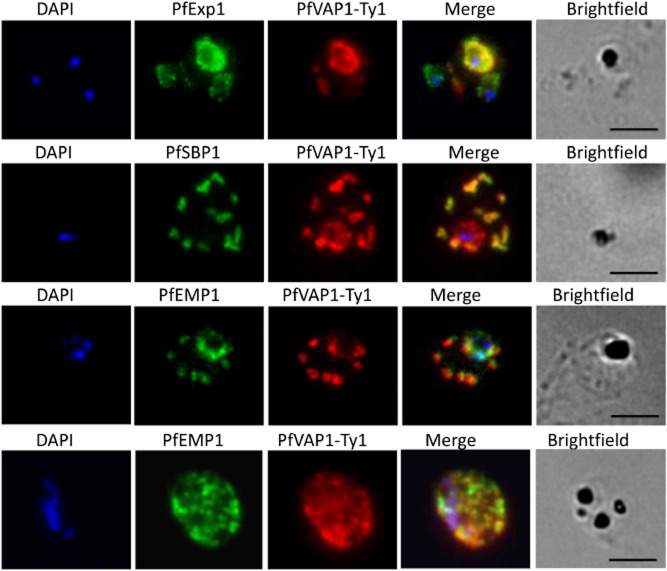
We generated D10 parasites complemented with Ty1-tagged PfVAP1 to investigate the subcellular localization and temporal expression profile. PfVAP1 is synthesized by ring stage parasites (from 8–11 h post-invasion) onwards. In D10Pfvap1 early trophozoites PfVAP1 is present in the parasitophorous vacuole (PV) where it partially co-localizes with PfExp1 (Fig. 3). PfVAP1 was observed in the PV until trophozoite stages and then the protein associates with the Maurer's clefts (24–36 h) as shown by labelling with PfSBP1 antibodies. PfEMP1 and PfVAP1 transiently co-localize in the Maurer's clefts suggesting that they interact in this particular compartment or that they are co-trafficked (Fig. 3; Supporting Information Fig. S1), they then appear to ‘dissociate’ when they reach the RBC membrane as very little overlap is evident in schizonts (Fig. 3). Similar to PfEMP1, PfVAP1 gives a punctate labelling pattern at the RBC membrane in schizonts (Fig. 3).
Fig. 3.
Localization of PfVAP1 in complemented D10 parasites. Ty1-tagged D10Pfvap1 parasites were labelled with anti-Ty1 monoclonal antibody to determine the localization of the protein. In early trophozoites PfVAP1 is associated with the parasitophorous vacuole (PV) as determined by double labelling with PfEXP1. It is then trafficked via the Maurer's clefts (PfSBP1) to the infected erythrocyte membrane. In the Maurer's clefts, PfVAP1 co-localizes with PfEMP1 (third row) and appears to localize in distinct regions, separate from PfEMP1 at the erythrocyte membrane (fourth row). Scale bars = 5 μm.
PfVAP1 plays a major role in cytoadhesion of FCR3-IE
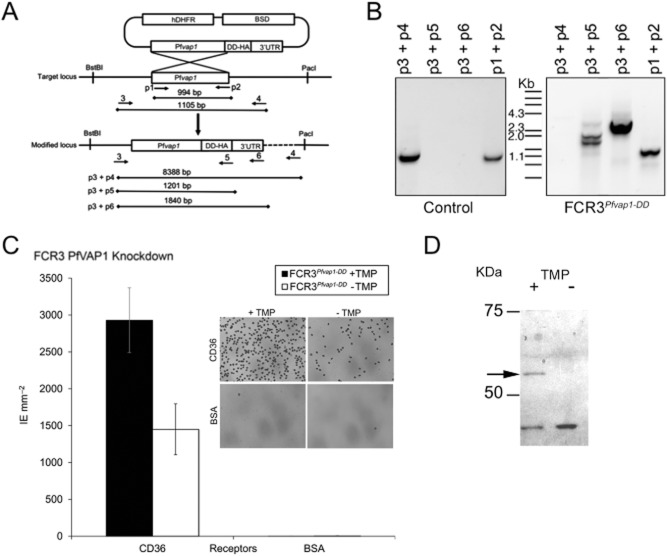
To directly assess the role of PfVAP1 in adhesion in the FCR3 reference strain, we used a protein knockdown approach in which Pfvap1 was cloned in frame with an E. coli dihydrofolate reductase (DHFR) destabilization domain (Muralidharan et al., 2011). The original pGDB2 vector was modified by the insertion of a human DHFR (hDHFR) cassette to transfect parasites without prior resistance to WR9220, necessary for the maintenance of the stable protein with trimethoprim (TMP; Fig. 4A; Supporting Information Table S1). In addition, we replaced GFP by a haemagglutinin A tag (HA-tag). Mutant parasites were selected with blasticidin-S (BSD) and following integration by single crossover homologous recombination, clones were obtained by limiting dilution and validated using specific PCR primer combinations to show integration into the genome (Fig. 4B; Supporting Information Table S1). In recombinant parasites, binding to CD36 was reduced by at least 50% upon removal of the stabilizing drug TMP indicating a significant role of PfVAP1 in this process (Fig. 4C). FCR3 transfected with the empty vector in the presence of TMP was comparable with the wild type, indicating no effects of drug treatment on binding (data not shown).
Fig. 4.
Knockdown of PfVAP1 reduces binding to CD36.A. Schematic representation of the knockdown (KD) strategy. Primer locations are indicated by arrows and numbered (e.g. p1). Expected amplicons are shown for the different primer pairs used.B. Integration of the PfVAP1-DD-HA in FCR3. Genomic DNA was used for PCR with the indicated primer pairs. Genomic DNA from parasites obtained from a control transfection with empty plasmid is shown.C. Binding of FCR3PfVAP1-DD KD. FCR3PfVAP1-DD KD parasites were cultured in the presence or absence of the drug TMP that stabilizes the degradation domain. A 50% reduction in binding was observed in FCR3PfVAP1-DD parasites without TMP. Mean binding of infected erythrocytes (IE) to purified CD36 and BSA is shown. Error bars = r standard deviation, n = 4 replicates). No effect of TMP on binding was detected using vector-transfected FCR3 parasites (data not shown). The inset shows Giemsa staining of FCR3PfVAP1-DD (+ /− TMP) bound to CD36 or the BSA negative control showing the effect of PfVAP1 in adhesion.D. Western blot of FCR3PfVAP1-DD parasites cultured in the presence (+) or absence (−) of TMP probed with anti-HA. A band of the expected molecular weight (52 kDA) corresponding the PfVAP1+HA fusion protein is observed in the presence of TMP that is lacking in the absence of TMP, confirming the degradation of the protein.
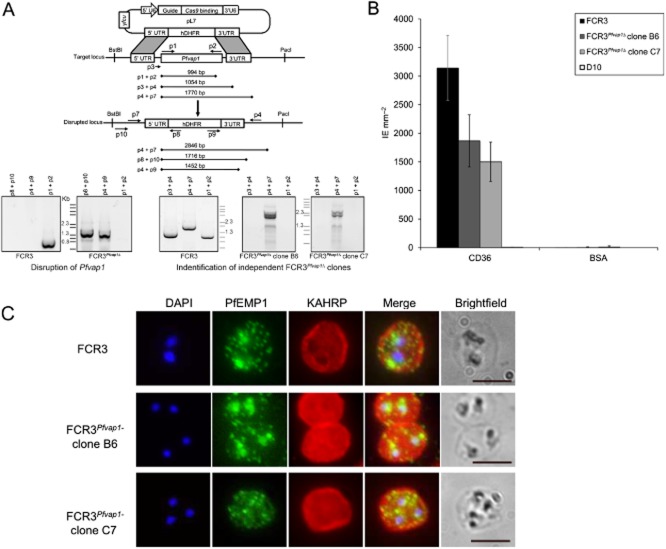
Western blot analysis of the HA-tagged FCR3Pfvap1-DD in the presence or absence of TMP using anti-HA antibodies shows that protein expression was abrogated in the conditional protein knockdown (Fig. 4D). However, since minimal amounts of protein still may mediate residual adhesion, we generated knockout FCR3Pfvap1Δ parasites by double homologous recombination (Fig. 5A; Supporting Information Table S1) replacing the entire genomic sequence of Pfvap1 with hDHFR using the recently developed CRISPR-Cas9 system (Ghorbal et al., 2014). Knockout parasites emerged 3 weeks after transfection. Cultures were treated with Ancotil to remove episomes and two independent clones (B6 and C7) were selected for further analysis by PCR and sequencing to confirm the integration of the hDHFR cassette and the absence of the gene (Fig. 5A; Supporting Information Table S1). Binding to CD36 was reduced by 55% (B6) and 40% (C7). After two rounds of panning on CD36, both clones retained similar adhesion levels (40% for B6 and 50% for C7; Fig. 5B), suggesting that maximal binding inhibition attributable to PfVAP1 was reached and had been obtained in the FCR3 PfVAP1 knockdown parasites.
Fig. 5.
FCR3Pfvap1Δ knockout parasites show a reduction in cytoadhesion.A. Diagram depicting the cas9/CRISPR strategy (Ghorbal et al., 2014) used for disruption of Pfvap1 in FCR3 parasites. Only the pL7 plasmid carrying the Pfvap1-specific guide RNA and homology boxes is shown. The position of primers is shown (arrows) and each primer (p) is numbered. The expected amplicon sizes for PCR verification are shown. Gene replacement with hDHFR was verified by PCR (left). In FCR3Pfvap1Δ parasites, part of the 5′UTR sequence is lost (p3), the absence of this sequence used to select two KO clones (right).B. Binding of FCR3Pfvap1Δ knockout clones panned on CD36. A 40% reduction in binding was observed in FCR3Pfvap1Δ clone B6 (dark grey). Binding to CD36 was reduced by 50% in the C7 clone (light grey). Mean binding of IE mm−2 to purified CD36 and BSA is shown. Error bars = o standard deviation, n = 3 replicates.C. FCR3Pfvap1Δ parasites export PfEMP1 and KAHRP in a manner comparable with wild-type FCR3 parasites, suggesting that there is no impairment or retention of these proteins in mutant parasites. Scale bar = 5 μm.
To ascertain that the observed decrease in binding levels in FCR3Pfvap1Δwere not due to off-target events, such as a concomitant deletion of KAHRP as has been reported for the complete rex1 knockouts in 3D7 (Dixon et al., 2011), we verified targeting of key proteins to different cellular compartments. Immunolabelling of FCR3Pfvap1Δ indicated that these parasites have KAHRP at the IE membrane and trafficking of PfEMP1 was comparable with FCR3 (Fig. 5C).
Trypsin resistance of PfEMP1 was reported in non-cytoadherent D10 parasites (Nacer et al., 2011). We investigated whether mutant FCR3Pfvap1Δ parasites show changes in trypsin sensitivity compared wild-type FCR3. The PfEMP1 of both B6 and C7 FCR3Pfvap1Δ clones were sensitive to trypsin digestion in a manner comparable with FCR3 as judged by the appearance of a shortened intracellular PfEMP1 fragment in Western blot analysis (Fig. 6).
Fig. 6.
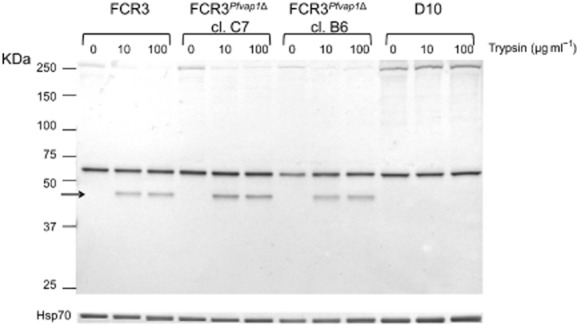
Trypsin sensitivity of PfEMP1 in FCR3Pfvap1Δ parasites. Synchronized FCR3 wild type and FCR3Pfvap1Δ parasites (30–35 h) were selected by gel floatation prior to trypsin. Parasites were treated with either 0, 10 or 100 μg ml−1 trypsin. The Triton X-100 insoluble/SDS soluble protein fraction was collected and separated by SDS-PAGE. A guinea pig anti-ATS antibody was used to detect PfEMP1. Treatment with trypsin results in the detection of an additional band (arrow) that represents the trypsin-resistant intracellular ATS domain. The membrane was then probed with the Hsp70 monoclonal antibody MAb1C11 as a loading control (bottom lane).
Discussion
We have identified a novel gene implicated in the major virulence process of P. falciparum, the adhesion of IE to host endothelial cells. Previously, we had observed the complete loss of parasite adhesion despite apparent normal expression levels of PfEMP1 at the IE surface. Since this phenotype is linked to the deletion of a large subtelomeric fragment on chromosome 9 in several distinct parasites lines, our screen for candidate molecules focused on genes that were lost on the truncated chromosome 9 of D10 parasites. Our complementation strategy and screening for restoration of adhesion led to the discovery of a single candidate gene out of 12 tested, which showed a clear increase in the levels of adhesion (D10 = 12.3 IE mm−2, D10Pfvap1-Ty1 = 255 IE mm−2 and D10Pfvap1 = 293 IE mm−2, i.e. 20-fold and 25-fold increase, respectively, for D10Pfvap1-Ty1 and D10Pfvap1). The screen revealed a protein that has never before been investigated or associated to parasite adhesion. Comparative genome analysis showed that Pfvap1 is only found in P. falciparum and despite its subtelomeric location adjacent to highly variable gene families, it is highly conserved between different parasite lines. The fact that this protein is exported together with PfEMP1 via Maurer's clefts to the host membrane suggests that it has evolved to contribute in the process of functional cytoadhesion at the surface of IE.
PfEMP1 trafficking to the surface does not depend on the deleted genes in D10. Our observation that PfVAP1 complementation does restore the adhesion phenotype adds an important new piece to this complex mechanism. How PfVAP1 contributes to this process is still unknown but we speculate that it may occur at different levels. PfEMP1, after having been exported to the IE cytoplasm via the Plasmodium translocon of exported proteins, may need chaperones to refold into a functional adhesion molecule (Elsworth et al., 2014). Although the amino acid sequence of PfVAP1 does not reveal any homology to chaperones, it may interact directly with PfEMP1 to allow proper refolding. Alternatively, PfVAP1 and PfEMP1 may form a heterodimer at the surface of IE that stabilizes a PfEMP1 conformation needed for functional display to host receptors. A third possibility is that posttranslational modifications such as phosphorylation, which have been shown to reduce cytoadhesion, have been modified in PfVAP1 mutant parasites (Hora et al., 2009). Further experiments are needed to obtain in-depth insight into the role of PfVAP1 in surface adhesion. The possibility that PfVAP1 is exposed on the IE surface could not be confirmed experimentally as surface labelling with the Ty-1 antibodies did not result in surface signals using immunofluorescence assay (IFA) on live IE. Failure of these antibodies to label surface-exposed regions of PfVAP1 may be due to the location of the C-terminus at the inner IE membrane.
Our protein knockdown and gene knockout data demonstrate that mutant PfVAP1 FCR3 parasites show reduced binding to CD36 (between 50% and 60% reduction). This suggests that other genes located on chromosome 9 modulate the binding capacity in adherent parasites. A promising candidate is rex1, which encodes for a Maurer's cleft resident protein, and which is also located on chromosome 9. Indeed, disruption of REX1 results in a partial loss of binding to CD36 (Dixon et al., 2011). Unlike the rex1 knockout or Hsp101 protein knockdown (Beck et al., 2014), PfEMP1 appears to be correctly trafficked in FCR3Pfvap1Δ knockout parasites.
Genes located on the right arm of chromosome 9 have also been implicated in gametocytogenesis and a number of them are predicted to be expressed in gametocytes, a stage that loses the expression of PfEMP1. Various transcriptomic data sets (http://www.plasmodb.org) suggests that Pfvap1 is also transcribed in gametocytes suggesting that the protein could have distinct functions depending on the life-cycle stage during which it is expressed.
In conclusion, we have identified a novel and unique P. falciparum protein with a major role in cytoadhesion named PfVAP1. Further characterization of PfVAP1 is warranted to determine exactly how this protein functions and interacts with PfEMP1 to clearly elucidate its role in binding.
Experimental procedures
Parasite cultures and transfections
Plasmodium falciparum D10 (derived from FC27), FCR3 and 3D7 were cultured in RPMI 1640 (GibcoBRL, Life Technologies), 5% Albumax I (GibcoBRL, Life Technologies) supplemented with 0.2 mM Hypoxathine (C.C.pro GmbH) as described (Cranmer et al., 1997). All parasite lines were regularly selected by gelatine flotation (Fresenius Kabi, France; Pasvol et al., 1978) and tested for the presence of mycoplasma (Rowe et al., 1998). Transfections were performed by electroporation of normal RBCs (Spalding et al., 2010) and the plasmid-loaded erythrocytes used for culturing plasmion-selected parasites. Drugs BSD (2.5 μg ml−1) or WR99210 (20 μg ml−1) were added 48h following transfections to select for mutant parasites. Several rounds of drug on/off cycling were performed for integration of the plasmid by single crossover.
Immunodetection
A pool of human hyperimmune sera (PIAG, Bouharoun-Tayoun and Druilhe, 1992), guinea-pig anti-PfEMP1 acidic transmembrane segment (ATS) (Wickert et al., 2003), mAb89 anti-KAHRP (Taylor et al., 1987), mAb1C11 anti-PfHSP70 (Mattei et al., 1989), anti-PfSBP1 (Blisnick et al., 2000) and mouse anti-PfExp1 (Günther et al., 1991) were used to detect parasite antigens. Tag-specific antibodies were mouse hybridoma supernatant Ty1 mAb BB2 (Bastin et al., 1996) and rabbit anti-HA (Abcam ab9110). Fluorochrome-conjugated secondary antibodies were purchased from Life Technologies. Alcaline phosphatase and horseradish peroxidase secondary antibodies were from Promega and GE Healthcare respectively.
Binding assays and panning
Parasites were enriched by gelatin flotation (Plasmion) and resuspended in binding medium (RPMI 1640 supplemented with 25 mM HEPES pH 6.8) at 5 × 107 IE ml−1. Assays were performed as described (Buffet et al., 1999) with 20 μl of purified CD36 (10 μg ml−1) immobilized on plastic Petri dishes or culture flasks, overnight at 4°C. For binding assays, control spots were coated with 1% BSA (fraction V, Sigma). Binding assays dishes were fixed with 2% glutaraldehyde, Giemsa stained for 10 min and quantified by microscopy.
DNA constructs
Complementation
The 6.4 kb expression vector was derived from pLN-ENR-GFP (Nkrumah et al., 2006) by replacing the ENR-GFP by selected genes (see Fig. 1). The genes were amplified by PCR from FCR3 genomic DNA (gDNA) and cloned into the FseI/AscI sites of the pComp-BSD plasmid. Construction with a Ty1- tag at the 3′ end of the Pfvap1 gene was also used for complementation and localization.
Conditional protein knockdown
The pGDB2-WR plasmid was generated from pGDB (Muralidharan et al., 2011) by insertion of a hDHFR cassette to use with parasites susceptible to WR99210 or TMP. FCR3 gDNA amplified by PCR was cloned into the AflII/XhoI sites and transfected FCR3 parasites selected by adding BSD. Integration was verified by PCR and clones obtained by limiting dilution.
Knockout of Pfvap1
The cas9/CRISPR genome editing system recently adapted to P. falciparum was used as described (Ghorbal et al., 2014) to knockout the Pfvap1 gene. In brief, homology regions in the 5′ and 3′ UTRs were selected to ensure complete replacement of the coding sequence and cloned into the pL7 plasmid on either side of the hDHFR selectable marker. The 555 bp 5′ UTR homology region was amplified from FCR3 DNA and cloned between the SpeI/AflII sites of the pL6 plasmid by In-Fusion reaction (In-Fusion HD cloning kit Clontech Laboratories). Similarly, the 477 bp 3′ UTR homology region was amplified by PCR and cloned into the EcoRI/NcoI sites. Single gRNA sequences were selected generating two plasmids. The first sequence corresponded to 20 bp in Pfvap1 coding sequence (5′-ACATATTAGATTTCTAACCC-3′) located at the beginning of the second exon (ATG + 327 bp), approximately halfway between the two homology regions. The second gRNA sequence (5′-TTTGTAGGTGTAACATATAC-3′) was selected towards centre of the coding sequence (ATG + 539 bp). After transfection with the two plasmids, parasites were obtained in 16 days, in two independent transfections. Parasites were treated with Ancotil to remove residual episomes, and were cloned by limiting dilution. After PCR verification, two FCR3 Pfvap1 clones (B6 and C7) were selected for further phenotypic studies.
Northern and Southern blots
Total RNA from 3D7, D10 and FCR3 was isolated and processed as previously described (Kyes et al., 1999; 2000,). TRIzol (TRIzol reagent, Ambion Life Technologies) was added to harvested ring-, trophozoite- and schizont-IE. Parasite DNA was extracted (Gene Elute Mammalian Genomic DNA miniprep kit, Sigma-Aldrich) digested with DraI/XmnI (BioLabs), and processed (Viebig et al., 2005). Nucleic acids were transferred onto Hybond N + (Amersham) and hybridized with a 32P-labelled Pfvap1 probe (Megaprime Labelling System, GE Healthcare). Membranes were washed with 2X sodium saline citrate (SSC), 0.1% sodium dodecyl sulfate (SDS) at 60°C and exposed with BioMax MS films and Transcreen HE (Kodak) at −80°C.
Parasite protein preparation
For the detection of tagged PfVAP1, IE were collected and saponin-lysed. Parasites were washed three times in PBS containing protease inhibitors and the pellet resuspended to obtain 106 IE μl−1. Samples were run on 10% Tris-Bis gels (BioRad) in MOPS Buffer (BioRad) and transferred onto nitrocellulose membranes (iBlot, Life Technologies). Membranes were blocked in 5% milk/0.05% tween 20 (Promega) in PBS (PBS/milk/tween) for 30 min at room temperature and incubated overnight with either mouse anti-Ty1 (1:100) or rabbit anti-HA (1:250). Following three washes with PBS/milk/tween, alkaline phosphate-conjugated secondary antibodies were added for 1 h at room temperature.
Trypsin cleavage assays
Infected RBCs were enriched by gelatin flotation and incubated for 15 min with either 0, 10 or 100 μg ml−1 of trypsin (Baruch et al., 1996). Following three washes in PBS containing protease inhibitors, cells were either incubated with pooled hyperimmune serum (1:5) or proteins extracted with 1% Triton X-100 (v/v) followed by 2% SDS. Extracts were resuspended in sample buffer, heated, separated on 10% Bis-Tris gels (BioRad), and transferred onto nitrocellulose membranes (iBlot, Life Technologies). Detection of PfEMP1 was performed by incubation with a guinea pig anti-ATS antibody (1:400) followed by anti-guinea pig alkaline phosphatase (AP)-conjugated secondary antibody. Equal protein loading was verified with the mAb1C11 anti-Hsp70 antibody (1:2000) and revealed with NBT/BCIP following incubation with an anti-mouse AP-conjugated secondary antibody.
qRT-PCR
To confirm the intraerythrocytic gene expression profile of Pfvap1, parasites were synchronized by plasmion and sorbitol to obtain a 5 h window. Parasites were harvested every 12 h during the course of a single cycle. Total blood was washed once in PBS and TRIzol immediately added. Samples were then stored at −80°C until further processing. RNA was extracted, precipitated, treated with DNAse (Ambion), reverse transcribed (SuperScript II Reverse Transcriptase – Life Technologies), and diluted to 300 ng μl−1 in nuclease-free water for qRT-PCR. Expression was analysed using the primers Exon1-F: 5′-GTTTCTTTATTTTCCCTATCC-3′; Exon1-R: 5′-GAAGTCTTTTTCCACCTTTC-3′; Intron-F: 5′-CCCTATCCAAAATGTGAAC-3′; Intron-R: 5′-GTCTATATTTTTTTGTGATG-3′. Control genes used were seryl transferase (Salanti et al., 2003), inositol (Mancio-Silva et al., 2013) and aldolase (Knapp et al., 1990).
Immunofluorescence
PfVAP1 was localized using anti-Ty1 or anti-HA tag antibodies. Parasites were spotted onto multiwell slides and air-dried (Hinterberg et al., 1994). To further define the localization of PfVAP1, double labelling IFA with PfEMP1 anti-ATS or rat anti-PfSBP1 Maurer's clefts markers were performed as already described (Nacer et al., 2011).
Flow cytometry
Parasites were synchronized (within a 5 h window) by plasmion and sorbitol and 30–35 h post-invasion schizonts were incubated with the human sera for 1 h at room temperature. After three washes with PBS, samples were reacted 1 h with anti-human Alexa 488 (1:100) (Life Technologies). Nuclei were stained 10 min with DAPI, washed, and the samples were diluted to 500 cells μl−1 for reading by flow cytometry (Fortesa). For the quantification of CD36 labelling, parasites were obtained as described above and incubated for 1 h at RT with 10 μg ml−1 chimeric CD36-human IgG in suspension. Cells were washed in PBS prior to the addition of mouse anti-human IgG Alexa 488-conjugated secondary antibody. Nuclei were stained with SYTO 61 (Life Technologies) and samples were analysed on a Guava flow cytometer (BD Biosciences). Data were compiled and analysed using FlowJo (version 10).
Acknowledgments
We thank A. Scherf for helpful discussions during the entire project. We also thank R. Miyazawa Martins for advice and assistance in flow cytometry data collection and analysis and N. Malmquist for advice on flow cytometry. In addition, we thank S. Vembar and J. Guizetti for their insight and advice in the course of the project. This work was supported by an ERC AdG (PlasmoEscape 250320), the French Parasitology Consortium ParaFrap (ANR-11-LABX0024) and ANR 11 JSV3 00401 PlasmoPiggyBac.
Conflict of interest
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. PfVAP1 and PfEMP1 co-localize in the Maurer's clefts in complemented D10 parasites. Ty1-tagged D10Pfvap1 parasites were labelled with anti-Ty1 monoclonal antibody (red), guinea pig anti-PfEMP1 (green), and rat anti-PfSBP1 (magenta), nuclei were stained with DAPI (blue). PfVAP1 associates with PfEMP1 in Maurer's clefts (PfSBP1). Scale bar = 5 μm.
Table S1. List of oligonucleotides used in this study.
References
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Gormley JA, Ma C, Howard RJ, Pasloske BL. Plasmodium falciparum erythrocyte membrane protein 1 is a parasitized erythrocyte receptor for adherence to CD36, thrombospondin, and intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1996;93:3497–3502. doi: 10.1073/pnas.93.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh A, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Beck JR, Muralidharan V, Oksman A, Goldberg DE. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature. 2014;511:592–595. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blisnick T, Betoulle MEM, Barale J-C, Uzureau P, Berry L, Desroses S, et al. Pfsbp1, a Maurer's cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Mol Biochem Parasitol. 2000;111:107–121. doi: 10.1016/s0166-6851(00)00301-7. [DOI] [PubMed] [Google Scholar]
- Bouharoun-Tayoun H, Druilhe P. Plasmodium falciparum malaria: evidence for an isotype imbalance which may be responsible for delayed acquisition of protective immunity. Infect Immun. 1992;60:1473–1481. doi: 10.1128/iai.60.4.1473-1481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet PA, Gamain B, Scheidig C, Baruch D, Smith JD, Hernandez-Rivas R, et al. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc Natl Acad Sci USA. 1999;96:12743–12748. doi: 10.1073/pnas.96.22.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyaroj SC, Coppel RL, Magowan C, Brown GV. A Plasmodium falciparum isolate with a chromosome 9 deletion expresses a trypsin-resistant cytoadherence molecule. Mol Biochem Parasitol. 1994;67:21–30. doi: 10.1016/0166-6851(94)90092-2. [DOI] [PubMed] [Google Scholar]
- Chan J-A, Howell KB, Reiling L, Ataide R, Mackintosh CL, Fowkes FJI, et al. Targets of antibodies against Plasmodium falciparum-infected erythrocytes in malaria immunity. J Clin Invest. 2012;122:3227–3228. doi: 10.1172/JCI62182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranmer SL, Magowan C, Liang J, Coppel RL, Cooke BM. An alternative to serum for cultivation of Plasmodium falciparum in vitro. Trans R Soc Trop Med Hyg. 1997;91:363–365. doi: 10.1016/s0035-9203(97)90110-3. [DOI] [PubMed] [Google Scholar]
- Crompton PD, Moebius J, Portugal S, Waisberg M, Hart G, Garver LS, et al. Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Ann Rev Immunol. 2014;32:157–187. doi: 10.1146/annurev-immunol-032713-120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day KP, Karamalis F, Thompson J, Barnes DA, Peterson C, Brown H, et al. Genes necessary for expression of a virulence determinant and for transmission of Plasmodium falciparum are located on a 0.3-megabase region of chromosome 9. Proc Natl Acad Sci USA. 1993;90:8292–8296. doi: 10.1073/pnas.90.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MWA, Kenny S, McMillan PJ, Hanssen E, Trenholme KR, Gardiner DL, Tilley L. Genetic ablation of a Maurer's cleft protein prevents assembly of the Plasmodium falciparum virulence complex. Mol Microbiol. 2011;81:982–993. doi: 10.1111/j.1365-2958.2011.07740.x. [DOI] [PubMed] [Google Scholar]
- Eastman RT, Fidock DA. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat Rev Microbiol. 2009;7:864–874. doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsworth B, Matthews K, Nie CQ, Kalanon M, Charnaud SC, Sanders PR, et al. PTEX is an essential nexus for protein export in malaria parasites. Nature. 2014;511:587–591. doi: 10.1038/nature13555. [DOI] [PubMed] [Google Scholar]
- Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- Günther K, Tümmler M, Arnold H-H, Ridley R, Goman M, Scaife JG, Lingelbach K. An exported protein of Plasmodium falciparum is synthesized as an integral membrane protein. Mol Biochem Parasitol. 1991;46:149–158. doi: 10.1016/0166-6851(91)90208-n. [DOI] [PubMed] [Google Scholar]
- Hinterberg K, Scherf A, Gysin J, Toyoshima T, Aikawa M, Mazie J-C, et al. Plasmodium falciparum: the Pf332 antigen is secreted from the parasite by a brefeldin A-dependent pathway and is translocated to the erythrocyte membrane via the Maurer's clefts. Exp Parasitol. 1994;79:279–291. doi: 10.1006/expr.1994.1091. [DOI] [PubMed] [Google Scholar]
- Hora R, Bridges DJ, Craig A, Sharma A. Erythrocytic casein kinase II regulates cytoadherence of Plasmodium falciparum-infected red blood cells. J Biol Chem. 2009;284:6260–6269. doi: 10.1074/jbc.M809756200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp B, Hundt E, Küpper HA. Plasmodium falciparum aldolase: gene structure and localization. Mol Biochem Parasitol. 1990;40:1–12. doi: 10.1016/0166-6851(90)90074-v. [DOI] [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Rowe JA, Kriek N, Newbold CI. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc Natl Acad Sci USA. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Mancio-Silva L, Lopez-Rubio JJ, Claes A, Scherf A. Si2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat Commun. 2013;4:1530. doi: 10.1038/ncomms2539. doi: 10.1038/ncomms2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Spielmann T. Protein export in malaria parasites: many membranes to cross. Curr Opin Microbiol. 2013;16:445–451. doi: 10.1016/j.mib.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei D, Scherf A, Bensaude O, Pereira da Silva L. A heat shock-like protein from the human malaria parasite Plasmodium falciparum induces autoantibodies. Eur J Immunol. 1989;19:1823–1828. doi: 10.1002/eji.1830191010. [DOI] [PubMed] [Google Scholar]
- Miller LH, Ackerman HC, Su XZ, Wellems TE. Malaria biology and disease pathogenesis: insights for new treatments. Nat Med. 2013;19:156–167. doi: 10.1038/nm.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci USA. 2011;108:4411–4416. doi: 10.1073/pnas.1018449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacer A, Roux E, Pomel S, Scheidig-Benatar C, Sakamoto H, Lafont F, et al. clag9 is not essential for PfEMP1 surface expression in non-cytoadherent Plasmodium falciparum parasites with a chromosome 9 deletion. PLoS ONE. 2011;6:e29039. doi: 10.1371/journal.pone.0029039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Böhme U, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasvol G, Wilson RJM, Smalley ME, Brown J. Separation of viable schizont-infected red cells of Plasmodium falciparum from human blood. Ann Trop Med Parasitol. 1978;72:87–88. doi: 10.1080/00034983.1978.11719283. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Scragg IA, Kwiatkowski D, Ferguson DJP, Carucci DJ, Newbold CI. Implications of mycoplasma contamination in Plasmodium falciparum cultures and methods for its detection and eradication. Mol Biochem Parasitol. 1998;92:177–180. doi: 10.1016/s0166-6851(97)00237-5. [DOI] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Sargeant TJ, Marti M, Caler E, Carlton JM, Simpson K, Speed TP, Cowman AF. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- Smith JD, Rowe JA, Higgins MK, Lavstsen T. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum infected erythrocytes. Cell Microbiol. 2013;15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Allary M, Gallagher JR, Prigge ST. Validation of a modified method for Bxb1 mycobacteriophage integrase-mediated recombination in Plasmodium falciparum by localization of the H-protein of the glycine cleavage complex to the mitochondrion. Mol Biochem Parasitol. 2010;172:156–160. doi: 10.1016/j.molbiopara.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DW, Parra M, Chapman GB, Stearns ME, Rener J, Aikawa M, et al. Localization of Plasmodium falciparum histidine-rich protein 1 in the erythrocyte skeleton under knobs. Mol Biochem Parasitol. 1987;25:165–174. doi: 10.1016/0166-6851(87)90005-3. [DOI] [PubMed] [Google Scholar]
- Tembo DL, Nyoni B, Murikoli RV, Mukaka M, Milner DA, Berriman M, et al. Differential PfEMP1 expression is associated with cerebral malaria pathology. PLoS Pathog. 2014;10:e1004537. doi: 10.1371/journal.ppat.1004537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholme KR, Gardiner DL, Holt DC, Thomas EA, Cowman AF, Kemp DJ. clag9: a cytoadherence gene in Plasmodium falciparum essential for binding of parasitized erythrocytes to CD36. Proc Natl Acad Sci USA. 2000;97:4029–4033. doi: 10.1073/pnas.040561197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viebig NK, Gamain B, Scheidig C, Lepolard C, Przyborski J, Lanzer M, et al. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep. 2005;6:775–781. doi: 10.1038/sj.embor.7400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Malaria Report Geneva. Geneva, Switzerland: WHO Press; 2013. [Google Scholar]
- Wickert H, Wissing F, Andrews KT, Stich A, Krohne G, Lanzer M. Evidence for trafficking of PfEMP1 to the surface of P. falciparum-infected erythrocytes via a complex membrane network. Eur J Cell Biol. 2003;82:271–284. doi: 10.1078/0171-9335-00319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. PfVAP1 and PfEMP1 co-localize in the Maurer's clefts in complemented D10 parasites. Ty1-tagged D10Pfvap1 parasites were labelled with anti-Ty1 monoclonal antibody (red), guinea pig anti-PfEMP1 (green), and rat anti-PfSBP1 (magenta), nuclei were stained with DAPI (blue). PfVAP1 associates with PfEMP1 in Maurer's clefts (PfSBP1). Scale bar = 5 μm.
Table S1. List of oligonucleotides used in this study.