Abstract
The urokinase receptor (uPAR) is a GPI-anchored cell surface receptor that is at the center of an intricate network of protein-protein interactions. Its immediate binding partners are the serine proteinase urokinase (uPA), and vitronectin (VTN), a component of the extracellular matrix. uPA and VTN bind at distinct sites on uPAR to promote extracellular matrix degradation and integrin signaling, respectively. Here, we report the discovery of a new class of pyrrolone small-molecule inhibitors of the tight ∼1 nM uPAR•uPA protein-protein interaction. These compounds were designed to bind to the uPA pocket on uPAR. The highest affinity compound, namely 7, displaced a fluorescently-labeled α-helical peptide (AE147-FAM) with an inhibition constant Ki of 0.7 µM and inhibited the tight uPAR•uPAATF interaction with an IC50 of 18 µM. Biophysical studies with surface plasmon resonance showed that VTN binding is highly dependent on uPA. This cooperative binding was confirmed as 7, which binds at the uPAR•uPA interface, also inhibited the distal VTN•uPAR interaction. In cell culture, 7 blocked the uPAR•uPA interaction in uPAR-expressing human embryonic kidney (HEK-293) cells, and impaired cell adhesion to VTN, a process that is mediated by integrins. As a result, 7 inhibited integrin signaling in MDA-MB-231 cancer cells as evidenced by a decrease in focal adhesion kinase (FAK) phosphorylation and Rac1 GTPase activation. Consistent with these results, 7 blocked breast MDA-MB-231 cancer cell invasion with IC50 values similar to those observed in ELISA and surface plasmon resonance competition studies. Explicit-solvent molecular dynamics simulations show that the cooperativity between uPA and VTN is attributed to stabilization of uPAR motion by uPA. In addition, free energy calculations revealed that uPA stabilizes the VTN•uPARSMB interaction through more favorable electrostatics and entropy. Disruption of the uPAR•VTNSMB interaction by 7 is consistent with the cooperative binding to uPAR by uPA and VTN. Interestingly, the VTNSMB•uPAR interaction was less favorable in the VTNSMB•uPAR•7 complex suggesting potential cooperativity between 7 and VTN. Compound 7 provides an excellent starting point for the development of more potent derivatives to explore uPAR biology.
INTRODUCTION
The urokinase receptor (uPAR) is a glycosyl-phosphatidylinositol (GPI)-anchored protein that is at the center of a network of more than 50 protein-protein interactions (1). Biochemical and biophysical studies reveal that its immediate binding partners are the serine proteinase urokinase-type plasminogen activator (uPA) (2) and the somatomedin B (SMB) domain of the extracellular matrix glycoprotein vitronectin (VTN) (3–5). uPA binding to uPAR is mediated by a 25-residue β-turn growth factor-like (EGF) domain that is ensconced in a large hydrophobic cavity lined with several residues known as hot-spots that contribute more than an order of magnitude to the binding (6). As a result, the uPAR•uPA interaction is a tight ∼1 nM and long-lived complex that occurs over a large protein-protein interface of more than 1,000 A2 (6–9). In contrast, the uPAR•VTN interaction occurs over a smaller interface of 75 Å (3–5), driven primarily by a by an electrostatic interaction between Arg91 on uPAR and Asp22 on VTNSMB (3). The binding of the SMB domain is a transient interaction with micromolar affinity that is uPA-dependent (10,11) despite the lack of direct interaction between uPA and VTN. Studies have shown that VTN binds to uPAR but only in the presence of uPA (12) suggesting a potential allosteric mechanism (10,13,14). Crystal structures of the VTNSMB•uPAR•uPAATF multiprotein complex reveals that the VTN and uPA binding interfaces occur at distal sites (5).
In cancer cells, uPAR-bound uPA and VTN engender different biological responses. uPA promotes proteolysis in the cellular milieu following its activation upon binding to uPAR (15,16). Active uPA is highly specific to plasminogen, turning it into a non-specific protease that triggers a cascade of proteolytic events such as activation of matrix metalloproteinases (MMPs) (17) and extracellular matrix breakdown (18–22). Plasmin and MMPs activate latent growth factors to release them from extracellular matrix sequestration (23,24). Therefore, inhibition of the uPAR•uPA interaction is expected to reduce degradation of the extracellular matrix and therefore impair cancer cell invasion. VTN, on the other hand, mediates uPAR interaction with integrins through an RGD motif located near its N-terminal somatomedin (SMB) domain (25,26). Binding of integrins to the RGD motif on VTN triggers integrin signaling, cell migration, adhesion, lamellipodia formation, and modification of focal adhesion sites (4,27–29). uPAR interaction with the fibronectin receptor α5β1 integrin promotes ERK-dependent tumor cell proliferation, adhesion, migration, induction of MMPs, pericellular proteolysis, and cancer cell invasion (30–32). uPAR binding to β1 integrin leads to downstream activation of FAK and ERK signaling pathways (32–38). uPAR-β3 integrin interaction is associated with activation of Rac1 (39,40). Src signaling is activated by uPAR downstream of both α5β1 and α3β1 integrins (33,41). In sum, inhibition of uPA and VTN binding to uPAR is expected to impair an array of cellular processes that include adhesion, cell spreading, and invasion.
Small molecules that bind to uPAR can be used to disentangle its protein interactions and define their role in cancer invasion and metastasis and other pathological processes associated with the receptor. Despite the tight and stable uPAR•uPA complex, the presence of a large number of hot-spot residues at the protein-protein interface, and the well-defined hydrophobic binding pocket of uPA, makes it suitable for the design of small-molecule antagonists (42). Our previous efforts have led to the first small molecule that inhibited the uPAR•uPA interaction (43,44). The compound IPR-803 was discovered by molecular docking of multiple structures obtained from explicit-solvent molecular dynamics simulations of uPAR. IPR-803 inhibited cancer cell invasion and blocked cancer metastasis in vivo (43). Our work has shown that compounds that bind to uPAR share common structural features and occupy specific pockets in uPAR that accommodate critical hot-spot residues of uPA (45).
Here, we explore the effect of the uPAR•uPA interaction on the distant uPAR•VTN. First, we study the cooperative binding between uPA and VTN using surface plasmon resonance and other biochemical assays. We then use our small molecules to probe the uPAR•VTN interaction in cultured cells with uPAR-expressing HEK-293 cells. We further probe the effect of our compound on uPAR signaling through integrins using MDA-MB-231 breast cancer cells. Finally, we employ explicit-solvent molecular dynamics simulations and free energy calculations to explore the structural basis for the cooperative binding between uPA and VTN.
MATERIALS AND METHODS
Microtiter-based ELISA for uPAR•uPA
uPAR without the GPI anchor was obtained by a purification process as previously described (46). High-binding microplates (Greiner Bio-One) were incubated for 2 h at 4°C with 100 µL of 2 µg·mL−1 of uPAATF in PBS for immobilization as previously described (44). The plate was washed with 0.05% Tween-20 in PBS buffer between each step. A 1:1 mixture of Superblock® buffer in PBS (Thermo Fisher Scientific, Inc. Waltham, MA) with 0.04 M NaH2PO4 and 0.3 M NaCl buffer was used for blocking at room temperature for 1 h. 75 nM uPAR in PBS with 0.025% triton X-100 was added with indicated concentrations of compounds. Compounds were screened initially at 50 µM. For concentration-dependent studies, a range of compound concentrations from 100 µM to 0.4 µM was used. Final DMSO concentration was 1%. Following incubation for 30 minutes and subsequent washing steps, human uPAR biotinylated antibody (1:3000 dilution of 0.2 mg·mL−1 BAF807, R&D Systems, Minneapolis, MN) in PBS containing 1% BSA was added to the wells (100 µL/well) and incubated for 1 h to allow for the detection of bound uPAR. Following washing, streptavidin-horseradish-peroxidase in PBS containing 1% BSA was added for 20 min. The signal obtained in the presence of TMB in phosphate-citrate buffer (pH = 5) and hydrogen peroxide was stopped by adding H2SO4 solution and detected using a SpectraMax M5e (Molecular Devices, Sunnyvale, CA).
Microtiter-based ELISA for uPAR•VTN
High-binding microplates (Greiner Bio-One) were incubated for 12 h at 4°C with 100 µL of 5 µg·mL−1 of VTN (2349-VN-100, R&D Systems, Minneapolis, MN) in carbonate buffer (PH=9.6) for immobilization. The plate was washed with 0.05% Tween-20 in PBS buffer between each step. A 1:1 mixture of Superblock® buffer in PBS (Thermo Fisher Scientific, Inc. Waltham, MA) with 0.04 M NaH2PO4 and 0.3 M NaCl buffer was used for blocking at room temperature for 1 h. 120 nM uPAR.uPAATF in PBS with 0.01% triton X-100 was added with indicated concentrations of compounds. Compounds were screened initially at 50 µM. For concentration-dependent studies, a range of compound concentrations from 100 µM to 0.4 µM was used. Final DMSO concentration was 1%. Following incubation for 60 minutes and subsequent washing steps, human uPAR biotinylated antibody (1:3000 dilution of 0.2 mg·mL−1 BAF807, R&D Systems, Minneapolis, MN) in PBS containing 1% BSA was added to the wells (100 µL/well) and incubated for 1 h to allow for the detection of bound uPAR. Following washing, streptavidin-horseradish-peroxidase in PBS containing 1% BSA was added for 20 min. The signal obtained in the presence of TMB in phosphate-citrate buffer (pH = 5) and hydrogen peroxide was stopped by adding H2SO4 solution and detected using a SpectraMax M5e (Molecular Devices, Sunnyvale, CA).
Fluorescence Polarization
Polarized fluorescence intensities were measured using EnVision® Multilabel plate readers (PerkinElmer) with excitation and emission wavelengths of 485 and 530 nm, respectively (44). Samples were prepared in Thermo Scientific Nunc 384-well black microplate with a final volume of 50 µL in duplicates. First, the compounds were serially diluted in DMSO and further diluted in 1 x PBS buffer with 0.01% Triton X-100 for a final concentration of 100 µM to 0.046 µM. Triton X-100 was added in the buffer to avoid compound aggregation. 35 µL of the compound solution and 10 µL of PBS containing uPAR was added to the wells and incubated for at least 15 minutes to allow the compound to bind to the protein. Finally 5 µL of fluorescent AE147-FAM peptide was added for a total volume of 50 µL in each well resulting in final uPAR and peptide concentrations of 320 nM and 100 nM respectively. The final DMSO concentration was 2%, which had no effect on the binding of the peptide. Controls included wells containing only the peptide and wells containing both protein and peptide each in quadruplicates to ensure the validity of the reaction assay. A unit of millipolarization (mP) was used for calculating percentage inhibition of the compounds. Inhibition constants were measured using the Ki calculator available at http://sw16.im.med.umich.edu/software/calc_ki/.
Surface Plasmon Resonance
A Biacore 3000 instrument (GE Healthcare Life Sciences, Uppsala, Sweden) was used to study the effect of small molecules on the uPAR• uPAATF interaction. uPAATF was immobilized on a CM5 sensor chip (Biacore, Piscataway, NJ) using standard amine coupling chemistry recommended by the manufacturer. Surface densities of 179 RU of uPAATF were reached. The running buffer was HBS-EP and was obtained from GE (GE healthcare Bio-sciences, Piscataway, NJ). For the injection of compounds that included DMSO, the buffer was supplemented with 1% DMSO. Protein controls, that included DMSO but no compounds, were injected following two to three injections with compounds to ensure that the regeneration or compounds were not affecting the levels of immobilized uPAATF. All samples were prepared from 10 mM stock solutions in final concentrations ranging from 0.7 µM to 100 µM in 1% DMSO. Injections of uPAR (60 µL) in the presence and absence of compounds were done at 20 µL/min with 180 s association and 120 s dissociation time. The concentration of uPAR for the inhibition studies with compound was selected from a curve following several injections at different concentration of uPAR to immobilized uPAATF. Bound uPAR response values were selected from the equilibration binding region of the curve 120 s after injection. Regeneration was done using a single injection of 10 µL of 5 mM HCl.
Two approaches were followed to study the binding between VTN to uPAR, uPAATF, and their complex uPAR•uPAATF. uPAATF or uPAR was immobilized on a CM5 sensor chip (Biacore, Piscataway, NJ) using standard amine coupling chemistry recommended by the manufacturer. Surface densities of 1,600 RU of uPAATF or 1,900 RU of suPAR were reached. The running buffer used in the experiment was HBS-EP (GE healthcare Bio-sciences, Piscataway, NJ). For the chip immobilized with uPAATF, 60 µL of uPAR at a concentration of 100 nM was injected for 180 s with 120 s dissociation time. Then 60 µL VTN was injected at a concentration of 100 nM for 180 s with 120 s dissociation time. In the case of immobilized suPAR, 25 nM of uPAATF was injected with the kinject for 180 s (60 µL total volume) with 120 s dissociation time. VTN was injected with the kinject option at a concentration of 100 nM VTN (60 µL, 180 s association time and 120 s dissociation time).
Isothermal Titration Calorimetry
Experiments were carried on an ITC200 microcalorimeter from MicroCal, LLC (GE Healthcare). The experiments were conducted at 25 °C. uPAR was concentrated and dialyzed against 150 mM NaCl, 10 mM potassium phosphate at pH 7.4. DMSO was added to the protein for a solution with 6% DMSO. Samples were degassed for 3 min. The microsyringe was loaded with a solution of 0.6 mM 7 in PBS with 6% DMSO and was inserted into the 0.2003 mL microcalorimeter cell, which was filled with the uPAR solution (0.0167 mM, PBS, 6% DMSO). The system was equilibrated to 25 °C. Titrations were conducted using an initial control injection of 0.2 µL followed by another 19 identical injections at 120 s intervals. The data was corrected for protein heats of dilution based on separate measurements by titrating protein into buffer. MicroCal Origin 7.0 software was used to process the data yielding the binding enthalpy (ΔH) and dissociation constant (KD). A single binding mode model was used, supplied with the MicroCal Origin software. Thermodynamic parameters ΔG and ΔS were determined using standard thermodynamic equations.
Saturation Transfer Difference (STD) NMR
To further characterize and confirm the binding of 7 to uPAR, saturation transfer difference (STD) nuclear magnetic resonance (NMR) was used (47). Selective pulses were applied at −0.2 ppm to irradiate the protein methyl groups (where no ligand peaks appear) while the off-resonance frequency was positioned at 30 ppm. Saturation was carried out with a total 2 s pulse train composed of a repeated 50 ms gauss shaped pulse and 0.1 ms inter-pulse delay. The STD experiment of the small molecule with protein was acquired with 2,048 scans. The same experiment was collected on small molecule alone using exactly the same acquisition parameters except with 1,024 scans. To ensure that we only observed magnetization from the small molecule, uPAR was added at a concentration of 50 fold less than the small molecule. 7 was dissolved in deuterated DMSO at a concentration of 20 mM; it was then diluted in phosphate buffer to a concentration of 438 µM for the NMR experiment. NMR experiments were acquired at 298 K on a VNMRS 600MHz NMR spectrometer operating at a magnetic field strength of 18.8 T and equipped with a cold probe (Agilent Technologies, Santa Clara CA). 5 mm NMR tubes were used for the NMR experiments. Water suppression was done using Varian water sculpting pulse sequence.
Reagents
Biotinylated anti-human uPAR antibody (BAF807) was purchased from R&D Systems (Minneapolis, MN).
Cell Culture
MDA-MB-231 cells were cultured in Dulbecco's Modified Eagle Medium (Cellgro, Manassas, VA) supplemented with 10% FBS, 1% penicillin/streptomycin in a 5% CO2 atmosphere at 37°C.
Invasion
Invasion assays were performed using BD Biocoat Matrigel invasion chambers (BD Biosciences, San Jose, CA) as previously described (43,44,48,49). The undersurface of the inserts was coated with 30 ng µl−1 of fibronectin at 4 °C overnight. The inserts were equilibrated with 0.5 mL of serum-free medium in the upper and lower chamber separately for 2 h at 37 °C. After 4 h of serum starvation, cells were harvested and 5 × 104 cells in 500 µL medium containing 0.1% FBS and the compounds at the indicated concentrations or 1% DMSO control were plated onto the upper chamber. As a control of cell viability, 104 cells at the above conditions were plated in 100 µl in each well of 96-well plates. 500 µL of 10% FBS medium containing the same amount of compounds or DMSO control was added to the lower chamber. After a 16 h incubation at 37°C in 5% CO2, non-invaded cells were removed from the upper chamber with a cotton swab, and the invaded cells were fixed in methanol for 30 min at room temperature and stained with Hematoxylin Stain Harris Modified Method (Fisher Scientific, Waltham, MA) for 1 h at room temperature. We washed the filters with water 3 times. Filters were air-dried, and the number of invaded cells was counted in ten separate 200 × fields; meanwhile, 20 µl 5 mg/ml MTT (Sigma-Aldrich, St. Louis, Missouri) were added to each well, cells were incubated at 37°C in 5% CO2 for 2 h, viable cells were quantified at absorbance of 570 nm and 630 nm (reference background) as previously described (48,50,51).
Cell Viability Assay
104 cells MDA-MB-231 were plated overnight in 100 µl in each well of 96 well plates. Cells were treated with DMSO (control) or compounds at the indicated concentrations for 3 days. 20 µl 5 mg/ml MTT (Sigma-Aldrich, St. Louis, Missouri) were added to each well, cells were incubated at 37°C in 5% CO2 for 2 h, viable cells were quantified at absorbance of 570 nm and 630 nm (reference background) as previously described (48,50,51).
Fluorescence-Activated Cell Sorting (FACS) Analysis
A confluent flask of HEK-293-uPAR cells was treated with PMA at 100 µM for 48 hours, and then harvested with trypsin. Harvested cells were treated with 50 mM glycine-HCL, 0.1 M NaCl, at pH 3.0 and room temperature for 1 min. Subsequently, the acidic buffer was neutralized by 10 equivalents of 0.5 M HEPES and 100 mM NaCl at a pH of 7.5. The cells were washed and re-suspended in 1 x PBS and 0.1 BSA and then adjusted to 106 cells ml−1. HEK-293-uPAR cells were pre-incubated with different concentration of 7 (45 mins, room temperature) prior to addition of 40 nM of FITC-conjugated HMW-uPA (Innovative Research, Novi, MI) and allowed to incubate for 30 mins at room temperature. Stained cells were washed and re-suspended with 1xPBS and 0.1 BSA, 1 µl of 100 µg·ml−1 propodium iodide (Sigma-Aldrich, St. Louis, Missouri) was added to the samples and the cells were incubated for 15 min at room temperature. 105 cells of each sample were analyzed with a BD Biosciences FACSCalibur cytometer.
Adhesion Assay
96-well plates were coated with 5 ng•µl−1 vitronectin (R&D, Minneapolis, MN, USA) at 4 °C overnight, and then blocked with 2 % bovine serum albumin (BSA) in PBS for 1 h. After starving with serum-free medium for 4 h, 293-uPAR and HEK293 cells (2.5×104 cells mL−1) were suspended in 100 µL of DMEM containing 0.1 % FBS with indicated concentrations of IPR1110, or DMSO control in the presence or absence of 500pM uPA-ATF at 37 °C for 90 min. Medium was then carefully suctioned out from each well. Each well was washed three times with PBS. After washing, the adherent cells were fixed, stained with crystal violet, and quantified by measuring the absorbance at 540 nm.
Wound Healing Assay
MDA-MB-231 cells were allowed to grow confluently in 12-well plates. A linear wound was created by scraping the wells with a micropipette tip. The floating cells were removed by gentle washes in culture medium, and then the cells were cultured with completed media in the presence of 7 at 100 µM or 1% DMSO. The degree of wound closure was assessed using a Nikon Diaphot 300 microscope in three randomly chosen regions by measuring the distance between the wound edges just after wounding and after 16 h.
Western Blot Analysis
Total cell lysates were prepared in standard RIPA extraction buffer containing protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, Missouri). Thirty micrograms of protein were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (Amersham, Arlington Heights, IL). The membranes were immunoprobed with FAK, Phospho-FAK (Y397) (Cell Signaling, Danvers, MA) or Actin (C-2) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C overnight separately. Next, membranes were incubated with IRDye 800-conjugated goat anti-mouse IgG (Rockland, Immunochemicals, Gibertsville, PA) or Alexa Fluor 680 goat anti-rabbit IgG (Life Technologies, Grand Island, NY) as secondary antibodies. Bands were detected using Li-Cor Odyssey Imaging System (Li-Cor, Lincoln, NE).
Rac1 Pull-Down Assay
Rac1 pulldown kits (Thermo Scientific) were used to assay the active GTP-bound Rac1. MDA-MB-231 cells were seeded in 10-cm plates at a density of 2.2 × 106 cells in DMEM with 10% FBS and 1% of P/S till 80–90% confluence. The cells were then treated with 25 µM 7 or 1% DMSO for 24 hours. After rinsing with ice-cold PBS, cell samples were collected with 0.5 ml of lysis/binding/wash Buffer and transferred to a microcentrifuge tube. After incubating on ice for 5 minutes, the supernatant of cell samples (total lysate) were collected through centrifuging at 16000 x g for 15 minutes. 500 µg of total lysate was added to the spin cup containing the glutathione resin and 20 µg of GST-human Pak1-PBD. Sealed spin cups were incubated at 4°C for 1 hour with gentle rocking for pulling down active RAC1. The pulled down active RAC1 was then eluted and subjected to western blot analysis. 20 µg of cell lysate was analyzed by SDS-PAGE and Western Blot to determine the level of total Rac1 protein.
Cheminformatics Search
The ChemDiv library containing 969,572 compounds in its SMILES string format was obtained from the ZINC (v12) website. The compound library was pre-processed with the RDKit (31) cheminformatics tool to eliminate the ones that are not able to be processed by the software, which resulted in a new set of 802530 compounds. Duplicates were removed using ZINC codes, leading to a set of 716113 with unique ZINC codes. A substructure search was carried out on the core structure of pyrazole, piperidinone, and pyrrolidinone compounds that were previously shown to bind to uPAR with inhibition constants ∼ 25 µM (45,50). The fragments that were used for the search are shown in Scheme S1. The search was done with RDKit against the processed ChemDiv compounds, which gave rise to 6213, 3370, and 636 compounds from the pyrazole, pyrrolidinone, and piperidinone cores respectively. Based on the binding mode predicted for the parental structures (45,50), we further filtered these compounds for molecules that possessed three or more substituents attached to the core, of which at least three were required to contain a ring structure (ring size greater or equal to 5). Finally a molecular weight filter (300 ∼ 650 Dalton) was applied to eliminate fragments for large compounds, resulting in a set of 2196 molecules. The compounds were imported into the Canvas program (Schrödinger, Inc., New York, NY, 2009) for further clustering. Atom pair fingerprints (52) and K-mean clustering algorithm were employed. The program was specified to cluster each set of compounds into 100 subsets, resulting in the selection of 72 compounds that were purchased and tested for activity as described below.
Compound Docking and Clustering
Three-dimensional structures of the commercial compounds were downloaded from ZINC Web site in mol2 file format. The compound structures were imported into Maestro (version 9.5, Schrödinger, LLC, New York, NY, 2013) and prepared by LigPrep (version 2.6, Schrödinger, LLC, New York, NY, 2013). The prepared compounds were docked into uPAR protein (3BT1) using Glide (version 5.9, Schrödinger, LLC, New York, NY, 2013) in standard precision (SP). 100 poses per ligand were included in the post-docking minimization. 20 poses per ligand were collected in the output file. All the binding poses generated by Glide were visually checked and clustered into different groups.
Molecular Dynamics Simulations
The initial coordinates of uPAR, uPA and vitronectin (VTN) were obtained from the Protein Data Bank (PDB). The PDB file (PDB code 3BT1) was imported into Maestro (version 9.4, Schrödinger, LLC, New York, NY, 2013) and prepared using the Protein Preparation Wizard. Bond orders were assigned, hydrogen atoms were added, disulfide bonds were created. Three separate simulations were carried out: (i) VTN·uPAR·uPA; (ii) VTN·uPAR (uPA chain subtracted manually); (iii) VTN·uPAR·7. A mol2 file for 7 was downloaded from the ZINC Web site (http://zinc.docking.org), imported into Maestro and prepared by LigPrep (version 2.6, Schrödinger, LLC, New York, NY, 2013). The compound was docked to uPAR protein (3BT1) using Glide (version 5.9, Schrödinger, LLC, New York, NY, 2013) in standard precision (SP). The compound was assigned AM1-BCC (53) charges and gaff (54) atom types using the antechamber program from the AMBER12 package (55). Complexes were immersed in a box of TIP3P (56) water molecules to perform molecular dynamics simulations. No atom on the complex was within 14 Å from any side of the box. The solvated box was further neutralized with Na+ or Cl− counterions using the leap program from the AMBER12 package.
Simulations were carried out using the pmemd program in AMBER12 with ff10 (57) and gaff (54) force field in periodic boundary conditions. All bonds involving hydrogen atoms were constrained by using the SHAKE algorithm (58), and a 2 fs time step was used in the simulation. The particle mesh Ewald (59) (PME) method was used to treat long-range electrostatics. Simulations were run at 298 K under 1 atm in NPT ensemble employing Langevin thermostat and Berendsen barostat. Water molecules were first energy-minimized and equilibrated by running a short simulation with the complex fixed using Cartesian restraints. This was followed by a series of energy minimizations in which the Cartesian restraints were gradually relaxed from 500 kcal·Å−2 to 0 kcal·Å−2, and the system was subsequently gradually heated to 298 K via a 48 ps molecular dynamics run. By assigning different initial velocities, 10 independent simulations 10 ns in length were carried out for each complex.
Molecular Dynamics-Based Free Energy Calculations
For each trajectory among the 10 conducted for each complex (10 ns in length), the first 2 ns were discarded for equilibration. Molecular dynamics simulation snapshots were saved every 1 ps yielding 8000 structures per trajectory. A total of 80000 snapshots were generated per 100 ns of simulation. 100 snapshots were selected at regular intervals from the 80000 snapshots for free energy calculations. Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) (60) method were used to calculate the free energy using the sander program in the AMBER12 suite. The entropy is determined by normal mode analysis with the nmode module from AMBER (61). In normal mode entropy calculation, distance-dependent dielectric constant was set to 4. Maximum number of cycles of minimization was set to 10000. The convergence criterion for the energy gradient to stop minimization was 0.5.
The MM-GBSA binding free energy is expressed as:
where ΔEGBTOT is the combined internal and solvation energies, T is system temperature. ΔSNM is entropy determined by normal mode analysis. The internal energy is determined using the Lennard-Jones and Coulomb potentials in the AMBER force-field (ΔEGAS). The solvation energy is determined using Generalized-Born solvation models (ΔEGBSOL):
where ΔEGBSOL is the solvation free energy calculated with GB model, and ΔEGAS:
where ΔEELE is the non-bonded electrostatic energy, ΔEVDW is non-bonded van der Waals energy, and ΔEINT is the internal energies composed of bond, angle, and dihedral energies.
The GB solvation free energy is expressed by:
where ΔEGBSUR is hydrophobic contribution to desolvation energy, ΔEGB is reaction field energy (62).
The total electrostatic energy ΔEelectrostatic is the combination of ΔEELE and ΔEGB. Total non-polar energy ΔEnon-polar is the combination of ΔEVDW and ΔEGBSUR. The final energy is:
where ECOM, EREC and ELIG are total energies corresponding to complex, receptor and ligand, respectively. In these three system, complexes are VTN·uPAR·uPA, VTN·uPAR and VTN·uPAR·7; receptors are uPAR·uPA, uPAR and uPAR·7; ligands are all VTN.
Residue-compound distance, RMSD, correlation matrix for dynamic cross-correlation map (DCCM) and atomic fluctuation of the complexes were determined using the ptraj program in AMBER12.
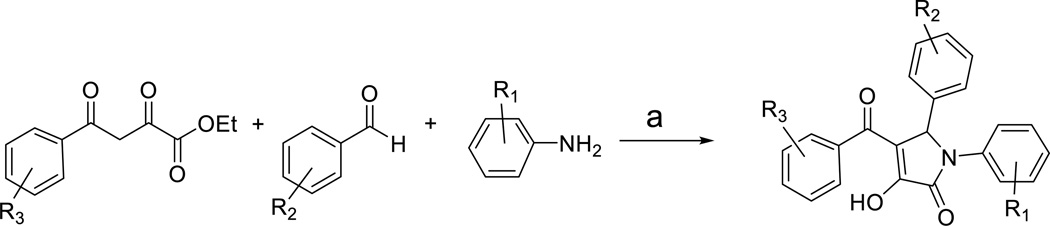
Chemistry
β-diketoesters were synthesized by Claisen condensation of ketones with diethyl oxalate in the presence of sodium ethoxide. The subsequent room temperature three-component condensation of β-diketoesters, amines, and aldehydes in acetonitrile gave the desired 2-pyrrolinones.
All chemicals were purchased from commercially available sources and used as received. Column chromatography was carried out with silica gel (25–63µ). High-Res Mass Spectra were measured on an Agilent 6520 Accurate Mass Q-TOF instrument. 1H NMR was recorded in CDCl3 or DMSO-d6 on a Bruker 500 MHz spectrometer. RP-LCMS was carried out on a Agilent 1100 LC/MSD fitted with a Eclipse XBD-C18 (4.6 × 150 mm) column eluting at 1.0 ml/min employing a gradient of (acetonitrile:methanol):water (each containing 5mM NH4OAc) from 70% to 100% acetonitrile:methanol over 15 min and holding at 100% acetonitrile:methanol for 2 min. Chemical shifts are reported in ppm using either residual CHCl3 or DMSO-d6 as internal references. All compounds are more than 95% purity unless otherwise stated. Synthesis of 3-(hexyloxy)aniline and 3-(hexyloxy)-4-methylaniline were carried out using a protocol from Marco et al. (63) β-diketoesters were synthesized with modification according to Milagre et al. (64). IPR-993 derivatives were synthesized by a modified procedure from Rose et al (65).
General Procedure for Synthesis
To a solution of β-diketoester (1 eq) in acetonitrile (1 mL) was added the appropriate amine or aniline (1 eq) and stirred for 10 min whereupon the aldehyde (1 eq) was added. The reaction was left to stir for 20 h at ambient temperature. The solvent was removed in vacuo and the product was isolated either by filtration (the precipitate was washed with cold diethyl ether) or flash chromatography (2–5% MeOH/DCM).
4-benzoyl-5-(4-ethylphenyl)-1-(3-(hexyloxy)phenyl)-3-hydroxy-1H-pyrrol-2(5H)-one (1)
(yellow solid, 7 mg, 15%); 1H NMR (500 MHz, DMSO-d6) δ 7.70 (d, J=7.0 Hz, 2H), 7.39 (m, 1H), 7.25-7.20 (m, 3H), 7.189-7.15 (m, 1H), 7.12 (t, J=8.5 Hz, 2H), 7.11-7.05 (m, 2H), 6.94-6.85 (m, 2H), 6.61 (s, 1H), 3.93-3.81 (m, 2H), 2.41 (q, J=7.5 Hz, 2H), 1.63 (q, J=6.5 Hz, 2H), 1.40-1.33 (m, 2H), 1.31-1.24 (m, 4H), 1.04 (t, J= 7.5 Hz, 3H), 0.87 (t, J=6.5 Hz, 3H); 13C NMR (126 MHz, DMSO-d6) δ 188.7, 164.2, 162.9, 160.8, 158.6, 141.8, 138.3, 136.6, 135.3, 129.8, 129.2, 128.4, 127.3, 127.2, 126.9, 114.0, 112.6, 110.9, 67.4, 61.2, 31.0, 28.5, 27.6, 25.2, 22.1, 15.1, 13.9. HRMS m/z calcd for C31H34NO4 [M+H]+: 484.2482, found 484.2495.
1-(3-bromo-4-methylphenyl)-4-(4-chlorobenzoyl)-3-hydroxy-5-(3-methoxyphenyl)-1H-pyrrol-2(5H)-one (6)
(white solid, 20 mg, 20%); 1H NMR (500 MHz, DMSO-d6) δ 7.94 (s, 1H), 7.73 (d, J=8.0 Hz, 2H), 7.53 (d, J=8.0 Hz, 2H), 7.50 (d, J=8.0 Hz, 1H), 7.28 (d, J=8.0 Hz, 1H), 7.12, (t, J=7.5Hz, 1H), 7.02–6.89 (m, 2H), 6.69 (d, J=7.5 Hz, 1H), 6.28 (s, 1H), 3.65 (s, 3H), 2.24 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 188.0, 164.4, 159.1, 150.2, 137.7, 137.6, 136.5, 135.3, 134.3, 130.9, 130.7, 129.5, 128.4, 125.7, 123.7, 121.6, 119.8, 119.7, 113.9, 113.2, 61.0, 55.0, 21.8. HRMS m/z calcd for C25H20BrClNO3 [M+H]+: 512.0259, found 512.0267.
1-(3-bromo-4-methylphenyl)-4-(4-chlorobenzoyl)-5-(3-fluorophenyl)-3-hydroxy-1H-pyrrol-2(5H)-one (7)
(white solid, 11 mg, 11%); 1H NMR (500 MHz, DMSO-d6) δ 7.94 (s, 1H), 7.73 (d, J=8.5 Hz, 2H), 7.52 (d, J=8.5 Hz, 2H), 7.49 (d, J=8.5 Hz, 1H), 7.32 (d, J=10.0 Hz, 1H), 7.30-7.22 (m, 3H), 6.99-6.92 (m, 1H), 6.34 (s, 1H), 2.24 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 188.0, 164.4, 162.9, 161.0, 150.7, 139.3, 139.2, 137.6, 136.5, 135.2, 134.4, 130.9, 130.6, 130.3, 128.4, 125.8, 123.9, 123.7, 121.6, 119.3, 115.1, 114.9, 60.5, 21.8. HRMS m/z calcd for C24H17BrClFNO3 [M+H]+: 500.0059, found 500.0054.
RESULTS
Computational Search for uPAR·uPA Small-Molecule Antagonists
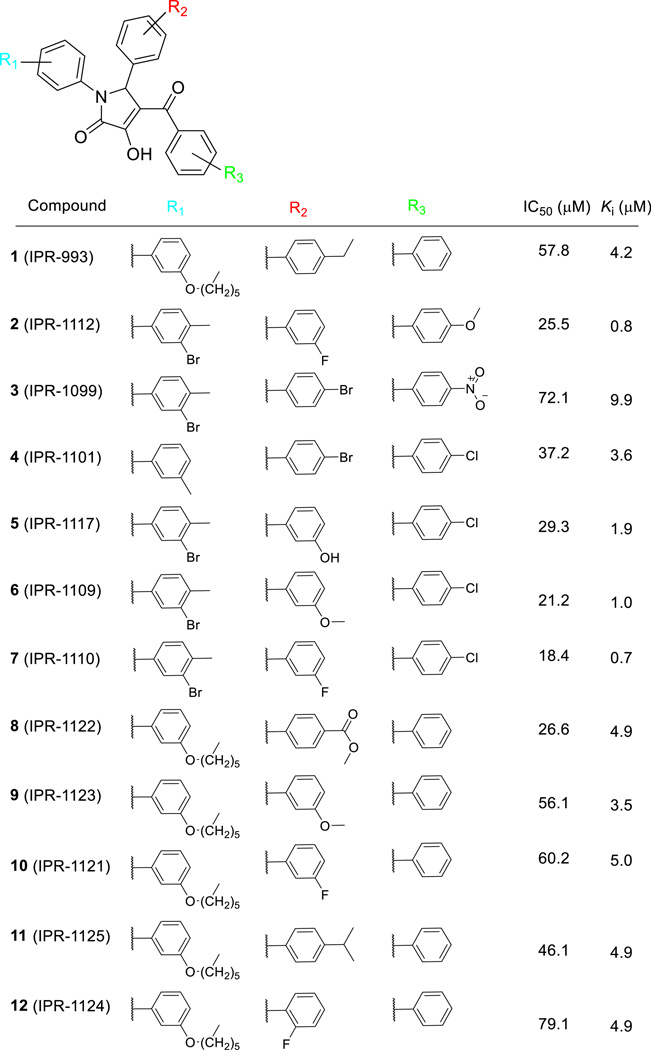
We had previously identified through structure-based virtual screening a set of small molecules (pyrazole, piperidinone, pyrrolidinone, and propylamine) that bind to uPAR (45,50). The compounds displaced a fluorescently-labeled small peptide (AE147-FAM) in the micromolar range with Ki ∼ 25 µM. Here, we search for small molecules that not only bind to uPAR but also disrupt the tight uPAR•uPA interaction. To that end, we used these small molecules as a starting point to conduct a sub-substructure search using 800,000 compounds in the ChemDiv library. A total of 2,196 compounds were identified and were further filtered to only include compounds with 3 or 4 substituents attached to the central ring. We also excluded fragments or larger compounds. Small molecules were clustered into 100 subsets, leading to the selection of 72 candidates. Compounds were purchased and screened for activity at an initial concentration of 50 µM. Among the 72 candidates that were screened, those that inhibited 50 percent or more were selected for a follow-up concentration-dependent study (Fig. S1A). One compound, namely 1 (IPR-993), inhibited AE147-FAM binding to uPAR with a Ki of 4.2 µM (Table 1 and Fig. 2a).
Table 1.
Compounds and Their Activity in ELISA and FP Assays
Figure 2.
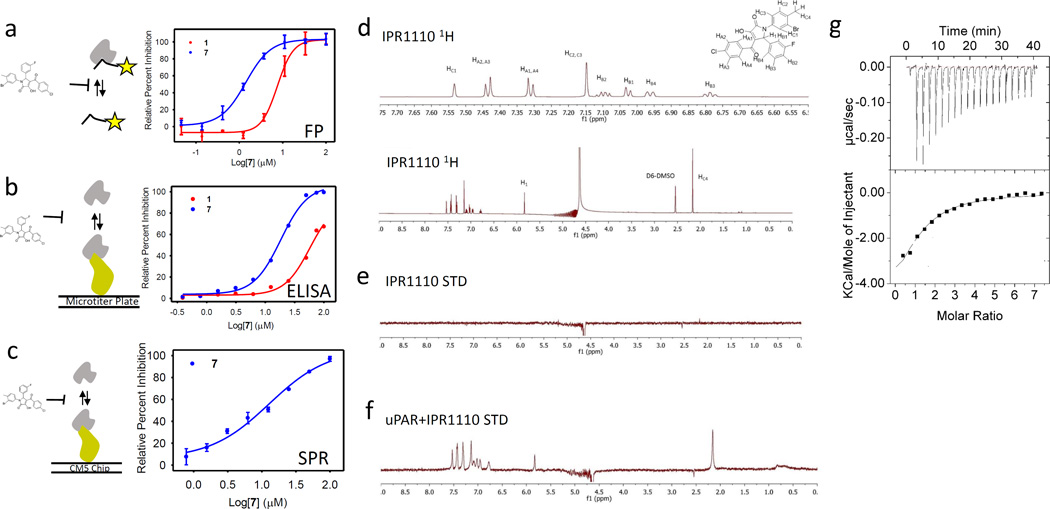
(a) Fluorescence polarization (mP) for FAM-AE147 and uPAR measured at increasing concentration of 7. Data are shown as mean +/− S.D. (n = 2). (b) An ELISA was used to measure inhibition of uPAR binding to uPAATF-coated microtiter plate by serial dilutions of compounds. Data are shown as mean +/−S.D. (n = 2). (c) A competition assay using surface plasmon resonance with uPAR injected along with increasing concentration of compound on immobilized uPAATF. Data are shown as mean +/− S.D. (n = 2). (d)1H NMR spectrum of 7. (e) STD NMR spectrum of 7 only showing no ligand peaks arising. (f) STD NMR spectrum of 7 in the presence of uPAR. (g) Isothermal titration calorimetry (ITC) data as a result of titration of 7 to uPAR.
The nearly one order of magnitude increase in potency of 1 over previously-identified compounds prompted us to assess whether it inhibits the full uPAR•uPA protein-protein interaction. A microtiter-based ELISA that we have previously developed was used for this purpose (44). The ELISA uses uPAR as well as the amino-terminal fragment of uPA (uPAATF), which contains the entire binding interface of the protein-protein interaction (Fig. 1) (7). The assay is particularly suitable since washing steps following addition of uPAR and compound rules out any interference of the compound with components of the assay. The compound inhibited the uPAR•uPAATF interaction with an IC50 of 58 µM (Table 1 and Fig. 2b). The difference between IC50 in the FP and ELISA is not unexpected, since the uPAR·uPAATF interface is significantly larger than the uPAR•AE147 interface, and higher concentration of the compound is required to shift the equilibrium towards dissociation.
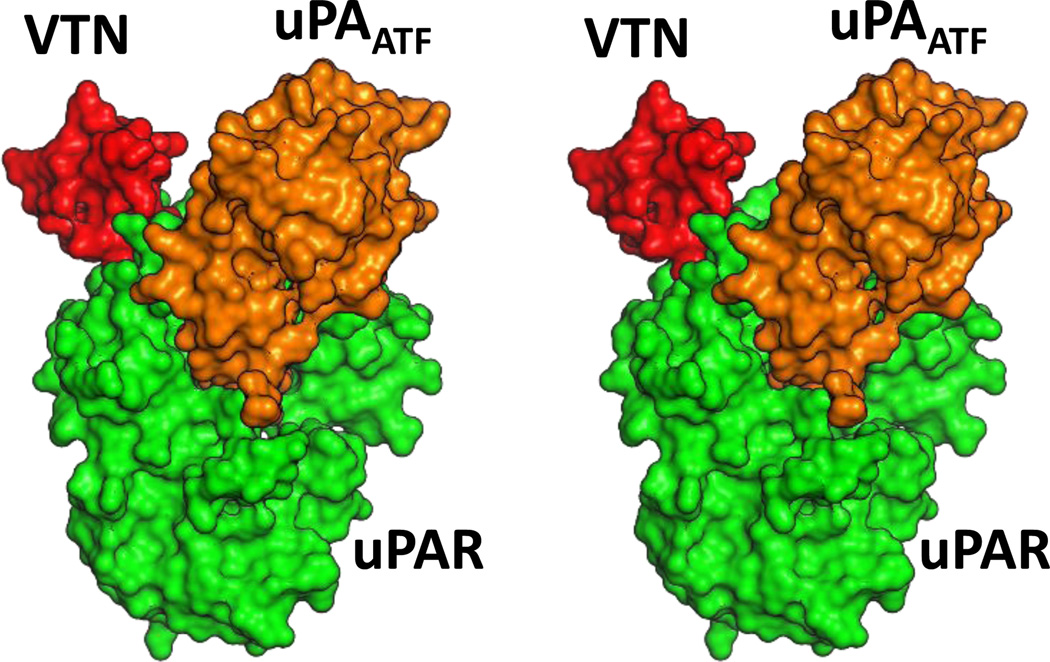
Figure 1.
Stereoview of the three-dimensional structure of uPAATF in complex with uPAR (PDB ID: 3BT2). uPAATF is shown in orange surface, while uPAR is depicted in green surface representation. VTNSMB is shown in red surface rendering.
The activity of 1 prompted us to conduct another computational search of the Chemdiv chemical library for additional compounds. Forty-eight derivatives were identified, purchased and tested for inhibition of uPAR•uPAATF initially at 50 µM. The 12 compounds that inhibited uPAR·uPAATF binding by more than fifty percent were evaluated in a concentration-dependent manner (Table 1 and Fig. S1B). The compound with highest affinity was 7 (IPR-1110). It inhibited fluorescently-labeled AE147-FAM binding with a sub-micromolar Ki of 0.7 µM (Fig. 2a) and inhibited uPAR•uPAATF interaction with an IC50 of 18.4 µM (Fig. 2b). Among all active compounds, the inhibition constant using FP with the AE147-FAM probe ranged from 0.7 µM for 7, to 10 µM for 3 (Table 1 and Fig. S1A). The IC50 values for inhibition of the full uPAR·uPAATF interaction of these compounds ranged from 18 µM for 7 (Fig. 2b) to 79 µM for 12 (IPR-1124) (Table 1 and Fig. S1B). To confirm the ELISA and FP results we resorted to the use of surface plasmon resonance in competition mode. uPAATF was immobilized on a dextran layer while uPAR was injected with increasing concentration of compound (Fig. 2c). The resulting IC50 of 12.9 µM for 7 is consistent with the ELISA results of 18 µM.
Direct Binding Studies
Saturation transfer difference (STD) NMR (66) was used to probe direct binding of 7 to uPAR. STD NMR is a ligand-based method. Measurement of the binding of a small molecule to a protein is done by monitoring the transfer of magnetization by fast chemical exchange. 1H NMR spectra were recorded for 7 and uPAR (Fig. 2d). STD spectra for 7 alone (Fig. 2e) and in the presence of uPAR (Fig. 2f) were subsequently collected. As expected, no saturation transfer is seen for compound only, as evidenced by the complete absence of peaks in Fig. 2e. Saturation transfer can be seen particularly in the aromatic range (6.5–8.0 ppm), 5.83 ppm and 2.17 ppm. These peaks occur only when spin diffusion signal (originating from the protein) is transferred to the small molecule.
Isothermal titration calorimetry (ITC) was used to further probe the direct binding of 7 to uPAR. Titration of 7 against purified uPAR led to protein binding site saturation (Fig. 2g). A fit of the data resulted in a dissociation constant Kd of 17.2 µM and an enthalpy (ΔH) of −6.4 kcal/mol (N = 1.1). Based on this data, the entropy (ΔS) was 0.01 kcal/mol. The binding is entirely driven by enthalpy. The negligible entropy suggests that (i) conformational changes in the receptor or (ii) displacement of water molecules likely compensate for the loss of entropy that is typically associated with ligand binding. A conformational change would be consistent with the highly dynamic nature of uPAR (10). It may also explain the difference between the inhibition constants Ki obtained in the FP studies (0.7 µM) and the ITC dissociation constant Kd (17.2 µM). The large AE147 peptide in the FP study likely promotes different conformational states of the protein that may not exist in the apo structure of uPAR.
Structure-Activity Relationships
Like 1, several derivatives had a hexyloxy moiety at R1. These compounds had relatively weaker potency with Ki values greater than 3 µM for inhibition of AE147-FAM binding to uPAR. These compounds also inhibited the uPAR•uPAATF interaction more weakly than other compounds as evidenced by ELISA IC50 values that were higher than 25 µM. In the remaining derivatives, the R1 group is a di-substituted benzene with a methyl group at the para-position and a meta-bromine 2 (IPR-1112), 3 (IPR-1099), 5 (IPR-1117), 6 (IPR-1109) and 7 (IPR-1110). Compound 4 (IPR-1101) is the exception as it bears a methyl group instead of bromine and has a hydrogen atom at the para-position.
At the R2 position, there was greater diversity in the structure and properties of the substituents. Two compounds (3 and 4) had a bromine atom instead of the ethyl group of 1, while other compounds such as 8 (IPR-1122) and 11 bear a methyl ester and an isopropyl group, respectively. It is worth noting that all compounds with a substituent at the para position had relatively weaker affinity with a Ki for inhibition of AE147-FAM binding to uPAR ranging from 3.6 µM for 4 to 4.9 µM for 8 and 11. Similarly, IC50 for blocking the uPAR•uPAATF interaction were higher among these compounds (37 µM for 4 and 46 µM for 11). Despite the similar Ki values for 8 and 11, compound 8 exhibited slightly greater potency in blocking the uPAR•uPAATF interaction suggesting that the compounds engage different residues on uPAR. When the benzene ring at R2 had a substituent at the meta position, higher affinity was observed with FP Ki values that were 1 µM or lower. Comparison of 5 and 7 shows that replacement of a hydroxyl moiety of 5 with fluorine resulted in nearly two-fold increase in the FP Ki, and a reduction of the ELISA IC50 by 10 µM. Compound 7 showed the highest affinity in both FP and ELISA.
In contrast to R1 and R2, the R3 substituent did not vary significantly. It consisted mostly of a benzene ring (1, 8, and 10–12) or a benzene ring with a chlorine atom at the para position (4–7, or 13). One compound, namely 3, had a nitro group at the para position and had the weakest affinity to uPAR.
Compound 7 Impairs the Distal VTN•uPAR Interaction in a uPA-dependent Manner
Previous studies have shown that uPA is required for VTN to bind to uPAR despite the large distance in the binding sites of the two ligands (3,10). Since 7 inhibits the uPAR•uPA interaction, we wondered whether the compound also affected the distant VTN•uPAR interaction. To explore this, we first investigate the cooperative binding of uPA and VTN to uPAR using surface plasmon resonance. Subsequently, we develop an ELISA to probe the effect of compound 7 on VTN binding to uPAR.
uPAR is well-known to bind to uPA in a tight and long-lived complex. We confirmed by surface plasmon resonance. uPAR was immobilized on the dextran surface of a CM5 chip and injection of uPAATF onto the immobilized protein resulted in a Kd of 2.2 nM (Fig. S2). Sensorgrams shown in Fig. S2 reveal a stable complex as evidenced by the lack of appreciable dissociation detected during the time period of the run. We performed the reverse experiment to ensure that we detect binding when uPAATF is immobilized on the surface. A similar tight and stable interaction with a Kd of 25.1 nM is observed (Fig. S2).
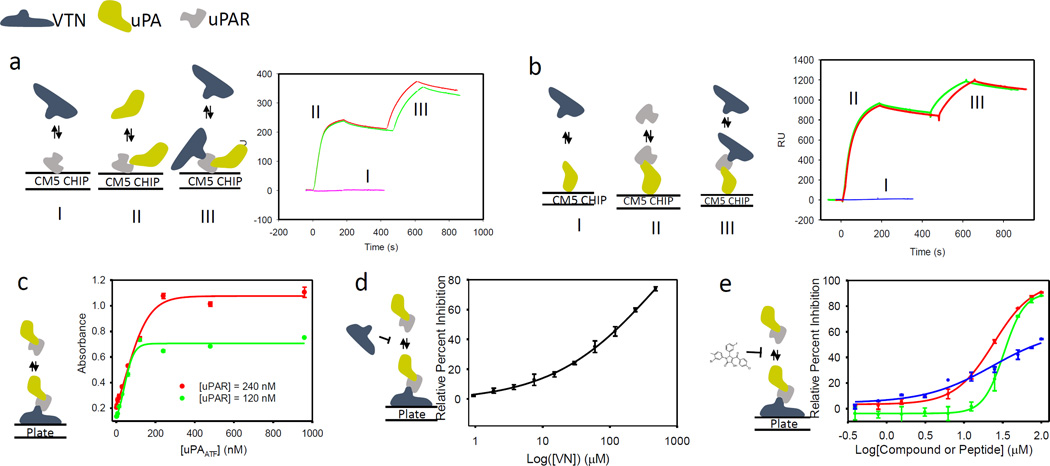
We subsequently investigated the binding of full-length VTN to uPAR. uPAR is immobilized on the dextran surface of the CM5 chip. First, VTN was injected onto immobilized uPAR. No binding between VTN and uPAR was detected (Fig. 3a). Next, uPAATF is injected on the immobilized uPAR resulting in strong binding as expected (Fig. 3a). The uPAATF injection was stopped after 180 s, and, 120 s later, VTN was injected. Strong binding of VTN to the uPAR•uPAATF complex was detected confirming that VTN binds to uPAR only in the presence of uPA (Fig. 3a). To rule out that VTN is binding to uPAATF, we immobilized uPAATF on the CM5 chip and injected VTN. No binding to uPAATF was detected (Fig. 3b). uPAR was subsequently injected resulting in tight binding as expected (Fig. 3b). After 120 seconds post uPAR injection, VTN was injected to the uPAR•uPAATF complex resulting in robust binding (Fig. 3b).
Figure 3.
(a) Direct binding assay using surface plasmon resonance with uPAR immobilized on the surface of the CM5 chip. First, VTN is injected onto uPAR as illustrated by the schematic and the curve labeled I; then uPAATF is injected as illustrated by the schematic and curve labeled II; this is followed by injection of VTN as illustrated by schematic and curves labeled III. The injection of uPAATF and VTN is carried out in duplicate as illustrated by the red and green curves. (b) Direct binding assay using surface plasmon resonance with uPAATF immobilized on the surface of the CM5 chip. First, VTN is injected onto uPAATF as illustrated by the schematic and the curve labeled I; then uPAR is injected as illustrated by the schematic and curve labeled II; this is followed by injection of VTN as illustrated by schematic and curves labeled III. The injection of uPAR and VTN is carried out in duplicate as illustrated by the red and green curves. (c) An ELISA was used to measure binding of uPAR to VTN in the presence of serial dilutions of uPAATF. Data are shown as mean +/− S.D. (n = 2). (d) An ELISA was used to measure inhibition of uPAR•uPAATF binding to VTN coated microtiter plate by serial dilutions of VTN. (c) An ELISA was used to measure inhibition of uPAR•uPAATF binding to VTN coated microtiter plate by serial dilutions of compounds or peptides. Data are shown as mean +/− S.D. (n = 2).
To test for the effect of 7 on the uPAR•VTN interaction, a microtiter-based ELISA was developed (Fig. 3c). First, wells were coated with full-length VTN followed by titration of uPAR at increasing concentration. As expected, no binding uPAR alone to VTN only was detected. This process was repeated in the presence of uPAATF. uPAR concentration was held fixed at 120 nM and an increase in uPAATF concentration showed a gradual increase of binding to VTN that eventually plateaued at 120 nM (Fig. 3c). Even more binding is observed as a result of an increase in uPAR concentration to 240 nM, confirming that the process is uPAR-dependent (Fig. 3c). To confirm that the binding of the uPAR•uPAATF complex was to VTN, we tested whether soluble VTN can compete with immobilized VTN. To that end, uPAR and uPAATF were pre-incubated with VTN and the complex was added to VTN immobilized on the microtiter plate. As expected, soluble VTN inhibited uPAR•uPAATF binding to immobilized VTN (Fig. 3d). Finally, the ELISA was used to test the effect of 7 on the VTN•uPAR•uPAATF interaction. The compound was found to inhibit the interaction with an IC50 of 32.8 µM (Fig. 3e). We show that the AE147 peptide, which also binds to the uPA pocket on uPAR, also inhibited uPAR binding to VTN albeit with weaker potency with an IC50 close to 100 µM.
Effect of 7 on Cellular Processes and Signaling Mediated by uPAR
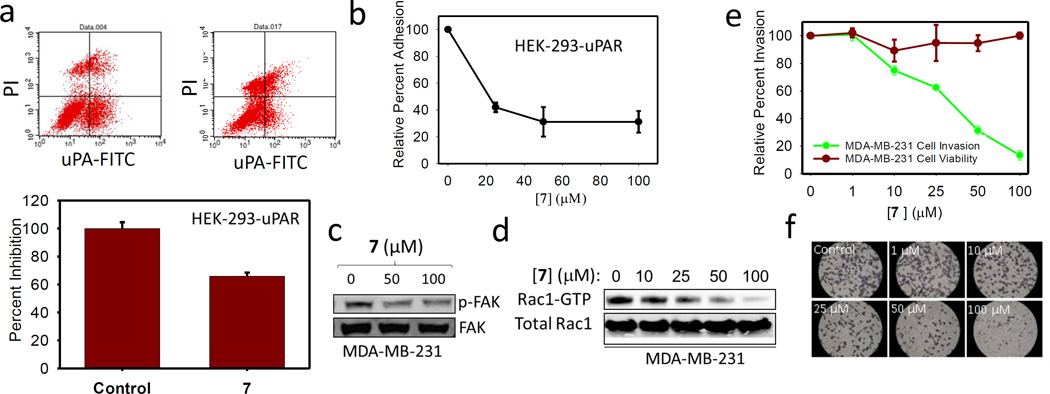
The ability of 7 to inhibit the uPAR•uPA and uPAR•VTN interaction at the cell surface was investigated by flow cytometry. HEK-293 cells that stably expressed membrane-associated uPAR were treated with fluorescein isothiocyanate-conjugated uPA (uPA-FITC) in the presence and absence of compound. Binding of uPA-FITC to uPAR was detected by fluorescence-activated cell sorting (FACS) analysis. As shown in Fig. 4a, 7 inhibited uPA binding to uPAR by nearly 40 percent.
Figure 4.
(a) Flow cytometry analysis using FITC-conjugated HMW-uPA and PI staining. 100 µM of 7 and control analyzed for uPAR•uPA binding. (b) Effects of 7 on HEK-293-uPAR adhesion to VTN; data are shown as mean +/− S.D. (n = 3). (c) Immunoblot showing the expression levels of FAK, p-FAK in MDA-MB-231 cell lines. (d) GST-Rac1 pull-down assay. The protein complexes were subjected to immunoblot analysis to detect active Rac1. Rac1 from total cell lysates was used as a control. (e) Cell invasion studies with the Boyden Matrigel Invasion Chamber and MTT assay to characterize the role of 7 in cell proliferation and cell Invasion; data are shown as mean +/− S.D. (n = 3). (f) Representative experimental cells from control and in the presence of 7 were photographed (×400) to illustrate the effect of 7 on invasion.
To probe the effect of 7 on the uPAR•uPA and uPAR•VTN interaction at the cell surface, we employed a microtiter-based adhesion assay that has been previously developed (4). The assay consists of using uPAR-expressing HEK-293 cells that are added to wells containing full-length VTN. We find that addition of exogenous uPA consistently enhanced uPAR-expressing HEK-293 cell adhesion to VTN as previously reported. This increase in adhesion was abrogated by compound 7 in a concentration-dependent manner (Fig. 4b). The compound inhibited uPAR-mediated HEK-923 cell adhesion to VTN by 60 percent at 25 µM. This is consistent with IC50 values obtained from the ELISA and surface plasmon resonance competition assay. The fact that uPA causes an increase in adhesion confirms our biophysical studies that VTN binding to uPAR is a uPA-dependent process.
It is well-established that uPAR promotes signaling by interaction with integrins (4,37,38,67). One mechanism by which uPAR activates integrins is through uPAR-bound VTN, a ligand of β3 integrins. An RGD motif located a few residues away from the SMB domain of VTN recruits β3 integrins to promote signaling. This interaction leads to FAK phosphorylation (34,68,69) as well as downstream activation of Rho family small GTPase Rac1 (39,70). Immunoblot analysis for phospho-FAK and pull-down assays for active Rac1 were conducted in MDA-MB-231 breast cancer cells treated with 7. As a result, FAK phosphorylation at Y397 was significantly inhibited by the compound (Fig. 4c). We also conducted a study to determine the effect of 7 on Rac1 activity in MDA-MB-231 cancer cells. This was done using a pull-down approach where the Rac1 effector Pak1 was used to test for Rac1 activity in MDA-MB-231. Pak1 only binds to GTP-bound (active) Rac1. As shown in Fig. 4d, 7 inhibited Rac1 activation in a concentration-dependent manner.
The inhibition of uPAR•uPA binding at the cell surface prompted a study of the effect of the compound on uPA-mediated cell invasion. A Boyden chamber apparatus that is pre-coated with a reconstituted basement membrane known as Matrigel was used for these studies (49). Using this approach, we have previously shown that siRNA knockdown of uPAR significantly inhibited invasion of breast MDA-MB-231 cells (44) and non-small cell lung cancer (NSCLC) cells A549, H460 and H1299 (48). We find that 7 inhibited MDA-MB-231 cell invasion with an IC50 of 35 µM (Fig. 4e and 4f). This IC50 is consistent with the IC50 for inhibition of the uPAR•uPA interaction measured in the ELISA and surface plasmon resonance competition assays (Fig. 2). 7 had little cytotoxicity in MDA-MB-231 cells with over 80 percent of cells remaining viable even after 3 days of treatment at 100 µM (Fig. 4e and 4f). This confirms that the effects on invasion are not due to cell killing.
uPA Promotes VTN binding by Stabilization of uPAR
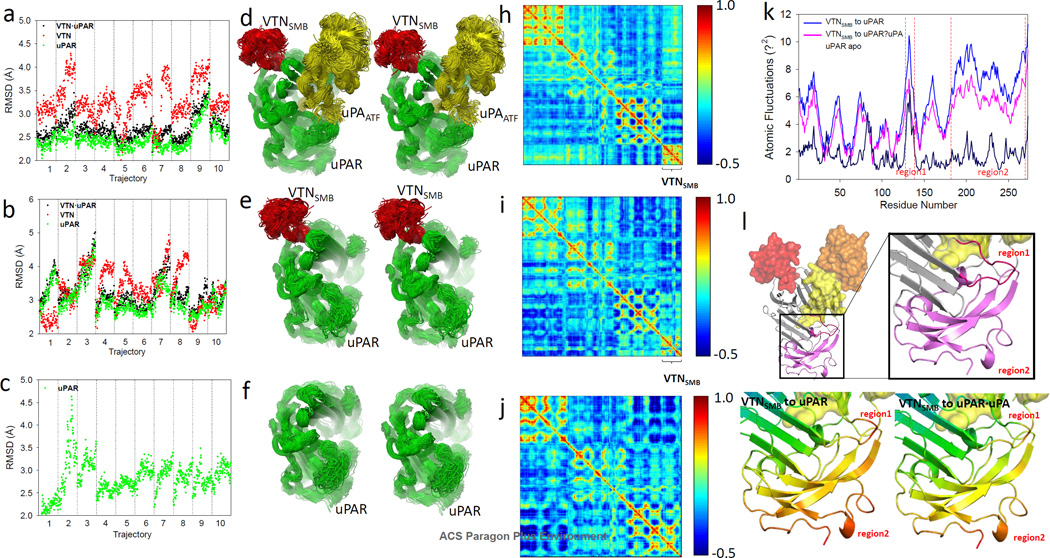
To understand the structural basis by which 7 modulates the distal VTN•uPAR interaction, we resorted to explicit-solvent molecular dynamics simulations (71–73). Molecular dynamics simulations use Newton’s equations of motion to follow the trajectory of each atom with respect to time. Molecular dynamics simulations can thus provide a detailed account of the motion of a macromolecule with respect to time. Here, we are particularly interested in understanding how the binding of uPA and 7 affects uPAR and its interaction with VTN. To that end, we carried out separate molecular dynamics simulations for (i) VTNSMB•uPAR•uPAATF (Fig. 5a and 5d); (ii) VTNSMB•uPAR (Fig. 5b and 5e); (iii) uPAR (Fig. 5c and 5f); and (iv) uPAR•7 complexes. In each case a total of 100 ns of simulation is conducted (10 trajectories that are 10 ns in length). The structural changes over the course of each trajectory of each of the proteins in the multiprotein complexes is quantified with the root-mean-squared deviation (RMSD) (Fig. 5). Larger RMSDs correspond to greater deviation from the crystal structure. In the VTNSMB•uPAR•uPAATF complex, VTN experiences RMSD fluctuations that range between 2 and 4.5 Å (Fig. 5a). uPAR experiences fluctuations in RMSDs between 2 and 2.5 Å except for trajectory number 9 where the RMSD reaches 3.5 Å. In the uPAR•VTNSMB complex, VTNSMB experiences similar structural changes with RMSDs ranging between 2 and 4.5 Å (Fig. 5b). uPAR structures, on the other hand, show significantly greater deviation from the crystal structure with uPAR RMSDs that range from 3 to 5 Å (Fig. 5b). An illustration of the structural changes experienced by uPAR in the VTNSMB•uPAR•uPAATF and VTNSMB•uPAR complexes is shown in Fig. 5c and 5d.
Figure 5.
RMSD for 10 × 10 molecular dynamics simulation of (a) VTNSMB·uPAR·uPA complex, (b) VTNSMB·uPAR complex and (c) uPAR apo structure. Stereoview of the superimposed structure of 100 snapshots selected at regular intervals from molecular dynamics simulations of (d) VTNSMB·uPAR·uPA complex, (e) VTNSMB·uPAR complex and (f) uPAR apo structure, respectively. VTN, uPA, and uPAR are shown in red, yellow and green ribbon representation, respectively. Dynamic cross-correlation map (DCCM) for (h) VTNSMB·uPAR·uPA, (i) VTNSMB·uPAR, and (j) uPAR. (k) Atomic fluctuations of the VTNSMB·uPAR·uPA (magenta), VTNSMB·uPAR (blue), and uPAR (dark blue). (l) Three-dimensional structure of VTNSMB·uPAR·uPA to illustrate region1 and region2 that undergo significant level of atomic fluctuation.
The dynamics of uPAR in the VTNSMB•uPAR•uPAATF and VTNSMB•uPAR complexes is further investigated using a dynamical cross-correlated map (normalized covariance matrix) of the proteins (73). The map is a two-dimensional matrix that provides correlation coefficients between among all residues within a three-dimensional structure. These coefficients range from −1 (highly anti-correlated motion) to 1 (highly correlated motion). Negative values correspond to anti-correlated motion, where residues are generally moving in opposite directions, and positive values correspond to correlated motions, during which residues are generally moving in the same direction. The maps in the presence (Fig. 5h) and absence (Fig. 5i and 5j) of uPA show correlations that range from highly anti-correlated (blue) to highly correlated (red). Comparison of the two maps confirms that uPA significantly alters the motion of uPAR. The most pronounced change in the correlations occurs on VTNSMB (Fig. 5h and 5i). In particular, the correlation of motion between the residues of VTNSMB and residues 1 and 150 on uPAR are significantly altered and switch from slightly anti-correlated motion to strongly anti-correlated motion.
To gain insight into the flexibility of uPAR in the various multiprotein complexes, the trajectories are used to measure the atomic fluctuations for individual residues over the course of the entire trajectory. Both VTNSMB•uPAR•uPAATF and VTNSMB•uPAR trajectories were analyzed (Fig. 5k). Overall, uPAR residues experience much larger fluctuations in the absence of uPAATF confirming previous results. Two regions on uPAR in particular experience the largest fluctuations. The first occurs between residues 125 and 150 (Region1) and residues 180 and 273 (Region2). These two regions are mapped onto the structure of uPAR (Fig. 5l).
Finally, we resorted to free energy calculations to better understand how uPA and 7 are able to affect binding of VTNSMB to uPAR despite the large distance between the ligands. Using the trajectories of VTNSMB •uPAR•uPAATF and VTNSMB •uPAR•7, we determined the free energy of binding of VTN to uPAR•uPAATF and uPAR•7 using the MM-GBSA approach (71–74). We find that the binding affinity of VTNSMB is significantly more favorable in the VTN•uPAR•uPA complex (ΔGMMGBSA = −19.8 kcal•mol−1) than in the VTNSMB •uPAR•7 complex (ΔGMMGBSA = −12.4 kcal•mol−1). Analysis of the components of the free energy reveals that the electrostatics and non-polar components ΔEelectrostatics and ΔEnon-polar are more favorable in the presence of uPA, while the entropy is more favorable when 7 is bound. These results are fully consistent with the ELISA, FP and surface plasmon resonance studies that reveal that VTNSMB binds more favorably to uPAR in the presence of uPA.
A question of interest is how uPA and 7 contribute to the free energy of binding of the distal VTN interaction. To address this, we used the same snapshots from the VTNSMB •uPAR•uPAATF and VTN•uPAR•7 trajectories to determine the VTNSMB •uPAR binding free energy in the presence or absence of uPA or 7. This approach removes the contributions of conformational change caused by uPA or 7 binding and provides information strictly on the direct effects of uPA and 7 to VTNSMB binding to uPAR. Interestingly, we find that the presence of uPA resulted in more favorable free energy of binding with a ΔGMMGBSA of −19.8 kcal•mol−1 compared to ΔGMMGBSA of −16.9 kcal•mol−1 in the absence of uPA (Fig. 5i). Inspection of the components of the free energy reveals that the absence of uPA led to less favorable contributions of the electrostatics and entropy to the binding of VTN, but no effect on the non-polar components. The less favorable entropy is consistent with uPA’s ability to stabilize the structure of uPAR and also consistent with our observations from the analysis of uPAR structures. In contrast to uPA, the removal of 7 from the structures resulted in negligible effect on the uPAR• VTNSMB free energy by 0.7 kcal mol−1 (Fig. 5h). This was primarily due to an unfavorable entropy of 0.7 kcal mol−1 (Fig. 5i).
DISCUSSION
Small-molecule inhibitors of uPAR protein interactions could provide valuable chemical probes to further explore the role of the receptor in a range of pathological processes including cancer metastasis particularly in vivo. Here, we report the discovery of new pyrrolone compounds that inhibit the tight and highly stable uPAR•uPAATF interaction. Among them, compound 7 was the most potent. The compound displaced an α-helical peptide AE147-FAM in the sub-micromolar range (Ki = 0.7 µM) using an FP assay, and inhibited the uPAR•uPAATF interaction with an IC50 of 18.4 µM based on an ELISA and surface plasmon resonance. The difference between the FP and ELISA are not unexpected, since the FP assay probe is a small α-helical peptide with a much smaller footprint than uPA used in the ELISA. STD-NMR and ITC were conducted to confirm that 7 directly binds directly to uPAR. The ITC study revealed that the binding of 7 was primarily driven by enthalpy. The favorable enthalpy change suggests good complementarity between small molecule and target. The lack of entropy change in the binding suggests that the displacement of water molecules from the hydrophobic binding cavity on uPAR as well as changes in conformations of uPAR likely compensate for the loss in entropy associated with binding of the small molecule.
Multiprotein complexes often involve cooperativity as binding events tend to lead to stabilization of the receptor and invariably affect the subsequent binding of other ligands at distal sites (75). The VTN•uPAR•uPAATF multiprotein complex occurs gradually as uPA first binds to uPAR leading to a series of proteolytic events that lead to the release of VTN from the extracellular matrix and its binding to uPAR (4,12,76). X-ray structures show that there is no contact between uPA and VTN (Fig. 1) (5,7,77,78). We conducted biophysical studies with surface plasmon resonance to explore the cooperative binding between uPA and VTN. We show that VTN and uPA binding to uPAR is highly cooperative confirming previous studies (3,10). While uPA binds to uPAR in a high-affinity and highly stable complex, VTN binds to uPAR only in the presence of uPA. This was elegantly illustrated by Ploug and co-workers when they showed that constraining uPAR using an engineered disulfide bond to resemble its structure in complex with uPA led to similar phenotype on vitronectin that is observed as a result of vitronectin binding (79). Our study found no binding of VTN to uPA confirming that the cooperative binding to uPAR is not due to direct contact of these two proteins in the VTN•uPAR•uPAATF multiprotein complex. The cooperative uPA and VTN binding was further supported by the fact that 7 also led to the inhibition of the uPAR•VTN interaction with an IC50 of 35.6 µM. These results show that the inhibition of the uPAR•uPA is expected to shut down all biological processes mediated by uPAR through uPA and VTN.
We use compound 7 to probe uPAR protein-protein interactions at the cell surface. The compound inhibited the uPAR•uPAATF interaction in uPAR-expressing HEK-293 cell line as evidenced by FACS analysis. This is consistent with our observed inhibition of breast MDA-MB-231 cancer cell invasion by compound 7 with an IC50 of 35 µM. To probe the effect of 7 on the uPAR•VTN interaction, we used an adhesion assay with uPAR-expressing HEK-293 cells and VTN coated on the surface of wells. Addition of uPA causes a significant increase in adhesion of these cells to VTN as expected since uPAR is the VTN receptor at the cell surface. Compound 7 inhibited uPAR-expressing HEK-293 cells adhesion to VTN in a concentration-dependent manner with an IC50 similar to the ELISA and surface plasmon resonance studies. Since VTN contains an RGD motif that is recognized by β3 integrins, it is expected that the compound should also impair the recruitment of these integrins by uPAR. Signaling through these integrins usually occurs through FAK and Rac1 activation (34,39,68–70). Immunoblotting analyses revealed that 7 blocked phosphorylation of FAK in MDA-MB-231 cells. The compound also impaired Rho GTPase Rac1 activation confirming the role of uPAR as a cell surface receptor of VTN that mediates signaling through integrins to promote invasion and metastasis. It was encouraging that the compound was not cytotoxic to cells even at 100 µM suggesting that the effects on invasion are not due to cell killing.
We resorted to explicit-solvent molecular dynamics simulations to gain more insight into the structural basis by which 7 modulates the distal uPAR•VTN interaction. Analysis of molecular dynamics simulation structures for the VTN•uPAR•uPA and VTN•uPAR•7 complexes showed that uPA considerably dampened uPAR motion as evidenced by reduced RMSDs and residue-based atomic fluctuations for uPAR in complex with uPA. A principal component analysis of the complex confirmed that uPA binding to uPAR had a significant effect on the correlation of the motion of VTN and uPAR. Free energy calculations using the MM-GBSA approach further shed light into the allosteric modulation of VTN by uPA. The calculations showed that the VTN•uPAR binding free energy is significantly more favorable in the presence of uPA compared with 7. We repeated these calculations using molecular dynamics structures of VTN•uPAR•uPAATF and VTN•uPAR•7 complexes except that uPA and 7 were deleted from these structures. Comparison of VTN•uPAR•uPAATF and VTN•uPAR free energy calculations show that the allosteric effects of uPA originates from the electrostatic and entropy component by making both more favorable. But the VTN•uPAR•7 and VTN•uPAR calculations show that the allosteric effects of 7 are confined to the entropy component by making it slightly unfavorable. This suggests that the compound works not only by displacing uPA and promoting conformational states of uPAR that are unsuitable for VTN binding, but may also contributes directly to the allosteric inhibition of VTN by affecting its entropy of binding to uPAR. Further experimental investigations will be required to confirm this effect.
In sum, our work demonstrates that inhibition of the uPAR•uPA protein-protein interaction with a small molecule also leads to the inhibition of the distal uPAR•VTN interaction. By inhibiting uPA binding to uPAR, compound 7 causes uPAR to become more flexible, weakening its interaction with VTN. Hence, blocking the uPAR•uPA interaction should have the dual effect of inhibiting uPA-mediated proteolysis at the cell surface and VTN-mediated integrin signaling. Compound 7 provides a scaffold for the development of higher affinity derivative compounds that will likely possess significantly greater in vivo efficacy for further probing of uPAR signaling.
Supplementary Material
Scheme 1.
Reagents and Conditions: a) MeCN, r.t.
Table 2.
Free energy Calculations Conducted using the MM-GBSA Approach Based on Snapshots Collected from the MD Simulations
| Binding Event | ΔG MMGBSA (kcal·mol−1) |
ΔE electrostatic (kcal·mol−1) |
ΔE non-polar (kcal·mol−1) |
−TΔS (kcal·mol−1) |
|---|---|---|---|---|
| VTNSMB→uPAR·uPA(I) | −19.8±1.3 | 21.2±0.3 | −71.8±0.1 | 30.9±1.0 |
| VTNSMB→uPAR(II) | −16.9±1.3 | 22.2±0.4 | −71.8±0.1 | 32.7±1.0 |
| ΔΔG (I-II) | −2.9±1.3 | −1.0±0.4 | 0.0±0.0 | −1.8±1.0 |
| VTNSMB→uPAR·7 (I) | −12.4±1.6 | 25.4±0.6 | −66.4±0.1 | 28.6±1.1 |
| VTNSMB→uPAR(II) | −13.1±1.8 | 25.5±0.6 | −66.5±0.1 | 27.9±1.0 |
| ΔΔG (I-II) | 0.7±1.7 | −0.1±0.6 | −0.1±0.1 | 0.7±1.0 |
ACKNOWLEDGMENT
The research was supported by the National Institutes of Health (CA135380) (SOM), the American Cancer Society Research Scholar Grant RSG-12-092-01-CDD (SOM), and by the 100 Voices of Hope (SOM).
Footnotes
Supporting Information Available: Chemical structure of compounds that emerged from virtual screening against uPAR crystal structure and multiple conformers is provided. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Eden G, Archinti M, Furlan F, Murphy R, Degryse B. The urokinase receptor interactome. Curr. Pharm. Des. 2011;17:1874–1889. doi: 10.2174/138161211796718215. [DOI] [PubMed] [Google Scholar]
- 2.Magdolen V, Rettenberger P, Koppitz M, Goretzki L, Kessler H, Weidle UH, Konig B, Graeff H, Schmitt M, Wilhelm O. Systematic mutational analysis of the receptor-binding region of the human urokinase-type plasminogen activator. Eur. J. Biochem. 1996;237:743–751. doi: 10.1111/j.1432-1033.1996.0743p.x. [DOI] [PubMed] [Google Scholar]
- 3.Gardsvoll H, Ploug M. Mapping of the vitronectin-binding site on the urokinase receptor: involvement of a coherent receptor interface consisting of residues from both domain I and the flanking interdomain linker region. J. Biol. Chem. 2007;282:13561–13572. doi: 10.1074/jbc.M610184200. [DOI] [PubMed] [Google Scholar]
- 4.Madsen CD, Ferraris GM, Andolfo A, Cunningham O, Sidenius N. uPAR-induced cell adhesion and migration: vitronectin provides the key. J. Cell. Biol. 2007;177:927–939. doi: 10.1083/jcb.200612058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huai Q, Zhou A, Lin L, Mazar AP, Parry GC, Callahan J, Shaw DE, Furie B, Furie BC, Huang M. Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes. Nat. Struct. Mol. Biol. 2008;15:422–423. doi: 10.1038/nsmb.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardsvoll H, Dano K, Ploug M. Mapping part of the functional epitope for ligand binding on the receptor for urokinase-type plasminogen activator by site-directed mutagenesis. J. Biol. Chem. 1999;274:37995–38003. doi: 10.1074/jbc.274.53.37995. [DOI] [PubMed] [Google Scholar]
- 7.Huai Q, Mazar AP, Kuo A, Parry GC, Shaw DE, Callahan J, Li YD, Yuan C, Bian CB, Chen LQ, Furie B, Furie BC, Cines DB, Huang MD. Structure of human urokinase plasminogen activator in complex with its receptor. Science. 2006;311:656–659. doi: 10.1126/science.1121143. [DOI] [PubMed] [Google Scholar]
- 8.Llinas P, Le Du MH, Gardsvoll H, Dano K, Ploug M, Gilquin B, Stura EA, Menez A. Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J. 2005;24:1655–1663. doi: 10.1038/sj.emboj.7600635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardsvoll H, Gilquin B, Le Du MH, Menez A, Jorgensen TJ, Ploug M. Characterization of the functional epitope on the urokinase receptor. Complete alanine scanning mutagenesis supplemented by chemical cross-linking. J. Biol. Chem. 2006;281:19260–19272. doi: 10.1074/jbc.M513583200. [DOI] [PubMed] [Google Scholar]
- 10.Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M. A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR) J. Biol. Chem. 2012;287:34304–34315. doi: 10.1074/jbc.M112.398404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardsvoll H, Hansen LV, Jorgensen TJ, Ploug M. A new tagging system for production of recombinant proteins in Drosophila S2 cells using the third domain of the urokinase receptor. Protein Expr. Purif. 2007;52:384–394. doi: 10.1016/j.pep.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Sidenius N, Andolfo A, Fesce R, Blasi F. Urokinase regulates vitronectin binding by controlling urokinase receptor oligomerization. J. Biol. Chem. 2002;277:27982–27990. doi: 10.1074/jbc.M111736200. [DOI] [PubMed] [Google Scholar]
- 13.Nussinov R, Tsai C-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Nussinov R, Tsai C-J. The different ways through which specificity works in orthosteric and allosteric drugs. Curr. Pharm. Des. 2012;18:1311–1316. doi: 10.2174/138161212799436377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis V, Scully MF, Kakkar VV. Plasminogen activation initiated by single-chain urokinase-type plasminogen activator. Potentiation by U937 monocytes. J. Biol. Chem. 1989;264:2185–2188. [PubMed] [Google Scholar]
- 16.Ellis V, Behrendt N, Dano K. Plasminogen activation by receptor-bound urokinase. A kinetic study with both cell-associated and isolated receptor. J. Biol. Chem. 1991;266:12752–12758. [PubMed] [Google Scholar]
- 17.Carmeliet P, Moons L, Lijnen R, Baes M, Lemaitre V, Tipping P, Drew A, Eeckhout Y, Shapiro S, Lupu F, Collen D. Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat. Genet. 1997;17:439–444. doi: 10.1038/ng1297-439. [DOI] [PubMed] [Google Scholar]
- 18.Dano K, Andreasen PA, Grondahl-Hansen J, Kristensen P, Nielsen LS, Skriver L. Plasminogen activators, tissue degradation, and cancer. Adv. Cancer Res. 1985;44:139–266. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- 19.Castellino FJ, Ploplis VA. Structure and function of the plasminogen/plasmin system. Thromb. Haemost. 2005;93:647–654. doi: 10.1160/TH04-12-0842. [DOI] [PubMed] [Google Scholar]
- 20.Andreasen PA, Kjoller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: A review. Int. J. Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Fridman R, Toth M, Chvyrkova I, Meroueh SO, Mobashery S. Cell surface association of matrix metalloproteinase-9 (gelatinase B) Cancer and Metast. Rev. 2003;22:153–166. doi: 10.1023/a:1023091214123. [DOI] [PubMed] [Google Scholar]
- 22.Brown S, Meroueh SO, Fridman R, Mobashery S. Quest for selectivity in inhibition of matrix metalloproteinases. Curr. Top. Med. Chem. 2004;4:1227–1238. doi: 10.2174/1568026043387854. [DOI] [PubMed] [Google Scholar]
- 23.Lyons RM, Gentry LE, Purchio AF, Moses HL. Mechanism of activation of latent recombinant transforming growth factor beta 1 by plasmin. J. Cell. Biol. 1990;110:1361–1367. doi: 10.1083/jcb.110.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houck KA, Leung DW, Rowland AM, Winer J, Ferrara N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 1992;267:26031–26037. [PubMed] [Google Scholar]
- 25.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 26.Tang ML, Vararattanavech A, Tan SM. Urokinase-type plasminogen activator receptor induces conformational changes in the integrin alphaMbeta2 headpiece and reorientation of its transmembrane domains. J. Biol. Chem. 2008;283:25392–25403. doi: 10.1074/jbc.M802311200. [DOI] [PubMed] [Google Scholar]
- 27.Salasznyk RM, Zappala M, Zheng M, Yu L, Wilkins-Port C, McKeown-Longo PJ. The uPA receptor and the somatomedin B region of vitronectin direct the localization of uPA to focal adhesions in microvessel endothelial cells. Matrix Biol. 2007;26:359–370. doi: 10.1016/j.matbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Kjoller L, Hall A. Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J. Cell. Biol. 2001;152:1145–1157. doi: 10.1083/jcb.152.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillig T, Engelholm LH, Ingvarsen S, Madsen DH, Gardsvoll H, Larsen JK, Ploug M, Dano K, Kjoller L, Behrendt N. A composite role of vitronectin and urokinase in the modulation of cell morphology upon expression of the urokinase receptor. J. Biol. Chem. 2008;283:15217–15223. doi: 10.1074/jbc.C700214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarui T, Andronicos N, Czekay RP, Mazar AP, Bdeir K, Parry GC, Kuo A, Loskutoff DJ, Cines DB, Takada Y. Critical role of integrin alpha 5 beta 1 in urokinase (uPA)/urokinase receptor (uPAR, CD87) signaling. J. Biol. Chem. 2003;278:29863–29872. doi: 10.1074/jbc.M304694200. [DOI] [PubMed] [Google Scholar]
- 31.Bass R, Ellis V. Regulation of urokinase receptor function and pericellular proteolysis by the integrin alpha(5)beta(1) Thromb. Haemost. 2009;101:954–962. [PubMed] [Google Scholar]
- 32.Liu D, Aguirre Ghiso J, Estrada Y, Ossowski L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer cell. 2002;1:445–457. doi: 10.1016/s1535-6108(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 33.Aguirre Ghiso JA. Inhibition of FAK signaling activated by urokinase receptor induces dormancy in human carcinoma cells in vivo. Oncogene. 2002;21:2513–2524. doi: 10.1038/sj.onc.1205342. [DOI] [PubMed] [Google Scholar]
- 34.Chaurasia P, Aguirre-Ghiso JA, Liang OD, Gardsvoll H, Ploug M, Ossowski L. A region in urokinase plasminogen receptor domain III controlling a functional association with α5β1 integrin and tumor growth. J. Biol. Chem. 2006;281:14852–14863. doi: 10.1074/jbc.M512311200. [DOI] [PubMed] [Google Scholar]
- 35.Aguirre-Ghiso JA, Liu D, Mignatti A, Kovalski K, Ossowski L. Urokinase receptor and fibronectin regulate the ERK(MAPK) to p38(MAPK) activity ratios that determine carcinoma cell proliferation or dormancy in vivo. Mol. Biol. Cell. 2001;12:863–879. doi: 10.1091/mbc.12.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y, Tang CH, Kim Y, Robillard L, Zhang F, Kugler MC, Chapman HA. Urokinase receptors are required for alpha 5 beta 1 integrin-mediated signaling in tumor cells. J. Biol. Chem. 2007;282:3929–3939. doi: 10.1074/jbc.M607989200. [DOI] [PubMed] [Google Scholar]
- 37.Wei Y, Eble JA, Wang Z, Kreidberg JA, Chapman HA. Urokinase receptors promote beta1 integrin function through interactions with integrin alpha3beta1. Mol. Biol. Cell. 2001;12:2975–2986. doi: 10.1091/mbc.12.10.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang CH, Hill ML, Brumwell AN, Chapman HA, Wei Y. Signaling through urokinase and urokinase receptor in lung cancer cells requires interactions with beta1 integrins. J. Cell Sci. 2008;121:3747–3756. doi: 10.1242/jcs.029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei C, Möller CC, Altintas MM, Li J, Schwarz K, Zacchigna S, Xie L, Henger A, Schmid H, Rastaldi MP. Modification of kidney barrier function by the urokinase receptor. Nat. Med. 2008;14:55–63. doi: 10.1038/nm1696. [DOI] [PubMed] [Google Scholar]
- 40.Smith SC, Theodorescu D. The Ral GTPase pathway in metastatic bladder cancer: Key mediator and therapeutic target. Urol Oncol-Semin O I. 2009;27:42–47. doi: 10.1016/j.urolonc.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F, Tom CC, Kugler MC, Ching TT, Kreidberg JA, Wei Y, Chapman HA. Distinct ligand binding sites in integrin alpha3beta1 regulate matrix adhesion and cell-cell contact. J. Cell Biol. 2003;163:177–188. doi: 10.1083/jcb.200304065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z, Lin L, Huai Q, Huang M. Challenges for drug discovery-a case study of urokinase receptor inhibition. Comb. Chem. High T. Scr. 2009;12:961. doi: 10.2174/138620709789824727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mani T, Wang F, Knabe WE, Sinn AL, Khanna M, Jo I, Sandusky GE, Sledge GW, Jr, Jones DR, Khanna R, Pollok KE, Meroueh SO. Small-molecule inhibition of the uPAR.uPA interaction: synthesis, biochemical, cellular, in vivo pharmacokinetics and efficacy studies in breast cancer metastasis. Bioorg. Med. Chem. 2013;21:2145–2155. doi: 10.1016/j.bmc.2012.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khanna M, Wang F, Jo I, Knabe WE, Wilson SM, Li L, Bum-Erdene K, Li J, G WS, Khanna R, Meroueh SO. Targeting multiple conformations leads to small molecule inhibitors of the uPAR.uPA protein-protein interaction that block cancer cell invasion. ACS Chem. Biol. 2011;6:1232–1243. doi: 10.1021/cb200180m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mani T, Liu D, Zhou D, Li L, Knabe WE, Wang F, Oh K, Meroueh SO. Probing binding and cellular activity of pyrrolidinone and piperidinone small molecules targeting the urokinase receptor. ChemMedChem. 2013;8:1963–1977. doi: 10.1002/cmdc.201300340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobsen B, Gårdsvoll H, Juhl Funch G, Ostergaard S, Barkholt V, Ploug M. One-step affinity purification of recombinant urokinase-type plasminogen activator receptor using a synthetic peptide developed by combinatorial chemistry. Potein Expres Purif. 2007;52:286–296. doi: 10.1016/j.pep.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 48.Wang F, Eric Knabe W, Li L, Jo I, Mani T, Roehm H, Oh K, Li J, Khanna M, Meroueh SO. Design, synthesis, biochemical studies, cellular characterization, and structure-based computational studies of small molecules targeting the urokinase receptor. Bioorg. Med. Chem. 2012;20:4760–4773. doi: 10.1016/j.bmc.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- 50.Wang F, Li J, Sinn AL, Knabe WE, Khanna M, Jo I, Silver JM, Oh K, Li L, Sandusky GE, Sledge GW, Nakshatri H, Jones DR, Pollok KE, Meroueh SO. Virtual screening targeting the urokinase receptor, biochemical and cell-based studies, synthesis, pharmacokinetic characterization, and effect on breast tumor metastasis. J. Med. Chem. 2011;54:7193–7205. doi: 10.1021/jm200782y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br. J. Cancer. 1987;56:279–285. doi: 10.1038/bjc.1987.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheridan RP. The most common chemical replacements in drug-like compounds. J. Chem. Inf. Comput. Sci. 2002;42:103–108. doi: 10.1021/ci0100806. [DOI] [PubMed] [Google Scholar]
- 53.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comp. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 54.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comp. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 55.Case DA, Darden TA, TE Cheatham I, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. San Fransico: University of California; 2012. [Google Scholar]
- 56.Jorgensen WL, Chandraseklar J, Maduar JD, Impey RW, Klein ML. TIP3P water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 57.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryckaert JP, Ciccotti G, Berendsen JJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:15. [Google Scholar]
- 59.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 60.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical Treatment of Solvation for Molecular Mechanics and Dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 61.Karplus M, Kushick JN. Method for estimating the configurational entropy of macromolecules. Macromolecules. 1981;14:325–332. [Google Scholar]
- 62.Hecht JL, Honig B, Shin YK, Hubbell WL. Electrostatic Potentials near-the-Surface of DNA - Comparing Theory and Experiment. J. Phys. Chem. 1995;99:7782–7786. [Google Scholar]
- 63.Marco ADCMD, Carotti A, Cellamare S, Candia ED, Altomare C. Eur. J. Pharm. Sci. 2004;22:153–164. doi: 10.1016/j.ejps.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Milagre CDFM, HMS, Moran PJS, Rodrigues JAR. J. Org. Chem. 2010;75:1410–1418. doi: 10.1021/jo902227f. [DOI] [PubMed] [Google Scholar]
- 65.Rose ROC, Richter A, Hedberg C, Waldmann H. Chem. Eur. J. 2012;18:6520–6527. doi: 10.1002/chem.201103761. [DOI] [PubMed] [Google Scholar]
- 66.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Edit. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 67.Yebra M, Parry GC, Stromblad S, Mackman N, Rosenberg S, Mueller BM, Cheresh DA. Requirement of receptor-bound urokinase-type plasminogen activator for integrin alphavbeta5-directed cell migration. J. Biol. Chem. 1996;271:29393–29399. doi: 10.1074/jbc.271.46.29393. [DOI] [PubMed] [Google Scholar]
- 68.Liu D, Ghiso JAA, Estrada Y, Ossowski L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer cell. 2002;1:445–457. doi: 10.1016/s1535-6108(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 69.Aguirre GJ. Inhibition of FAK signaling activated by urokinase receptor induces dormancy in human carcinoma cells in vivo. Oncogene. 2002;21:2513–2524. doi: 10.1038/sj.onc.1205342. [DOI] [PubMed] [Google Scholar]
- 70.Smith HW, Marra P, Marshall CJ. uPAR promotes formation of the p130Cas-Crk complex to activate Rac through DOCK180. J. Cell. Biol. 2008;182:777–790. doi: 10.1083/jcb.200712050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang B, Li L, Hurley TD, Meroueh SO. Molecular recognition in a diverse set of protein-ligand interactions studied with molecular dynamics simulations and end-point free energy calculations. J. Chem. Inf. Model. 2013;53:2659–2670. doi: 10.1021/ci400312v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Liang S, Pilcher MM, Meroueh SO. Incorporating receptor flexibility in the molecular design of protein interfaces. Protein Eng. Des. Sel. 2009;22:575–586. doi: 10.1093/protein/gzp042. [DOI] [PubMed] [Google Scholar]
- 73.Li L, Uversky VN, Dunker AK, Meroueh SO. A computational investigation of allostery in the catabolite activator protein. J. Am. Chem. Soc. 2007;129:15668–15676. doi: 10.1021/ja076046a. [DOI] [PubMed] [Google Scholar]
- 74.Liang S, Li L, Hsu WL, Pilcher MN, Uversky V, Zhou Y, Dunker AK, Meroueh SO. Exploring the molecular design of protein interaction sites with molecular dynamics simulations and free energy calculations. Biochemistry. 2009;48:399–414. doi: 10.1021/bi8017043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson AD, Dugan A, Gestwicki JE, Mapp AK. Fine-tuning multiprotein complexes using small molecules. ACS Chem. Biol. 2012;7:1311–1320. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cunningham O, Andolfo A, Santovito ML, Iuzzolino L, Blasi F, Sidenius N. Dimerization controls the lipid raft partitioning of uPAR/CD87 and regulates its biological functions. EMBO J. 2003;22:5994–6003. doi: 10.1093/emboj/cdg588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu X, Gårdsvoll H, Yuan C, Lin L, Ploug M, Huang M. Crystal structure of the urokinase receptor in a ligand-free form. J. Mol. Biol. 2012;416:629–641. doi: 10.1016/j.jmb.2011.12.058. [DOI] [PubMed] [Google Scholar]
- 78.Lin L, Gardsvoll H, Huai Q, Huang M, Ploug M. Structure-based engineering of species selectivity in the uPA·uPAR interaction. Implication for preclinical cancer therapy. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.093492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gardsvoll H, Kjaergaard M, Jacobsen B, Kriegbaum MC, Huang M, Ploug M. Mimicry of the regulatory role of urokinase in lamellipodia formation by introduction of a non-native interdomain disulfide bond in its receptor. J. Biol. Chem. 2011;286:43515–43526. doi: 10.1074/jbc.M111.300020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.