Abstract
The DNA wrapping model of transcription stipulates that DNA bending and wrapping around RNA polymerase causes an unwinding of the DNA helix at the enzyme catalytic center that stimulates strand separation prior to initiation and during transcript elongation. Recent experiments with mammalian RNA polymerase II indicate the significance of DNA bending and wrapping in transcriptional mechanisms. These findings have important implications in our understanding of the role of the general transcription factors in transcriptional initiation and the mechanisms underlying transcriptional regulation.
Keywords: mRNA synthesis, transcription initiation, RNA polymerase II, DNA wrapping, general transcription factors
The RNA polymerase II general transcription machinery
Eukaryotic mRNA is synthesized by RNA polymerase II (Pol II). In vitro the accurate initiation of transcription by Pol II at most promoters requires a set of five general transcription factors (GTFs) (Orphanides et al. 1996; Hampsey 1998). The mammalian GTFs include the TATA box-binding protein (TBP), and transcription factors (TF) IIB, TFIIE (TFIIE56 and TFIIE34 subunits), TFIIF (RAP74 and RAP30 subunits), and TFIIH. TFIIH comprises nine subunits including a kinase and two ATP-dependent, single-stranded DNA helicases, one of which is essential for transcription initiation (Tirode et al. 1999). In mammalian cell extracts, TBP is tightly associated with 8–11 TAFIIs (TBP-associating factors) to form the TFIID complex (Mr = ~700 kDa) (Orphanides et al. 1996; Hampsey 1998). TFIIA is a trimeric factor that is not essential for transcription but which stimulates the interaction of TFIID with some promoters (Coulombe et al. 1992). Early models of preinitiation complex formation have proposed that the GTFs and Pol II assemble in a sequential order onto promoter DNA (Fig. 1). This notion has recently been challenged by the discovery of large multisubunit complexes, called Pol II holoenzymes, which contain a number of GTFs, core Pol II, the products of a set of SRB genes (Suppressors of RNA polymerase B), and several additional polypeptides (Greenblatt 1997). Some of these Pol II holoenzymes contain all the necessary factors for accurate transcriptional initiation in vitro in addition to those required for response to transcriptional activators. Preinitiation complex formation with the Pol II holoenzyme can be achieved in either one or a limited number of steps in vitro (Fig. 1); however, it is generally agreed that the single-step recruitment of the holoenzyme is the mechanism of preinitiation complex formation in vivo.
Fig. 1.
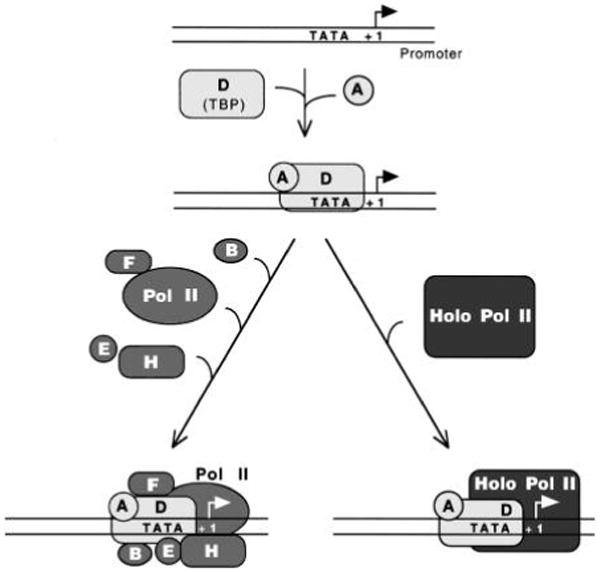
Preinitiation complex formation on a class II promoter. The GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) and Pol II can assemble sequentially onto the promoter DNA in vitro. Alternatively, the Pol II holoenzyme, which typically contains core Pol II, a number of GTFs, and several additional polypeptides, can bind to the promoter in a single step. TFIID (TBP and TAFIIs) or TBP alone bind to the TATA box of promoters to begin preinitiation complex formation close to the transcriptional initiation site (+1).
Core Pol II is a ~550 kDa enzyme composed of 8–12 subunits depending of the species of origin (Orphanides et al. 1996; Hampsey 1998). The two largest subunits, Rpb1 (~220 kDa) and Rpb2 (~180 kDa), are the most highly conserved subunits and are homologous to the β′ and β subunits, respectively, of bacterial RNA polymerase. Rpb1 contains a C-terminal domain (CTD) composed of an heptapeptide tandemly repeated 26–27 times in yeast and 52 times in humans (Dahmus 1996). Two different forms of Pol II exist in vivo, Pol IIA and Pol IIO. Pol IIA possesses an unphosphorylated CTD and preferentially enters the preinitiation complex, while Pol IIO possesses an extensively phosphorylated CTD and is found in the elongation complex. A number of kinases and one phosphatase that regulate CTD phosphorylation have been identified (Dahmus 1996; Orphanides 1997; Hampsey 1998). The structure of core Pol II in solution has been determined, and indicates the presence of finger-like projections that close to form a channel large enough to accommodate DNA (Darst et al. 1991). See Fig. 2A for a schematic representation. Cross-linking studies using E. coli RNA polymerase, which has a solution structure strikingly similar to that of yeast Pol II, indicate that the finger-like projections constitute the catalytic center of the enzyme and can grasp DNA at the transcriptional initiation site in the preinitiation complex and at the site of ribonucleotide addition in the elongation complex (Mustaev et al. 1997; Nudler et al. 1998) (see Fig. 2B). In solution, the CTD of Pol II has been shown to attach to the body of the enzyme opposite to the catalytic center (Meredith et al. 1996) (see Fig. 2A). Recently, the position of the CTD in the initiation complex has been determined (Douziech et al. 1999).
Fig. 2.
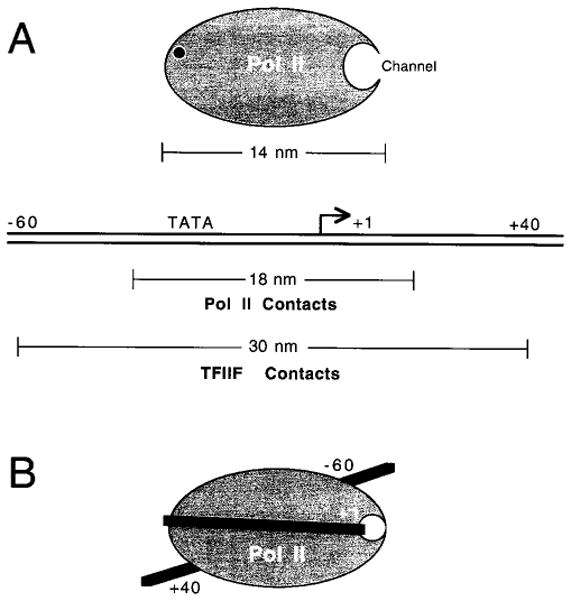
Promoter DNA is wrapped around core Pol II in the initiation complex. (A) Eukaryotic core Pol II is represented with the 2.5-nm channel in its open conformation and the point of attachment of the CTD of Rpb1 (black dot). The longest dimension of Pol II is indicated (14 nm). The contact points of both Pol II and TFIIF with promoter DNA are as determined by the photo-cross-linking studies and comprise 18 and 30 nm of B-form DNA, respectively. (B) The paradox described in (A) (e.g., the “footprint” is longer than the “foot”) is resolved if the promoter DNA is wrapped around core Pol II. After DNA binding, the 2.5-nm channel, which contains the catalytic center of the enzyme, is in the closed conformation.
Promoter DNA is wrapped around Pol II in the initiation complex
Evidence for the wrapping of promoter DNA around Pol II in the preinitiation complex is emerging from site-specific DNA–protein photo-cross-linking studies using the adenovirus 2 major late promoter (Forget et al. 1997; Kim et al. 1997; Robert et al. 1998). Various intermediate complexes have been analyzed using different cross-linking reagents located along the promoter DNA. These studies indicate extensive bending and wrapping of the DNA in the initiation complex (Coulombe and Burton 1999).
In studying a preinitiation complex containing TBP, TFIIB, TFIIF, TFIIE, and Pol IIB (a form of Pol II lacking the CTD), we found that Pol II closely approaches promoter DNA in several regions between nucleotides −40 to +13 (Forget et al. 1997; Robert et al. 1998). This 53-bp region corresponds to ~18 nm of B-form DNA and is longer than the longest dimension of Pol II (14 nm; see Fig. 2A). Thus, the “footprint” is longer than the “foot.” This apparent paradox can be explained if promoter DNA is wrapped around Pol II in the preinitiation complex (Fig. 2B). In addition, the TFIIF subunits have been detected along promoter DNA from nucleotides −56 to +34, a 89-bp-long region that corresponds to ~30 nm of B-form DNA (Coulombe et al. 1994; Robert et al. 1996; Forget et al. 1997; Robert et al. 1998). This finding alone argues against promoter DNA being in a linear conformation. The cross-linking data also indicates that single molecules of RAP74, RAP30, and TFIIE34 simultaneously approach the DNA both downstream of the transcriptional initiation site (nucleotides +13 and +26) and upstream of the TATA element (nucleotides −39/−40 and −56/−61) (Robert et al. 1998), demonstrating that these sequences are juxtaposed (see Fig. 3, BACK view1 and Fig. 4A). In addition, electron microscopy images of the initiation complex on promoter DNA allow the direct visualization of DNA wrapping in the complex (Forget et al. 1997), providing conclusive support for the cross-linking data. Measurements obtained from electron microscopy images indicate that ~50–70 bp of DNA are consumed by the protein core of the complex (Forget et al. 1997; Kim et al. 1997). Given the dimensions of Pol II (14 × 13.6 × 11 nm), and the existence of a number of channels and grooves on the enzyme surface that can accommodate DNA, a 70–100 bp DNA sequence should be sufficient to wrap completely around the enzyme. This would imply that ~50–80 bp of B-form DNA are effectively consumed by the complex, in good agreement with electron microscopy measurements.
Fig. 3.
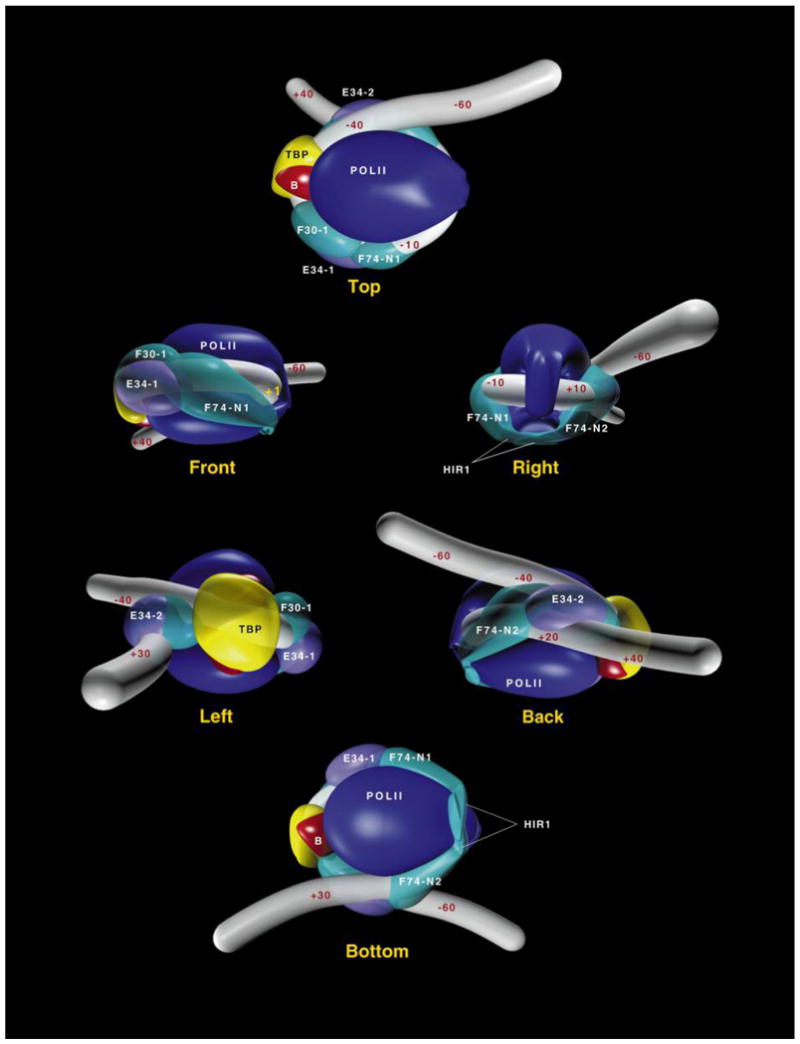
Proposed structure for a TBP–TFIIB–TFIIF–Pol II–TFIIE–promoter complex. Six views are presented. In the FRONT view, the complex is shown with the promoter between the TATA element and the transcriptional initiation site being placed in the direction of the view. The positions of the transcriptional initiation site (+1), TBP, TFIIB (B), Pol II, and each of two molecules of TFIIE34 (E34-1 and E34-2), RAP74 (F74-1 and F74-2), and RAP30 (F30-1 and F30-2) are shown.
Fig. 4.
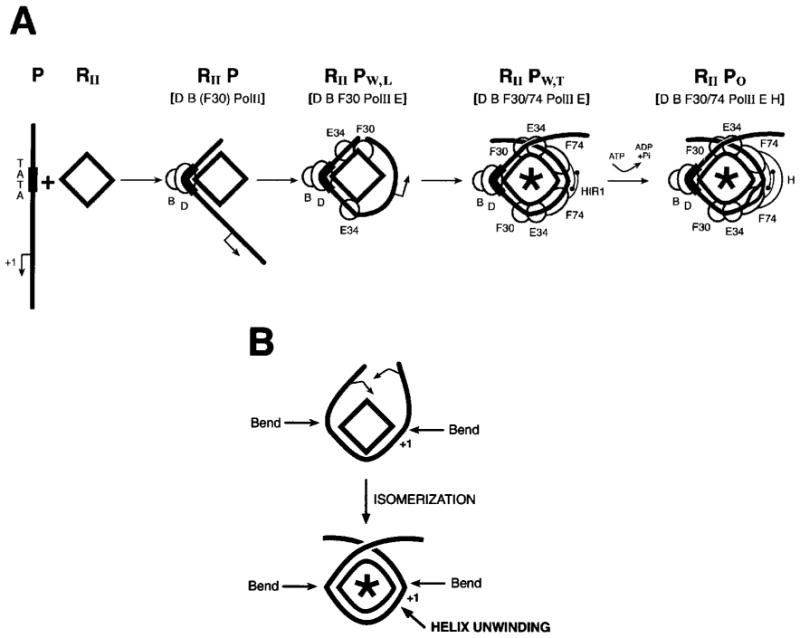
Wrapping of promoter DNA around Pol II accompanies a conformational isomerization of the preinitiation complex. (A) During preinitiation complex formation, the promoter DNA (P) is progressively wrapped around Pol II (RII) through the action of the various GTFs including TBP (D), TFIIB (B), TFIIE (E), TFIIF (F), and TFIIH (H). The HIR1 of RAP74, which is important for isomerization, is shown. Isomerization refers to the progression of specific conformational changes in protein and DNA that occurs during preinitiation complex formation. In the open complex (RIIPo), the melting of the DNA helix is completed through the action of TFIIH in the presence of ATP. In RIIPW,T (RIIPWRAPPED,TIGHT), tight wrapping is expected to induce a torsional strain in the DNA helix which facilitates local helix unwinding. RIIPW,L (RIIPWRAPPED,LOOSE) and RIIP are intermediate complexes that have been analyzed in photo-cross-linking experiments. The asterisk (*) indicates a conformational change in core Pol II. (B) DNA bending and wrapping around Pol II (isomerization) induces unwinding of the DNA helix in the vicinity of the transcriptional initiation site.
DNA wrapping around RNA polymerase is an evolutionarily conserved mechanism
DNA wrapping during preinitiation complex formation was first proposed for E. coli RNA polymerase (Cowing et al. 1989; Schickor et al. 1990; Craig et al. 1995; Polyakov et al. 1995). In a manner reminiscent to that described above for Pol II, the footprint of the E. coli RNA polymerase holoenzyme (the α2ββ′σ complex) extends from either −70 or −50 to +20, depending on the promoter. This corresponds to a ~31 nm-long region of B-form DNA, and is longer than the longest dimension (16 nm) of E. coli RNA polymerase (Darst et al. 1989). Preinitiation complex formation using bacterial holoenzyme has been studied at various temperatures (0, 15, and 30–37°C), thereby permitting characterization of the intermediates in the formation of the holoenzyme–promoter complex. An important difference in the structures of these complexes is the extent of the DNA region that is protected by RNA polymerase. At low temperatures (0–15°C) the footprint lies between −70 or −50 and −10, whereas at higher temperatures (30–37°C), the footprint extends from −70 or −50 to +20. In a dynamic perspective, this observation indicates that promoter DNA is progressively wrapped around E. coli RNA polymerase during preinitiation complex formation.
In the Pol II system, we have also been able to characterize intermediates in the formation of the preinitiation complex. Instead of using different temperatures, we took advantage of a set of RAP74 deletion mutants in our photo-cross-linking experiments (Forget et al. 1997; Robert et al. 1998). The presence of a fragment of RAP74 containing the first 172 amino acids (e.g., one containing the RAP30-binding domain) resulted in promoter contacts by Pol II between positions −19 and −5. In contrast, RAP74(1–205) and other longer mutants extend the promoter contacts by Pol II from −39/−40 to +13. These results indicate that RAP74 supports the progressive wrapping of the promoter DNA around Pol II. The progression of specific conformational changes in protein and DNA that occurs during preinitiation complex formation is referred to as complex isomerization (Coulombe and Burton 1999) and is summarized in Fig. 4A.
Together, these observations indicate that DNA is wrapped around both prokaryotic and eukaryotic RNA polymerases and suggest that DNA wrapping around RNA polymerase may be a fundamental transcriptional mechanism that is evolutionarily conserved.
A function of the GTFs is to help the wrapping of promoter DNA around Pol II
The DNA wrapping model suggests that a function of the GTFs is to help promoter DNA wrap around Pol II (Figs. 3 and 4A). X-ray crystallography and NMR studies have revealed that the binding of TBP and TFIIB to the TATA box induces a ~80° bend in the DNA (Kim et al. 1993a, 1993b; Werner and Burley 1997). This observation indicates that the binding of TBP and TFIIB to the promoter initiates DNA wrapping. Cross-linking experiments with the TBP–TFIIB–TFIIF–Pol II–TFIIE-promoter complex have revealed that the DNA is bent in the region of the TATA box, thereby allowing promoter contacts by Pol II in the −39/−40 region, and that a second bend in the +1 region is responsible for promoter contacts by Pol II downstream of the transcriptional initiation site (Forget et al. 1997; Robert et al. 1998). See Fig. 4B. Such a bend in the region of the transcriptional initiation site has been described by Ebright and colleagues (Kim et al. 1997). Interestingly, Pol II contacts both upstream of the TATA box and downstream of the initiation site require the presence of RAP74, minimally a fragment containing amino acids 1–205. RAP74(1–205) contains both the RAP30-binding domain (amino acids 1–172) and a homomeric interaction region (HIR1; amino acids 172–205) required for the maintenance of TFIIF in a α2 β 2 arrangement in the preinitiation complex assembled with recombinant polypeptides (Robert et al. 1998). This observation indicates that the association of a TFIIF α 2 β 2 heterotetramer is required for tight DNA wrapping around Pol II. The presence of TFIIE, which also enters the preinitiation complex as a α 2 β 2 heterotetramer, favors DNA wrapping (Robert et al. 1998). Interestingly, the cross-linking data further indicate that TFIIH tightens the DNA loop around the enzyme (Robert et al. 1998). This observation defines a novel function for TFIIH in transcriptional initiation.
In conclusion, the GTFs are all nvolved in the wrapping of the DNA around Pol II. In principle, a role for the GTFs in DNA wrapping may exist regardless of whether the preinitiation complex is assembled sequentially or through a single-step recruitment of the Pol II holoenzyme.
DNA wrapping induces helix unwinding which facilitates transcriptional initiation
Transcriptional initiation requires the melting of promoter DNA immediately upstream of the transcriptional initiation site, leading to the formation of the open complex (see Fig. 4A). Open complex formation is necessary for the proper access of ribonucleotides to the catalytic center of the enzyme. In the Pol II system, open complex formation requires both ATP hydrolysis and one of the TFIIH helicases (Orphanides et al. 1996; Hampsey et al. 1998). The TFIIH helicases use single-stranded DNA as substrate, implying that the promoter DNA must be at least partly melted prior to helicase action. A possible explanation for this observation is that the tight wrapping of promoter DNA around Pol II in the wrapping model induces a torsional strain in the DNA helix that may create just enough stand separation so as to allow the TFIIH helicase to latch onto single-stranded DNA and catalyze the formation of the open complex in the presence of ATP (Fig. 4). This idea has yet to be supported by direct experimental results but is consistent with a number of observations. For example, TFIIF and TFIIE have been shown to facilitate promoter melting prior to initiation (Pan and Greenblatt 1994; Holstege et al. 1995). The effects of TFIIF and TFIIE may be the consequence of the torsional strain that follows DNA wrapping in the preinitiation complex. Interestingly, the domain of RAP74 that is minimally required to support tight DNA wrapping around Pol II, HIR1, is important for transcriptional initiation in vitro (Wang and Burton 1995; Lei et al. 1998). DNA wrapping during transcriptional elongation may also play a role in facilitating the access of ribonucleotides to the catalytic center of Pol II for transcript elongation (Coulombe and Burton 1999).
Promoter DNA is wrapped around TFIID: replacement of wrapped structures?
The role of TFIID in transcriptional initiation has been extensively studied and is still a subject of debate (Burley and Roeder 1996; Hahn 1998). Depending on the context, TAFIIs act as general promoter selectivity factors, co-activators or co-repressors of initiation, or are not required at all for transcription. Thus, it appears that the requirement for TAFIIs is promoter specific and may operate at various levels. Recent experiments have indicated that some TAFIIs are subunits of other complexes, such as a histone acetyltransferase complex called SAGA, adding to the complexity of their function (Hahn 1998).
Photo-cross-linking experiments have revealed that promoter DNA wraps around the TFIID–(TFIIA) complex (Oelgeschlager et al. 1996; Hoffmann et al. 1997) in a structure that is very similar to that proposed for the preinitiation complex formed in the presence of TBP, TFIIB, TFIIF, Pol II, and TFIIE. Both wrapped structures occupy the same region of the promoter, extending from about position −30 to +40 (Burke and Kadonaga 1996, 1997; Oelgeschlager et al. 1996). This suggests that these two promoter complexes cannot co-exist. Because of this incompatibility, preinitiation complex assembly requires that the TAFIIs release their DNA contacts within the core promoter (Fig. 5). In many cases, a promoter may be a nucleoprotein complex of TBP, TAFIIs, and DNA, rather than a linear DNA sequence, and Pol II and the GTFs may recognize both DNA and TFIID in promoter binding. Evidence supporting this hypothesis comes from the identification of protein–protein interactions between TBP and TFIIB (Ha et al. 1993), TAFII250 and RAP74 (Diskstein et al. 1996), TFIIA and TFIIE (Yokomori et al. 1998; Rojas, A., and Coulombe, B., unpublished data), and TFIIA and TFIIF (A. Rojas, M.-F. Langelier, D. Forget, and B. Coulombe, unpublished data). These data provide a rational explanation for the ability of TAFIIs to act as promoter selectivity factors or to be completely unrequired for transcriptional initiation depending on the particular promoter. The role of TAFs as co-activators/co-repressors is addressed below.
Fig. 5.
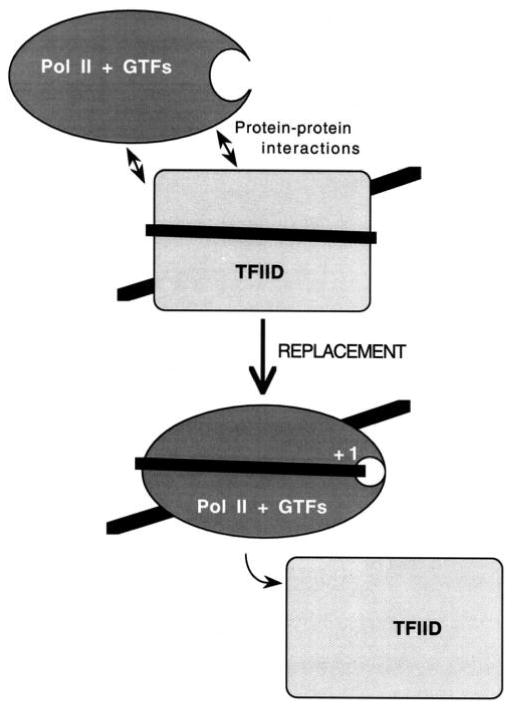
A proposed additional step in preinitiation complex assembly involving the replacement of TAFIIs by the GTFs and Pol II on core promoter sequences. Protein–protein interactions between TAFIIs/TBP/TFIIA and the GTFs/Pol II may facilitate this replacement step.
DNA wrapping as a mechanism of transcriptional control
Several reports have indicated that direct protein–protein interactions between the activation domain of DNA-bound transcriptional activators and components of the Pol II holoenzyme are both necessary and sufficient for activation both in vitro and in vivo (Treisenberg 1995; Ptashne and Gann 1997). These results have been interpreted as an indication that activator proteins recruit the basal transcriptional machinery on core promoters. Activation-domain bypass experiments appear to support this notion (Ptashne and Gann 1997). In these experiments, the DNA-binding domain of an activator (lacking its activation domain) is fused to a component of the general transcription machinery. The fusion protein is expressed in cells and tested for its capability to activate transcription of a gene containing the appropriate binding site. When general factors such as TBP have been used and found to cause activation, the conclusion has been drawn that the binding of TBP, and therefore recruitment of Pol II holoenzyme to the promoter, has been enhanced (Ptashne and Gann 1997).
According to the DNA wrapping model, however, another interpretation of this activation data is possible. Activation may result not only from Pol II recruitment, but also from stimulation of open complex formation resulting from the DNA wrapping that must be induced by looping the DNA between the upstream DNA element and the core promoter (Fig. 6, top part). In the bacterial system, the CAP protein (Crothers and Steitz 1992; Lavigne et al. 1992; Niu et al. 1996) and the λ repressor (Hawley and McClure 1982) have been shown to stimulate not only holoenzyme-promoter binding, but also isomerization of the preinitiation complex.
Fig. 6.
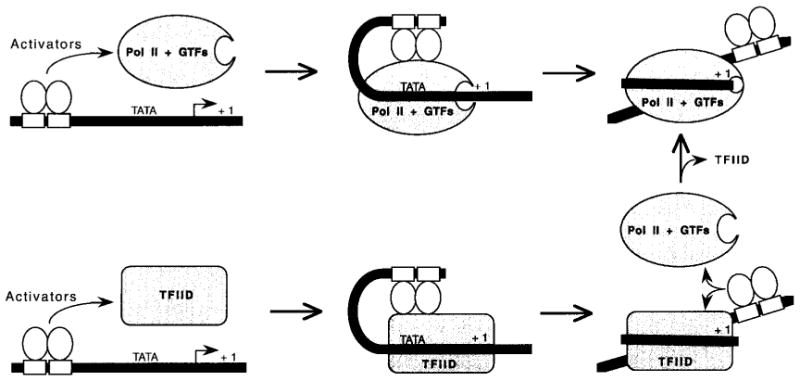
A model for the proposed role of transcriptional activators in the isomerization of the preinitiation complex. Protein–protein interactions between the activation domain of a DNA-bound activator and the Pol II transcription machinery induce the formation of a loop that initiates DNA wrapping around Pol II. For activators requiring TFIID as a co-activator, the promoter DNA is first wrapped around the TFIID complex. In such cases, the activator can work by either helping the formation of a wrapped TFIID-promoter structure, or by facilitating the displacement of TAFIIs and their replacement by Pol II/GTFs.
In the DNA wrapping model, transcriptional regulation by activation (or repression) may also occur at the level of the wrapped structure of the TFIID–(TFIIA)–promoter complex. Activators requiring TFIID as a co-activator may help in the binding of TFIID to the promoter DNA and, consequently, the formation of the wrapped promoter structure that is ready to accommodate Pol II and the GTFs (Fig. 6, bottom part). Alternatively, these TFIID-dependent activators may facilitate the displacement of TFIID from the promoter, a mechanism analogous to active anti-repression. Transcriptional repressors requiring TFIID as cofactor may work by interfering with the positive role of TFIID.
The DNA wrapping model may also help to explain the synergetic effect of transcriptional activators. Synergy may result from activators making a number of relatively weak contacts with the Pol II holoenzyme. Formation of the DNA wrap is an inherently cooperative process, because as the DNA is docked into grooves on the RNA polymerase holoenzyme surface many favorable binding contacts are simultaneously developed along an extended protein-DNA interface. Transcriptional activators can affect this process cooperatively through multiple distinct protein–protein and protein–DNA contacts. Cooperativity between activators and the natural DNA wrapping process is expected to stimulate both the recruitment of Pol II and isomerization of the holoenzyme and promoter DNA in much the same way that CAP and λ repressor are thought to activate transcription in E. coli.
Acknowledgments
I wish to thank the following people without whom these ideas would not have been developed: Zach Burton, Maxime Douziech, Diane Forget, Jack Greenblatt, François Robert, and Pierre Vandenberghe. I also thank Will Home and Maxime Douziech for help in preparing the manuscript. My work is supported by grants from the Medical Research Council of Canada and the Cancer Research Society Inc., and I hold a Scholarship from the Fonds de la Recherche en Santé du Québec.
Abbreviations
- CTD
C-terminal domain
- GTFs
general transcription factors
- HIR
homomeric interaction region
- Pol II
RNA polymerase II
- TBP
TATA box-binding protein
- TAFIIs
TBP-associating factors
- TF
transcription factor
Footnotes
Some of our molecular models are available on the Internet at http://www.ircm.qc.ca/coulombe.
References
- 1.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 2.Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 3.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to human and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: a revolutionary model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulombe B, Killeen M, Liljelund P, Honda B, Xiao H, Ingles CJ, Greenblatt J. Identification of three mammalian proteins that bind to the yeast TATA box protein TFIID. Gene Expr. 1992;2:99–110. [PMC free article] [PubMed] [Google Scholar]
- 6.Coulombe B, Li J, Greenblatt J. Topological localization of the human transcription factors IIA, IIB, TATA box-binding protein, and RNA polymerase II-associated protein 30 on a class II promoter. J Biol Chem. 1994;269:19 962–19 967. [PubMed] [Google Scholar]
- 7.Cowing DW, Mecsas J, Record MT, Jr, Gross CA. Intermediates in the formation of the open complex by RNA polymerase holoenzyme containing the sigma factor σ32 at the groE promoter. J Mol Biol. 1989;210:521–530. doi: 10.1016/0022-2836(89)90128-9. [DOI] [PubMed] [Google Scholar]
- 8.Craig ML, Suh WC, Record MT., Jr HO., and DNase I probing of Eσ70 RNA polymerase-λPR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15 624–15 632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 9.Crothers DM, Steitz TA. Transcriptional regulation. Vol. 1. Cold Spring Harbor Press; Cold Spring Harbor, New York: 1992. Transcriptional activation by E. coli CAP protein; pp. 501–534. [Google Scholar]
- 10.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19 009–19 012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 11.Darst SA, Kubalek EW, Kornberg RD. Three-dimensional Esherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature (London) 1989;340:730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- 12.Darst SA, Edwards AM, Kubalek EW, Kornberg RD. Three-dimensional structure of yeast RNA polymerase II at 16 Å resolution. Cell. 1991;66:121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- 13.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 14.Douziech M, Forget D, Greenblatt J, Coulombe B. Topological localization of the carboxyl-terminal domain of RNA polymerase II in the initiation complex. J Biol Chem. 1999;274:19 868–19 873. doi: 10.1074/jbc.274.28.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douziech M, Rorget D, Greenblatt J, Coulombe B. Topological localization of the carboxy-terminal domain of RNA polymerase II in the initiation complex. J Biol Chem. 1999;274:19 868–19 873. doi: 10.1074/jbc.274.28.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forget D, Robert F, Grondin G, Burton ZF, Greenblatt J, Coulombe B. RAP74 induces promoter contact by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenblatt J. RNA polymerase II holoenzyme and transcriptional regulation. Curr Opin Cell Biol. 1997;9:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 18.Ha I, Roberts S, Maldonado E, Sun X, Kim LU, Green M, Reinberg D. Multiple functional domains of human transcription factor IIB: Distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 19.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 20.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley DK, McClure WR. Mechanism of activation of transcription initiation from the λ PRM promoter. J Mol Biol. 1982;157:493–525. doi: 10.1016/0022-2836(82)90473-9. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann A, Oelgeschlager T, Roeder RG. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege FC, Tantin D, Carey M, van der Vliet PC, Timmers HT. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of the TATA element. Nature (London) 1993a;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature (London) 1993b;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 26.Kim TK, Lagrange T, Wang YH, Griffith JD, Reinberg D, Ebright RH. Trajectory of DNA in the RNA polymerase II transcription pre-initiation complex. Proc Natl Acad Sci USA. 1997;94:12 268–12 273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavigne M, Herbert M, Kolb A, Buc H. Upstream curved sequences influence the initiation of transcription at the E. coli galactose operon. J Mol Biol. 1992;224:293–306. doi: 10.1016/0022-2836(92)90995-v. [DOI] [PubMed] [Google Scholar]
- 28.Lei L, Ren D, Finkelstein A, Burton ZF. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meredith GD, Chang WH, Li Y, Bushnell DA, Darst SA, Kornberg R. The C-terminal domain revealed in the structure of RNA polymerase II. J Mol Biol. 1996;258:413–419. doi: 10.1006/jmbi.1996.0258. [DOI] [PubMed] [Google Scholar]
- 30.Mustaev A, Kozlov M, Markovtsov V, Zaychikov E, Denissova L, Goldfarb A. Modular organization of the catalytic center of RNA polymerase. Proc Natl Acad Sci USA. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Spatial organization of transcription elongation complex in Escherichia coli. Science (Washington, DC) 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
- 33.Oelgeschlager T, Chiang CM, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 34.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 35.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30 101–30 104. [PubMed] [Google Scholar]
- 36.Polyakov A, Severinova E, Darst SA. Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 37.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 38.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 39.Robert F, Douziech M, Forget D, Egly JM, Greenblatt J, Burton ZF, Coulombe B. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: Assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 42.Treisenberg SJ. Structure and function of transcriptional activation domains. Curr Opin Gen Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 43.Wang BQ, Burton ZF. Functional domains of human RAP74 including a masked polymerase binding domain. J Biol Chem. 1995;270:27 035–27 044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 44.Werner MH, Burley SK. Architectural transcription factors: Proteins that remodel DNA. Cell. 1997;88:733–736. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- 45.Yokomori K, Verrijzer CP, Tjian R. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc Natl Acad Sci USA. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
This article has been cited by
- 46.Petro Yakovchuk Benjamin Gilman, Goodrich James A, Kugel Jennifer F. RNA Polymerase II and TAFs Undergo a Slow Isomerization after the Polymerase Is Recruited to Promoter-Bound TFIID. Journal of Molecular Biology. 2010;397:57–68. doi: 10.1016/j.jmb.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 47.Brenowitz M, Erie DA, Chance MR. Catching RNA polymerase in the act of binding: Intermediates in transcription illuminated by synchrotron footprinting. Proceedings of the National Academy of Sciences. 2005;102:4659–4660. doi: 10.1073/pnas.0501152102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tornøe Jens, Kusk Philip, Johansen Teit E, Jensen Peter R. Generation of a synthetic mammalian promoter library by modification of sequences spacing transcription factor binding sites. Gene. 2002;297:21–32. doi: 10.1016/s0378-1119(02)00878-8. [DOI] [PubMed] [Google Scholar]
- 49.Kretz-Remy Carole, Michaud Sébastien, Robert M Tanguay. The nuclear chronicles: gene transcription and molecular traveling. Biochemistry and Cell Biology. 1999;77:4. [PubMed] [Google Scholar]


