Abstract
The comprehensive MS analysis of the peptidome, the intracellular and intercellular products of protein degradation, has the potential to provide novel insights on endogenous proteolytic processing and their utility in disease diagnosis and prognosis. Along with the advances in MS instrumentation and related platforms, a plethora of proteomics data analysis tools have been applied for direct use in peptidomics; however an evaluation of the currently available informatics pipelines for peptidomics data analysis has yet to be reported. In this study, we begin by evaluating the results of several popular MS/MS database search engines including MS-GF+, SEQUEST and MS-Align+ for peptidomics data analysis, followed by identification and label-free quantification using the well-established accurate mass and time (AMT) tag and newly developed informed quantification (IQ) approaches, both based on direct LC-MS analysis. Our results demonstrate that MS-GF+ out-performed both SEQUEST and MS-Align+ in identifying peptidome peptides. Using a database established from MS-GF+ peptide identifications, both the AMT tag and IQ approaches provided significantly deeper peptidome coverage and less missing data for each individual data set than the MS/MS methods, while achieving robust label-free quantification. Besides having an excellent correlation with the AMT tag quantification results, IQ also provided slightly higher peptidome coverage. Taken together, we propose an optimized informatics pipeline combining MS-GF+ for initial database searching with IQ (or AMT tag) approaches for identification and label-free quantification for high-throughput, comprehensive and quantitative peptidomics analysis.
Keywords: peptidomics, identification, quantification, MS-GF+, accurate mass and time tag, informed quantitation
Introduction
Peptidomics is defined as the comprehensive characterization of “native” peptides in a biological sample [1]. Without digesting proteins into peptides using trypsin or other proteases as applied in conventional bottom-up proteomics, peptidomics is able to preserve the endogenous information of the peptidome peptides from a biological sample, including post-translational modifications (PTMs) and proteolytic products revealing the natural proteases participated in the proteolytic processes [2]. In contrast to limited study of small neuropeptides [3-6], broad and large-scale studies have been increasingly conducted to characterize endogenous peptides in different biological samples, including cell lines [7, 8], body fluids [9, 10], as well as tissues [11], for biomarker screening or clinically-related studies.
The peptidome coverage and effectiveness for quantification provided by a peptidomics pipeline is dependent on each step of the pipeline, including peptide extraction and/or enrichment/fractionation, separation, LC-MS data acquisition, as well as the subsequent informatics analysis [12]. Despite many new advances in both sample preparation [13, 14] and instrumentation [15], very limited progress has been made toward improved data analysis for peptidomics studies. Many proteomics software tools have been applied directly to peptidomics studies, such as MS-GF+ [16], SEQUEST [17], Mascot [18], MS-Align+ [19], which has been reviewed recently [20]. However, to the best of our knowledge, there is no detailed study reported for comparing the performance of these proteomics software tools for peptide identification for peptidomics analysis.
Different stable isotope labeling strategies such as 18O-labeling [21] and other chemical labeling [8] have been applied for quantification of peptidomic changes. Although the labeling approaches can provide accurate quantification, they are typically associated with increased cost, sample losses and increased sample processing time. In contrast, label-free quantification, a simple yet effective method, can be highly reliable in well-controlled experiments [22]. In addition, there are also well-established methods, such as the accurate mass and time (AMT) tag approach [23], that provide both improved measurement throughput and reliable label-free quantification. More recently, we have developed a new software tool, informed quantification (IQ), which capitalizes on peptide LC elution and high-accuracy mass information, and is capable of accurate de-isotoping, peak matching as well as label-free quantification, independent of MS/MS data [11]. Such MS/MS-independent proteomics analysis strategies have the potential of providing both increased coverage and reliable quantification in large-scale peptidomics studies.
In this study, we applied both the AMT tag and the augmented IQ informatics pipelines for analyzing data sets from our recent peptidomics study on potential ischemia effects in ovarian cancer tumors [11], and evaluated their performance in greater detail. Both the AMT tag and IQ analyses use a database consisting of peptides identified from conventional database searching of the MS/MS data from each individual peptidomics data set from the entire study, and the study included evaluation of the performance of different search engines, including MS-GF+, SEQUEST and MS-Align+, for effectiveness in peptidomic peptide identification. The results showed that MS-GF+ could identify many more unique peptidome peptides than SEQUEST and MS-Align+. Both AMT tag and IQ approaches were shown to provide more unique peptide identification than the database searching methods, which greatly reduced missing data across the entire data sets. In addition to the good correlation that was observed between AMT tag and IQ quantification results, IQ also provided slightly higher peptidome coverage and less missing data than AMT tag approach. Taken together, our results demonstrate that integration of MS-GF+ database research and IQ analysis for label-free quantification drastically improves peptidome coverage and reduces missing data, and represents an optimized informatics pipeline for large-scale, comprehensive and quantitative peptidomics analysis.
Experimental
Tumor samples, peptidomics sample preparation and LC-MS/MS analysis
The ovarian tumor samples, sample preparation methods and LC-MS/MS instrument analysis methods used for generating the peptidomics data sets have been described in details previously [11]. Briefly, tumor tissues collected from three patients with high-grade serous ovarian carcinoma (A, B and C) were rapidly dissected into four contiguous and adjacent specimens strips, and placed into cryovials and frozen in liquid nitrogen at 4 different time points (0, 5, 30, and 60 min, at room temperature). The ovarian cancer tumor samples were further processed by cryopulverization and acid extraction (using 0.25% acetic acid and protease inhibitor cocktail) for peptidomic peptides, followed by LC-MS/MS analysis using nanoACQUITY UPLC® system (Waters Corporation, Milford, MA) coupled on-line to a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA). A 110 cm × 75 μm i.d. (flow rate 200 nL/min) fused-silica capillary column packed with 3 μm Jupiter C18 bonded particles (Phenomenex, Torrance, CA) was used for analysis of the ovarian tumor samples. Mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid acetonitrile (B) were operated with effective gradient profiles as follow (min:%B): 0:1, 6:8, 60:12, 225:35, 291:45, 300: 95. The LTQ Orbitrap Velos mass spectrometer was operated in the data-dependent mode acquiring high-resolution CID scans (R=15,000, 5 × 104 target ions) after each full MS scan (R=60,000, 1 × 106 target ions) for the top six most abundant ions within the mass range of 400 to 2000 m/z. An isolation window of 2 Th and a normalized collision energy of 35 were used for CID. The dynamic exclusion time was 60 s.
Mass spectrometry data analysis
The resulting MS data was first subject to DTARefinery [24] to correct overall mass measurement deviation before database searching. After that, the corrected spectrum was further searched against human protein sequences from UniProt (UniProt Knowledgebase release 2013_09) using MS-GF+ [16], with the following parameters: no enzyme digestion, precursor mass tolerance 50 ppm, methionine oxidation as variable modification, and target-decoy strategy was adopted for false discovery rate (FDR) calculation. The database searching result was finally filtered by spectrum level FDR less than 1% and precursor mass error less than 10 ppm, and only these confidently identified peptides were kept for next step analysis.
After the initial database searching, the data sets were further analyzed using the PNNL-developed AMT tag approach [25]. Confidently identified peptides from all the individual data sets were assembled into an AMT tag database containing both theoretical masses (calculated from the peptide sequence) and LC elution times of those peptides. LC-MS features in each individual data set were then matched against the AMT tags in the database for peptide identification using VIPER [26] with a mass error tolerance of <10 ppm and a normalized elution time (NET) error tolerance of <2.5%. The AMT tag matching result was further filtered by uniqueness probability (UP) higher than 0.5 to reduce ambiguous match to multiple AMT tags, and finally by Statistical Tools for AMT tag Confidence (STAC) value filtering to ensure FDR less than 2.5% [27]. Integrated MS peak area was used to derive changes in abundance for the peptide identifications.
The MS data sets were also analyzed using a recently in-house developed IQ approach [11], a derivative and improved approach, which provides better de-isotoping and peak selection. Briefly, in IQ the m/z values of the theoretical isotopic profile (derived based on the peptide sequences that were included in the AMT tag database) are used to guide the extraction of the observed isotopic profile from the summed mass spectra. Least-squares fitting of the theoretical isotopic profile on the observed profile is then performed [28], providing a measure of how well the observed isotopic profile matches the theoretical isotopic profile. This metric is called the “fit score” and is a key metric for resolving correct vs. incorrect features. A key step in IQ, as with the AMT tag approach, is the alignment of observed mass and LC elution times to database values in order to correct for variations in mass and elution time measurements taken across multiple datasets. Alignment of mass and the LC elution time makes it possible to narrow the mass tolerance used in generating extracted ion chromatograms (XICs) and the elution time window for selecting the correct chromatographic peak. Currently, VIPER is also used in a first-pass analysis to output mass and NET alignment information, which is then loaded into IQ and used for mass and NET correction during subsequent processing. Data processed by IQ approach was initially filtered by fit score (<0.1), NET tolerance (<2.5%) and mass accuracy (<10 ppm), and followed by manual validation to eliminate false positives [29] (development on computing FDR for IQ is currently in progress). If a chromatographic peak has been selected for a given peptide/charge state target, IQ then performs a final step of extracting the abundance information. Currently, this is comprised of summing a total of 5 mass spectra, centering around the apex scan of the elution profile. The abundance from different charge states is then added up for the specific peptide for quantification.
The peptide to protein mapping was performed using IDPicker3 [30]. All the quantification result from AMT tag and IQ analyses were imported into DanteR program [31] for processing and plotting: the data was first log10 transformed followed by median normalization, and used for further analysis; hierarchical clustering analysis was performed with Euclidean distance as distance metrics and average linkage for clustering; principal component analysis was performed with default parameters.
Results and Discussion
MS-GF+ outperforms SEQUEST and MS-Align+ in peptide identification for peptidomics analysis
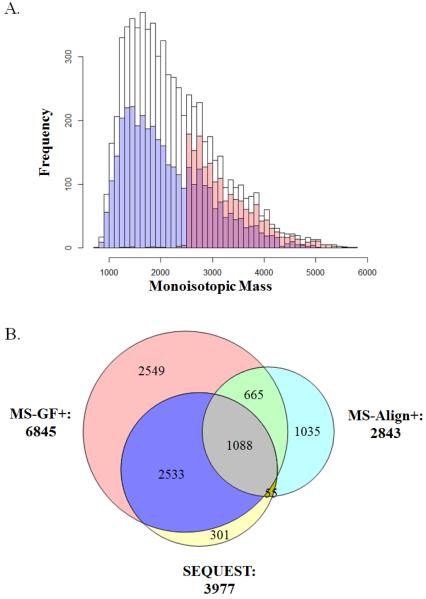
Totally 6845 unique peptides were confidently identified using MS-GF+ after 1% FDR filtering. As shown in Figure 1A (white), the distribution of precursor monoisotopic mass ranged from 785 ~ 6000 Da with a median of 2059.3 Da. These peptides were further mapped to 1136 non-redundant protein groups using IDPicker3 (Supplementary Table S1). In our previous study [11], both SEQUEST and MS-Align+ were used for database search for the same peptidomics data, resulting in 3977 and 2843 unique peptides after filtering (FDR <1%), respectively. MS-GF+ was able to identify many more unique peptides than SEQUEST and MS-Align+ (Figure 1B). Similar results were also obtained for the breast tumor peptidomics data in our previous study (data not shown). Furthermore, 91.05% of SEQUEST identified peptides (3621) were overlapped with MS-GF+ identifications, and the distribution of which was depicted in Figure 1A(Blue); similarly, 61.66% of MS-Align+ identified peptides (1753) were covered by MS-GF+, and the distribution of which is depicted in Figure 1A(Red). In agreement with previous report [19], SEQUEST appears to better identify peptides with relatively lower molecular weight, whereas MS-Align+ has a preference for peptides with higher molecular weight (Figure 1A). In comparison, peptide identifications from MS-GF+ covered a larger dynamic range of molecular weight distribution, almost as large as the combination of those from the other two methods. To confirm the quality of the MS-GF+ search results, we manually inspected the spectra with the worst scores for 50 peptides that were identified only by MS-GF+. The results suggest that the majority of the spectra are of acceptable quality to us for the peptide identification (Supplementary Figure 1). Taken together, our data suggested that MS-GF+ outperformed SEQUEST and MS-Align+ in terms of peptide identification for peptidomics analysis by providing significantly more unique peptide identifications and better molecular weight range coverage.
Figure 1.
Comparison of the peptidome peptide identification results from MS-GF+, SEQUEST and MS-Align+. (A) Distribution of precursor monoisotopic mass for MS-GF+ (white), SEQUEST (blue) and MS-Align+ (red). (B) Venn diagram showing the overlap of the identifications resulted from MS-GF+ (red), SEQUEST (yellow) and MS-Align+ (blue) analyses.
AMT tag and IQ provide significantly higher peptidome coverage and fewer missing data than MS-GF+
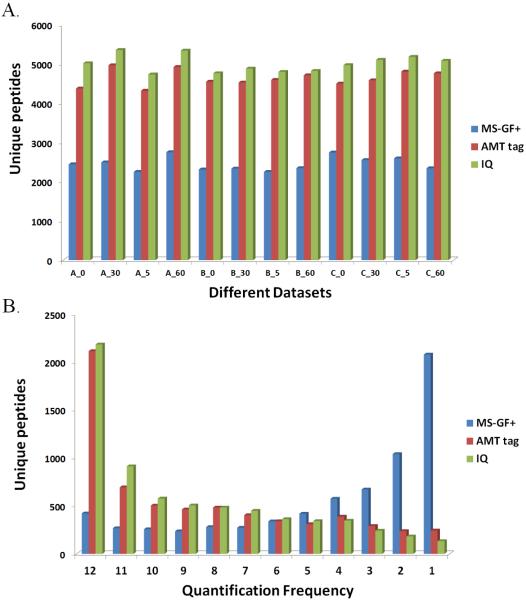
Due to the low stoichiometry of peptidome as well as under-sampling nature of typical data-dependent acquisition of MS/MS data, very low consistency is usually observed in MS/MS based peptide identification from run to run, leading to poor peptidome coverage and significant amount of missing data in label-free quantification [32]. Indeed, although there were a total of 6845 unique peptides identified by MS-GF+ from all 12 datasets, on average only approximately 2500 unique peptides were detected for each data set (Blue in Figure 2A; also see Table 1). Furthermore, only about 500 peptides were consistently identified and quantifiable across all 12 samples, whereas more than 2000 peptides were only detected from one sample (Figure 2B, Blue). This led to much smaller peptidome coverage and a much larger number of missing data in the individual sample analysis, both well known and significant issues for label-free quantification.
Figure 2.
Comparison of identification and quantification results from MS-GF+, AMT tag and IQ analyses. (A) Comparison of number of identified unique peptides in each sample for MS-GF+ (blue), AMT (red) and IQ (green). (B) Distribution of quantification frequency among all samples for MS-GF+ (blue), AMT tag (red) and IQ (green).
Table 1.
Number of unique peptidome peptides identified by different informatics approaches in each tumor sample.
| Tumor samples | MS-GF+ | AMT tag | IQ |
|---|---|---|---|
| A_0 | 2440 | 4370 | 5012 |
|
| |||
| A_5 | 2248 | 4315 | 4729 |
|
| |||
| A_30 | 2489 | 4963 | 5351 |
|
| |||
| A_60 | 2748 | 4919 | 5336 |
|
| |||
| B_0 | 2309 | 4544 | 4759 |
|
| |||
| B_5 | 2247 | 4590 | 4791 |
|
| |||
| B_30 | 2332 | 4523 | 4878 |
|
| |||
| B_60 | 2344 | 4707 | 4817 |
|
| |||
| C_0 | 2741 | 4496 | 4966 |
|
| |||
| C_5 | 2590 | 4802 | 5176 |
|
| |||
| C_30 | 2549 | 4579 | 5100 |
|
| |||
| C_60 | 2341 | 4757 | 5075 |
In order to improve data quality in both identification and quantification for each individual analysis, we next utilized both the AMT tag and IQ approaches for analysis of the same 12 data sets, taking advantage of a LC-MS database created using the MS-GF+ peptide identifications. Both approaches are expected to obtain more comprehensive quantification results due to the LC elution time alignment, and hence effective LC-MS peak matching. As depicted in Figure 2A, the average numbers of unique peptides identified from individual dataset via AMT tag and IQ approaches were much larger than that from MS-GF+: 4630 and 5000, respectively, as opposed to 2500. Moreover, the number of unique peptides detected across all 12 datasets via AMT tag (2112) and IQ (2182) approaches, was significantly increased compared to MS-GF+ (421), and peptides only detected in one sample greatly decreased (243 for AMT tag and 133 for IQ), significantly reducing missing data (Figure 2Band Table 2). The peptide identifications resulted from MS-GF+, AMT tag and IQ analysis for each individual peptidomic dataset are provided in Supplementary Tables 2-4.
Table 2.
The distribution of quantification frequency (number of unique peptide identifications common across the different samples) for the three different informatics approaches.
| Quantification Frequency |
MS-GF+ | AMT tag | IQ |
|---|---|---|---|
| 12 | 421 | 2112 | 2182 |
|
| |||
| 11 | 267 | 693 | 911 |
|
| |||
| 10 | 256 | 503 | 576 |
|
| |||
| 9 | 235 | 463 | 505 |
|
| |||
| 8 | 279 | 482 | 482 |
|
| |||
| 7 | 272 | 403 | 449 |
|
| |||
| 6 | 338 | 340 | 362 |
|
| |||
| 5 | 418 | 309 | 341 |
|
| |||
| 4 | 574 | 388 | 346 |
|
| |||
| 3 | 670 | 290 | 241 |
|
| |||
| 2 | 1039 | 237 | 182 |
|
| |||
| 1 | 2076 | 243 | 133 |
|
| |||
| Total | 6845 | 6463 | 6710 |
When the performance of the two MS/MS-independent LC-MS analysis approaches were compared, IQ provided slightly more unique peptide identifications (2.34% ~ 14.69%, average 8.02%), and thus less missing data than did the AMT tag approach. This is likely because IQ employs all isotopic peaks whereas the AMT tag approach uses individually deisotoped spectra, resulting in improved sensitivity, better distinguished overlapping features, and better reproducibility. Taken together, our results demonstrate that benefiting from significantly reduced undersampling, the direct LC-MS analysis pipelines including the AMT tag and IQ approaches provide significantly higher peptidome coverage and more importantly, less missing data across the entire peptidomics data sets comparing to even the best performing MS/MS-dependent analysis approaches such as MS-GF+.
Both the AMT tag and IQ approaches provide more robust label-free quantification for peptidomics
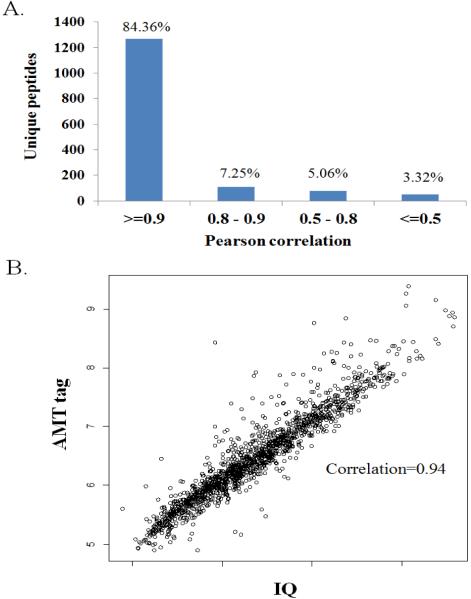
The AMT tag approach is a well-established approach for label-free quantification [23, 33]; it is interesting to compare its performance in quantification with that of the relatively newer IQ approach. Altogether, there were 1503 unique peptides overlapped between IQ and AMT tag analyses with no missing data across all 12 samples. Pearson correlation was first calculated for all 1503 unique peptides to assess the consistency between the quantification results of AMT tag and IQ analyses. As shown in Figure 3A, 91.62% of peptides displayed a correlation coefficient no less than 0.8, whereas only 50 peptides (3.32%) had a correlation lower than 0.5. Pearson correlations between the AMT tag and IQ quantification results for each sample were also calculated. With no Pearson correlation coefficient less than 0.93, the quantification results from both AMT tag and IQ analyses were well-correlated (Table 3). As an example, the AMT tag and IQ quantification results of one sample (A_0) shown in Figure 3B displayed excellent consistency, with most of the data points aligned along the diagonal line with an overall correlation of 0.94.
Figure 3.
Comparison of the label-free quantification results from AMT tag and IQ analyses. (A) Distribution of the Pearson correlation values for peptides common in all samples quantified by AMT tag and IQ approaches. (B) Scatter plot showing the correlation of all peptides quantified by AMT tag and IQ approaches from sample A_0.
Table 3.
Pearson correlation of the AMT tag and IQ label-free quantification results for each tumor sample.
| Sample | Correlation |
|---|---|
| A_0 | 0.94 |
|
| |
| A_5 | 0.95 |
|
| |
| A_30 | 0.94 |
|
| |
| A_60 | 0.94 |
|
| |
| B_0 | 0.94 |
|
| |
| B_5 | 0.95 |
|
| |
| B_30 | 0.94 |
|
| |
| B_60 | 0.95 |
|
| |
| B_0 | 0.94 |
|
| |
| B_5 | 0.94 |
|
| |
| B_30 | 0.93 |
|
| |
| B_60 | 0.94 |
The IQ and AMT tag label-free quantitation results also produced very similar hierarchical clustering analysis (HCA) and principle component analysis (PCA) plots (Supplementary Figure 2). Consistent with previously reported results [11], in both IQ and AMT tag analyses of the same peptidomics data sets the HCA heatmaps and PCA plots showed that the peptidomic profiles from the four time points of the same patient sample were clustered together. This indicated that potential changes in the peptidomes due to post-excision delay (up to one hour) were much smaller than that from patient heterogeneity, and that both IQ and AMT tag informatics pipelines provide robust quantitation for peptidomics analysis.
Conclusions
We describe an improved LC-MS based informatics workflow for comprehensive and quantitative peptidomics analysis, which consists of MS-GF+ for initial database searching and IQ (or AMT tag) approach for improved identification and more robust label-free quantification. MS-GF+ provides significantly more peptide identifications spanning a broader molecular weight range than the frequently used SEQUEST and MS-Align+ MS/MS search engines for identifying peptidome peptides. Owing to the direct LC-MS analysis strategy employed, both the AMT tag and IQ approaches significantly alleviate the under-sampling issue and provide much better peptidome coverage and much less missing data for each sample in comparison to even the best MS/MS-based analysis methods such as MS-GF+. In addition to the excellent correlation with the quantification results provided by the AMT tag approach, the IQ approach showed further improvement of peptidome coverage and reduced missing data across the entire data sets, likely due to better peak picking and retention time alignment. We believe that the powerful combination of MS-GF+ and IQ (or AMT tag) approach represent an optimal peptidomics informatics pipeline and expect broad application of this pipeline for large-scale and highly effective and robust peptidomics analysis.
Supplementary Material
Supplementary Figure 1. Manual inspection of spectra with the worst scores for 50 peptides that were identified only by MS-GF+ (see separate PDF file).
Acknowledgements
Portions of this work were supported by the grant U24CA160019, from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), National Institutes of Health Research Resource grant P41GM103493, and Department of Defense Interagency Agreement MIPR2DO89M2058. The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
References
- 1.Schrader M, Schulz-Knappe P. Peptidomics technologies for human body fluids. Trends Biotechnol. 2001;19:S55–60. doi: 10.1016/S0167-7799(01)01800-5. [DOI] [PubMed] [Google Scholar]
- 2.Gelman JS, Fricker LD. Hemopressin and other bioactive peptides from cytosolic proteins: are these non-classical neuropeptides? AAPS J. 2010;12:279–289. doi: 10.1208/s12248-010-9186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 4.Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 6.Ma M, Chen R, Ge Y, He H, Marshall AG, Li L. Combining bottom-up and top-down mass spectrometric strategies for de novo sequencing of the crustacean hyperglycemic hormone from Cancer borealis. Anal Chem. 2009;81:240–247. doi: 10.1021/ac801910g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Peptidomic analysis of human cell lines. J Proteome Res. 2011;10:1583–1592. doi: 10.1021/pr100952f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fricker LD, Gelman JS, Castro LM, Gozzo FC, Ferro ES. Peptidomic analysis of HEK293T cells: effect of the proteasome inhibitor epoxomicin on intracellular peptides. J Proteome Res. 2012;11:1981–1990. doi: 10.1021/pr2012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, Sanchez-Carbayo M, Holland EC, Cordon-Cardo C, Scher HI, Tempst P. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiedler GM, Baumann S, Leichtle A, Oltmann A, Kase J, Thiery J, Ceglarek U. Standardized peptidome profiling of human urine by magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem. 2007;53:421–428. doi: 10.1373/clinchem.2006.077834. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z, Wu C, Xie F, Slysz GW, Tolic N, Monroe ME, Petyuk VA, Payne SH, Fujimoto GM, Moore RJ, Fillmore TL, Schepmoes AA, Levine DA, Townsend RR, Davies SR, Li S, Ellis M, Boja E, Rivers R, Rodriguez H, Rodland KD, Liu T, Smith RD. Comprehensive Quantitative Analysis of Ovarian and Breast Cancer Tumor Peptidomes. J Proteome Res. 2015;14:422–433. doi: 10.1021/pr500840w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tinoco AD, Saghatelian A. Investigating endogenous peptides and peptidases using peptidomics. Biochemistry. 2011;50:7447–7461. doi: 10.1021/bi200417k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Dijck A, Hayakawa E, Landuyt B, Baggerman G, Van Dam D, Luyten W, Schoofs L, De Deyn PP. Comparison of extraction methods for peptidomics analysis of mouse brain tissue. J Neurosci Methods. 2011;197:231–237. doi: 10.1016/j.jneumeth.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Finoulst I, Pinkse M, Van Dongen W, Verhaert P. Sample preparation techniques for the untargeted LC-MS-based discovery of peptides in complex biological matrices. J Biomed Biotechnol. 20112011:245291. doi: 10.1155/2011/245291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 16.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mommen GP, Frese CK, Meiring HD, van Gaans-van den Brink J, de Jong AP, van Els CA, Heck AJ. Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD) Proc Natl Acad Sci U S A. 2014;111:4507–4512. doi: 10.1073/pnas.1321458111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dasgupta S, Castro LM, Dulman R, Yang C, Schmidt M, Ferro ES, Fricker LD. Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PLoS One. 2014;9:e103604. doi: 10.1371/journal.pone.0103604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Sirotkin Y, Shen Y, Anderson G, Tsai YS, Ting YS, Goodlett DR, Smith RD, Bafna V, Pevzner PA. Protein identification using top-down. Mol Cell Proteomics. 2012;11(M111):008524. doi: 10.1074/mcp.M111.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menschaert G, Vandekerckhove TT, Baggerman G, Schoofs L, Luyten W, Van Criekinge W. Peptidomics coming of age: a review of contributions from a bioinformatics angle. J Proteome Res. 2010;9:2051–2061. doi: 10.1021/pr900929m. [DOI] [PubMed] [Google Scholar]
- 21.Stingl C, Soderquist M, Karlsson O, Boren M, Luider TM. Uncovering effects of ex vivo protease activity during proteomics and peptidomics sample extraction in rat brain tissue by oxygen-18 labeling. J Proteome Res. 2014;13:2807–2817. doi: 10.1021/pr401232e. [DOI] [PubMed] [Google Scholar]
- 22.Quintana LF, Campistol JM, Alcolea MP, Banon-Maneus E, Sol-Gonzalez A, Cutillas PR. Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Mol Cell Proteomics. 2009;8:1658–1673. doi: 10.1074/mcp.M900059-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Petyuk VA, Mayampurath AM, Monroe ME, Polpitiya AD, Purvine SO, Anderson GA, Camp DG, 2nd, Smith RD. DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Mol Cell Proteomics. 2010;9:486–496. doi: 10.1074/mcp.M900217-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23:2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 27.Stanley JR, Adkins JN, Slysz GW, Monroe ME, Purvine SO, Karpievitch YV, Anderson GA, Smith RD, Dabney AR. A statistical method for assessing peptide identification confidence in accurate mass and time tag proteomics. Anal Chem. 2011;83:6135–6140. doi: 10.1021/ac2009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slysz GW, Steinke L, Ward DM, Klatt CG, Clauss TR, Purvine SO, Payne SH, Anderson GA, Smith RD, Lipton MS. Automated data extraction from in situ protein-stable isotope probing studies. J Proteome Res. 2014;13:1200–1210. doi: 10.1021/pr400633j. [DOI] [PubMed] [Google Scholar]
- 30.Holman JD, Ma ZQ, Tabb DL. Identifying proteomic LC-MS/MS data sets with Bumbershoot and IDPicker. Curr Protoc Bioinformatics. 2012:17. doi: 10.1002/0471250953.bi1317s37. Chapter 13, Unit13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taverner T, Karpievitch YV, Polpitiya AD, Brown JN, Dabney AR, Anderson GA, Smith RD. DanteR: an extensible R-based tool for quantitative analysis of -omics data. Bioinformatics. 2012;28:2404–2406. doi: 10.1093/bioinformatics/bts449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie F, Liu T, Qian WJ, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. J Biol Chem. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci U S A. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Manual inspection of spectra with the worst scores for 50 peptides that were identified only by MS-GF+ (see separate PDF file).