Abstract
A relationship between the actions of TSH and IGF-1 was first recognized several decades ago. The close physical and functional associations between their respective receptors (TSHR and IGF-1R) has been described more recently in thyroid epithelium and human orbital fibroblasts as has the noncanonical behavior of IGF-1R. Here we report studies conducted in lung fibroblasts from female wild-type C57/B6 (TSHR+/+) mice and their littermates in which TSHR has been knocked out (TSHR−/−). Flow cytometric analysis revealed that cell surface IGF-1R levels are substantially lower in TSHR−/− fibroblasts compared with TSHR+/+ fibroblasts. Confocal immunofluorescence microscopy revealed similar divergence with regard to both cytoplasmic and nuclear IGF-1R. Western blot analysis demonstrated both intact IGF-1R and receptor fragments in both cellular compartments. In contrast, IGF-1R mRNA levels were similar in fibroblasts from mice without and with intact TSHR expression. IGF-1 treatment of TSHR+/+ fibroblasts resulted in reduced nuclear and cytoplasmic staining for IGF-1Rα, whereas it enhanced the nuclear signal in TSHR−/− cells. In contrast, IGF-1 enhanced cytoplasmic IGF-1Rβ in TSHR−/− fibroblasts while increasing the nuclear signal in TSHR+/+ cells. These findings indicate the intimate relationship between TSHR and IGF-1R found earlier in human orbital fibroblasts also exists in mouse lung fibroblasts. Furthermore, the presence of TSHR in these fibroblasts influenced not only the levels of IGF-1R protein but also its subcellular distribution and response to IGF-1. They suggest that the mouse might serve as a suitable model for delineating the molecular mechanisms overarching these two receptors.
TSH binds to and activates the TSH receptor (TSHR), which in turn functionally couples multiple G protein subtypes (1–3). A complex pattern of cell signaling is initiated, depending in large part on the cell type being examined (1, 4). TSHR represents the dominant self-antigen targeted uniquely in Graves' disease (GD) by thyroid-stimulating immunoglobulins (5). In addition to its well-established and characterized role in the development and function of the thyroid, TSHR has been detected more recently in tissues and cell types peripheral to the gland (6, 7) in which its signaling characteristics have been partially characterized. Furthermore, a potential role for the receptor expressed by orbital fibroblasts in the pathogenesis of the ocular manifestation of GD, called thyroid-associated ophthalmopathy (TAO), has been proposed (7, 8).
Unlike TSHR, the IGF-1 receptor (IGF-1R) is a ubiquitous membrane-spanning tyrosine kinase receptor with two dissimilar subunits (9). It plays important roles in growth and cell survival. Several pieces of evidence have suggested a close physical and functional relationship between the TSHR and IGF-1R (10). For instance, IGF-1 enhances the actions of TSH in thyroid epithelial cells and alters the balance between levels of TSH and thyroid hormones in serum in vivo (11, 12). Thus, there exists reason to further investigate the nature of this putative functional relationship between these two receptors.
We have reported previously that TSHR and IGF-1R colocalize in human thyroid epithelial cells, orbital fibroblasts, and orbital fat derived from individuals with TAO (10). Furthermore, when treated with either IgGs from patients with GD (GD-IgG) or IGF-1, a fragment of IGF-1R containing an epitope located in the IGF-1Rα subunit translocates from the cell membrane to the nucleus, a process requiring the activities of the protease, a disintegrin and metalloproteinase domain 17 (13). This translocation of IGF-1R was absent in orbital fibroblasts from healthy donors. Inhibition of IGF-1R activity with blocking monoclonal antibodies dampens not only signaling initiated at IGF-1R but also the induction by TSH of several cytokines in human orbital fibroblasts and CD34+ fibrocytes (10, 14). That property has led to an ongoing multinational, multicenter therapeutic trial examining the benefits of the blocking anti-IGF-1R monoclonal antibody, teprotumumab (http://clinicaltrials.gov/show/NCT01868997) in treating severe, active TAO.
In the current study, lung fibroblasts isolated from homozygous TSHR−/− C57/B6 female mice and their wild-type TSHR+/+ littermates were examined for IGF-1R expression, subcellular localization, and responses in vitro to IGF-1. Our aim was to examine the impact of the presence and absence of TSHR on the expression and cellular distribution of IGF-1R, a topic that has yet to be clarified in the literature. We report here that untreated TSHR−/− mouse lung fibroblasts express equivalent levels of IGF-1R mRNA but display substantially lower levels of IGF-1R protein than do their TSHR+/+ counterparts. The cellular distribution of the protein after treatment with IGF-1 differs in fibroblasts, depending on whether they express TSHR. In TSHR−/− fibroblasts, IGF-1 enhances nuclear IGF-1Rα levels while diminishing both cytoplasmic and nuclear signal in TSHR+/+ cells. In contrast, IGF-1 enhances cytoplasmic IGF-1Rβ in TSHR−/− fibroblasts while increasing its nuclear content in TSHR+/+ cells. These findings support the general concept that a functional relationship between TSHR and IGF-1R exists in mouse fibroblasts. Furthermore, TSHR appears to determine the subcellular distribution of IGF-1R and its response to IGF-1.
Materials and Methods
Materials
DMEM containing 4.5 g/L D-glucose and L-glutamine (catalog number 11965-092), fetal bovine serum (FBS) (catalog number 16000-044), penicillin/streptomycin (catalog number 15140-122), trypsin-EDTA (catalog number 25200-056), and IGF-1 (catalog number PHC0815) were supplied by Life Technologies-Invitrogen.
Animals
TSHR-knockout (KO) mice were purchased from Jackson Labs (strain B6;129S1-TSHRtm1Rmar/J, catalog number 004858, http://jaxmice.jax.org/strain/004858.html) (15). TSHR+/+ and TSHR−/− C57/B6 mice derived from common breeding pairs. All animals were genotyped and RNA typed for assignment confirmation. TSHR+/+ mice were weaned at 21 days, whereas TSHR−/− mice were weaned at 28–30 days as described (15). All mice were maintained on Mod PicolLab Rod containing 100 ppm powdered thyroid hormone (catalog number 44099; Frontier). A total of 17 TSHR+/+ and 19 TSHR−/− female mice were used in the studies described. They were between 7 and 9 weeks of age and weighed 18–22 g at the time of the tissue harvest. They were anesthetized and/or euthanized according to the guidelines of the University of Michigan University Committee on the Use and Care of Animals.
Animal genotyping
All primers used in these studies were designed with P3 Primer Design online software (http://bioinfo.ut.ee/primer3/) and DNAstar software (DNAstar Inc) and are listed in Table 1. They were synthesized by Life Technologies. Reactions were optimized with temperature gradients from 55°C to 70°C. They were performed with iQ SYBR Green Supermix (catalog number 170-8884; Bio-Rad Laboratories).
Table 1.
Primers Used
| Reaction Type | Target | Primers | Sequence (5′–3′) | Annealing Temperature, ºC | Product Size, bp |
|---|---|---|---|---|---|
| Real-time PCR | IGF-R1 α | IGF-R1 forward | CAAGCTGTGTGTCTCCGAAA | 56 | 161 |
| IGF-R1 reverse | TGATTCGGTTCTTCCAGGTC | ||||
| GAPDH | GAPDH forward | CATGGCCTTCCGTGTTCCTA | 56–63.4 | 111 | |
| GAPDH reverse | GCGGCACGTCAGATCCA | ||||
| TSHR (exons 6–7) | TSHR exon 6 forward | TCCCTGAAAACGCATTCCA | 63.4 | 111 | |
| TSHR exon 6 reverse | GCATCCAGCTTTGTTCCATTG | ||||
| Genotyping | WT primers 1 | TSHR exon 1 forward 1 | TGCACCCGGGTCTCTTCCAGC | 63.4 | 241 |
| TSHR exon 1 reverse 2 | AGGCTGGGGATTCGGTGGAGC | ||||
| WT primers 2 | TSHR exon 1 forward 2 | CAGGGCACTGAGAATGTGGCGA | 63.4 | 212 | |
| TSHR exon 1 reverse 2 | AGGCTGGGGATTCGGTGGAGC | ||||
| TSHR-KO primers | KO forward | AAGTTCATCTGCACCACCG | 63.4 | 170 | |
| KO reverse | TCCTTGAAGAAGATGGTGCG |
Thyroid and lung isolation and fibroblast cultivation
Thyroid was isolated by placing the animal on the dorsum and an incision was made exposing the soft tissues of the neck. Thyroids were removed from the trachea and placed in Bio-Rad lysis buffer (Bio-Rad Laboratories), sonicated, and RNA extracted. Lungs were blanched with sterile PBS and excised in sterile Hanks' balanced salt solution (catalog number 14025-076; Life Technologies). Tissue was minced and digested enzymatically in Hanks' balanced salt solution containing collagenase type III (49.2 mg per lung, catalog number LS004208; Worthington), deoxyribonuclease (375 μg per lung, catalog number LS002058; Worthington), and 2% FBS for 90 minutes at 37°C with gentle agitation. Cell suspensions were filtered through 100-μm filters (catalog number 352360; BD Biosciences), centrifuged at 670 × g, and the resultant pellet resuspended in DMEM and used to inoculate T-175 flasks. Monolayers were covered with DMEM supplemented with 20% FBS and antibiotics, which was replaced every 3–4 days. These were serially passaged with trypsin/EDTA and cryopreserved in liquid nitrogen until used in experiments.
RNA preparation and real-time PCR
Cellular RNA was extracted using Aurum Total RNA mini kit (catalog number 732-6820; Bio-Rad Laboratories) and reverse transcribed using a QuantiTect reverse transcription kit (catalog number 205314; QIAGEN). RT-PCR was performed with either a CFX96 or a 384-well real-time PCR detection system (Bio-Rad Laboratories). Data were analyzed using Bio-Rad CFX software (Bio-Rad Laboratories). Sample values were generated against a standard curve and normalized to their respective glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signals.
Flow cytometry
Cell monolayers were washed, mechanically disrupted, and stained with the relevant primary antibodies (Table 2). They were suspended in PBS containing 2% FBS. Incubations with antibodies were conducted at 4°C for 30 minutes followed by extensive washing. For collagen I staining, cells were treated with CytoFix/CytoPerm (catalog number 51-2090KZ; BD Biosciences) and incubated with a biotinylated goat antihuman collagen I polyclonal antibody (catalog number AB758B; Millipore). Flow cytometry was performed using an LSRII instrument (BD Biosciences). At least 5 × 105 events were collected. Mean fluorescent intensity (MFI) was calculated as a ratio of mean fluorescence sample per isotype fluorescence. Percentage positive expression was defined as the fraction of cells with increased fluorescent intensity compared with an isotype control.
Table 2.
Primary Antibodies
| Antibody | Company | Catalog Number | Dilution |
|---|---|---|---|
| Flow cytometry | |||
| Goat α-collagen 1 | Millipore | AB758B | 1:100 |
| FITC streptavidin | BD Biosciences | 554060 | 1:100 |
| IGF-1Rα (3B7) PE | Santa Cruz | SC-462 | 1:40 |
| Immunofluorescence | |||
| Rabbit anti-IGF-1Rα | Santa Cruz | sc-712 | 1:100 |
| Rabbit anti-IGF-1Rβ | Santa Cruz | sc713 | 1:100 |
| Donkey anti-rabbit IgG Alexa fluor 488 | Life Technologies | A21206 | 1:1200 |
Abbreviation: FITC, fluorescein isothiocyanate.
Immunofluorescence confocal microscopy
Fibroblasts were inoculated on 3-cm glass coverslips in six-chamber arrays and covered with DMEM with 20% FBS. These were incubated for at least 3 days and were then shifted to DMEM with 10% FBS. Cells were fixed in 4% paraformaldehyde and blocked with PBS containing 3% goat serum and 0.2% Triton X-100 for 30 minutes at room temperature. Cells were then incubated with primary antibodies in 1% goat serum and 0.2% Triton X-100 overnight at 4°C. After the washes, they were incubated with secondary antibodies for 60 minutes at room temperature. They were then mounted with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (catalog number P36931; Life Technologies) or propidium iodide (PI) (Vectashield with PI, catalog number H-1300; Vector Labs) and subjected to confocal microscopy (Leica SP5 confocal microscope; Leica).
Image analysis
IGF-1R-α and IGF-1R-β fluorescent signals were quantified as follows: confocal z-stack images were opened with Volocity software (version 4.4; PerkinElmer), and individual fibroblasts were isolated from neighboring cells with an imbedded selection and cropping tool. The cropped cell images were exported from Volocity as OME TIFF files and imported into Imaris software (version 7.7.2; Bitplane USA). Imaris was used to isolate nuclear and cytosolic-specific IGF-1R-α and IGF-1Rβ expression, allowing quantification and comparison of the mean IGF-1R signal intensity between nuclei and cytosol. For each cell, three separate, 3-dimensional surfaces were made, with the surface of the nucleus being created first. Local contrast was used (background subtraction tool) to set the threshold for the nuclear specific PI staining to accurately mark the nucleus. The nuclear surface was then used to isolate nuclear and cytosolic-specific IGF-1R/PI signals. The nuclear IGF-1R signal was isolated by setting voxels outside the nuclear PI surface to zero, whereas voxels inside the nucleus maintained their fluorescence intensity values. The cytosolic IGF-1R signal was isolated by setting voxels inside the nuclear PI surface to zero, whereas voxels throughout the cytosol maintained their fluorescence intensity values. Surfaces were then rendered around the newly created fluorescent channels of the nuclear and cytosolic IGF-1R signals. Local contrast was used to define the surfaces and the intensity. Threshold was set to accurately encompass the IGF-1R signals. The same threshold settings for PI and IGF-1R surfaces were used for all cells analyzed. The MFI values were exported from Imaris and into an Excel spreadsheet for all IGF-1R surfaces and used to compare expression levels between the treatment groups. Data are expressed as mean ± SD.
Subcellular fractionation and Western blotting
Confluent fibroblasts were rinsed in PBS and subjected to cytosolic and nuclear protein extractions using the NE-PER extraction kit (catalog number 78833; Thermo Fisher Scientific). These were quantified using a DC Protein Assay Kit I (catalog number 500-0111; Bio-Rad Laboratories) and boiled in Laemmli buffer. Protein (50 μg) was loaded on 4%–20% triglycine gels and subjected to SDS-PAGE and transferred to a polyvinyl difluoride membrane (catalog number IPFL00010\; Millipore). Membranes were blocked with 5% milk for 3 hours, washed, and incubated with primary antibody rabbit anti-IGF-1Rα (1:1000, catalog number sc-712) and rabbit anti-IGF-1Rβ (1:1000, catalog number sc-713; Santa Cruz Biotechnology) overnight 4°C in 5% milk. Washed membranes were incubated with horseradish peroxidase-conjugated antirabbit secondary antibody (1:2000 dilution, catalog number 7047; Cell signaling). For the pAkt time course, confluent fibroblast monolayers were shifted to medium with 1% FBS and stimulated with IGF-1 (10 nM) for the graded interval indicated (see Figure 5). Monolayers were extracted with buffer (FNN0011; Invitrogen/Thermo Fisher), quantified, and electrophoresed as above. They were probed with antiphospho-Akt (Thr308) (1:1000, catalog number 9275; Cell Signaling). An enhanced chemiluminescence reagent (catalog number 32106; Thermo Fisher) was used for signal development. Anti-β-actin (1:1000, catalog number 3700; Cell Signaling) and anti-c-Jun (1:1000, catalog number sc-44; Santa Cruz Biotechnology) were used as loading controls (Table 2).
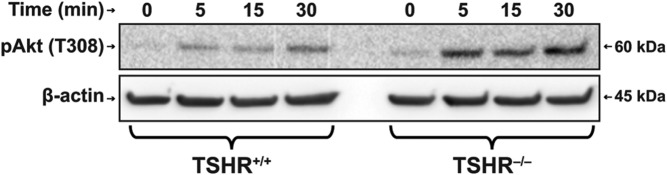
Figure 5.
Time course of Akt phosphorylation in response to IGF-1 in TSHR+/+ and TSHR−/− fibroblasts. Fibroblasts were grown to confluence and treated as described in Materials and Methods with IGF-1 (100 ng/mL) for the times indicated along the abscissas. Monolayers were collected and subjected to Western blot analysis for pAkt. Densities were corrected for their β-actin controls. Relative densities were as follows: time 0, TSHR+/+ 0.23, TSHR−/− 0.22; time 5 minutes, TSHR+/+ 0.70, TSHR−/− 1.62; 15 minutes, TSHR+/+ 0.61, TSHR−/− 1.46; and 30 minutes, TSHR+/+ 0.86, TSHR−/− 2.50.
Statistics
The results are expressed as the mean ± SD. A value of P < .05 was considered significant. ANOVA and post hoc Holm-Šídák (16–18) were used to test for a trend of change over time or to test for differences among groups. Plots and statistical calculations were performed in GraphPad Prism 6 (GraphPad Software, www.graphpad.com) and JMP Pro version 10.0.1 (SAS Institute Inc, 1989–2007).
Results
Lung fibroblasts express TSHR and IGF-1R mRNA
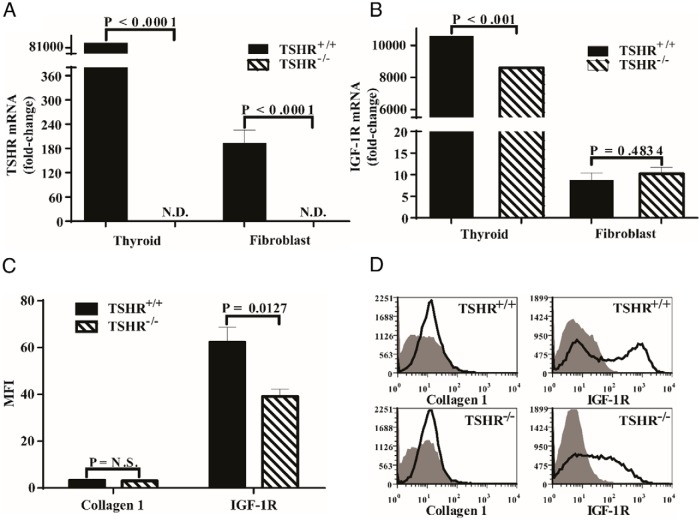
Initial studies focused on determining the expression of TSHR and IGF-1R mRNA in lung fibroblasts. As the data in Figure 1A demonstrate, TSHR mRNA was substantially more abundant in thyroid tissue from wild-type animals than in fibroblasts from those mice (425-fold). The transcript was undetectable in both thyroid and fibroblasts from TSHR−/− mice, as anticipated. IGF-1R mRNA was also expressed at dramatically higher levels in thyroid tissue than in lung fibroblasts (>1000-fold, Figure 1B). It was significantly lower in the thyroids from TSHR−/− mice than from their wild-type littermates (19%, P < .001). In contrast, IGF-1R mRNA was expressed at equivalent levels in lung fibroblasts from TSHR+/+ and TSHR−/− animals (Figure 1B).
Figure 1.
IGF-1R expression in TSHR+/+ and TSHR−/− female mouse thyroid and lung fibroblasts. A and B, Thyroid tissues were harvested from mice and RNA immediately extracted as described in Materials and Methods. Lung fibroblasts were allowed to proliferate to confluence, harvested, processed, and cellular RNA extracted as described in Materials and Methods. Each sample was reversed transcribed and subjected to real-time PCR for TSHR (A) or IGF-1R (B) mRNA. Data are expressed as the mean ± SD of three replicates. Data from lung fibroblasts represent those from five mice in each group. ND, not detected. C and D, Flow cytometric analysis of cell surface IGF-1R and intracellular collagen 1 in TSHR+/+ and TSHR−/− lung fibroblasts. C, Aggregate results, expressed as MFI from five fibroblast strains, each from a separate animal. Data are expressed as the mean ± SD. D, Individual examples of cytometric gating strategies used in analyses the results from which comprise panel C.
Unlike IGF-1R mRNA levels, the fibroblast surface display of IGF-1R was significantly lower in those from TSHR−/− mice compared with fibroblasts from TSHR+/+ animals as determined by flow cytometry (Figure 1C). The levels of IGF-1R density, expressed as MFI, were found to differ substantially in fibroblasts with the two genotypes. Receptor density was 1.6-fold lower in TSHR−/− fibroblasts compared with their wild-type counterparts (P < .02). The gating strategy for this interrogation is demonstrated in Figure 1D. Thus, despite similar levels of IGF-1R mRNA levels, surface receptor levels are significantly lower in TSHR−/− fibroblasts.
Cellular distribution of IGF-1R in TSHR+/+ and TSHR−/− lung fibroblasts
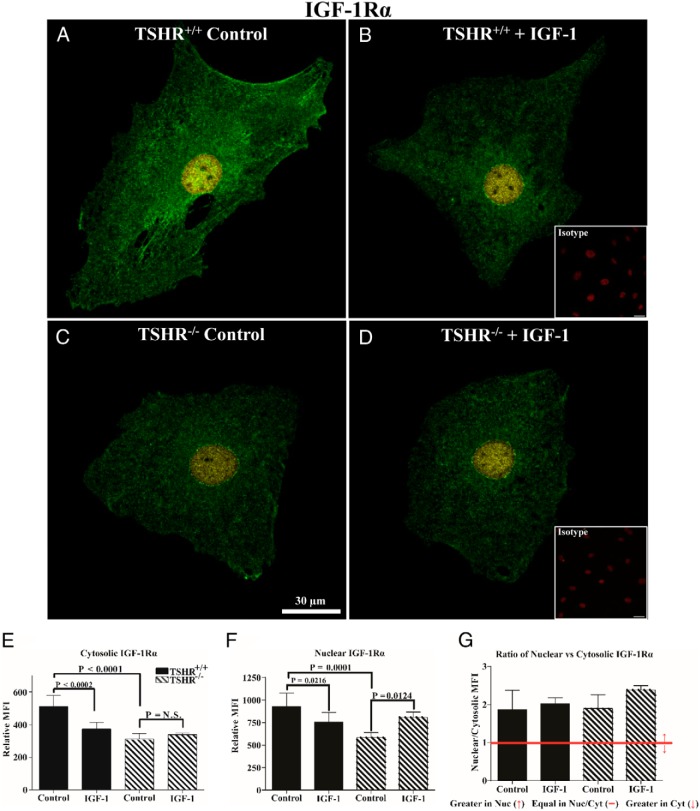
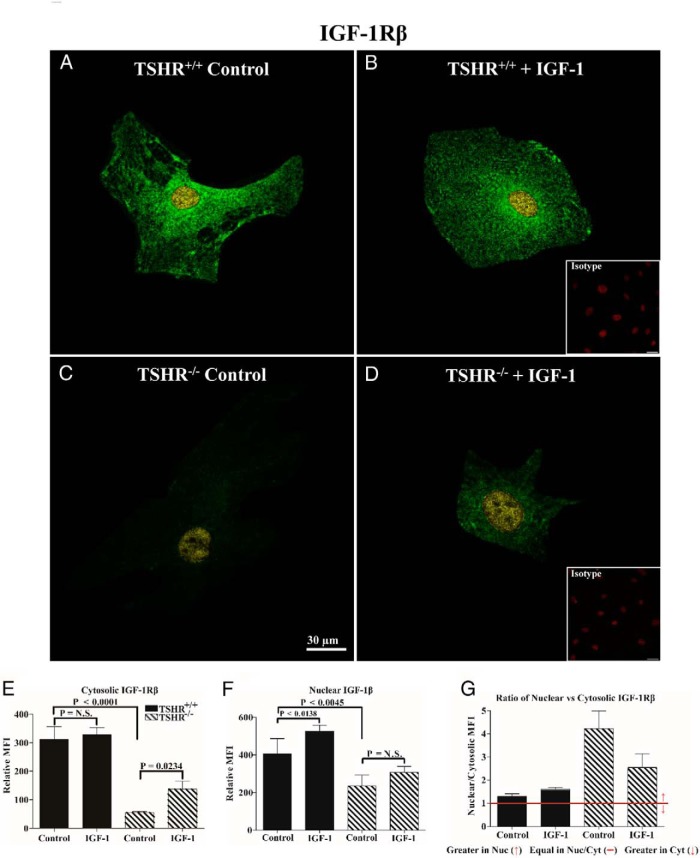
Given the divergence in surface display of IGF-1R, we next examined the intensity and subcellular distribution using monoclonal antibodies (Abs) targeting epitopes on IGF-1Rα and IGF-1Rβ subunits. The overall intensities of immunoflorescent signals from anti-IGF-1Rα (Figure 2) and anti-IGF-1Rβ Abs (Figure 3) are substantially lower in TSHR−/− fibroblasts compared with cells expressing TSHR. This divergence is apparent in both cytoplasmic and nuclear compartments. In untreated TSHR+/+ and TSHR−/− fibroblasts, IGF-1Rα can be detected in both the nucleus and cytoplasm, with considerably higher levels found in the former compartment. IGF-1 (100 ng/mL) for 16 hours reduced IGF-1Rα MFI in both the nuclei and cytoplasm of TSHR+/+ fibroblasts (Figure 2, P = .0216 and P < .0002, respectively). In contrast, IGF-1 increased the nuclear IGF-1Rα signal in TSHR−/− fibroblasts, whereas the levels in the cytosol remained unchanged. With regard to IGF-1Rβ, untreated TSHR+/+ fibroblast cytoplasm stains far more intensely than does that of TSHR−/− fibroblasts (Figure 3, 82%, P < .0001). Nuclear staining in the two untreated fibroblast types is more equivalent. IGF-1 enhances the cytoplasmic IGF-1Rβ staining in TSHR−/− fibroblasts (60%, P = .0234), whereas a smaller increase in the nuclei of TSHR−/− fibroblasts failed to reach statistical significance (23%, P = .1983). In contrast, IGF-1 enhances IGF-1Rβ nuclear staining in TSHR+/+ fibroblasts (23%, P < .0138), whereas a small increase in the cytoplasmic staining was insignificant (5%, P < .9989). It would thus appear that the expression of TSHR exerts a substantial impact on the subcellular distribution and levels of IGF-1R in lung fibroblasts. Furthermore, TSHR also determines the pattern of IGF-1R after exposure to IGF-1.
Figure 2.
Analysis of IGF-1Rα subcellular distribution by immunofluorescence in TSHR+/+ and TSHR−/− mouse lung fibroblasts. A–D, Immunofluorescence staining for IGF-1Rα (green and yellow) and PI (red, insets) demonstrates cellular localization of the receptor protein by confocal microscopy in lung fibroblasts isolated from TSHR+/+ (panels A and B) and TSHR−/− mice (panels C and D). Fibroblasts were inoculated on coverslips as described in Materials and Methods. Cells were either left untreated (panels A and C, control) or were treated with 100 ng/mL IGF-1 for 16 hours (panels B and D, IGF-1). Yellow signal represents nuclear IGF-1Rα colocalizing with PI and green represents cytosolic localization. Isotype control Abs (panels B and D, insets) demonstrate an absence of background labeling. Fluorescence intensities of the cytosolic (E) and nuclear (F) compartments and the ratios (G) of the signal intensities in the two cellular compartments were quantified. Intensity measurements from five different cells from each group were used in the analysis. Data are expressed as mean ± SD. Bar, 30 μm.
Figure 3.
Analysis of IGF-1Rβ cellular distribution by immunofluorescence in TSHR+/+ and TSHR−/− mouse lung fibroblasts. A–D, Immunofluorescence staining for IGF-1Rβ (green and yellow) and PI (red, insets) demonstrates cellular localization of the receptor protein by confocal microscopy in lung fibroblasts isolated from TSHR+/+ (panels A and B) and TSHR−/− mice (panels C and D). Fibroblasts were inoculated on coverslips as described in Materials and Methods. Cells were either left untreated (panels A and C, control) or were treated with 100 ng/mL IGF-1 for 16 hours (panels B and D, IGF-1). Yellow signal represents nuclear IGF-1Rβ colocalizing with PI and green represents cytosolic localization. Isotype control Abs (panels B and D, insets) demonstrate an absence of background labeling. Fluorescence intensities of the cytosolic (E) and nuclear (F) compartments and the ratios (G) of the signal intensities in the two cellular compartments were quantified. Intensity measurements from five different cells were used in the analysis in the TSHR+/+ group, whereas four were used from the TSHR−/− cultures. Data are expressed as mean ± SD. Bar, 30 μm.
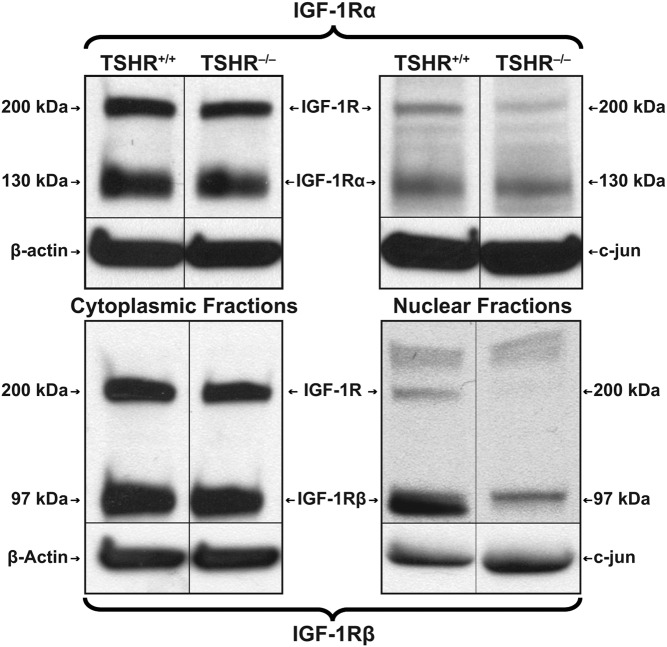
Cell fractionation studies were next conducted to determine whether the cytoplasmic and nuclear compartments contained intact or fragmented IGF-1R, as had been detected in human orbital fibroblasts (Figure 4) (13). In both TSHR+/+ and TSHR−/− fibroblasts, evidence of intact receptor migrating at 200 kDa as well as IGF-1Rα (130 kDa) could be detected in the cytoplasm and at somewhat lower levels in the nucleus. In contrast, IGF-1Rβ (97 kDa) was abundant in the cytoplasm but barely detectable in the nucleus of either fibroblast type. This pattern of IGF-1Rβ is consistent with the very low nuclear levels found earlier in human orbital fibroblasts (13).
Figure 4.
Western blot analysis of IGF-1R content in cytoplasm and nucleus of TSHR+/+ and TSHR−/− fibroblasts. Fibroblasts were subjected to subcellular fractionation and proteins extracted, as described in Materials and Methods. Proteins were loaded on 4%–20% triglycine gels and subjected to SDS-PAGE. Separated proteins were then probed by Western blotting with anti-IGF-1Rα and anti-IGF-1Rβ Abs.
Impact of TSHR on IGF-1R signaling
The effects of IGF-1 on Akt phosphorylation were next compared in TSHR+/+ and TSHR−/− fibroblasts (Figure 5). As the Western blots demonstrate, levels of pAkt were increased in both fibroblast types after exposure to IGF-1 (100 ng/mL) in a time-dependent manner. Levels achieved at each time point were substantially lower in TSHR+/+ fibroblasts than in their TSHR−/− counterparts.
Discussion
The array of biological functions attributed to TSHR continues to expand as mounting evidence suggests that its expression outside the thyroid may have biological consequence (6, 19, 20). Detail concerning the molecular context in which TSHR is expressed and initiates signals and how this might vary in different cell types and tissues is emerging. For instance, regulation of TSHR expression by TSH in 3T3 cells appears distinct from that in FRTL-5 thyroid cells (21). The apparently close relationship between TSHR and other signaling proteins suggests that TSH may exert influence on signaling pathways not traditionally associated with thyroid economy. Furthermore, these peripheral pathways intersecting with those downstream from the TSHR may be necessary for a subset of TSH actions. With specific regard to connective tissue, it remains uncertain what physiological roles TSHR might play. In fibrocytes and orbital fibroblasts, TSH and thyroid-stimulating immunoglobulins induce several cytokines (22, 23). Given their participation in tissue reactivity, remodeling, and repair, it seems reasonable to speculate that TSHR on fibrocytes may represent an important link between immunity and endocrine regulation.
The repertoire of recognized cellular processes attributed to IGF-1R regulation has also widened as examination of its pattern of expression and signaling characteristics has been undertaken. Its functions while membrane anchored are dependent on its tyrosine kinase activities, which set into motion signaling down multiple pathways involved in cell differentiation, survival, growth, and immune function (24, 25). Several reports have appeared recently demonstrating that IGF-1R can localize to the cell nucleus (26). Among these, evidence has surfaced indicating that the receptor might regulate specific target gene expression. One study identified IGF-1R translocation to the perinuclear compartment and then to the nucleus (27). The proposed model involves IGF-1 provoking SUMOylation of IGF-1R by small ubiquitin-like modifier protein-1 in which the receptor protein associates with enhancer elements and up-regulates target gene transcription (27). Warsito et al (28) demonstrated nuclear IGF-1R could associate with the transcription factor lymphoid enhancer binding factor 1 and in so doing enhanced cyclin D1 and axin2. In breast cancer cells, nuclear IGF-1R autoregulates expression of its own gene (29). We have also reported that IGF-1 and circulating Abs from patients with GD can provoke translocation of membrane-associated IGF-1R to the nucleus in TAO orbital fibroblasts but not in cells from healthy donors (13). The functional consequences of nuclear accumulation of the entire receptor or its fragments remain to be fully determined.
Among the first reports of a functional relationship existing between TSHR and IGF-1R were those of Ingbar and colleagues (30). They recognized that IGF-1 enhances TSH actions in cultured FRTL-5 thyroid cells in which both insulin and IGF-1 synergistically enhance the effects of TSH on cell proliferation (30). Deficiency of IGF-1R in thyroid epithelium results in reduced sensitivity to TSH in vivo (12) but can be compensated for by higher serum levels of TSH (31). In contrast, overexpression in the thyroid of both IGF-1 and IGF-1R reduces the requirement for TSH (11). It is currently unknown whether the canonical tyrosine kinase functions of IGF-1R or those activities arising from its intracellular redistribution and nuclear accumulation underlie the influences the receptor exerts on TSH actions. None of these earlier studies examined the impact of an absence of TSHR on the expression or subcellular localization of IGF-1R. Therein lies the rationale for the current studies, which are now extended to mouse cells. Besides their informing on receptor distribution, the current studies also suggest that the effects of IGF-1in fibroblasts might be enhanced in the absence of TSHR (Figure 5). Further studies will be required to determine the mechanisms underlying the more robust activation of Akt in TSHR−/− fibroblasts and whether this influences the downstream genes responsive to IGF-1.
The findings we report here suggest that IGF-1R protein expression, subcellular distribution, and response to IGF-1 in mouse lung fibroblasts are influenced by the mere presence or absence TSHR. Furthermore, the divergent abundance of IGF-1R in TSHR+/+ and TSHR−/− fibroblasts appears to be orchestrated through an as-yet-unidentified posttranslational mechanism. These studies demonstrate that IGF-1 diminishes cytoplasmic and nuclear IGF-1Rα staining in TSHR+/+ fibroblasts, whereas nuclear IGF-1Rβ signal is enhanced in these cells. In contrast, IGF-1 enhances nuclear IGF-1Rα staining in TSHR−/− fibroblasts. Thus, TSHR acts as a determinant of IGF-1R levels and subcellular distribution in mouse fibroblasts. In some respects, these findings are congruent with findings reported earlier in human orbital fibroblasts. Pritchard et al (32) found IGF-1R overexpression in TAO orbital fibroblasts compared with fibroblasts from healthy tissues. Subsequent studies identified the physical association between IGF-1R and TSHR and that inhibition of the former results in the attenuation of TSH-provoked signaling to target genes such as proinflammatory cytokines (10, 14). A later report by Hoa et al (13) described the cellular redistribution of IGF-1R after exposure to IGF-1 in TAO orbital fibroblasts. But those earlier studies did not examine the influence of TSHR on the behavior of IGF-1R. This as-yet-unexplored aspect of TSHR/IGF-1R interactions has thus prompted our current studies in which TSHR is knocked out. Taken in aggregate with findings in human fibroblasts (10), it appears that the interplay occurring between the two receptors may carry complex functional consequences. Identification of the molecular events downstream from the redistribution of IGF-1R and whether the set of IGF-1 targeted genes is altered by an absence of TSHR will require additional studies.
Acknowledgments
This work was supported in part by National Institutes of Health Grant EY08976 and unrestricted grants from the Research to Prevent Blindness and the Bell Family Charitable Foundation. These studies used the core Center for Vision by Grant EY007003 from the National Eye Institute.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Ab
- antibody
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GD
- Graves' disease
- IGF-1R
- IGF-1 receptor
- KO
- knockout
- MFI
- mean fluorescent intensity
- PI
- propidium iodide
- TAO
- thyroid-associated ophthalmopathy
- TSHR
- TSH receptor.
References
- 1. Kleinau G, Neumann S, Gruters A, Krude H, Biebermann H. Novel insights on thyroid-stimulating hormone receptor signal transduction. Endocr Rev. 2013;34:691–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Parmentier M, Libert F, Maenhaut C, et al. Molecular cloning of the thyrotropin receptor. Science 1989;246:1620–1622. [DOI] [PubMed] [Google Scholar]
- 3. Laugwitz K-L, Allgeier A, Offermanns S, et al. The human thyrotropin receptor: a heptahelical receptor capable of stimulating members of all four G protein families. Proc Natl Acad Sci USA. 1996;93:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kleinau G, Krause G. Thyrotropin and homologous glycoprotein hormone receptors: structural and functional aspects of extracellular signaling mechanisms. Endocr Rev. 2009;30:133–151. [DOI] [PubMed] [Google Scholar]
- 5. Chazenbalk GD, Pichurin P, Chen C-R, et al. Thyroid-stimulating autoantibodies in Graves disease preferentially recognize the free A subunit, not the thyrotropin holoreceptor. J Clin Invest. 2002;110:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shimura H, Miyazaki A, Haraguchi K, Endo T, Onaya T. Analysis of differentiation-induced expression mechanisms of thyrotropin receptor gene in adipocytes. Mol Endocrinol. 1998;12:1473–1486. [DOI] [PubMed] [Google Scholar]
- 7. Agretti P, Chiovato L, De Marco G, et al. Real-time PCR provides evidence for thyrotropin receptor mRNA expression in orbital as well as in extraorbital tissues. Eur J Endocrinol. 2002;147:733–739. [DOI] [PubMed] [Google Scholar]
- 8. Paschke R, Vassart G, Ludgate M. Current evidence for and against the TSH receptor being the common antigen in Graves' disease and thyroid associated ophthalmopathy. Clin Endocrinol (Oxf). 1995;42:565–569. [DOI] [PubMed] [Google Scholar]
- 9. De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov. 2002;1:769–783. [DOI] [PubMed] [Google Scholar]
- 10. Tsui S, Naik V, Hoa N, et al. Evidence for an association between thyroid stimulating hormone and insulin-like growth factor 1 receptors: a tale of two antigens implicated in Graves' disease. J Immunol. 2008;181:4397–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clement S, Refetoff S, Robaye B, Dumont JE, Schurmans S. Low TSH requirement and goiter in transgenic mice overexpressing IGF-I and IGF-Ir receptor in the thyroid gland. Endocrinology. 2001;142:5131–5139. [DOI] [PubMed] [Google Scholar]
- 12. Ock S, Ahn J, Lee SH, et al. IGF-1 receptor deficiency in thyrocytes impairs thyroid hormone secretion and completely inhibits TSH-stimulated goiter. FASEB J. 2013;27:4899–4908. [DOI] [PubMed] [Google Scholar]
- 13. Hoa N, Tsui S, Afifiyan NF, et al. Nuclear targeting of IGF-1 receptor in orbital fibroblasts from Graves' disease: apparent role of ADAM17. PLoS One. 2012;7:e34173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen H, Mester T, Raychaudhuri N, et al. Teprotumumab, an IGF-1R blocking monoclonal antibody inhibits TSH and IGF-1 action in fibrocytes. J Clin Endocrinol Metab. 2014;99:E1635–E1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marians RC, Ng L, Blair HC, Unger P, Graves PN, Davies TF. Defining thyrotropin-dependent and -independent steps of thyroid hormone synthesis by using thyrotropin receptor-null mice. Proc Natl Acad Sci USA. 2002;99:15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 17. Aickin M, Gensler H. Adjusting for multiple testing when reporting research results: the Bonferroni vs Holm methods. Am J Public Health. 1996;86:726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seaman MA, Levin JR, Serlin RC. New developments in pairwise multiple comparisons: some powerful and practicable procedures. Psychol Bull. 1991;110:577–586. [Google Scholar]
- 19. Feliciello A, Porcellini A, Ciullo I, Bonavolonta G, Avvedimento EV, Fenzi G. Expression of thyrotropin-receptor mRNA in healthy and Graves' disease retro-orbital tissue. Lancet. 1993;342:337–338. [DOI] [PubMed] [Google Scholar]
- 20. Cianfarani F, Baldini E, Cavalli A, et al. TSH receptor and thyroid-specific gene expression in human skin. J Invest Dermatol. 2010;130(1):93–101. [DOI] [PubMed] [Google Scholar]
- 21. Shimura H, Haraguchi K, Endo T, Onaya T. Regulation of thyrotropin receptor gene expression in 3T3-L1 adipose cells is distinct from its regulation in FRTL-5 thyroid cells. Endocrinology. 1997;138(4):1483–90. [DOI] [PubMed] [Google Scholar]
- 22. Raychaudhuri N, Fernando R, Smith TJ. Thyrotropin regulates IL-6 expression in CD34+ fibrocytes: clear delineation of its cAMP-independent actions. PLoS One. 2013;8:e75100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gillespie EF, Papageorgiou KI, Fernando R, et al. Increased expression of TSH receptor by fibrocytes in thyroid-associated ophthalmopathy leads to chemokine production. J Clin Endocrinol Metab. 2012;97:E740–E746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer. 2012;12(3):159–169. [DOI] [PubMed] [Google Scholar]
- 25. Smith TJ. Insulin-like growth factor-I regulation of immune function: a potential therapeutic target in autoimmune diseases? Pharmacol Rev. 2010;62(2):199–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarfstein R, Werner H. Minireview: nuclear insulin and insulin-like growth factor-1 receptors: a novel paradigm in signal transduction. Endocrinology. 2013;154:1672–1679. [DOI] [PubMed] [Google Scholar]
- 27. Sehat B, Tofigh A, Lin Y, et al. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal 2010;3:ra10. [DOI] [PubMed] [Google Scholar]
- 28. Warsito D, Sjostrom S, Andersson S, Larsson O, Sehat B. Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF. EMBO Rep. 2012;13:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarfstein R, Pasmanik-Chor M, Yeheskel A, et al. Insulin-like growth factor-I receptor (IGF-IR) translocates to nucleus and autoregulates IGF-IR gene expression in breast cancer cells. J Biol Chem. 2012;287:2766–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tramontano D, Cushing GW, Moses AC, Ingbar SH. Insulin-like growth factor-I stimulates the growth of rat thyroid cells in culture and synergizes the stimulation of DNA synthesis induced by TSH and Graves'-IgG. Endocrinology. 1986;119:940–942. [DOI] [PubMed] [Google Scholar]
- 31. Muller K, Fuhrer D, Mittag J, et al. TSH compensates thyroid-specific IGF-I receptor knockout and causes papillary thyroid hyperplasia. Mol Endocrinol. 2011;25:1867–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pritchard J, Han R, Horst N, Cruikshank WW, Smith TJ. Immunoglobulin activation of T cell chemoattractant expression in fibroblasts from patients with Graves' disease is mediated through the IGF-1 receptor pathway. J Immunol. 2003;170:6348–6354. [DOI] [PubMed] [Google Scholar]