Significance
Non-Hodgkin lymphomas (NHLs) are associated with HIV-1 infection, but the HIV1 genome is not detectable in malignant B cells. Here we show that variants of the HIV-1 matrix protein p17 (vp17s) are detected in the NHL specimens of HIV+ patients. These vp17s are more frequently detected in HIV+ patients with NHL than in patients without NHL. These vp17s display a potent B-cell growth-promoting activity, which is exerted by activating the Akt signaling pathway. Results obtained by CD spectroscopy and thermal denaturation suggest that mutation-induced protein destabilization may lead to a conformational change potentially responsible for the viral protein to promote B-cell growth. Our results suggest that vp17s may have a role in sustaining lymphomagenesis, thus offering new opportunities to prevent and/or treat HIV-associated NHLs.
Keywords: non-Hodgkin lymphoma, HIV-1 matrix protein p17, AIDS, p17 variants, B-cell clonogenicity
Abstract
Although in decline after successful anti-HIV therapy, B-cell lymphomas are still elevated in HIV-1-seropositive (HIV+) persons, and the mechanisms are obscure. The HIV-1 matrix protein p17 persists in germinal centers long after HIV-1 drug suppression, and some p17 variants (vp17s) activate Akt signaling and promote growth of transformed B cells. Here we show that vp17s derived from four of five non-Hodgkin lymphoma (NHL) tissues from HIV+ subjects display potent B-cell growth-promoting activity. They are characterized by amino acid insertions at position 117–118 (Ala–Ala) or 125–126 (Gly–Asn or Gly–Gln–Ala–Asn–Gln–Asn) among some other mutations throughout the sequence. Identical dominant vp17s are found in both tumor and plasma. Three of seven plasma samples from an independent set of NHL cases manifested multiple Ala insertions at position 117–118, and one with the Ala–Ala profile also promoted B-cell growth and activated Akt signaling. Ultradeep pyrosequencing showed that vp17s with C-terminal insertions are more frequently detected in plasma of HIV+ subjects with than without NHL. Insertion of Ala–Ala at position 117–118 into reference p17 (refp17) was sufficient to confer B-cell growth-promoting activity. In contrast, refp17 bearing the Gly–Asn insertion at position 125–126 did not, suggesting that mutations not restricted to the C terminus can also account for this activity. Biophysical analysis revealed that the Ala–Ala insertion mutant is destabilized compared with refp17, whereas the Gly–Asn form is stabilized. This finding provides an avenue for further exploration of structure function relationships and new treatment strategies in combating HIV-1–related NHL.
Although AIDS-defining cancers have declined following the introduction of combined antiretroviral therapy (cART), non-Hodgkin lymphomas (NHLs) still comprise more than 50% of all AIDS-defining cancers (1) and are the most frequent cause of death in these patients (2). Most HIV-1–related NHLs (HIV–NHLs) are high-grade B-cell lymphomas such as diffuse large B-cell lymphoma (DLBCL) and Burkitt lymphoma (BL). HIV–NHLs derived from either germinal or postgerminal center B lymphocytes are often characterized by clinical aggressiveness and display a predilection for extranodal sites (3–5). To date, the usual reason for the increase of HIV–NHL is said to be an HIV-1–driven immune dysfunction (6). The onset of HIV–NHL is often preceded by an overproduction of B-cell stimulatory cytokines, which further sustains B-cell activation and possibly drives the generation of activation-induced DNA modification errors and oncogenic translocations, conferring the transformed phenotype to B cells (7). On the other hand, the loss of immune control promotes the reactivation of potentially oncogenic herpesviruses, such as Epstein–Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), which are causally involved in a significant fraction of HIV–NHLs (8). Lymphomas associated with EBV or KSHV show a mainly latent infection by these oncogenic herpesviruses with a pattern of viral antigen expression that varies according to the degree of HIV1-related immunosuppression (4).
A possible direct role of HIV-1 and its gene products in lymphomagenesis has been less considered. Although the HIV-1 genome is not clonally integrated in the malignant B cells as seen for oncogenic retroviruses such as HTLV-1, some HIV-1 structural (gp120) and regulatory proteins (Tat) induce chronic inflammation, leading to B-cell activation and proliferation within lymph nodes, fostering polyclonal B-cell expansion (9, 10).
The HIV-1 matrix protein p17 is the N-terminal domain of a larger precursor polyprotein encoded by the HIV-1 gag gene, which directs, via its N-terminal myristoyl group, HIV-1 Gag polyproteins to the host membrane, where assembly and budding of virions occur (11). In mature virions, p17 forms a protective shell lining the inner leaflet of the viral membrane and can dissociate from it in the early stages of the HIV-1 life cycle to direct the preintegration complex to the host cell nucleus. We have recently highlighted a strong extracellular presence of p17 in mice transgenic for a partial HIV-1 provirus in which B-cell lymphoma develops in a significant percent of animals (12). Moreover, we have provided evidence that genetic variation occurring in p17 during the natural course of HIV-1 infection may differentially impact signaling pathways and clonogenic B-cell growth and contribute to B-cell lymphomagenesis (13). In fact, in contrast to a prototype p17 (reference p17, refp17; from clone BH10 of the clade B isolate), which does not promote B-cell growth, a vp17 derived from a HIV-1 strain A1-infected Ugandan patient whose clinical status is unknown, named S75X, was found to activate the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway in B cells and increase B-cell proliferation and clonogenicity in soft agar (14). Recent data indicate that the intrapatient diversity of amino acid sequences of the p17 protein is much higher than expected (15), thus suggesting that p17 variants (vp17s) potentially endowed with different biologic effects may spontaneously originate in HIV-1–infected cells. Considering that p17 may accumulate and persist within lymphoid tissues, even in patients treated with cART (16), the possibility that vp17s with lymphomagenic properties may be enriched in some HIV+ patients with possible predilection for lymphomas is a testable hypothesis. Here we report on the amino acid sequence associated with lymphomas and the structural impacts of some, which may result in biologic effects of potential pathogenic relevance for the development of HIV–NHL.
Results
Detection of vp17s in HIV–NHL Specimens.
We sought to evaluate the occurrence of dominant species of p17 in HIV-1–infected cellular reservoirs within lymphoma biopsies obtained from five patients with HIV–NHL (group A patients) (Table S1). Considering that all these patients displayed low-level viremia, a nested PCR approach was used to obtain p17-specific amplicons in a setting in which a low amount of proviral DNA was expected. Amplification of the same target DNA was repeated at least 20 times, and HIV–DNA was successfully detected in all biopsies. These findings are consistent with the presence of HIV-1–infected cellular reservoirs within lymphoma biopsies, even in those obtained from patients with a very low level of circulating virus. Data obtained from repeated amplifications of p17 target sequences demonstrated the reproducibility of the amplification and confirmed that the sequence variability observed was not due to lack of polymerase fidelity. Analysis of p17 sequences obtained from each DNA sample showed that, within each lymphoma sample, the proviral population always contained a dominant vp17 sequence (Fig. 1A) and paired plasma virus shared this same sequence.
Table S1.
Clinical and virological features of patients
| Patient group | Pt | Sex | Age, y | CD4, cells/μL | HIV viral load, log 10 cp/mL | NHL histology | cART* |
| A | a101 | F | 36 | 80 | <1.70 | DLBCL | Experienced |
| a102 | M | 39 | 243 | 2.46 | DLBCL | Experienced | |
| a103 | M | 44 | 89 | 2.18 | DLBCL | Experienced | |
| a104 | M | 44 | 166 | <1.70 | DLBCL | Experienced | |
| a105 | F | 42 | 460 | 3.54 | Burkitt | naïve | |
| Median (range) | 42 (36–44) | 166 (80–460) | 2.18 (<1.70–3.54) | 20.0; 80.0† | |||
| B | b101 | M | 39 | n/a | n/a | DLBCL | Experienced |
| b102 | M | 48 | 54 | 4.63 | DLBCL | Experienced | |
| b103 | M | 40 | 125 | n/a | LCL | Experienced | |
| b104 | M | 41 | n/a | 4.40 | LBCL | Experienced | |
| b105 | M | 44 | n/a | n/a | LBCL | n/a | |
| b106 | M | 45 | 10 | <1.70 | DLBCL | n/a | |
| b107 | M | 57 | n/a | n/a | DLBCL | n/a | |
| Median (range) | 44 (39–57) | 54 (10–125) | 4.40 (<1.70–4.63) | 0; 57.14 | |||
| p (B vs. A) | 0.29‡ | 0.14‡ | 0.36‡ | NA | NA | ||
| C | c103 | M | 45 | 148 | 5.06 | DLBCL | Naive |
| c104 | F | 31 | 73 | 4.48 | DLBCL | Experienced | |
| c105 | F | 43 | 157 | 5.29 | DLBCL | Naive | |
| c106 | M | 35 | 246 | 5.35 | DLBCL | Experienced | |
| c107 | M | 50 | 32 | 4.73 | Burkitt | Experienced | |
| c108 | M | 63 | 56 | 4.76 | DLBCL | Experienced | |
| c109 | M | 38 | 247 | 4.87 | DLBCL | Naive | |
| c110 | M | 43 | 50 | 4.49 | DLBCL | Naive | |
| c112 | M | 34 | 16 | 4.93 | DLBCL | Experienced | |
| c113 | M | 36 | 186 | 4.11 | DLBCL | Experienced | |
| c114 | M | 60 | 148 | 5.70 | DLBCL | Naive | |
| c115 | M | 45 | 3 | 4.18 | DLBCL | Experienced | |
| c116 | M | 34 | 161 | 4.20 | n/a | Experienced | |
| c117 | M | 34 | 72 | 4.98 | Burkitt | Naive | |
| c118 | M | 53 | 16 | 5.44 | Burkitt | Naive | |
| c119 | M | 42 | 123 | 4.01 | DLBCL | Naive | |
| c120 | M | 40 | 560 | 4.60 | DLBCL | n/a | |
| c121 | F | 32 | 10 | 5.26 | DLBCL | Naive | |
| Median (range) | 41 (31–63) | 98 (3–560) | 4.82 (4.01–5.70) | 50.0; 44.44 | |||
| D | d104 | M | 31 | 375 | 4.52 | NA | Experienced |
| d105 | M | 31 | 285 | 4.32 | NA | Experienced | |
| d106 | M | 27 | 611 | 4.21 | NA | Naive | |
| d107 | M | 30 | 510 | 4.54 | NA | Naive | |
| d108 | M | 43 | 422 | 4.47 | NA | Experienced | |
| d110 | M | 32 | 252 | 4.71 | NA | Experienced | |
| d112 | F | 32 | 447 | 4.62 | NA | Experienced | |
| d113 | F | 21 | 99 | 4.94 | NA | Experienced | |
| d114 | M | 50 | 39 | 4.61 | NA | Experienced | |
| d115 | F | 41 | 486 | 4.30 | NA | Experienced | |
| d117 | F | 49 | 417 | 4.98 | NA | Experienced | |
| d118 | M | 45 | 44 | 5.44 | NA | Experienced | |
| d120 | M | 43 | 217 | 6.12 | NA | Experienced | |
| d121 | M | 44 | 252 | 4.66 | NA | Experienced | |
| d122 | M | 44 | 454 | 4.62 | NA | Experienced | |
| d123 | F | 27 | 643 | 4.05 | NA | Naive | |
| d125 | M | 44 | 406 | 4.83 | NA | Experienced | |
| d126 | M | 37 | 652 | 4.63 | NA | Naive | |
| Median (range) | 39 (21–50) | 411.5 (39–652) | 4.62 (4.05–6.12) | 22.22; 77.78 | |||
| p (D vs. C) | 0.18 | 0.0004 | 0.44 | NA | 0.086§ | ||
DLBCL, diffuse large B-cell lymphoma; F, female; LBCL, large B-cell lymphoma; LCL, large cell lymphoma; M, male; n/a, not available; NA, not applicable; NHL, non-Hodgkin lymphoma.
Combined antiretroviral therapy.
% naive; % experienced.
B vs. A and D vs. C, Mann–Whitney test.
D vs. C, Fisher exact test.
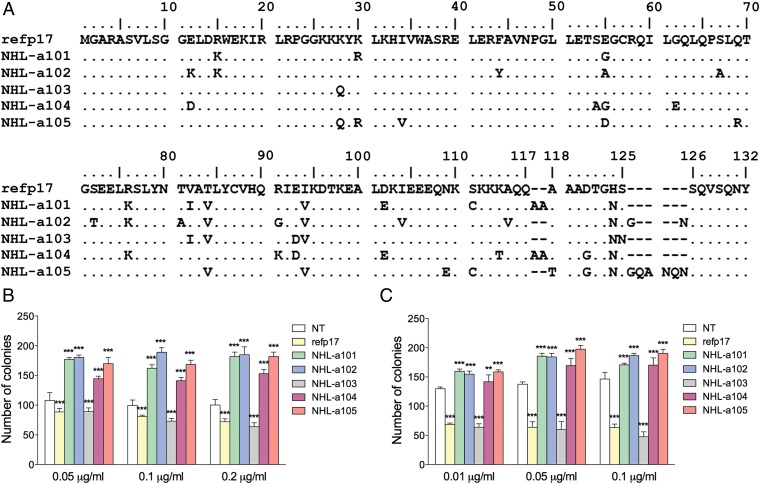
Fig. 1.
Alignment and comparison among amino acid sequences of refp17 and vp17s isolated from HIV–NHL patients and effects of different recombinant p17s on B-cell clonogenicity. (A) Sequences are represented by the single-letter amino acid code. Amino acid positions are referred to the prototype genotype B strain BH10 (UniProtKB P04585; refp17), adopted as reference for this analysis. Each amino acid residue of NHL-derived vp17s not differing from the refp17 sequence is represented by a dot. (B) Raji and (C) BJAB cells were plated in 12-well plates, and after 4 d, medium was replaced by fresh medium with various concentrations, 0.01, 0.05, 0.1, and 0.2 μg/mL of refp17 and lymphoma-associated variants, as indicated. Cells not treated (NT) were used as negative control. The cell growth was analyzed by using 3-[4, 5-Dimethylthiazol-2-y1]-2, 5-diphenyltetrazolium bromide (MTT). Data represent the average number of colonies ± SD from three independent experiments performed in triplicate. The statistical significance between control and treated cultures was calculated using one-way ANOVA performed separately for each concentration of p17 variants, across the three groups. Bonferroni’s posttest was used to compare data. *P < 0.05, **P < 0.01, ***P < 0.001.
HIV–NHL-Derived vp17s Promote Lymphoma B-Cell Colony Formation.
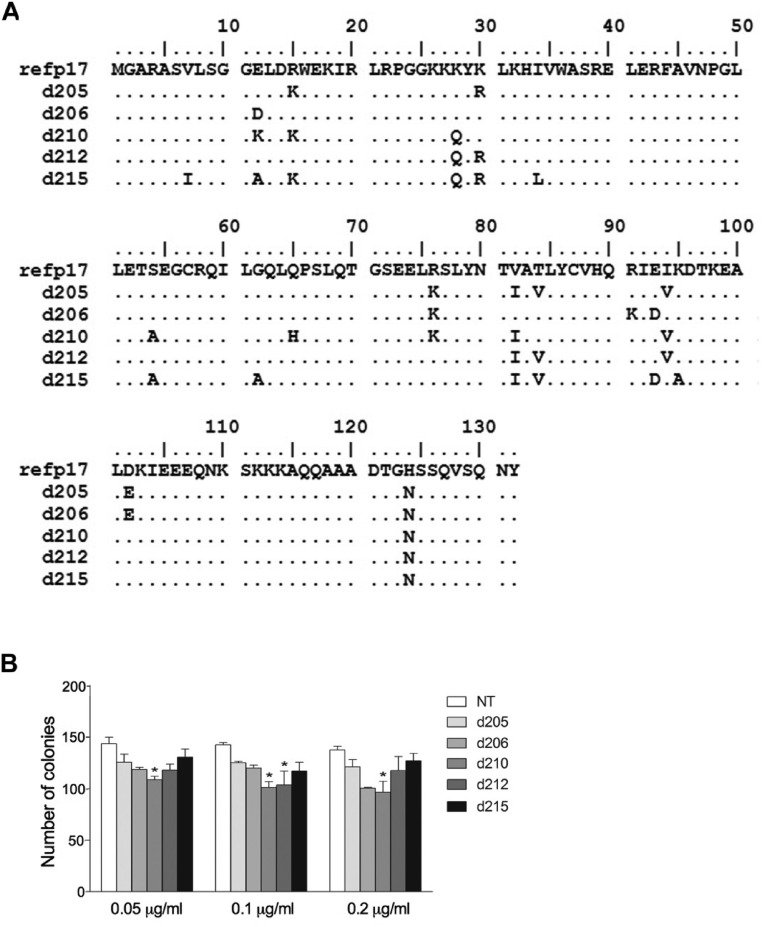
Recombinant vp17s obtained from the five dominant sequences detected in lymphoma tissues of group A patients (NHL-a101, -a102, -a103, -a104, and -a105) were then investigated for their ability to enhance clonogenic activity of the Raji lymphoma B-cell line. As shown in Fig. 1B, NHL-a101, NHL-a102, NHL-a104, and NHL-a105, at a concentration ranging from 0.05 to 0.2 µg/mL, significantly increased the number of Raji cell colonies in soft agar compared with untreated cells. By contrast, NHL-a103 was unable to increase the colony-forming ability of Raji cells. Its activity was superimposable to that exerted by refp17. For comparison, dominant p17 sequences were detected as proviral DNA in blood from five HIV+ patients without lymphoma, and recombinant proteins were produced and tested in the Raji clonogenic assay. Similar to refp17, all of the five p17s derived from lymphoma-negative HIV+ patients did not show any capability to increase the clonogenic activity of Raji cells (Fig. S1).
Fig. S1.
Alignment and comparison among amino acid sequences of refp17 and p17s isolated from HIV+ non-NHL patients and effects of different recombinant p17s on B-cell clonogenicity. (A) Sequences are represented by the single-letter amino acid code. Amino acid positions are referred to the prototype genotype B strain BH10 (UniProtKB P04585; refp17), adopted as the reference for this analysis. Each amino acid residue of p17s not differing from the refp17 sequence is represented by a dot. (B) Raji cells were plated in 12-well plates, and after 4 d, medium was replaced by fresh medium with various concentrations, 0.05, 0.1, and 0.2 μg/mL, of refp17 and p17 derived from blood samples of HIV+ non-NHL patients, namely d205, d206, d210, d212, and d215, as indicated. Cells not treated (NT) were used as negative control, whereas cells treated with vp17 NHL-a101 and NHL-a102 were used as the positive clonogenic control. The cell growth was analyzed by using MTT. Data represent the average number of colonies ± SD from three independent experiments performed in triplicate. The statistical significance between control and treated cultures was calculated using one-way ANOVA performed separately for each concentration of p17 across the three groups. Bonferroni’s posttest was used to compare data. *P < 0.05, **P < 0.01, ***P < 0.001.
Raji is an EBV-infected lymphoma B-cell line. To verify that vp17s operate independently of EBV, we tested the clonogenic activity of these variants in the EBV-negative BJAB human lymphoma B-cell line. As for Raji, NHL-a101, -a102, -a104, and -a105 significantly increased, whereas refp17 and NHL-a103 significantly reduced, the number of colonies of BJAB cells in soft agar, compared with untreated control cultures (Fig. 1C). These results confirm the capacity of distinct lymphoma-associated vp17s to increase B-cell growth and clonogenicity irrespective of the presence of EBV.
HIV–NHL-Derived vp17 Signaling.
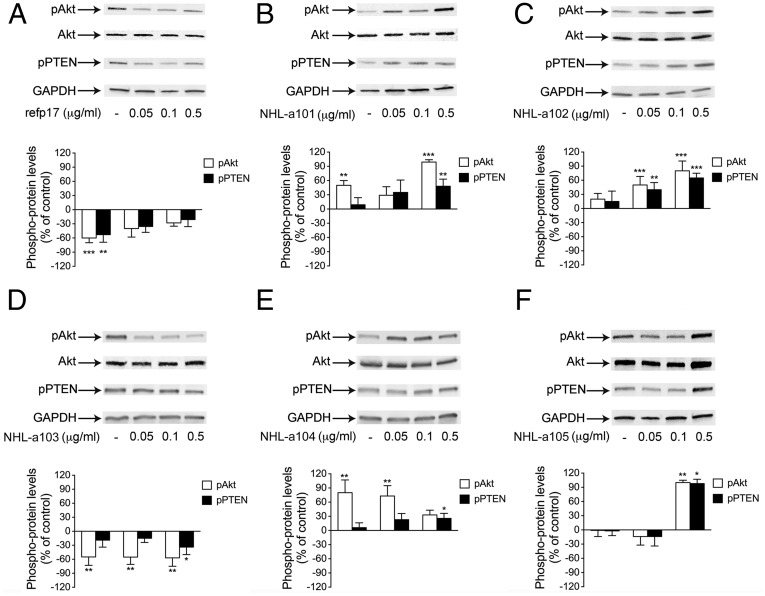
The PI3K/Akt signaling pathway plays a crucial role in the development and malignant progression of several tumor histotypes, including lymphomas (17). Because NHL-a101, -102, -104, and -105 promote B-cell clonogenicity, we explored their capability to modulate the phosphorylation status of PI3K/Akt, compared with the nonclonogenic refp17. As shown in Fig. 2, Raji cells stimulated with refp17 or NHL-a103 showed a significant inhibition of Akt activation, as evidenced by down-regulation of phosphorylated Akt. By contrast, all lymphoma-associated vp17s with clonogenic activity significantly increased the active phosphorylation of the Akt kinase. Considering that Akt phosphorylation can be modulated by phosphatase and tensin homolog (PTEN) (18), we also investigated the same samples for Ser/Thr phosphorylation status of PTEN. Treatment of cells with clonogenic vp17s led to increased, whereas refp17 and the nonclonogenic p17 NHL-a103 reduced, levels of PTEN Ser/Thr phosphorylation (Fig. 2). These findings are consistent with the potential link between heightened clonogenicity of lymphoma-derived vp17s and activation of the Akt–PTEN pathways.
Fig. 2.
Effects of refp17 and lymphoma-associated vp17 stimulation on Akt and PTEN activity in Raji cells. Cells were treated for 5 min with 0.05, 0.1, and 0.5 μg/mL of refp17 (A) and lymphoma-associated p17 variants NHL-a101 (B), NHL-a102 (C), NHL-a103 (D), NHL-a104 (E), and NHL-a105 (F). Untreated cells were used as control (lane 1). Western blot analysis of Raji lysates shows that refp17 and NHL-a103 inhibit the activation of Akt and maintain PTEN in an active state (A), as shown by the respective phosphorylation state, verified by densitometric analysis and plotting of the pAkt/Akt and pPTEN/GAPDH. On the contrary, NHL-a101, NHL-a102, NHL-a104, and NHLa105 induce the activation of Akt and maintain PTEN in an inactive state, as shown by the increased phosphorylation, verified by densitometric analysis and plotting of the pAkt/Akt and pPTEN/GAPDH. In the Upper panels, blots from one representative experiment of three with similar results are shown. In the Lower panels, values reported for phosphorylation of Akt and PTEN are the mean ± SD of three independent experiments. Statistical analysis was performed by one-way ANOVA, and the Bonferroni’s posttest was used to compare data. *P < 0.05, **P < 0.01, ***P < 0.001.
Sequence Patterns Related to Clonogenic Activity.
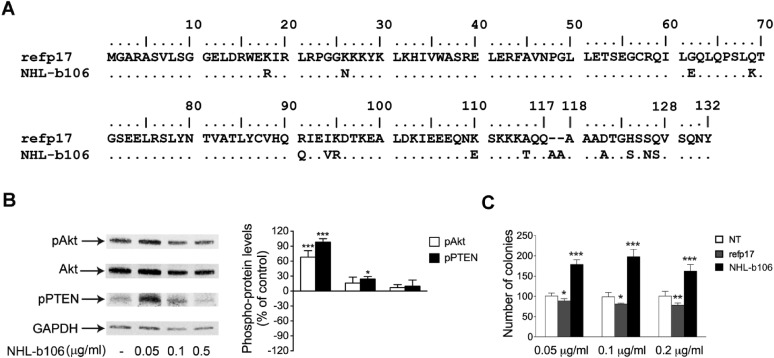
Analysis of the dominant sequences of vp17s derived from group A patients showed that four out of five clonogenic vp17s (NHL-a101, -a102, -a104, and -a105) shared the presence of amino acid insertions at positions 117–118 or 125–126. In particular, NHL-a101 and NHL-a104 showed the insertion of an Ala–Ala stretch between amino acids 117 and 118, whereas NHL-a102 and NHL-a105 had the insertion of a Gly–Asn or of a GlyGln–Ala–Asn–Gln–Asn stretch between amino acids 125 and 126 (Fig. 1A). Considering that amino acid insertions at position 117–118 or 125–126 are a common feature of all HIV–NHL-derived clonogenic vp17s from group A patients, we therefore evaluated if these molecular signatures could be frequently detected in samples obtained from other patients with HIV–NHL belonging to different cohorts. One group represented a series of seven plasmas obtained from AIDS and Cancer Specimen Resource (ACSR), funded by the National Cancer Institute (group B in Table S1). Data obtained showed that three out of seven group B-derived plasma samples expressed vp17s characterized by amino acid insertions at position 117–118. One vp17, similar to NHL-a101 and -a104, showed an Ala–Ala insertion. The other two displayed more complex insertions at the same position, containing Ala–Ala–Ala or Ala–Ala–Ala–Ala–Gln stretches. When examined for B-cell growth-promoting activity, the vp17 displaying the shortest insertion (Ala–Ala), named NHLb106 (Fig. S2A), was found to activate the Akt signaling pathway, down-modulate PTEN (Fig. S2B), and promote a potent clonogenic activity (Fig. S2C). Although based on one case, this result indicates the potential predictive value of certain amino acid substitutions associated with a B-cell growth-promoting phenotype.
Fig. S2.
Effect of NHL-b106 stimulation on the PTEN/Akt signaling pathway and on B-cell clonogenicity. (A) Alignment and comparison among p17 and NHL-b106 amino acid sequences. Sequences are represented by the single-letter amino acid code. Each amino acid residue of NHL-b106 not differing from refp17 is represented by a dot. (B) Raji cells were treated for 5 min with 0.05, 0.1, and 0.5 μg/mL of NHL-b106. Cells not treated (NT) were used as the control. Western blot analysis of lysates shows that NHL-b106 induces the activation of Akt and maintains PTEN in an inactive phosphorylated state, verified by densitometric analysis and plotting of pAkt/Akt and pPTEN/GAPDH. In the Left panels, blots from one representative experiment of three with similar results are shown. In the Right panels, values reported for pAkt and pPTEN are the mean ± SD of three independent experiments. Statistical analysis was performed by one-way ANOVA, and the Bonferroni.s posttest was used to compare data. *P < 0.05, ***P < 0.001. (C) Raji were plated in 12-well plates, and after 4 d, medium was replaced by fresh medium with various concentrations, 0.05, 0.1, and 0.2 μg/mL, of refp17 and NHL-b106. Cells not treated (NT) were used as the negative control. The cell growth was analyzed by using MTT. Data represent the average number of colonies ± SD from three independent experiments performed in triplicate. The statistical significance between control and treated cultures was calculated using one-way ANOVA performed separately for each concentration of p17 variant across the three groups. Bonferroni’s posttest was used to compare data. *P < 0.05, **P < 0.01, ***P < 0.001.
Vp17s With Insertions at Their C Terminus Are More Frequently Detected in Plasma of Patients With HIV–NHL Than in Plasma of Patients Without NHL.
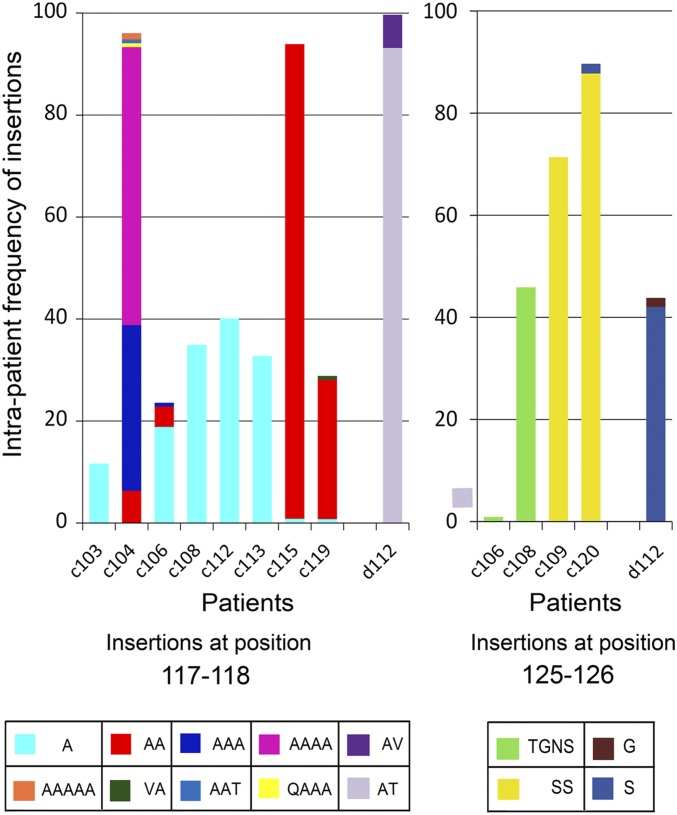
To gain more information about the frequency of detection of vp17s showing insertions at position 117–118 and 125–126 among the circulating quasispecies detected in NHL patients, two further series of plasma samples from viremic HIV+ patients with (group C) or without NHL (group D) (Table S1) were analyzed by ultradeep pyrosequencing (UDPS). Ten out of 18 (55.56%) patients belonging to group C had vp17s characterized by amino acid insertions at position 117–118 or at position 125–126 in their plasma. As shown in Fig. 3, eight of them displayed from one to five Ala insertions at position 117–118, sometimes including other amino acids such as Thr, Val, or Gln. Two out of 10 (c106 and c108) also showed vp17s with Tyr–Gly–Asn–Ser insertions at position 125–126, whereas c109 and c120 have an insertion of Ser or Ser–Ser solely at position 125–126. By contrast, only 1 out of 18 group D patients (5.5%; P = 0.0027 vs. group C in Fisher exact test) showed an insertion both at position 117–118 and 125–126, characterized by the presence of Ala–Thr/Ala–Val or Ser/Gly stretches, respectively. Beside the higher frequency of insertions detected in the vp17 C terminus, it is worth noting that the intrasample diversity was significantly higher in group C than in group D patients [median (range), 251.30 (50.21–445.60) × 10−4 vs. 141.10 (28.69–366.70) × 10−4 substitutions per site (P = 0.05)], even if it was calculated only on the stretch encompassing residues 5–113 due to the quasispecies simplification introduced by the additional clustering. Taken together, these results show that vp17s derived from tumor tissue and blood of HIV–NHL patients are mostly characterized by amino acid insertions occurring at well-defined amino acid positions within the viral protein C terminus. Moreover, they show that insertions of different Ala stretches at position 117–118 are much more represented in the blood of HIV–NHL patients than insertions of different amino acids at position 125–126.
Fig. 3.
Frequency of amino acid insertions at positions 117–118 and 125–126 in vp17 of HIV+ patients with or without NHL. Bars represent the frequency of amino acid insertions detected at positions 117–118 (Left panel) and 125–126 (Right panel) in patients belonging to groups C and D. Each insertion is identified by a color code (see table at the bottom of the histogram). *Amino acid positions are referred to the prototype genotype B strain BH10 (UniProtKB P04585; refp17), adopted as the reference for this analysis.
To evaluate the frequency of the Ala–Ala insertion at position 117–118 of the matrix protein p17, all available subtype B sequences at the Los Alamos HIV Database were examined. The initial query resulted in a total of 9,750 sequences; after selecting only one sequence per patient, removing all of the clones and problematic sequences, the final alignment contained 5,332 sequences from around the world. Out of the 5,332 sequences, 851 (16%) had one or more Ala insertions at position 117–118. One of the most frequent insertions was the Ala–Ala insertion, which was displayed by 304 (5.7%) of the sequences analyzed.
Insertion of Ala–Ala at Position 117–118 of the refp17 Backbone Is Sufficient to Confer Clonogenic Activity to the Viral Protein.
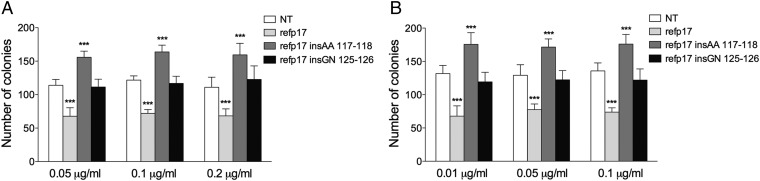
To better define the role of specific amino acid insertions in conferring a clonogenic property to HIV–NHL-derived vp17s, we engineered the inactive refp17 by insertions of Ala–Ala or Gly–Asn at position 117–118 and 125–126, respectively, and investigated their clonogenic activity. As shown in Fig. 4, the refp17 mutant bearing the Ala–Ala insertion at position 117–118 (refp17 insAA 117–118), similar to its natural vp17 counterparts NHL-a101 and -a104, was able to increase Raji (Fig. 4A) and BJAB (Fig. 4B) cell clonogenicity. On the other hand, the refp17 mutant bearing the Gly–Asn insertion at position 125–126 (refp17 insGN 125–126), unlike its NHL-a102 counterpart, showed no evidence of enhanced clonogenic activity. These data indicate that the Ala–Ala insertion at position 117–118 may be one of the possible signatures for vp17s characterized by enhanced B-cell clonogenic activity. On the other hand, the Gly–Asn insertion at position 125–126 is not involved, at least per se, in the NHL-a102 B-cell clonogenic activity. This finding suggests that amino acid mutations scattered throughout the vp17 NHL-a102, per se or in addition to the Gly–Asn insertion, may be responsible for the enhanced clonogenic activity of this vp17.
Fig. 4.
Effect of Ala–Ala or Gly–Asn insertion at position 117–118 and 125–126, respectively, of the refp17 backbone on B-cell clonogenicity. Raji (A) and BJAB (B) were plated in 12-well plates, and after 4 d, medium was replaced by fresh medium with various concentrations, 0.01, 0.05, 0.1, and 0.2 μg/mL of refp17, refp17 insAA 117–118, and refp17 insGN 125–126, as indicated. Cells not treated (NT) were used as the negative control. The cell growth was analyzed by using MTT. Data represent the average number of colonies SD from three independent experiments performed in triplicate. The statistical significance between control and treated cultures was calculated using one-way ANOVA performed separately for each concentration of p17 variants, across the three groups. Bonferroni’s posttest was used to compare data. ***P < 0.001.
The Ala–Ala Insertion at 117–118 Destabilizes refp17, Whereas the Gly–Asn Insertion at 125–126 Stabilizes It.
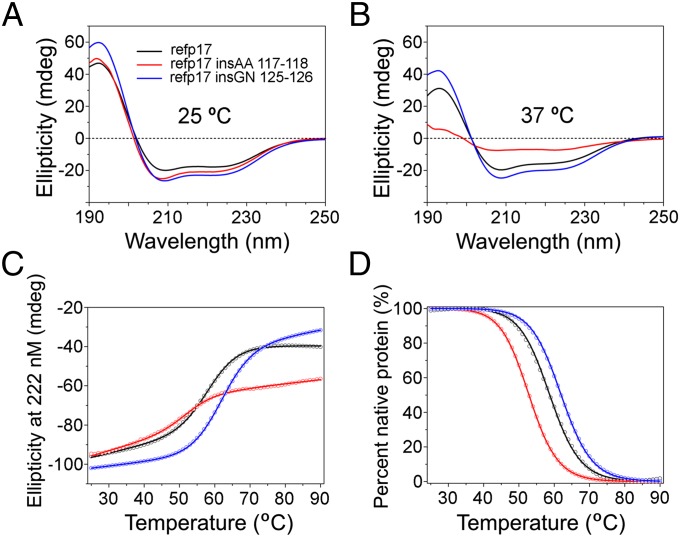
Various structural analyses by X-ray crystallography and NMR spectroscopy indicate that p17 comprises five major alpha helices connected primarily by short loops (19), where the C-terminal alpha helix H5, extending to Ala119, packs against H3 and the central alpha helix H4 to stabilize the protein. Although the Gly–Asn insertion in the disordered C-terminal region of p17 may be structurally tolerated, the outcome of the Ala–Ala insertion within the H5 helix remains difficult to predict, due in part to the fact that Ala is known to be of high helix propensity (20). To better understand the structural ramification of these amino acid insertions in refp17, we comparatively characterized recombinant refp17, refp17 insAA 117–118, and refp17 insGN 125–126 proteins using circular dichroism (CD) spectroscopy and protein thermal denaturation techniques. Shown in Fig. 5A are CD spectra of the three p17 proteins that displayed similar secondary structures of an alpha-helical nature at room temperature, as evidenced by a strong positive maximum at 195 nm and two negative minima at 208 and 222 nm, consistent with the known structural features of p17. Furthermore, the two insertion mutants were marginally more helical than refp17, suggesting that both insertions are well tolerated structurally at 25 °C. At 37 °C, however, refp17 insAA 117–118 was significantly less structured than either refp17 or refp17 insGN 125–126 (Fig. 5B), suggesting that the Ala–Ala insertion accentuates the protein to unfold at elevated temperature. To verify these findings, we subjected all three p17 proteins to heat-induced denaturation monitored at 222 nm by CD spectroscopy. As shown in Fig. 5C, refp17 exhibited a cooperative unfolding with a single transition as the temperature was raised from 25 °C to 90 °C, typical of a compact, single-domain globular protein. Normalization of the experimental data based on a two-state protein denaturation model (21) gave rise to a characteristic melting temperature (Tm at which 50% of protein is denatured or unfolded) of 58.7 ± 0.1 °C for refp17 (Fig. 5D). The Gly–Asn insertion stabilized the p17 protein (Tm of 62.0 ± 0.1 °C), whereas the Ala–Ala insertion noticeably destabilized it by ∼6 °C (Tm of 52.9 ± 0.3 °C). In sharp contrast to refp17 and refp17 insGN 125–126, massive protein precipitation of refp17 insAA 117–118 was observed during thermal denaturation, resulting in “artificially” increased ellipticity in its unfolded state. Taken together, these results strongly indicate that although the Gly–Asn insertion at 125–126 is structurally amenable to refp17, the Ala–Ala insertion at 117–118 is detrimental to its stability.
Fig. 5.
Effects of the Ala–Ala and Gly–Asn insertions in the C terminus of refp17 on protein conformation and stability. (A) CD spectra of refp17, refp17 insAA 117–118, and refp17 insGN 125–126, each at 2.5 µM, collected at 25 °C in 10 mM phosphate buffer, pH 7.4. (B) CD spectra of the three recombinant p17 proteins at 37 °C under otherwise identical conditions. (C) Thermal denaturation of refp17, refp17 insAA 117–118, and refp17 insGN 125–126, each at 10 µM in PBS, pH 7.4, as monitored at 222 nm by CD spectroscopy. (D) The experimental data normalized according to a two-state protein denaturation model from which Tm values were derived.
Discussion
Despite the improved control of HIV-1 infection achieved by cART, malignant lymphomas still remain a major cause of morbidity and mortality in this setting. In fact, even if NHLs are considered closely associated with HIV-1 infection, with the advent of cART, their incidence decreased to a markedly less extent compared with other HIV-1–related tumors and mainly affected the most immunogenic histotypes, such as EBV-associated immunoblastic DLBCL and primary central nervous system lymphomas. These lymphomas express the broadest pattern of EBV latent antigens similarly to posttransplant lymphoproliferative disorders that are almost invariably EBV-positive. In contrast, less immunogenic forms of HIV-1–related lymphomas are still relatively prevalent despite cART immune reconstitution. This is particularly true for BL, in which the expression of viral proteins is restricted to EBNA-1 (4). These findings strongly indicate that factors different from HIV-1–related immunosuppression are probably involved in the development of lymphomas still occurring in HIV+ patients. In light of the emerging role of the microenvironment in promoting and sustaining the growth and survival of tumor cells (22), the possibility that HIV-1–encoded proteins derived from infected cells and endowed with peculiar biologic properties contribute to lymphomagenesis was entertained. Indeed, a recent study has shown that a variant of p17 slightly increases cell proliferation of EBV-infected primary human B cells (23).
The current analysis provides direct evidence for a potential role of vp17s in prompting lymphoma in HIV+ patients and thus provides a rational explanation for the paradox of persistent risk for lymphoma among patients despite the immune-restorative effects of antiretroviral therapy. Popovic et al. previously demonstrated persistence of p17 but not viral RNA in lymph nodes of patients on cART who were virally suppressed (16), suggesting the presence of soluble p17 in these tissues. If so, the impact of p17 on B-cell growth is likely to persist. In the current report, we document for the first time, to our knowledge, evidence for the presence of vp17s in lymphoma tissue, mirrored in their plasma counterparts that are capable of augmenting clonogenic proliferation of both EBV and non–EBV-associated lymphoma cell lines.
Derived from the current analysis are fundamental insights into the molecular basis of how vp17s may play a role in lymphomagenesis. First is the finding that in four out of five NHL cases from Italy, vp17s derived from the lymphoma tissue augment clonogenic activity and mediate up-regulation of the Akt signaling pathway, whereas one of these shares the property of down-modulation of clonogenic and Akt signaling characteristic of the refp17 derived from the BH10 sequence. The four with growth-promoting effects all share a common pattern of amino acid substitutions at positions 117–118 and 125–126, particularly Ala–Ala insertions at the 117–118 that appear to predict this activity, insertions also observed in plasma-derived virus. Second, in an independent set of cases from the United States, several manifest similar amino acid substitutions, and one with such an Ala–Ala profile predicted to promote B-cell growth and signaling was functionally tested and showed the expected vp17 activity. Third, a survey of 18 cases of HIV–NHL documents a higher proportion of cases with the vp17 signatures compared with 18 cases of HIV+ patients without lymphoma. Additionally, these HIV–NHL cases demonstrate higher intrapatient p17 sequence diversity compared with non-HIV–NHL cases, suggesting a possible difference in biological properties in those at risk for lymphoma.
The most convincing evidence for this particular pattern of amino acid substitutions to predict vp17 activity comes from our data showing that insertion of the Ala–Ala substitution into the refp17 (BH10) backbone reproduced the observed up-regulation of clonogenicity. However, the refp17 mutant bearing the Gly–Asn insertion at position 125–126 did not reproduce the pattern of its native NHL-a102 counterpart, suggesting that other additional mutations or structural motifs account for this activity. This finding is not surprising, as recent data point to the C terminus of refp17 beyond residue 119 as fully disordered (24), suggesting that mutations in this region are expected to have little impact, if any, on protein folding and function. Because interactions between side chains are important for maintaining the stability of a protein as well as for enabling the correct folding of the molecule, comparative mutational studies of NHL-a102 and refp17 will help identify which amino acid mutations in the vp17 are associated with Akt activation and B-cell clonogenicity.
The finding that p17 variants with certain amino acid insertions in the C-terminal region promote B-cell growth initially makes it tempting to ascribe this biological activity to the C terminus of the protein interacting directly with its putative receptors. However, we have previously shown that the entire alpha helix H5 is dispensable with respect to vp17 functionality (13). In fact, truncation of the last 36 amino acid residues of refp17 (refp17 Δ36) resulted in a protein that enhanced B-cell clonogenicity through up-regulation of the Akt signaling pathway. Compared with refp17, the C-terminally truncated variant is partially unfolded, drastically destabilized, and extremely prone to precipitation in solution. These properties of refp17 Δ36 suggest that protein destabilization, induced by mutations (deletions and insertions included) not restricted to the C terminus of p17, may ensue a conformational change sufficient to endow the viral protein with B-cell growth-promoting activity presumably through better receptor recruitment and activation. Our findings on the positive Ala–Ala insertion variant and negative Gly–Asn insertion variant of refp17 appear consistent with this tenet.
According to our data, insertion of an Ala–Ala stretch at position 117–118 may be considered as one of the possible molecular signatures identifying vp17 proteins with enhanced B-cell growth-promoting and clonogenic activity. Analysis of subtype B sequences in the Los Alamos HIV Database documented that the Ala–Ala insertion reported here is frequent (5.7%), followed by the insertion of three Ala (5.1%) and a single Ala (4.6%) at this location. Extending the current analysis to determine the impacts of these different types of insertion on p17 stability and to further study their association with lymphoma in both a cross-sectional and longitudinal study will contribute to our understanding of the potential of such sequence analysis to predicting which p17 alterations help identify HIV+ patients at risk for lymphomas.
Recent data have highlighted the capability of p17 to also promote both angiogenesis (25, 26) and lymphangiogenesis (26, 27), which are essential in supporting proliferation and survival of lymphoma, as well as tumor cell dissemination (28). Altogether, these evidences corroborate the hypothesis that p17 may play a key role in producing a microenvironment that fosters lymphoma development, progression, and metastasis.
In conclusion, our results suggest that vp17s may have a key role in sustaining B-cell growth and transformation, thus offering new opportunities to develop preventive and/or treatment strategies in combating HIV–NHLs.
Materials and Methods
This project has been approved by the Institutional Review Board (IRB) of the Centro di Ricerca Oncologica of Aviano Pordenone (PN), Italy. An IRB exemption was obtained from the University of Maryland, Baltimore. Samples provided by the National Cancer Institute supported AIDS Malignancy Repository were collected with informed consent at various US cancer centers. For a complete description of the source of materials and our methods, see SI Materials and Methods. It includes description of patients and procedures for Western blot analysis, p17 sequencing, and UDPS (15, 29). It also includes description of different recombinant monomeric LPS-free HIV p17 protein production, soft agar anchorage-independent B-cell growth assay, CD spectroscopy and thermal denaturation assay, and the statistical analysis performed (21, 30–32).
SI Materials and Methods
Patients.
This project, dealing on the presence of vp17s in plasma and tumor tissue of HIV+ patients with NHL and on the effect of NHL-derived vp17s on B-cell clonogenicity, has been approved by the Institutional Review Board of the Centro di Ricerca Oncologica (CRO) of Aviano Pordenone (PN), Italy. Samples provided by the National Cancer Institute supported AIDS Malignancy Repository were collected with informed consent at various US cancer centers. P17 gene sequences were analyzed comparatively in tumor tissue and plasma of five patients with HIV–NHL (group A), in plasma of five patients without NHL treated at the CRO of Aviano, and also in plasma of seven patients with HIV–NHL (group B) obtained from ACSR, funded by the National Cancer Institute. To perform UDPS on genomes from circulating virions, plasma samples from HIV+ patients with HIV-1–RNA values of at least 10,000 copies per mL were selected. These included plasma of 18 HIV+ patients affected by NHL (group C) collected and stored at the CRO of Aviano and 18 HIV+ subjects without NHL (group D) we previously described for the expression of vp17 quasispecies in plasma (15). All patients were infected with HIV-1 subtype B, as determined by the sequence analysis of p17 region, according to the Los Alamos genotyping algorithm. Clinical and virological characteristics of patients belonging to the four different groups are reported in Table S1. It is worth noting that one limitation of the study is that patients are not matched for geographical origin, and some data on clinical and virological features of group B patients were not available.
P17 Gene Sequencing.
Because, at the time of sampling, all group A and B patients showed a low-level viremia, we used a nested PCR approach amplifying a 559-nucleotide (nt)-long amplicon (from nt 326 to nt 885) to evaluate the whole p17 coding sequence. RNA was extracted from plasma and tumor samples using the Qiamp viral RNA Kit (Qiagen). RNA was then retrotranscribed, and cDNAs, as well as genomic DNAs, were thermal-cycled using the GoTaq polymerase master mix (Promega) as follows: 4 µL of genomic DNA in a total volume of 50 µL, containing each external primer at 0.3 µM (UGF1, 5′-GTGCCCGTCTGTTGTGTG; p24R1, 5′-TAACATTTGCATAGCTGCTT), were subjected to a denaturation step (5 min) followed by 40 cycles of PCR (95 °C for 45 s, 56 °C for 45 s, and 72 °C for 60 s) and a final extension step (7 min). We subjected 5 µL of PCR products to a second round of amplification (95 °C for 45 s, 58 °C for 45 s, and 72 °C for 60 for 35 cycles) in a total volume of 60 µL containing each internal primer at 0.25 µM (p17F1, 5′-AAGGAGAGAGATGGGTGCGA; UGR1, 5′-AATCTTGTGGGGTGGCTCCTT). Amplicons were analyzed on a 1.5% agarose gel, purified using the PCR Clean-Up System (Promega), and sequenced by BMR Genomics.
Recombinant HIV-1 p17 Proteins.
Purified endotoxin (LPS)-free recombinant HIV-1-matrix protein p17s (in their monomeric form) were produced as previously described (30). The absence of LPS contamination (<0.25 endotoxin units per mL) in protein preparation was assessed by Limulus amoebocyte assay (Associates of Cape Cod Inc.).
UDPS and Data Analysis.
Plasma HIV-1 RNA extraction and quantification were performed as previously described (15). As regards bioinformatic analysis, reads shorter than 360 nts were excluded, short InDels were removed, and all correct reads were translated and then clusterized by cd-hit program (31). Amino acid sequences, aligned using muscle software, were trimmed to cover positions 5–125 (121 amino acids). Considering the corrected error rate that we had previously calculated for p17 amplicon (i.e., 0.04%), a sensitivity threshold of 0.4% (i.e., a value exceeding by at least 10-fold the error rate) was adopted. All residue changes detected with frequency below this threshold after the correction pipeline were ignored. For the stretch 5–113, the analysis was performed exactly as previously described (15). For the stretch 114–125, a further clusterization step was adopted to overcome the extreme variability observed in this portion of p17 that prevented correct alignment. After this additional clusterization, only clusters with frequency >0.4% were considered. After these steps, ambiguous reads were manually corrected. The intrapatient amino acid diversity was calculated as previously described (15) only on the stretch encompassing residues 5–113, whereas the stretch 114–125 was not considered in the calculation of sequence diversity, due to the quasispecies simplification introduced by the additional clustering.
Cell Cultures and Recombinant Proteins.
Human lymphoma B-cell lines Raji and BJAB were cultured in RPMI 1640 containing 10% (vol/vol) FBS. The coding sequence of HIV-1 isolate BH10 p17 (refp17; amino acids 1–132) was amplified by PCR with specific primers (30) that allowed us to clone the refp17 sequence into the BamH1 site of the prokaryotic expression vector pGEX-2T (Pharmacia). The vp17s obtained from the lymphoma biopsy of patient a101, a102, a103, a104, and a105 (NHL-a101, NHL-a102, NHL-a103, NHL-a104, and NHL-a105 vp17s, respectively); from the blood of patient b106 with HIV–NHL (NHL-b106); and from the blood of patients without NHL (d205, d206, d210, d212, and d215), were amplified by PCR and cloned into BamH1 and XhoI sites of the same vector. Specific stretches of Ala–Ala and Gly–Asn were inserted within the refp17 primary sequence between amino acids Gln and Ala at position 117–118 and between amino acids Ser–Ser at position 125–126, respectively, by using the Quick Change Site-Directed Mutagenesis Kit (Stratagene) and cloned in the expression vector as above. The recombinant proteins were further purified (>98%) by reverse-phase fast performance liquid chromatography. The absence of endotoxin contamination (<0.25 endotoxin U/mL) in the protein preparation was assessed by Limulus amoebocyte assay (Associates of Cape Cod Inc.).
Western Blot Analysis.
Raji cells were starved for 24 h by serum deprivation (RPMI containing 1 mM l-glutamine and 0.5% FBS). Raji cells (7 × 105/mL) were treated with recombinant refp17 or vp17 NHL-a101, NHL-a102, NHL-a103, NHL-a104, NHL-a105, and NHL-b106 at different concentrations (0.05, 0.1, and 0.5 μg/mL) and lysed in 200 μL of 10 mM Hepes (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM EDTA, and 0.6% Nonidet P-40, containing a mixture of protease inhibitors (Complete Mini Roche) and phosphatase inhibitors (sodium vanadate, PAO, and sodium fluoride). Equal amounts of total protein were resolved on a 12% SDS-polyacrylamide gel and then electroblotted onto a nitrocellulose membrane. The blots were incubated overnight at 4 °C with a mouse monoclonal antibody to pAkt (Cell Signaling Technology), a mouse monoclonal antibody to Akt (Cell Signaling Technology), a rabbit monoclonal antibody to pPTEN (Ser380/Thr382/383) (Cell Signaling Technology), or a mouse monoclonal antibody to GAPDH (Santa Cruz Biotechnology). The antigen–antibody complex was detected by incubation of the membranes for 1 h at room temperature with peroxidase-coupled goat anti-rabbit IgG or goat anti-mouse IgG (Thermo Scientific) and revealed using the ECL System.
Soft Agar Anchorage-Independent Growth Assay.
Raji cells (12,500 per well) and BJAB (2,000 per well) were plated in 12-well plates in 2 mL RPMI containing 5% FBS and 0.35% and 0.5% SeaPlaque agarose (Lonza), respectively, over a 0.7% agarose base. One day after plating, medium containing treatments, as indicated, was added to the top of the layer and replaced every 4 d. After 10 d, 300 μL of MTT (Sigma-Aldrich) were added to each well and allowed to incubate for 4 h at 37 °C. Plates were then placed overnight at 4 °C, and colonies >50 μm in diameter were counted.
CD Spectroscopy and Thermal Denaturation.
Recombinant proteins were quantified by UV absorbance measurements at 280 nm using calculated molar extinction coefficients as described (32). CD spectra of p17 proteins at 2.5 μM in 10 mM phosphate buffer (pH 7.4) were obtained at 25 °C and 37 °C on a Jasco J-810 spectropolorimeter using a 1-mm quartz cuvette. Protein thermal denaturation was carried out on the Jasco spectropolorimeter equipped with a temperature controller. We aliquoted 2.5 mL of protein solution (10 μM) prepared in PBS, pH 7.4, into a 3 mL cuvette. Under constant stirring, CD measurements at 222 nm were made at a 1° interval between 25 °C and 90 °C, at a heating rate of 1 °C per minute. After each 1-min heating, the solution in the cuvette waited for 20 s before signals were sampled over a 16-s period. Heating and data acquisition were fully automated with control software provided by Jasco. Denaturation data were fitted to a six-parameter equation derived from a two-state protein denaturation model (21).
Statistical Analysis.
Data obtained from multiple independent experiments are expressed as the mean ± SD. Data were analyzed for statistical significance using the Student’s two-tailed t test, one-way ANOVA, or two-way ANOVA when appropriate. Bonferroni’s posttest was used to compare data. The Mann–Whitney U test and χ2 tests (or Fisher’s exact test when applicable) were used for univariate analysis, as appropriate. To evaluate correlations, the Spearman’s rank correlation coefficient was calculated. Statistical significance threshold was set at *P < 0.05.
Acknowledgments
The authors thank Dr. Man Charurat, Dr. Alfredo Garzino-Demo, Ms. Joyce Johnson, and Dr. Fabio Romerio for careful reviews of the manuscript. The authors acknowledge the contribution of biological samples and clinical data by the ACSR, funded by the National Cancer Institute under Grant UM1CA181255. The authors thank the Dean, Dr. Albert E. Reece, University of Maryland School of Medicine for the support of this work through the Deans Challenge Award to Accelerate Innovation and Discovery in Medicine and the Director of the Greenebaum Cancer Center for funding support through the Cigarette Restitution Fund Pilot Grant Program. This study was supported in part by Associazione Italiana per la Ricerca sul Cancro Grant 14287 (to R.D.) and by the Italian Ministry of Health (Ricerca Corrente e Finalizzata). E.M. was supported by a Fondazione Umberto Veronesi Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514748112/-/DCSupplemental.
References
- 1.Shiels MS, et al. Cancer burden in the HIV-infected population in the United States. J Natl Cancer Inst. 2011;103(9):753–762. doi: 10.1093/jnci/djr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandenhende MA, et al. ANRS EN20 Mortalité 2010 study group Cancer-related causes of death among HIV-infected patients in France in 2010: Evolution since 2000. PLoS One. 2015;10(6):e0129550. doi: 10.1371/journal.pone.0129550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coté TR, et al. AIDS/Cancer Study Group Non-Hodgkin’s lymphoma among people with AIDS: Incidence, presentation and public health burden. Int J Cancer. 1997;73(5):645–650. doi: 10.1002/(sici)1097-0215(19971127)73:5<645::aid-ijc6>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 4.Gloghini A, Dolcetti R, Carbone A. Lymphomas occurring specifically in HIV-infected patients: From pathogenesis to pathology. Semin Cancer Biol. 2013;23(6):457–467. doi: 10.1016/j.semcancer.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Knowles DM. Etiology and pathogenesis of AIDS-related non-Hodgkin’s lymphoma. Hematol Oncol Clin North Am. 2003;17(3):785–820. doi: 10.1016/s0889-8588(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 6.Epeldegui M, Vendrame E, Martínez-Maza O. HIV-associated immune dysfunction and viral infection: Role in the pathogenesis of AIDS-related lymphoma. Immunol Res. 2010;48(1-3):72–83. doi: 10.1007/s12026-010-8168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez-Maza O, Breen EC. B-cell activation and lymphoma in patients with HIV. Curr Opin Oncol. 2002;14(5):528–532. doi: 10.1097/00001622-200209000-00009. [DOI] [PubMed] [Google Scholar]
- 8.da Silva SR, de Oliveira DE. HIV, EBV and KSHV: Viral cooperation in the pathogenesis of human malignancies. Cancer Lett. 2011;305(2):175–185. doi: 10.1016/j.canlet.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 9.He B, et al. HIV-1 envelope triggers polyclonal Ig class switch recombination through a CD40-independent mechanism involving BAFF and C-type lectin receptors. J Immunol. 2006;176(7):3931–3941. doi: 10.4049/jimmunol.176.7.3931. [DOI] [PubMed] [Google Scholar]
- 10.Kundu RK, et al. Expression of the human immunodeficiency virus-Tat gene in lymphoid tissues of transgenic mice is associated with B-cell lymphoma. Blood. 1999;94(1):275–282. [PubMed] [Google Scholar]
- 11.Freed EO. HIV-1 gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251(1):1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 12.Curreli S, et al. B cell lymphoma in HIV transgenic mice. Retrovirology. 2013;10:92. doi: 10.1186/1742-4690-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giagulli C, et al. Opposite effects of HIV-1 p17 variants on PTEN activation and cell growth in B cells. PLoS One. 2011;6(3):e17831. doi: 10.1371/journal.pone.0017831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caccuri F, et al. Simian immunodeficiency virus and human immunodeficiency virus type 1 matrix proteins specify different capabilities to modulate B cell growth. J Virol. 2014;88(10):5706–5717. doi: 10.1128/JVI.03142-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giombini E, et al. Detection of HIV-1 matrix protein p17 quasispecies variants in plasma of chronic HIV-1-infected patients by ultra-deep pyrosequencing. J Acquir Immune Defic Syndr. 2014;66(3):332–339. doi: 10.1097/QAI.0000000000000164. [DOI] [PubMed] [Google Scholar]
- 16.Popovic M, et al. Persistence of HIV-1 structural proteins and glycoproteins in lymph nodes of patients under highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2005;102(41):14807–14812. doi: 10.1073/pnas.0506857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim Biophys Acta. 2008;1784(1):150–158. doi: 10.1016/j.bbapap.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96(8):4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93(7):3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrabartty A, Kortemme T, Baldwin RL. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 1994;3(5):843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koepf EK, Petrassi HM, Sudol M, Kelly JW. WW: An isolated three-stranded antiparallel beta-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 1999;8(4):841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor JG, Liapis K, Gribben JG. The role of the tumor microenvironment in HIV-associated lymphomas. Biomarkers Med. 2015;9(5):473–482. doi: 10.2217/bmm.15.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martorelli D, et al. A natural HIV p17 protein variant up-regulates the LMP-1 EBV oncoprotein and promotes the growth of EBV-infected B-lymphocytes: Implications for EBV-driven lymphomagenesis in the HIV setting. Int J Cancer. 2015;137(6):1374–1385. doi: 10.1002/ijc.29494. [DOI] [PubMed] [Google Scholar]
- 24.Goh GK, Dunker AK, Uversky VN. A comparative analysis of viral matrix proteins using disorder predictors. Virol J. 2008;5:126. doi: 10.1186/1743-422X-5-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caccuri F, et al. HIV-1 matrix protein p17 promotes angiogenesis via chemokine receptors CXCR1 and CXCR2. Proc Natl Acad Sci USA. 2012;109(36):14580–14585. doi: 10.1073/pnas.1206605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basta D, et al. Angiogenic, lymphangiogenic and adipogenic effects of HIV-1 matrix protein p17. Pathog Dis. 2015;73(8):ftv062. doi: 10.1093/femspd/ftv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caccuri F, et al. HIV-1 matrix protein p17 promotes lymphangiogenesis and activates the endothelin-1/endothelin B receptor axis. Arterioscler Thromb Vasc Biol. 2014;34(4):846–856. doi: 10.1161/ATVBAHA.113.302478. [DOI] [PubMed] [Google Scholar]
- 28.Pepper MS, Tille JC, Nisato R, Skobe M. Lymphangiogenesis and tumor metastasis. Cell Tissue Res. 2003;314(1):167–177. doi: 10.1007/s00441-003-0748-7. [DOI] [PubMed] [Google Scholar]
- 29.Pratesi C, et al. γ-Herpesvirus load as surrogate marker of early death in HIV-1 lymphoma patients submitted to high dose chemotherapy and autologous peripheral blood stem cell transplantation. PLoS One. 2015;10(2):e0116887. doi: 10.1371/journal.pone.0116887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Francesco MA, et al. HIV-1 matrix protein p17 increases the production of proinflammatory cytokines and counteracts IL-4 activity by binding to a cellular receptor. Proc Natl Acad Sci USA. 2002;99(15):9972–9977. doi: 10.1073/pnas.142274699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 32.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4(11):2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]