Significance
The conduction of protons through the highly restricted paths of transmembrane proteins is an essential process of living systems and an intriguing problem in modern physical chemistry. The small size of the influenza M2 proton channel makes it an ideal system for the study of proton transport across a membrane. Additionally, the M2 channel has medical relevance as an anti-flu drug target. These high-resolution structures of the channel were obtained by crystallizing the protein in a membrane-like environment and reveal networks of hydrogen-bonded waters that change with temperature and pH. The locations of these waters, in conjunction with molecular dynamics simulations that predict their hydrogen bond orientations, provide insight into the mechanism of proton stabilization and transduction within the channel.
Keywords: proton channels, influenza M2 channel, membrane proteins
Abstract
The matrix 2 (M2) protein from influenza A virus is a proton channel that uses His37 as a selectivity filter. Here we report high-resolution (1.10 Å) cryogenic crystallographic structures of the transmembrane domain of M2 at low and high pH. These structures reveal that waters within the pore form hydrogen-bonded networks or “water wires” spanning 17 Å from the channel entrance to His37. Pore-lining carbonyl groups are well situated to stabilize hydronium via second-shell interactions involving bridging water molecules. In addition, room temperature crystallographic structures indicate that water becomes increasingly fluid with increasing temperature and decreasing pH, despite the higher electrostatic field. Complementary molecular dynamics simulations reveal a collective switch of hydrogen bond orientations that can contribute to the directionality of proton flux as His37 is dynamically protonated and deprotonated in the conduction cycle.
Proton transport and conduction is essential to life. Proteins conduct protons over long distances through membranes to facilitate proton-coupled electron transfer and the formation and utilization of proton gradients. The M2 proton channel from the influenza A virus (1) is not only a medically important protein but also a simple, well-defined system for studying proton transport through confined spaces (2–4). This channel is the target of the anti-flu drug amantadine. M2 is activated at low pH by protonation of His37, which also participates in proton conduction by shuttling protons into the interior of the virus (5–7). His37 lies near the center of the bilayer, where it is connected to the viral exterior by a water-filled pore through which protons must pass to gain access to the viral interior (8–13).
Visualizing the flow of protons within protein channels such as M2 is one of the long-standing challenges in molecular biophysics. Based on computational studies (9, 14–19) it has been suggested that protons reach His37 through “water wires” via the Grotthuss mechanism, but there is little high-resolution information concerning the path by which protons are conducted. A previously solved 1.65-Å crystal structure (9) showed six ordered waters immediately above the His37 tetrad, but ordered waters spanning the entire aqueous pore of M2 have not been observed until now. Previous MD simulations suggested a pore with mobile waters (12, 15), whereas the results of NMR and IR experiments are more consistent with an environment that is more similar to bulk water at low pH (13, 19, 20). However, it is difficult to deconvolute the changes in the water structure and dynamics when the protonation of His37 is raised from those induced indirectly via the conformation of the protein’s main chain.
The M2 channel is known to have at least two conformational states that are populated to differing extents at low versus high pH (1, 10, 12). One, seen primarily at high pH, has been characterized extensively by solution NMR (21, 22), solid-state NMR (SSNMR) (10, 12), and X-ray crystallography (9). A second form is observed in dynamic equilibrium at lower pH (21–23), as evidenced by a broadening of magnetic resonances that thus far has made it impractical to determine a high-resolution structure of the protein in this state by SSNMR or solution NMR. X-ray crystallographic studies, however, have provided structures of both states (8, 9), which differ primarily in the conformation of the C terminus where protons exit the channel. Here we have obtained crystals that diffract to high resolution (1.10 Å) at both low and high pH, allowing visualization of water wires leading to His37 as a function of pH. The conformations of the backbone at the two pH values are essentially identical, permitting us to isolate changes in the organization of the water without any confounding factors.
Results and Discussion
High-Resolution Structures of M2TM.
In this work, we use a minimal construct, M2TM, which is fully active as a proton channel (24). M2TM was crystallized using lipidic cubic phase (LCP) techniques, which provide a continuous lipid bilayer in which crystallization can occur (25). Indeed, the channel is surrounded by a layer of monoolein molecules, and very few protein–protein interactions are present in the crystal lattice (SI Appendix, Fig. S1). The conformation of the protein (Fig. 1A) lies between the C-terminally closed conformation previously seen in several crystal and NMR structures (9, 10, 12, 21, 22) and a conformation with a dilation of the C-terminal end of the pore seen in some crystal structures (8). The primary focus of the current study is on the water-filled pore leading to His37, whose backbone structure is very similar in all previously solved structures of M2, including a structure of a longer construct determined by SSNMR in phospholipid bilayers (12) (Fig. 1A). However, the current structures include a complete set of water molecules in the pore and were resolved at exceptional resolution (1.1 Å); fewer than 10 structures of membrane proteins have been defined to less than a 1.2-Å resolution, and the present structures are also, to our knowledge, among the first high-resolution structures of a membrane protein near room temperature. Because of their high resolution, these structures provide novel insights into the water molecules that work in concert with the protein macromolecule to affect biological proton transport.
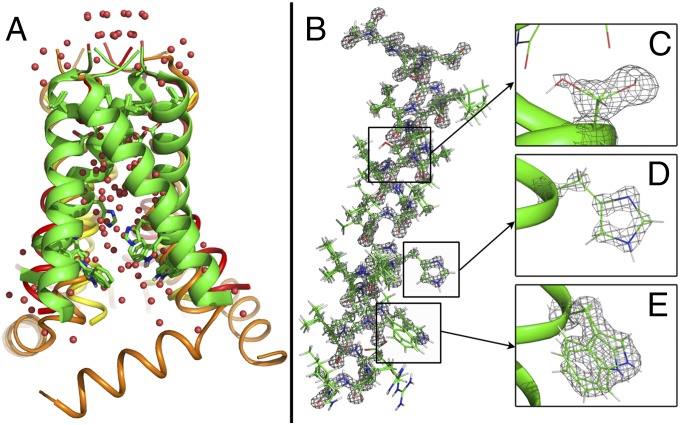
Fig. 1.
Crystal structure of M2TM at 1.10 Å at pH 8.0 (4QK7) under cryogenic conditions. (A) Superposition of this structure (green) with previously solved structures of M2 2LOJ (12) (orange), 3LBW (9) (yellow), and 3BKD (8) (red) shows that the conformation of the N-terminal half of the channel is relatively conserved whereas the conformation of the C terminus varies. Waters from the high-pH cryogenic structure (red spheres) are overlaid with waters from the low-pH cryogenic structure (pink spheres). (B) Density of backbone and side chains of a monomer of M2TM at a contour of 3.0 σ shows features typical of high-resolution structures. (C) Ser31 side chain (conformer A and B) at a contour of 1.0 σ. (D) His37 side chain at a contour of 3.0 σ. (E) Trp41 side chain (conformer A and B) at a contour of 1.5 σ.
Crystals were grown at pH 8.0 (“high pH”) and 5.5 (“low pH”) and crystallographic data were collected at cryogenic and room temperatures (SI Appendix, Table S1). Although the conformations of the backbone and even most side chains are essentially identical between the structures, there are marked differences in the distribution and occupancy of solvent molecules as a function of pH and temperature throughout the channel in the N-terminal pore leading to His37. The cryo-cooled structures were solved to a 1.10-Å resolution and show electron density maps typical of high-resolution structures, including features from the hydrogen atoms of CH bonds (Fig. 1 B–E). As in a previous crystallographic structure (9) and consistent with SSNMR (10, 11), water molecules bind to both the delta and epsilon nitrogens of His37. The N-terminal part of the pore is occluded by four Val27 residues, but it is possible for protons entering from the outside of the virus to gain access to water in the pore via low-energy fluctuations of the Val27 residues (19, 26).
A Multitude of Water Wires Lead to His37.
The cryo structures determined at high pH (4QK7) and low pH (4QKC) show multiple continuous paths of water molecules stretching ∼17 Å from Val27 to His37 (Fig. 2). Some water molecules are found at full occupancy, whereas others are found at one of two closely adjoining sites that together add up to full occupancy. The water wires have multiple branching points that create multiple hydrogen bond pathways by which a proton can pass through the pore en route to one of the four His37 residues. Thus, the low-temperature structures (∼100 K) identify a highly degenerate path of enthalpically stable (27) water wires.
Fig. 2.
Waters in the cryogenic crystal structures form ordered water wires whereas room temperature waters are less ordered. The front helix of each tetramer has been removed. (Top) Water wires inferred from crystallographic water positions from the 1.10-Å resolution cryo crystal structures at pH 8.0 (Left, PDB code 4QK7) and pH 5.5 (Right, PDB code 4QKC) form a continuous network of water wires within the N-terminal half of the pore leading to the gating His37 residue, with some positional changes in the waters between the two pH conditions. Alternate occupancy water-wire networks A (light blue) and B (dark blue) intersect at full occupancy waters (red). Continuous water wires span 16.8 Å of the channel pore. (Bottom) Water networks observed under room temperature conditions at pH 8.0 at a 1.71-Å resolution (Left, PDB code 4QKL) and pH 5.5 at a 1.44-Å resolution (Right, PDB code 4QKM) have fewer ordered waters and no longer form a continuous path leading to His37; in the low-pH structure, the waters do not form ordered networks at all and instead appear fluid-like.
Fluid Water Wires at Room Temperature.
To determine whether additional paths would be present near room temperature, two additional structures were determined at 273 K at high pH (4QKL) and low pH (4QKM) to 1.71- and 1.44-Å resolution, respectively. The electron density maps for the room temperature conditions versus the cryo temperature conditions, calculated to a common resolution, show a number of striking similarities and also differences in electron density originating from water in the pore (Fig. 3 A and B; SI Appendix, Fig. S2). Water molecules remain bound to the highly conserved His37 residue, but higher variability is seen across the other pore-lining water molecules. At pH 8, a clathrate-like structure is observed below Val27 and is anchored by hydrogen bonds to the main chain carbonyls facing the pore, but the water layer connecting to the hydrated His37 residues has become too diffuse to be seen clearly in the electron density. At pH 5.5, the water density in most of the pore is diffuse and a fully connected hydrogen bond network can not be observed. The water in this region is disordered, indicating that the wires seen at low temperature represent snapshots of the energetically most favorable pathways among a larger ensemble of dynamically exchanging sites populated at room temperature.
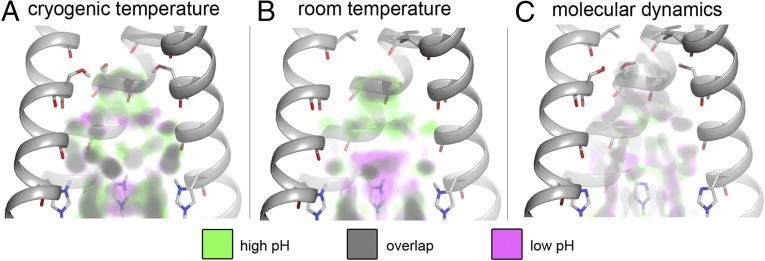
Fig. 3.
Electron density comparisons show differences and similarities between the solvent density at high and low pH. The front helices have been removed. Electron density at a contour of 1.5 sigma under (A) cryogenic conditions, (B) room temperature conditions, and (C) calculated electron density from molecular dynamics simulations is shown for both high (green) and low (magenta) pH experiments/simulations (high pH corresponds to a neutral His37 cluster, and low pH to a 4+ charge state). Areas where low- and high-pH densities overlap are shown in gray. Electron density from water binding to carbonyl groups and histidines is relatively conserved under all conditions whereas variations in density are seen elsewhere.
An Abundance of Hydrogen-Bond Donors Relative to Acceptors in the Channel.
The pore of the protein is richer overall in hydrogen bond acceptors than in donors. This presents an environment that should intrinsically stabilize a hydronium ion, which has three hydrogen bond donors and only one relatively weak acceptor. The water molecules in the channel form hydrogen bonds with the accepting carbonyl groups of Gly34, Ala30, and Val27. The hydroxyl group of Ser31 donates an intramolecular hydrogen bond to a carbonyl in the helix, leaving its two electron lone pairs free to accept hydrogen bonds from water molecules in the pore. At high pH, the δ-nitrogen of His37 also presents a hydrogen bond acceptor (7, 11), although this residue switches to being a donor with protonation at low pH. Thus, the channel would appear to be intrinsically suited to stabilize hydronium in the pore, particularly when His37 residues are in the neutral state. This environment is consistent with the idea that M2-blocking drugs, such as amantadine and rimantadine, act as substrate analogs of the hydronium ion due to their positively charged ammonium groups and lack of hydrogen bond acceptors (28).
Density of Water by Molecular Dynamics Simulations.
To gain further insight into the water network inside the M2 pore and its response to pH changes (i.e., the protonation state of His37), we performed force field-based molecular dynamics (MD) simulations of the hydrated protein crystals (SI Appendix, Figs. S3–S5) at a temperature of 310 K as in other simulations of M2TM (9). Computational studies of different proteins have shown that MD simulations in a crystal environment yield water densities in good agreement with experimentally determined electron densities (29–32). Additionally, MD simulations can complement the X-ray data by providing an atomic picture of the water hydrogens and their H-bonding connectivity and dynamics, as recently shown for aquaporin (4). The highest calculated density of water is near His37 and the pore-facing carbonyls of residue 34 (Fig. 3), similarly to what is indicated by the present structures and by the previous X-ray structure of M2TM (9). In addition to these, simulations also predict the presence of water molecules at other positions (SI Appendix, Fig. S2), including molecules that are not in direct contact with the protein’s side chains. Although the measured and calculated densities bear many similarities, it is possible that the mismatches between the two may affect the calculation of the hydrogen positions. To assess this potential issue, we also performed control simulations with all water molecules restrained at their crystallographic positions: these yield nearly identical distributions of donors and acceptors of H-bonds as when water molecules are left free to diffuse in the pore (SI Appendix, Fig. S10).
Hydrogen Bond Network Within the M2 Pore.
The M2 proton channel has the interesting property of being an asymmetric conductor: it conducts protons from the outside to the inside of the virus when the outside pH is low (as in an acidifying endosome), but does not as easily conduct protons outward when the pH gradient is reversed. The current crystallographic structures and MD simulations provide a detailed molecular description for this behavior. As the protonation state of the His37 changes, the polarity of the H-bonds between water molecules becomes increasingly aligned with the axis of the channel (SI Appendix, Fig. S6 and Table S2). The total number of H-bonds is higher in the charge states of the histidine tetrad accessible at high pH (0 and 1+) than in the higher charge states populated at low pH (3+ and 4+). Nevertheless, the H-bonds in the high pH states do not adopt a preferred orientation, whereas in the low-pH states all H-bonds are highly directional. In the 3+ and 4+ charge states (Fig. 4C and SI Appendix, Fig. S7H), the H-bonds are oriented outward in the extraviral half of the pore and are oriented inward in the intraviral half. The change of polarity occurs at His37 (SI Appendix, Fig. S8). The protonated His can donate two H-bonds, one upward to the water molecules in the extraviral side of the pore and one downward to the water molecules in the intraviral side. In contrast, in the 0 and 1+ states (Fig. 4B and SI Appendix, Fig. S7E), the water molecules can form both outward- and inward-oriented H-bonds with their neighbors regardless of their position along the pore, and the flipping between the two orientations occurs in the subnanosecond timescale. Except for the immediate vicinity of His37, the high-pH pore exhibits only mild imbalances between outward- and inward-oriented H-bonds (SI Appendix, Fig. S6). In other words, the H-bond network inside the M2 pore can be described as an assembly of “loopback circuits” at high pH and as a single “parallel circuit” at low pH. The switch between the two configurations takes place at the 2+ charge state (SI Appendix, Fig. S7 F and G), which is most populated at neutral pH (7, 24).
Fig. 4.
Hydrogen bond orientation calculated from MD simulations. (A) Schematic representation of the water wires and the hydrogen bond orientation. The oxygens of water molecules observed in the room temperature X-ray structures are shown as van der Waals spheres (orange color) with diameter proportional to the crystallographic B-factors and transparency reflecting the partial occupancy. Hydrogen bonds are displayed as sticks with a color scale denoting their orientation and the thickness proportional to their population. (B) Water wires for a neutral channel. (C) Water wires at the 4+ charge state.
We compared this behavior with analogous simulations of the two most prevalent M2 mutants, S31N-M2TM and D44N-M2TM (33), in the same unit cell [S31N-M2TM crystallizes in the LCP with unit-cell parameters within 1 Å from those shown here (22), and the backbone of D44N-M2TM modeled by MD and NMR (34) is at less than 2 Å rmsd from the current structures]. S31N-M2TM also exhibits “loopback circuits” at high pH (SI Appendix, Figs. S13–S15), but D44N-M2TM features a “parallel circuit” configuration at all pH levels (SI Appendix, Figs. S19–S21). Electrophysiology experiments showed that WT-M2 and S31N-M2 have similar pH/current profiles (33), but the proton conductance of D44N-M2 saturates below pH 5 (34). Thus, the “loopback circuits” appear to correlate with the ability to promote the addition of protons onto a charged His37 cluster. A similar H-bond network structure was also seen for the water molecules adjacent to His37 in the previous 1.65-Å crystal structure of M2TM (9).
Hydrogen-Bond Donor/Acceptor Water Molecules Along the M2 Pore.
As posited above, the M2 pore-lining residues are expected to act preferentially as H-bond acceptors, thus providing the ideal environment to stabilize a hydronium ion. Our MD simulations show that, in response, the water molecules behave preferentially as H-bond donors along the 17-Å stretch between Val27 and His37 (SI Appendix, Fig. S9). There is a larger net number of water molecules acting as H-bond donors on the extraviral side of the M2 pore, irrespective of the charge state of the histidine tetrad. In contrast, the H-bonding population in the C-terminal part of the pore (between Trp41 and Leu46) is highly sensitive to pH. In the neutral state, there is only a marginal preference for the water molecules to act as donors in the direction of proton influx. As the charge on the His37 tetrad increases, the water molecules switch into H-bond acceptors so that they can establish H-bonds with the protonated His37, which can donate two H-bonds (Fig. 4 and SI Appendix, Table S2). These interactions are also seen in the S31N- and D44N-M2TM simulations (SI Appendix, Tables S3 and S4 and Figs. S16 and S22), indicating that no significant changes in the H-bonds between protein and water are brought about by naturally occurring mutations.
Conclusions.
It is interesting to compare the present studies of the M2 channel with previous studies of the water channel (4) and ammonia channel (3), both structures that have been solved at moderate to high resolution and are responsible for diffusion of solvent molecules. In the case of the ammonia and water channels, the conduction pore allows for diffusion of single molecules of neutral H2O or NH3 while excluding ammonium or hydronium ions. In contrast, the selectivity filter of the M2 channel is formed by a general acid/base, His37, which shuttles protons through the channel. The conduction path leading to this selectivity filter in M2 is markedly different from the ammonia and water channels in that it is broader and able to conduct protons through partially occupied water wires. Although there are only ∼16 water molecules at a given time in the path leading to His37, they are at partial occupancy at low temperature, giving rise to hundreds of potential paths conducive to proton conduction. The apparent fluid nature of the pore becomes even more apparent near room temperature, where the density suggests greater disorder, particularly at low pH. Moreover, examination of the structures and the predominant protonation state of His37 suggests that the overall collection of individual water molecules, although likely mobile, are on average highly polarized to favor hydrogen bond orientations that encourage entry of protons when the His37 residues are deprotonated; similarly, as His37 becomes more protonated, diffusion of protons into the channel might be encouraged. This dipolar switching might work in concert with rotameric switching to create the asymmetric proton conductance of the channel. Finally, the water molecules in the channel are surrounded by hydrogen bond-donating carbonyl groups, which should serve to stabilize hydronium through second-shell effects via bridging water molecules. It is encouraging that the present MD simulations in a crystal environment are consistent with experimental observations and add quantitative detail to these qualitative conclusions. Moreover, given the small size and biological relevance of the M2 channel, these structures should be ideally suited for more detailed simulations based on valence bond (15, 35) quantum and quantum mechanics/molecular mechanics simulations (14, 36) in which the formation and cleavage of covalent bonds associated with proton transfer can be examined in atomic detail.
Materials and Methods
Experimental Methods.
The peptide construct used in this study is M2(22-46) from influenza A/Udorn/307/1972. The sequence, the molecular weight of which is 2682.268 Da (average), is as follows: Ac-SSDPLVVAASIIGILHLILWILDRL-NH3. The peptide was synthesized manually at high temperature using Fmoc chemistry, cleaved from the resin and then purified using reverse-phase HPLC. The molecular weight of the desired product was confirmed using mass spectrometry, and the purity of the product was tested using analytical HPLC. The purified peptide was dissolved in ethanol and stored at −80 °C. The lipidic cubic phase was prepared with some modifications to the protocol described by Caffrey and Cherezov (38), and then crystallization conditions were screened in 96-well plastic sandwich plates. Square-shaped crystals belonging to space group I4 formed after 2–4 wk of incubation at 10 °C. These conditions were optimized to grow 20- to 120-µm crystals. The crystals used for data collection at cryogenic conditions were grown in 96-well plates at 10 °C and then harvested into liquid nitrogen. The crystals used for room temperature data were grown in 96-well plates at 20 °C and then were transported to the beam line at ambient temperature for harvesting and data collection.
All crystallographic data were collected at the Advanced Light Source on beam 8.3.1. The beam size was 100 µm for all data sets; the detector used was a 3 × 3 CCD array (ADSC Q315r). Data from the two cryo condition crystals that diffracted to 1.1 Å were collected at a temperature of <100 K with a 13.0 keV beam and a detector-to-sample distance of 125 mm. The high-pH cryo condition crystal (PDB entry 4QK7) was exposed to the beam for 6 s per frame, and the low-pH cryo condition crystal (PDB entry 4QKC) was exposed for 4 s per frame; both crystals were oscillated 1° during data collection. The room temperature data were collected at 273 K using room temperature data collection techniques (38) with a 11.111-keV beam and an aluminum foil attenuator for both data sets. The crystals accumulated radiation damage faster under room temperature diffraction conditions, so larger crystals (50–120 µm) had to be used to obtain complete data sets.The low-pH room temperature condition crystal (PDB entry 4QKM) was exposed to the beam for 3 s per frame with a detector-to-sample distance of 125 mm; the high-pH room temperature condition crystal (PDB entry 4QKL) was exposed to the beam for 1 s per frame with a detector-to-sample distance of 150 mm; both crystals were oscillated 1° during data collection.
Data processing was done in iMosflm (39). Phasing was done by molecular replacement in the Phenix suite using Phaser MR (40) with chain A from PDB entry 3C9J (8) as a search model, and then refinement was carried out in Phenix Refine (17). Protein model manipulation and addition of water and ions were done in Coot (20), and monoolein molecules were manually fit into Fo-Fc density using both Coot and PyMol (41). Alternate conformers were predicted using the qFit web server (25) and Ringer (42) and were also manually added where positive Fo-Fc density indicated that they were present. See SI Appendix, Table S1, for data processing and refinement statistics. Anisotropic B-factors were used for both cryo conditions (PDB entries 4QK7 and 4QKC) and the low-pH room temperature condition (PDB entry 4QKM) but not the high-pH room temperature condition (PDB entry 4QKL).
Molecular Dynamics Simulations.
Classical MD simulations of the hydrated protein crystal were performed to study the network of water molecules inside the M2 pore and its response to pH changes (i.e., the protonation state of His37). The initial configurations were built using the two cryo X-ray structures of the transmembrane region of the M2 bundle solved either at low pH (PDB entry 4QKC) or high pH (PDB entry 4QK7). To model the I4 symmetry of the crystal, two staggered tetramers were included in the simulation box (SI Appendix, Fig. S3), along with the crystallographically resolved water molecules. Simulations were performed starting from either of the solved structures for each of the five possible protonation states of the His37 tetrad (0, +1, +2, +3, and +4). The neutral histidine residues were set in the ε-tautomeric state, and the histidine charge was increased by protonating one, two (nonadjacent), three, or all four histidines, respectively, for each of the two tetramers. Each of the 10 resulting systems consists of ∼7,000 atoms. The simulations were performed in the NVT ensemble at 310 K using the measured crystal lattice dimensions (29.310 × 29.310 × 67.310 Å for the low-pH structure and 29.536 × 29.536 × 66.853 Å for the high pH); periodic boundary conditions were applied to mimic the crystal environment. The protein was modeled using the CHARMM force field (29, 30) with correlation maps (CMAP) corrections (32), and the water molecules were described using the TIP3P model (43). All MD simulations were carried out with the NAMD program (44), and a production trajectory of ∼350 ns was sampled for each system.
Analysis of the MD trajectories was performed over the last ∼250 ns of the MD trajectories and enforcing fourfold rotational symmetry. Water densities (Fig. 3A) were obtained from the MD simulations using the Volmap plugin (45) in the VMD program (46). Populations of hydrogen-bonded water molecules (Fig. 4 and SI Appendix, Fig. S7) were calculated using the following clustering protocol. A hydrogen-bond vector was defined between a donor and an acceptor when the two oxygen atoms were at a distance less than 3.5 Å and the donor-hydrogen-acceptor angle was between 150° and 180°. The clusters of these vectors were calculated using g_cluster (47) with a 1.5-Å cutoff. The occupancy of a hydrogen bond represented by the centroid of one cluster was obtained by dividing the population of the cluster by the total number of MD frames and by 4 (to normalize upon symmetrization), such that the occupancy of a given hydrogen bond was at most 1. A similar protocol was used in Gianti et al. (48).
Supplementary Material
Acknowledgments
Experimental work was funded by NIH Grant R01-GM056423. Use of the lipidic cubic phase crystallization robot was made possible by National Center for Research Resources Grant 1S10RR027234-01. All experimental data were collected at the Advanced Light Source on beam 8.3.1: University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR‐15‐328599 and the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation. Molecular dynamics simulations were performed using the Temple University High-Performance Computing System Owl’s Nest [purchased in part with National Science Foundation (NSF) Grant MRI-R2 0958854]. J.S.F. is a Searle Scholar and a Pew Scholar and is supported by NIH Grants OD009180 and GM110580 and by NSF Grant STC-1231306. R.A.W. is supported by a NSF Graduate Research Fellowship. We also acknowledge partial support from the NSF through Grants DMR-1120901 (to M.A.-P.) and CHE-1212416 (to G.F.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4QK7, 4QKC, 4QKM, and 4QKL.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518493112/-/DCSupplemental.
References
- 1.Hong M, DeGrado WF. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci. 2012;21(11):1620–1633. doi: 10.1002/pro.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hummer G, Rasaiah JC, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414(6860):188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 3.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science. 2004;305(5690):1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 4.Kosinska Eriksson U, et al. Subangstrom resolution X-ray structure details aquaporin-water interactions. Science. 2013;340(6138):1346–1349. doi: 10.1126/science.1234306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: A role for the transmembrane domain histidine residue. Biophys J. 1995;69(4):1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chizhmakov IV, et al. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494(Pt 2):329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, et al. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci USA. 2006;103(18):6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451(7178):596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acharya R, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci USA. 2010;107(34):15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu F, Schmidt-Rohr K, Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J Am Chem Soc. 2012;134(8):3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330(6003):505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma M, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo W, Hong M. Conformational changes of an ion channel detected through water-protein interactions using solid-state NMR spectroscopy. J Am Chem Soc. 2010;132(7):2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carnevale V, Fiorin G, Levine BG, DeGrado WF, Klein ML. Multiple proton confinement in the M2 channel from the influenza A virus. J Phys Chem C. 2010;114(48):20856–20863. doi: 10.1021/jp107431g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Wu Y, Voth GA. Proton transport behavior through the influenza A M2 channel: Insights from molecular simulation. Biophys J. 2007;93(10):3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei C, Pohorille A. Activation and proton transport mechanism in influenza A M2 channel. Biophys J. 2013;105(9):2036–2045. doi: 10.1016/j.bpj.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi M, Cross TA, Zhou HX. A secondary gate as a mechanism for inhibition of the M2 proton channel by amantadine. J Phys Chem B. 2008;112(27):7977–7979. doi: 10.1021/jp800171m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc Natl Acad Sci USA. 2011;108(15):6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451(7178):591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, et al. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc Natl Acad Sci USA. 2013;110(4):1315–1320. doi: 10.1073/pnas.1216526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768(12):3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma C, et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2009;106(30):12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landau EM, Rosenbusch JP. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci USA. 1996;93(25):14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khurana E, et al. Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc Natl Acad Sci USA. 2009;106(4):1069–1074. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halle B. Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc Natl Acad Sci USA. 2004;101(14):4793–4798. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, et al. Molecular dynamics simulation directed rational design of inhibitors targeting drug-resistant mutants of influenza A virus M2. J Am Chem Soc. 2011;133(32):12834–12841. doi: 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vangunsteren WF, Berendsen HJC, Hermans J, Hol WGJ, Postma JPM. Computer-simulation of the dynamics of hydrated protein crystals and its comparison with X-ray data. Proc Natl Acad Sci USA. 1983;80(14):4315–4319. doi: 10.1073/pnas.80.14.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P, Huang X, Zhou R, Berne BJ. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature. 2005;437(7055):159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 31.Cerutti DS, Le Trong I, Stenkamp RE, Lybrand TP. Simulations of a protein crystal: Explicit treatment of crystallization conditions links theory and experiment in the streptavidin-biotin complex. Biochemistry. 2008;47(46):12065–12077. doi: 10.1021/bi800894u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, Jiang J. Molecular dynamics simulations for water and ions in protein crystals. Langmuir. 2008;24(8):4215–4223. doi: 10.1021/la703591e. [DOI] [PubMed] [Google Scholar]
- 33.Balannik V, et al. Functional studies and modeling of pore-lining residue mutants of the influenza A virus M2 ion channel. Biochemistry. 2010;49(4):696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma C, et al. Asp44 stabilizes the Trp41 gate of the M2 proton channel of influenza A virus. Structure. 2013;21(11):2033–2041. doi: 10.1016/j.str.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang R, Li H, Swanson JM, Voth GA. Multiscale simulation reveals a multifaceted mechanism of proton permeation through the influenza A M2 proton channel. Proc Natl Acad Sci USA. 2014;111(26):9396–9401. doi: 10.1073/pnas.1401997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong H, Fiorin G, Degrado WF, Klein ML. Exploring histidine conformations in the M2 channel lumen of the influenza A virus at neutral pH via molecular simulations. J Phys Chem Lett. 2013;4(18):3067–3071. doi: 10.1021/jz401672h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4(5):706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraser JS, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108(39):16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277(42):39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 40.Lang PT, et al. Automated electron-density sampling reveals widespread conformational polymorphism in proteins. Protein Sci. 2010;19(7):1420–1431. doi: 10.1002/pro.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3r1.
- 42.van den Bedem H, Dhanik A, Latombe JC, Deacon AM. Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 10):1107–1117. doi: 10.1107/S0907444909030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 44.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gianti E, Carnevale V, DeGrado WF, Klein ML, Fiorin G. Hydrogen-bonded water molecules in the M2 channel of the influenza A virus guide the binding preferences of ammonium-based inhibitors. J Phys Chem B. 2015;119(3):1173–1183. doi: 10.1021/jp506807y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.