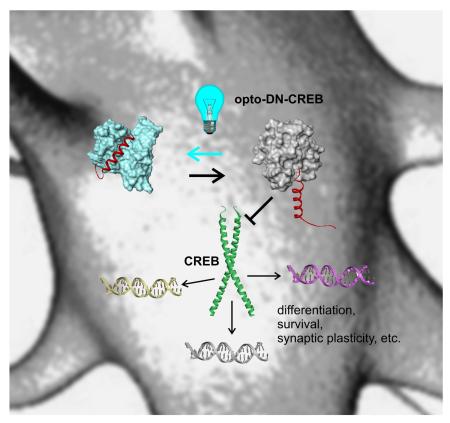
Summary
Current approaches for optogenetic control of transcription do not mimic the activity of endogenous transcription factors, which act at numerous sites in the genome in a complex interplay with other factors. Optogenetic control of dominant negative versions of endogenous transcription factors provides a mechanism for mimicking the natural regulation of gene expression. Here we describe opto-DN-CREB, a blue light controlled inhibitor of the transcription factor CREB created by fusing the dominant negative inhibitor A-CREB to photoactive yellow protein (PYP). A light driven conformational change in PYP prevents coiled-coil formation between A-CREB and CREB, thereby activating CREB. Optogenetic control of CREB function was characterized in vitro, in HEK293T cells, and in neurons where blue light enabled control of expression of the CREB targets NR4A2 and c-Fos. Dominant negative inhibitors exist for numerous transcription factors; linking these to optogenetic domains offers a general approach for spatiotemporal control of native transcriptional events.
Keywords: photo-control, photoactive yellow protein, PYP, optogenetics, photoisomerization, genetically encoded, CREB, endogenous, transcription factor, dominant negative
Graphical abstract
Introduction
The cyclic AMP response element binding protein (CREB) is a transcription factor that regulates the expression of genes involved in a diverse set of processes (Impey et al., 2004). A variety of extracellular signals trigger CREB activity via the well described cAMP signaling cascade (Altarejos and Montminy, 2011; Luo et al., 2012). CREB-dependent gene expression plays an important role in the central nervous system, where it regulates neuronal survival, memory consolidation, addiction, entrainment of the biological clock, and synaptic refinement (Impey et al., 2004). CREB has played a central role of the development of a molecular description of how memories are stored in the brain (Kandel, 2012; Zhou et al., 2009).
CREB activity is tightly regulated in time and space (Cowansage et al., 2013; Han et al., 2009; Lopez and Fan, 2013; Rogerson et al., 2014; Silva et al., 2009; Wu et al., 2001; Yiu et al., 2014; Zhou et al., 2009). For instance, spatiotemporal variations in CREB activity drive segmentation and posterior polarity specification in mammalian somitogenesis (Lopez and Fan, 2013). Cell to cell variations in CREB activity in the amygdala result in differential allocation of specific neurons to a fear memory engram (Cowansage et al., 2013; Han et al., 2009; Rogerson et al., 2014; Silva et al., 2009; Yiu et al., 2014; Zhou et al., 2009). The temporal dynamics of CREB activation may influence whether a fear memory is reconsolidated or undergoes extinction (Kim et al., 2014; Mamiya et al., 2009; Monfils et al., 2009).
Current optogenetic approaches for controlling transcription rely on targeting transcriptional activators or repressors to a specific gene loci chosen for external control (Konermann et al., 2013; Liu et al., 2012; Motta-Mena et al., 2014; Niopek et al., 2014; Polstein and Gersbach, 2015; Shimizu-Sato et al., 2002; Wang et al., 2012). While this approach can provide robust control of a given target gene, it does not adequately mimic transcriptional control by endogenous factors such as CREB. Approximately ~5000 functional CREB binding sites occur in promoter regions of human genes (Zhang et al., 2005) and CREB activation typically leads to transcriptional changes in ~100 of these in a given cell type (Altarejos and Montminy, 2011; Barco and Marie, 2011). The subset of responsive genes varies considerably between cell types (Zhang et al., 2005) and which genes becomes cAMP responsive in a given situation depends both on CREB and a family of co-activator proteins (Altarejos and Montminy, 2011). An optogenetic approach to probe CREB function, therefore needs to target the endogenous transcription factor directly.
A widely used approach for experimentally manipulating the activity of endogenous transcription factors with high specificity is the use of dominant negatives (Barco and Marie, 2011). For example, Vinson and colleagues developed acidic-CREB (A-CREB), a derivative of the CREB leucine zipper with an acidic N-terminal extension that traps the positively charged basic residues of CREB and prevents binding to DNA (Ahn et al., 1998). A-CREB is a highly specific dominant negative against the CREB family and has become a commonly used tool for studying CREB biology (Lopez and Fan, 2013; Zhang et al., 2005). Analogous dominant negatives have been engineered for other endogenous transcription factors including Myc/Max, (Krylov et al., 1997) AP-1, (Olive et al., 1997) C/EBP, (Chatterjee et al., 2011) and c-Jun/ATF2 (Yuan et al., 2009).
The challenge of conferring light-switchability on a given protein by fusing an optogenetic domain has been met in several cases with relatively little exploration of protein sequence space, implying that solutions to this protein engineering problem are relatively easy to find (Morgan et al., 2010; Motta-Mena et al., 2014; Strickland et al., 2008; Wu et al., 2009). We decided to use photoactive yellow protein (PYP), a small protein that functions in the phototactic swimming responses of the bacterium H. halophila, (Kumauchi et al., 2008) to confer light sensitivity on A-CREB. The N-terminal sequence of PYP shares rudimentary homology with leucine zipper sequences. Among optogenetic domains that have been structurally characterized, PYP undergoes relatively large changes in conformation and dynamics upon blue light irradiation. In addition, PYP is robust, reversibly folds and unfolds, and is tolerant of extensive mutagenesis (Kumar et al., 2013; Philip et al., 2010).
Here we report an A-CREB / PYP hybrid (designated opto-DN-CREB) that acts as a blue-light dependent CREB dominant negative. Using a combination of biochemical assays, we show that opto-DN-CREB provides effective optical control of CREB DNA binding in vitro, and of CREB signaling in HEK293T cells and primary cultured neurons.
Results
Protein Design
PYP senses light through the chromophore p-coumaric acid, which is covalently bound to the protein. Upon illumination with blue light, the p-coumaric acid chromophore in PYP undergoes trans to cis isomerization that leads to the formation of a highly dynamic, molten-globule-like, light-adapted state of the protein. The N-terminal cap (residues 1-25), in particular, undergoes significant rearrangement (Bernard et al., 2005; Ramachandran et al., 2011). How this large structural change is conveyed to an attached target protein depends on the amino acid sequence of the hybrid. Previously studied hybrids that merged the N-terminal region of PYP with the leucine zipper of GCN4 exhibited either enhanced or diminished target binding upon illumination, depending on the connection between the domains (Fan et al., 2011; Morgan et al., 2010; Morgan and Woolley, 2010). Here we used the dark state structure of PYP as a starting point to design a chimera in which the N-terminus of PYP was replaced by an A-CREB coiled-coil-like sequence, with the intent of producing a protein that was stable, folded, and underwent a normal photocycle, but had potential CREB-bZIP binding properties. To help define the amino acid sequence of the chimera, we employed the Rosetta software suite, (Leaver-Fay et al., 2011) which has been used extensively to guide the design of well-behaved proteins by avoiding amino acid sequences that would form internal cavities, buried polar interactions, or surface-exposed hydrophobic patches.
To maintain the interaction specificity of A-CREB toward CREB, it has been previously shown that keeping a/d/e/g positions of the coiled-coil is important; in contrast, the b/c/f positions can be varied (Grigoryan et al., 2009). The rudimentary sequence similarity between the N-terminal segment of PYP and leucine zippers suggested a unique register for the chimera (Fig. S1). Using Rosetta (see SI), we identified a new sequence that we designated opto-DN-CREB (Fig. 1, S1). This sequence differs from wild-type PYP at 29 positions, 20 of which are in the N-terminal region; the other residues are on the globular part of PYP, facing the N-terminal cap. The sequence is identical to A-CREB at all a/d/e/g positions of the coiled-coil, except for two, a Leu→Lys in an e position and a Phe→Leu in a g position; neither of these substitutions are expected to alter the interaction specificity (see SI)(Fong et al., 2004; Grigoryan et al., 2009; Newman and Keating, 2003). Mutations introduced in the globular core were designed to enhance packing or provide partners for buried polar sites.
Figure 1. Models of CREB bound to DNA and CREB inhibited by opto-DN-CREB.
(a) The bZIP domain of CREB (cyan ribbon) forms a dimeric parallel coiled-coil when bound to the DNA (solid surface) (PDBID: 1DH3). (b) A model of the designed protein opto-DN-CREB (shown in surface representation) bound to a single CREB-bZIP domain (cyan ribbon). Residues in the A-CREB-like N-terminal hybrid region of opto-DN-CREB that are critical for coiled-coil formation with the CREB zipper are colored cyan. Interactions between the acidic extension of A-CREB and the basic region of CREB are not modeled.
Characterization of opto-DN-CREB in vitro
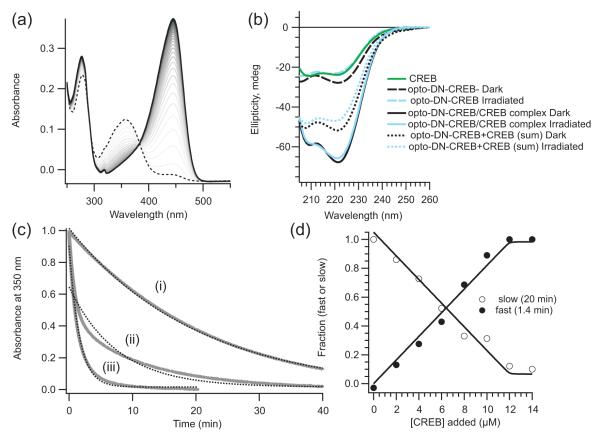
We expressed opto-DN-CREB in E. coli, purified it to homogeneity using nickel affinity followed by gel filtration chromatography, and characterized it using standard biophysical methods. The protein exhibited a UV-Vis spectrum characteristic of wild-type PYP (wtPYP) and underwent a normal photocycle upon exposure to blue light (Fig. 2a). The stability of the dark state of the protein was lower than wtPYP, as measured by its sensitivity to heat denaturation and the appearance of a band near 380 nm in the UV-Vis spectrum at elevated temperatures – the latter is typical of an altered protein conformation near the chromophore binding pocket (Joshi et al., 2009). The addition of an equimolar amount of CREB-bZIP stabilized the dark state of opto-DN-CREB according to both these measures, indicating that CREB-bZIP and opto-DN-CREB are forming a complex and that this interaction is sensed by the chromophore binding core of PYP. Circular dichroism spectra (Fig. 2b) indicated that the α-helical content is enhanced in the complex, consistent with coiled-coil formation. The addition of CREB-bZIP to opto-DN-CREB also affected the photocycle of opto-DN-CREB (Fig. 2c). After removal of the blue light source, opto-DN-CREB underwent thermal relaxation with a half-life of 20 min (at 20oC). The addition of CREB-bZIP produced a biexponential relaxation behavior with a faster component (1.4 min half-life) in addition to the 20 min process. The fraction of the faster process increased until CREB-bZIP was equimolar with opto-DN-CREB, at which point, only the fast relaxation process was observed (Fig. 2 c,d). This behavior is consistent with the formation of a tight 1:1 complex between opto-DN-CREB and CREB-bZIP. Enhanced thermal relaxation observed after the addition of the binding partner means that isomerization of the PYP chromophore and CREB-bZIP binding are energetically coupled and indicates that zipper formation lowers the barrier for reisomerization to the dark state (Morgan and Woolley, 2010).
Figure 2. Biophysical characterization of opto-DNA-CREB.
(a) UV-Vis spectra of opto-DN-CREB immediately after blue light irradiation (dotted line) then at 2 min intervals during the process of dark adaptation (gray lines). The dark-adapted spectrum is the solid black line. (b) CD spectra of opto-DN CREB and CREB-bZIP separately and together. The arithmetic sum of the separate spectra is also shown and exhibits significantly less negative ellipticity than the spectra obtained from the opto-DN CREB /CREB-bZIP mixture. (c) Time courses of thermal relaxation measured at 350 nm in the absence of CREB-bZIP (i), with 0.5 equivalents of CREB-bZIP (ii) and with 1.1 eq. of CREB-bZIP (iii). Solid lines are experimental data and dotted lines are single exponential fits to these data. In (ii), two components to the relaxation rate are clearly visible. (d) Relative fractions of fast and slow time constants extracted from global analysis of thermal relaxation data as a function of the concentration of added CREB-bZIP. The opto-DN-CREB concentration is 10 μM.
We conducted a series of experiments to gain further insight into the structural differences between opto-DN-CREB in dark-adapted and blue light irradiated states. At low concentrations (< 20 μM), both the light and dark states behaved as monomers in gel permeation chromatography (Fig. S2); a monomeric dark state for opto-DN-CREB at these lower concentrations was also confirmed by analytical ultracentrifugation (Fig S2). At higher concentrations (>100 μM), gel permeation chromatography revealed that opto-DN-CREB was self-associating (Fig S2). This is unsurprising given that the CREB zipper alone has been shown to self-associate at concentrations >100 μM (Santiago-Rivera et al., 1993). Notably, the self-association was more pronounced in the dark than under blue light illumination, suggesting that the A-CREB leucine zipper domain of opto-DN-CREB is more exposed in the dark state. (Fig. S2).
Chemical cross-linking experiments were conducted to test the hypothesis that the leucine zipper portion of opto-DN-CREB is more exposed in the dark state. Cross-linking opto-DN-CREB with bis (sulfosuccinimidyl) suberate (BS3) produced dimers and higher order oligomers in the dark, whereas very little cross-linking was observed under blue light (Fig. 3b). This result is consistent with self-association via a detached and helical conformation of the A-CREB domain that is adopted in the dark as depicted in Fig. 3a. Sequestration of the A-CREB portion of opto-DN-CREB by the PYP core in the light state reduces self-association and thereby cross-linking yield. Moreover, interaction with opto-DN CREB removes a stabilizing interaction between the A-CREB zipper and the light state, thereby enhancing thermal relaxation to the dark state (Fig. 2c,d).
Figure 3. Blue light dependent exposure of the zipper domain of opto-DN-CREB.
(a) Models of opto-DN-CREB derived from Rosetta design simulations. The PYP core is shown as a yellow surface. The A-CREB N-terminal cap is shown as a cyan ribbon. In the dark-adapted state (top), the A-CREB cap can be detached and helical. The detached helix can lead to a self-associated state that is trapped by chemical cross-linking. In the light state (lower model), the PYP core traps the A-CREB portion via an exposed hydrophobic patch. This state is shown schematically having the A-CREB N-terminal cap hidden. (b) Chemical cross-linking of opto-DN-CREB. SDS-PAGE gel showing oligomers (dimer, trimer, etc.) formed by crosslinking dark-adapted opto-DN-CREB (250 μM) with BS3 (5 mM). These species are formed to a much lesser extent when the reaction is carried out under blue light. (c) Schematic showing light dependent inhibition of CREB DNA binding by opto-DN-CREB. In the dark, the detached helical region A-CREB N-terminal cap can interact with the CREB-bZIP domain, thereby inhibiting full length CREB from binding to DNA. Full length CREB also contains glutamine rich domains (Q1, Q2) and a kinase inducible domain (KID). Blue light leads to sequestration of the A CREB region of opto-DN-CREB and permits normal CREB dimerization, DNA binding, and transcriptional regulation.
Rosetta simulations using the NMR-derived ensemble for the light state suggested that sequestration of A-CREB in the light state could be due to binding of the hydrophobic surface of the leucine zipper to a hydrophobic patch on the PYP domain, that is not available in the dark state (Fig. S1). Exposure of a hydrophobic surface in the light state of PYP has been detected previously by hydrophobic dye-binding studies (Hendriks et al., 2002).
Optical control of CREB-bZIP function in vitro
To assess the ability of opto-DN-CREB to bind CREB-bZIP and prevent CREB-bZIP from binding to DNA, we employed an electrophoretic mobility shift assay (EMSA) – a method commonly used to detect protein-DNA interactions. Initially, a fluorescently labeled DNA oligomer containing the CREB-bZIP (CRE) target binding site was electrophoresed in the presence or absence of CREB-bZIP. Mobility of the CREB-bZIP/DNA complex was slowed on the gel in comparison to free DNA. The apparent affinity of CREB-bZIP for the CRE oligo was 10 ± 2 nM (Fig. S3). The addition of increasing concentrations of opto-DN-CREB inhibited DNA binding by CREB-bZIP. In the dark, opto-DN-CREB exhibited an apparent Ki of 25 ± 10 nM (Fig. 4), similar to that of A-CREB alone (Fig. S3). Under blue light this inhibition was relieved – the apparent Ki was 500 ± 70 nM, which corresponds to a 20-fold increase in comparison to the Ki measured in the dark (Fig. 4). To confirm that the interaction with the CREB-bZIP protein was occurring through the leucine zipper of the A-CREB part of opto-DN-CREB, we tested the behavior of an opto-DN-CREB mutant in which two key Leu residues of the leucine zipper were mutated to Gly (Fig. 4, Fig. S1). This mutant showed a greatly reduced ability to inhibit CREB-bZIP DNA binding in the dark, and an even further reduced ability under blue light.
Figure 4. EMSA analysis of opto-DN-CREB inhibition of CREB-bZIP DNA binding.
(a) Increasing concentrations (indicated by the black wedge) of opto-DN-CREB (dark-adapted) prevent formation of the CREB-bZIP/CRE DNA complex (controls – free DNA, + CREB-bZIP bound DNA). (b) Much larger concentrations of opto-DN-CREB are required to inhibit CREB-bZIP DNA binding when the system is illuminated with blue light. Inhibition curves derived from 3 replicates of these data are shown at right (see SI for fitting). Solid dark line (dark-adapted, Ki 25 ± 10 nM), solid blue line (blue light illuminated, Ki 500 ± 70 nM), dashed line (thermally relaxed after blue light irradiation, Ki 30 ± 10 nM). (c) EMSA analysis using opto-DN-CREB (2Gly) under dark-adapted conditions and under blue light (d) shows much weaker inhibition. Solid dark line (dark-adapted, Ki 550 ± 90 nM), solid blue line (blue light illuminated, Ki 6 ± 1 μM).
Collectively our data support the model for opto-DN-CREB activity shown in Fig. 3c. In the dark, the A-CREB N-terminal cap of opto-DN-CREB can detach from the PYP core and can interact with the CREB-bZIP domain, thereby inhibiting CREB from binding to DNA. Blue light leads to sequestration of the A-CREB region of opto-DN-CREB and permits normal CREB dimerization and DNA binding
Optogenetic control of CREB signaling in living cells
In the bacterium H. halophila, the p-coumaric acid chromophore of PYP is produced biosynthetically from tyrosine through the action of tyrosine ammonia lyase (Kyndt et al., 2002). Activation of p-coumaric as a CoA thioester for loading onto PYP is catalyzed by p-coumaric acid ligase (Kyndt et al., 2003). We expected that the functional expression of this pair of enzymes in mammalian cells might require significant optimization. Since we were primarily interested in assaying the functionality of opto-DN-CREB in cell culture, we opted instead to form holo-PYP by adding pcoumaric acid thiophenyl ester to the culture medium. The compound is membrane permeable and reacts rapidly and selectively with apo-PYP both in vitro and when expressed in cell culture (Hori et al., 2013) and only hydrolyzes slowly (τ½ 6 h) in media.
We confirmed the functional reconstitution and photoswitching of the PYP optogenetic domain in mammalian cells using a reporter construct consisting of blue fluorescent protein (BFP) fused to PYP missing the N-terminal cap (BFP-Δ25PYP; Fig. S4). In the dark state, the blue fluorescence of BFP was quenched by PYP (Fig. S4). When PYP was switched to its light state, BFP quenching was relieved. Thus, light driven time-dependent changes in BFP fluorescence confirmed delivery of the p-coumaric acid thiophenylester and the resulting functionality of PYP (Fig. S4).
We systematically assayed the effects of added chromophore concentration and blue light intensity on cell viability using MTT assays (Cole et al., 2012) and morphological analysis to determine suitable working ranges for these parameters (Fig. S6).
To evaluate the ability of opto-DN-CREB to modulate CREB signaling in living cells we assessed CREB-mediated expression of the protein NR4A2 (nuclear receptor subfamily 4, group A, member 2), also known as nuclear receptor related 1 protein (NURR1). NR4A2 is important for the transcription-dependent consolidation process that converts a short-term memory into a long-term memory (Hawk et al., 2012). In addition, NR4A2 plays a key role in the maintenance of dopaminergic pathways in the brain (Sacchetti et al., 2006). NR4A2 is an ideal target because it is well established that CREB activation promotes its expression (summarized in the hESNET database (http://wanglab.ucsd.edu/star/hESnet/) (http://wanglab.ucsd.edu/star/hESnet/evidence.jsp?g1=CREB1&g2=NR4A2).
Opto-DN-CREB was expressed in HEK293T cells under control of the constitutive herpes simplex virus (HSV) - IE4/5 promoter via transfection with the pHSV vector. Approximately 40-48 h after transfection, activated chromophore was added and a subset of cells were irradiated with blue light for 60 min. Cells were then treated with forskolin for 2 h to activate cAMP signaling, and thereby CREB.
We directly assayed transcript levels of the CREB target gene nurr1 (coding for NR4A2) using quantitiative PCR using hprt1 (hypoxanthine-guanine phosphoribosyltransferase) as a control (Volpicelli et al., 2012). In control cells, forskolin treatment led to an increase in the level of nurr1, as expected (Fig. 5a). This increase occurred whether or not cells were irradiated with blue light (Fig. 5a). When cells were transfected with opto-DN-CREB and incubated with chromophore in the dark, the forskolin-mediated increase in nurr1 was strongly inhibited (Fig. 5a), indicating that opto-DN-CREB blocks CREB signaling in the dark. Under blue light irradiation (0.2 mW/cm2), this inhibition by opto-DN-CREB was completely relieved (Fig. 5a), exactly as occurred in vitro. These data confirm that opto-DN-CREB modulates CREB mediated transcription.
Figure 5.
(a) Forskolin treatment of HEK293T cells produces an increase in nurr1 that is blocked by opto-DN-CREB in the dark, but not under blue light. Changes in levels of nurr1 were determined by qPCR. Cells were transfected with control (empty) vector (empty bars) or with the opto-DN-CREB vector (filled bars). Data from cells illuminated with blue light are shown as blue bars; data from dark-adapted cells are shown as black bars. In control cells, forskolin treatment caused a large increase in nurr1 transcript levels (the hprt1 transcript was used as an internal control) (* p<0.05 using Student's t-test (unpaired) between dark and light condtions). In cells transfected with opto-DN-CREB and incubated in the dark, the forskolin-mediated increase seen in nurr1 levels seen with controls was prevented. Under blue light illumination (0.2 mW/cm2), this inhibition by opto-DN-CREB was relieved. Blue light alone did not affect levels of nurr1 under basal conditions or after forskolin stimulation.
(b) Forskolin treatment of HEK293T cells produces an increase in NR4A2 that is blocked by opto-DN-CREB in the dark, but not under blue light. Changes in levels of NR4A2 were determined by Western blots (see Fig. S6 for blots). Cells were transfected with control (empty) vector (empty bars) or with the opto-DN-CREB vector (filled bars). Data from cells illuminated with blue light are shown as blue bars; data from dark-adapted cells are shown as black bars. In cells transfected with opto-DN CREB and incubated in the dark, NR4A2 levels under basal conditions were decreased and the forskolin-mediated increase seen in controls was prevented. Under blue light illumination, this inhibition by opto-DN-CREB was relieved (* p<0.05 using Student's t-test (unpaired) between dark and light condtions). Blue light alone did not affect cellular levels of NR4A2 under basal conditions or after forskolin stimulation.
(c) Opto-DN-CREB inhibits forskolin and KCl mediated NR4A2 or c-Fos production in primary cortical neurons. This inhibition is relieved by blue light. Cells were treated with HSV-opto-DN CREB viral particles and exposed to blue light or dark-adapted (blue bars and black bars respectively). CREB signaling was stimulated in a subset of cells (stim) with forskolin and KCl. Cells were harvested and Western blots for NR4A2, c-Fos and β-actin (loading control) (Fig. S6) were quantified for cells without stimulation (unstim) and with stimulation (stim) (*, significant difference between S and US, p<0.05 using Student's t-test (unpaired), n=8-10 for each condition). With a control vector, increases in response to forskolin and KCl stimulation were observed as expected in both dark and under light illumination (not shown).
Optical modulation of nurr1 transcript levels by opto-DN-CREB are expected to lead to changes in levels of the NR4A2 protein. Following the same protocol described above, cells were collected, lysed, and the levels of NR4A2 were assayed via Western blotting (Fig. 5b). Again, in control cells forskolin treatment led to an increase in the level of NR4A2, as expected. This increase occurred whether or not the cells were irradiated with blue light (Fig. 5b). When cells were transfected with opto-DN-CREB and incubated with chromophore in the dark, NR4A2 levels under basal conditions were decreased and the forskolin-mediated increase was strongly inhibited, indicating that opto-DN-CREB blocks CREB signaling in the dark. Again, under blue light, this inhibition by opto-DN-CREB was completely relieved (Fig. 5b). The effect of opto-DN-CREB inhibition appears more pronounced on protein levels than on transcript levels.
To confirm that opto-DN-CREB was inhibiting CREB activity via the same mechanism as occurs in vitro, we tested the mutated version of opto-DN-CREB in which two Leu residues of the coiled-coil domain were mutated to Gly residues. When cells were transfected with this construct (opto-DN-CREB-2Gly), no inhibition of forskolin induced elevation of NR4A2 was observed, consistent with the much weaker interaction between opto-DN-CREB-2Gly and CREB (Fig. S6).
We then examined opto-DN-CREB function in cultured primary cortical neurons (Fig. 5c). Replication-deficient HSV viral vector amplicons were generated and used to infect primary cortical neurons obtained from E14 mice as previously described (Cole et al., 2012). After 8-10 days of growth in vitro, neurons were incubated with HSV-opto-DN-CREB viral particles. Five hours later the medium was replaced. One day later, cells were loaded with chromophore and then incubated for 1 h in the dark or under blue light before stimulating with 30 μM forskolin and 50 mM KCl. Cells were then harvested and processed for Western blotting as in the HEK293T cell experiments. In addition to assaying levels of NR4A2, we also measured effects on c-Fos, another well studied CREB target gene (Ahn et al., 1998). As observed in HEK293T cells, opto-DN-CREB inhibition of forskolin and KCl-mediated NR4A2 or c-Fos production is relieved by blue light (Fig. 5c). Collectively, these data demonstrate that opto-DNCREB provides a means for blue-light controlled dominant negative inhibition of endogenous CREB activity in cultured cells.
Discussion
This work demonstrates that coupling an optogenetic domain to a dominant negative inhibitor can produce a light-controlled tool for manipulating the activity of an endogenous transcription factor. The dominant negative used here (A-CREB) was designed to bind to the bZIP domain of CREB. Numerous analogous dominant negative inhibitors have been developed using the same design strategy against both bZIP-type and bHLH-type transcription factors (Krylov et al., 1997). In fact, the determinants of interaction specificity and affinity of coiled-coils are perhaps the best understood of any protein-protein interaction (Fletcher et al., 2013; Negron and Keating, 2014). Keating and colleagues have delineated the interaction specificities of the full complement of these factors in human cells (Newman and Keating, 2003). Thus, a similar approach to the one taken here could be used to produce light-controlled tools for manipulating a variety of endogenous transcription factors.
The choice of PYP as an optogenetic domain was made based on some rudimentary sequence homology to coiled-coils but also because PYP is a robust domain that undergoes large changes in conformational dynamics upon exposure to blue light. Our results indicate that a light state interaction between the PYP core and the leucine zipper of A-CREB is important for the function of opto-DNCREB. Rosetta simulations suggest that the light-adapted PYP core traps the A-CREB sequence folded as an α-helix and that the hydrophobic face of the helix interacts with an exposed hydrophobic patch on the PYP core. Rosetta simulations using NMR-derived light and dark state ensembles of the PYP domain may be used to guide the design of other light-switchable dominant negative inhibitors. Our experience here, and those of others, indicate that Rosetta protein design software is a powerful tool to guide the choice of sequences that produce folded, soluble proteins (Kaufmann et al., 2010). Based on the studies presented here, it appears that linking a binding sequence to PYP is likely to yield a functional switch in other cases as well. From the starting point of a folded, switchable protein with some affinity for its target, directed evolution or screening approaches could also be used to find improved switches (Guntas et al., 2014; Mazumder et al., 2015).
We have shown that opto-DN-CREB acts as a blue light-dependent modulator of CREB signaling in living neurons. Demonstration of blue light controlled activity on validated CREB targets implies that opto-DN-CREB could be used directly for experiments aimed at probing effects of spatiotemporal differences in CREB activity in cell culture or brain slice preparations. For example, the precise timing of CREB activation for chemically-induced long term potentiation (LTP) could be investigated in neuronal cell cultures. Incubation of cells in the dark would produce inactive opto-DNCREB / CREB complexes, similar to what occurs with A-CREB alone in neurons (Ahn et al., 1998). Blue light irradiation could then be used to release active CREB from this complex in targeted cells. Since conversion to the light state of opto-DN-CREB occurs in seconds at typical light intensities used for imaging (Fig. S4), the time frame at which CREB becomes functional in cells would then depend only on the time taken for free CREB to assemble into transcriptionally active complexes. If desired, opto-DN-CREB inhibition could be restored rapidly by a UV light pulse, or allowed to reoccur via thermal relaxation (in minutes per Fig. S4). As with channelrhodopsins, opto-DN-CREB, ideally when co-expressed with the genes required for chromophore biosynthesis, could also permit modulation of CREB activity in freely moving animals via delivery of blue light through fiber optic cables (Aravanis et al., 2007). This modality would enable an unprecedented level of spatiotemporal dissection of CREB mediated transcription in processes such as long term memory formation.
Experimental procedures
Design of opto-DN-CREB as guided by Rosetta modeling. Details of the sequence alignment performed and the overall design procedure are provided in the Supplemental Information (SI). To predict the specificity of coiled coil formation, we used the “bZIP Coiled-Coil Scoring Form” server (http://compbio.cs.princeton.edu/bzip/).
In vitro analysis of opto-DN-CREB
Opto-DN-CREB, the CREB bZIP domain and the A-CREB dominant negative were overexpressed and purified from bacteria using Ni-NTA affinity and size exclusion chromatography. Full details are provided in the SI. Spectral and kinetic measurements of optical switching of purified opto-DN-CREB were acquired using a PE Lambda 35 spectrophotometer with a Peltier temperature control unit. Circular dichroism was performed using an Olis RSM 1000 circular dichroism spectrophotometer. Self-association of purified opto-DN-CREB was assessed by size exclusion chromatography using a Superdex 75 10/300 GL column (GE Healthcare) running in tris-acetate-EDTA, 100 mM NaCl buffer (pH 7.5) at various protein concentrations and under dark and blue light irradiated conditions. Chemical cross-linking of purified opto-DN-CREB was carried out at a protein concentration of 250 μM in sodium phosphate buffer, pH 7.0. Solutions of dark-adapted or blue-light-irradiated opto-DN-CREB (~10 mW/cm2) were incubated in at 25oC in the presence of bis(sulfosuccinimidyl) suberate (BS3)(ThermoScientific) for 30 min. The reactions were quenched by adding 200 mM lysine. Samples were then analyzed by SDS-PAGE (10% polyacrylamide, 1 M Tris, 10% (w/v) glycerol).
Electrophilic Mobility Shift Assays (EMSA) of Opto-DN-CREB function
Binding of the CREB bZIP domain to a 28 base-pair target DNA duplex containing the CRE site was assayed using EMSA analysis as detailed in the SI. Protein and DNA samples were incubated then electrophoresed on an 8% polyacrylamide gel. The gels were scanned with a red laser on Pharos FX® plus molecular imager (Bio-Rad) and the images were recorded using QualityOne software (Bio-Rad). The extent of CREB bZIP CRE binding was quantified by analysis with Image Lab® software (Bio-Rad). Using Igor Pro software, three sets of data were averaged and fit to the Hill equation to determine the apparent Kd, i.e., the concentration of protein required for 50% binding to the CRE site. Inhibtion of CREB-bZIP binding by A-CREB or by opto-DN-CREB was assayed in a simailr manner. To assay under blue light irradiation, the sample preparation, incubation, and gel run were performed under cycles of 450 nm illumination composed of 3 minutes of irradiation followed by 2 minutes in the dark. An array of Luxeon III Star LED Royal Blue LEDS (455 nm), (overall ~50 mW/cm2), was used as the light source
Functional analysis of opto-DN-CREB in cells
The cDNAs for mammalian expression of A-CREB and opto-DN-CREB were synthesized (Biobasic, Inc.) and subcloned into the vector pHSV. Viability of transfected cells was assessed using Trypan blue assays, MTT assays and morpholgical analysis as descrined in the SI. Optical switching in of PYP in living HEK293 cells was initially assessed using a reporter construct in which BFP was fused to the N-terminus of 25-PYP via a flexible linker (Fig. S4). Excitation of BFP at 400 nm leads to fluorescence emission at 450 nm. Emission is partially quenched by dark-adapted PYP. The excitation wavelength also causes isomerization of PYP to its light state. This state does not quench BFP so effectively, leading to a time dependent increase in fluorescence emission when dark-adapted BFP-Δ25-PYP is exposed to blue light.
Modulation of Cre activity by opto-DN-CREB: qPCR
To examine modulation of Cre-mediated transcription by opto-DN-CREB in HEK 293T cells, quantitative PCR was performed to detect transcript levels of nurr1 (NR4A2) and hprt1 (hypoxanthine-guanine phosphoribosyltransferase) as a control. Full details are provided in the SI. HEK cells were grown in 96-well plates and transfected with either pHSV-opto-DN-CREB. Approximately 40-48 hrs after transfection, cells were washed and chromophore was added. All samples were incubated for 15 min in the dark and a subset of plates were subsequently moved to an incubator with blue light illumination (0.2 mW/cm2) for 60 min. Cells under both dark and illuminated conditions were then treated with forskolin (50 μM) for 2 hrs in serum free media. Media was aspirated and cells were flash frozen prior to RNA isolation. cDNA was made using a high capacity cDNA reverse transcriptase kit (Life Technologies Inc., cat#4374966). Quantitative real-time PCR (qPCR) was performed using the Evagreen mastermix (ABM, #mastermix-S) in a Biorad CFX96 real time detection system.
Modulation of Cre activity by opto-DN-CREB: Western blots
To assay protein levels of NR4A2, HEK293T cells were grown in 6 cm dishes to 80% confluency in DMEM with 10% FBS. Each dish was then transfected with 4 g of either pHSV-ACREB, pHSV-opto-DN-CREB or empty vector. Cells under both dark and illuminated conditions were then treated with forskolin (50 μM) for 2 h in serum free media before collection and lysis for Western blotting. To examine light dependent inhibition of CRE activity by opto-DN-CREB in neurons, replication-deficient Herpes virus amplicons were generated and used to infect primary cortical neurons. After 8-10 days of growth in vitro, neurons were incubated with HSV-ACREB-PYP or HSV-GFP (control vector) viral particles. Five hours later the medium was replaced. One day later, cells were loaded with chromophore (0.1 mg/mL) and then incubated for 1 hr in the dark or under light illumination (0.2 mW/cm2) before stimulating with 30 μM forskolin and 50 mM KCl for 2h at 37oC. Cells were then harvested and processed for Western blotting. For Western blotting, lysates were subjected to polyacrylamide gel electrophoresis and electroblotted onto PVDF membranes. Membranes were incubated with antibodies against NR4A2 (rabbit, 1:1000; Sigma-Aldrich, Cat# N6413) and β-actin (rabbit, 1:1000; Cell Signaling Inc., Cat# 4967) as a loading control. With neurons, blotting was also performed to detect protein levels of c-Fos (1:1000; rabbit polyclonal, sc-52, Santa Cruz). For quantification, optical densities of bands associated with NR4A2 or β-actin were measured using Image J software (NIH). NR4A2 levels were measured relative to β-actin levels in each lane and then normalized to controls indicated in each blot.
Supplementary Material
Significance.
CREB (cAMP response element-binding protein), a member of the bZIP family of cellular transcription factors, plays central roles in numerous cellular processes and has been a focus for studies on the molecular basis of learning and memory. Current optogenetic tools are not suitable for controlling spatial and temporal variations the activity of endogenous transcription factors like CREB. Here we describe the design and characterization a new type of optogenetic tool - opto-DN-CREB - a blue light controlled dominant negative inhibitor of CREB that can permit fine spatial and temporal control of CREB binding to cAMP response elements.
Highlights.
Specific dominant negative (DN) inhibitors exist for many transcription factors.
Fusing DNs to optogenetic domains permits optical control of native transcription.
Opto-DN-CREB was created by fusing a DN for CREB to the optogenetic domain PYP.
Optical control of CREB function in vitro, in HEK cells, and in cultured neurons.
Acknowledgements
This work was supported by the Human Frontiers Science Program and NIH R01 MH086379, and the National Science Foundation through XSEDE allocation MCB130049. NMR instrumentation at the Centre for Spectroscopic Investigation of Complex Organic Molecules and Polymers was supported by the Canada Foundation for Innovation (Project number: 19119) and the Ontario Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature reviews. Molecular cell biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, Schneider MB, Deisseroth K. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- Barco A, Marie H. Genetic approaches to investigate the role of CREB in neuronal plasticity and memory. Mol. Neurobiol. 2011;44:330–349. doi: 10.1007/s12035-011-8209-x. [DOI] [PubMed] [Google Scholar]
- Bernard C, Houben K, Derix NM, Marks D, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NA. The solution structure of a transient photoreceptor intermediate: Delta25 photoactive yellow protein. Structure. 2005;13:953–962. doi: 10.1016/j.str.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Chatterjee R, Bhattacharya P, Gavrilova O, Glass K, Moitra J, Myakishev M, Pack S, Jou W, Feigenbaum L, Eckhaus M, et al. Suppression of the C/EBP family of transcription factors in adipose tissue causes lipodystrophy. J. Mol. Endocrinol. 2011;46:175–192. doi: 10.1530/JME-10-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, Vetere G, Pekar T, Ross PJ, Neve RL, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat. Neurosci. 2012;15:1255–1264. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- Cowansage KK, Bush DE, Josselyn SA, Klann E, Ledoux JE. Basal variability in CREB phosphorylation predicts trait-like differences in amygdala-dependent memory. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16645–16650. doi: 10.1073/pnas.1304665110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Morgan SA, Brechun KE, Chen YY, Jaikaran AS, Woolley GA. Improving a designed photocontrolled DNA-binding protein. Biochemistry. 2011;50:1226–1237. doi: 10.1021/bi101432p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, et al. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, Josselyn SA. Selective erasure of a fear memory. Science. 2009;323:1492–1496. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, Kroener BT, Manglesdorf DJ, Abel T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J. Clin. Invest. 2012;122:3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks J, Gensch T, Hviid L, van Der Horst MA, Hellingwerf KJ, van Thor JJ. Transient exposure of hydrophobic surface in the photoactive yellow protein monitored with Nile Red. Biophys. J. 2002;82:1632–1643. doi: 10.1016/S0006-3495(02)75514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Norinobu T, Sato M, Arita K, Shirakawa M, Kikuchi K. Development of fluorogenic probes for quick no-wash live-cell imaging of intracellular proteins. J. Am. Chem. Soc. 2013;135:12360–12365. doi: 10.1021/ja405745v. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Joshi CP, Otto H, Hoersch D, Meyer TE, Cusanovich MA, Heyn MP. Strong hydrogen bond between glutamic acid 46 and chromophore leads to the intermediate spectral form and excited state proton transfer in the Y42F mutant of the photoreceptor photoactive yellow protein. Biochemistry. 2009;48:9980–9993. doi: 10.1021/bi9012897. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kwon JT, Kim HS, Josselyn SA, Han JH. Memory recall and modifications by activating neurons with elevated CREB. Nat. Neurosci. 2014;17:65–72. doi: 10.1038/nn.3592. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D, Kasai K, Echlin DR, Taparowsky EJ, Arnheiter H, Vinson C. A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12274–12279. doi: 10.1073/pnas.94.23.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Burns DC, Al-Abdul-Wahid MS, Woolley GA. A circularly permuted photoactive yellow protein as a scaffold for photoswitch design. Biochemistry. 2013;52:3320–3331. doi: 10.1021/bi400018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumauchi M, Hara MT, Stalcup P, Xie AH, Hoff WD. Identification of six new photoactive yellow proteins - Diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochem. Photobiol. 2008;84:956–969. doi: 10.1111/j.1751-1097.2008.00335.x. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Meyer TE, Cusanovich MA, Van Beeumen JJ. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 2002;512:240–244. doi: 10.1016/s0014-5793(02)02272-x. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Vanrobaeys F, Fitch JC, Devreese BV, Meyer TE, Cusanovich MA, Van Beeumen JJ. Heterologous production of Halorhodospira halophila holo-photoactive yellow protein through tandem expression of the postulated biosynthetic genes. Biochemistry. 2003;42:965–970. doi: 10.1021/bi027037b. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Gomez G, Lin S, Lin S, Lin C. Optogenetic control of transcription in zebrafish. PloS one. 2012;7:e50738. doi: 10.1371/journal.pone.0050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez TP, Fan CM. Dynamic CREB family activity drives segmentation and posterior polarity specification in mammalian somitogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E2019–2027. doi: 10.1073/pnas.1222115110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Viste K, Urday-Zaa JC, Senthil Kumar G, Tsai WW, Talai A, Mayo KE, Montminy M, Radhakrishnan I. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20865–20870. doi: 10.1073/pnas.1219028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N, Fukushima H, Suzuki A, Matsuyama Z, Homma S, Frankland PW, Kida S. Brain region-specific gene expression activation required for reconsolidation and extinction of contextual fear memory. J. Neurosci. 2009;29:402–413. doi: 10.1523/JNEUROSCI.4639-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder M, Brechun KE, Kim YB, Hoffmann SA, Chen YY, Keiski CL, Arndt KM, McMillen DR, Woolley GA. An Escherichia coli system for evolving improved light-controlled DNA-binding proteins. Protein Eng. Des. Sel. 2015;28:293–302. doi: 10.1093/protein/gzv033. [DOI] [PubMed] [Google Scholar]
- Monfils MH, Cowansage KK, Klann E, LeDoux JE. Extinction-reconsolidation boundaries: key to persistent attenuation of fear memories. Science. 2009;324:951–955. doi: 10.1126/science.1167975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SA, Al-Abdul-Wahid S, Woolley GA. Structure-based design of a photocontrolled DNA binding protein. J. Mol. Biol. 2010;399:94–112. doi: 10.1016/j.jmb.2010.03.053. [DOI] [PubMed] [Google Scholar]
- Morgan SA, Woolley GA. A photoswitchable DNA-binding protein based on a truncated GCN4-photoactive yellow protein chimera. Photochemical & photobiological sciences : Official journal of the European Photochemistry Association and the European Society for Photobiology. 2010;9:1320–1326. doi: 10.1039/c0pp00214c. [DOI] [PubMed] [Google Scholar]
- Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negron C, Keating AE. A set of computationally designed orthogonal antiparallel homodimers that expands the synthetic coiled-coil toolkit. J. Am. Chem. Soc. 2014;136:16544–16556. doi: 10.1021/ja507847t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- Niopek D, Benzinger D, Roensch J, Draebing T, Wehler P, Eils R, Di Ventura B. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat. Commun. 2014;5:4404. doi: 10.1038/ncomms5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- Philip AF, Kumauchi M, Hoff WD. Robustness and evolvability in the functional anatomy of a PER-ARNT-SIM (PAS) domain. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17986–17991. doi: 10.1073/pnas.1004823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran PL, Lovett JE, Carl PJ, Cammarata M, Lee JH, Jung YO, Ihee H, Timmel CR, van Thor JJ. The short-lived signaling state of the photoactive yellow protein photoreceptor revealed by combined structural probes. J. Am. Chem. Soc. 2011;133:9395–9404. doi: 10.1021/ja200617t. [DOI] [PubMed] [Google Scholar]
- Rogerson T, Cai DJ, Frank A, Sano Y, Shobe J, Lopez-Aranda MF, Silva AJ. Synaptic tagging during memory allocation. Nat. Rev. Neurosci. 2014;15:157–169. doi: 10.1038/nrn3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti P, Carpentier R, Segard P, Olive-Cren C, Lefebvre P. Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor NURR1. Nucleic Acids Res. 2006;34:5515–5527. doi: 10.1093/nar/gkl712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Rivera ZI, Williams JS, Gorenstein DG, Andrisani OM. Bacterial expression and characterization of the CREB bZip module: circular dichroism and 2D 1H-NMR studies. Protein science : a publication of the Protein Society. 1993;2:1461–1471. doi: 10.1002/pro.5560020910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat. Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. Molecular and cellular approaches to memory allocation in neural circuits. Science. 2009;326:391–395. doi: 10.1126/science.1174519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli F, De Gregorio R, Pulcrano S, Perrone-Capano C, di Porzio U, Bellenchi GC. Direct regulation of Pitx3 expression by Nurr1 in culture and in developing mouse midbrain. PloS one. 2012;7:e30661. doi: 10.1371/journal.pone.0030661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- Wu GY, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HL, Pressey J, Mahadevan V, Tran MM, Kushner SA, et al. Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron. 2014;83:722–735. doi: 10.1016/j.neuron.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gong S, Luo J, Zheng Z, Song B, Ma S, Guo J, Hu C, Thiel G, Vinson C, et al. Opposing roles for ATF2 and c-Fos in c-Jun-mediated neuronal apoptosis. Mol. Cell Biol. 2009;29:2431–2442. doi: 10.1128/MCB.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, Neve R, Poirazi P, Silva AJ. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat. Neurosci. 2009;12:1438–1443. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.