Summary
Type-2 innate lymphoid cells (ILC2s) promote anti-helminth responses and contribute to allergies. Here we report that Bcl11b, previously considered a T-cell lineage identity transcription factor, acts directly upstream of the key ILC2 transcription factor Gfi1 to maintain its expression in mature ILC2s. Consequently, Bcl11b−/− ILC2s downregulated Gata3 and downstream genes, including Il1rl1, encoding IL-33 receptor, and upregulated Rorc and type-3 ILC (ILC3) genes. Additionally, independent of Gfi1, Bcl11b directly repressed expression of the ILC3 transcription factor Ahr, further contributing to silencing of ILC3 genes in ILC2s. Thus, Bcl11b−/− ILC2s lost their functions and gained ILC3 functions, expanding in response to the protease allergen papain, while at the same time producing ILC3, and not ILC2 cytokines, and causing increased airway infiltration of neutrophils instead of eosinophils. Our results broaden Bcl11b's role from a T-cell only transcription factor, and establish that Bcl11b sustains mature ILC2 genetic and functional programs and lineage fidelity.
Introduction
Innate lymphoid cells (ILCs) reside at mucosal sites, produce T helper cell-like cytokines and play a critical role in maintaining mucosal homeostasis and preventing pathogen invasion. ILC2s respond to interleukins IL-33, IL-25 and thymic stromal lymphopoietin (TSLP) produced by damaged lung epithelial tissue, following viral and helminth infections or allergic reactions. ILC2s produce IL-13 and IL-5 and contribute to mucus secretion and accumulation of eosinophils in the airway and inflammation (Bartemes et al., 2012; Chang et al., 2011; Halim et al., 2012b). ILC2s also produce amphiregulin, promoting airway epithelial repair during viral infections (Monticelli et al., 2011). ILC2s are controlled by the transcription factors Gata3, Rorα, TCF-1 and Gfi1 (Halim et al., 2012b; Hoyler et al., 2012; Spooner et al., 2013; Wong et al., 2012; Yang et al., 2013). In contrast to ILC2s, ILC3s are normally excluded from the lung. They express the transcription factors Rorγt and Aryl hydrocarbon receptor (Ahr), absent in ILC2s, and produce the Th17 cell-associated cytokines IL-17 and IL-22 (Cella et al., 2009; Halim et al., 2012b; Kiss et al., 2011; Qiu et al., 2012). In the gut, ILC3-derived IL-22 induces repair of the gut epithelium and production of anti-microbial peptides (Luci et al., 2009; Sawa et al., 2011; Sonnenberg et al., 2012). ILC2s and ILC3s are dependent in their development on Id2 and Notch signaling (Cherrier et al., 2012; Lee et al., 2012; Possot et al., 2011), as well as on Gata3 and TCF-1 (Halim et al., 2012b; Hoyler et al., 2012; Mielke et al., 2013; Serafini et al., 2014; Wong et al., 2012; Yang et al., 2013). Gata3 is also responsible for the expression of the ILC2 cytokine genes IL-5 and IL-13, as well as for the IL-33 receptor (ST2), encoded by Il1rl1 gene (Hoyler et al., 2012; Yagi et al., 2014). TCF-1 is positioned upstream of Gata3 in the control of ILC2 lineage development (Yang et al., 2013). Another transcription factor, Gfi1, controls ILC2 development and its absence is associated with reduced Gata3, concomitant with derepression of the ILC3 transcription factor gene Rorc along with increased IL-17 production (Spooner et al., 2013).
Bcl11b is a C2H2 transcriptional regulator (Avram et al., 2000; Avram et al., 2002) which functions both as a transcriptional activator and repressor (Cismasiu et al., 2005; Cismasiu et al., 2006). Its expression is initiated at the thymic DN2 stage and it remains expressed in all mature T lymphocytes. Bcl11b is important for T lineage differentiation and T cell identity (Albu et al., 2007; Li et al., 2010a; Li et al., 2010b; Wakabayashi et al., 2003). Bcl11b also controls mature cytotoxic T lymphocyte (CTL) function, restricts T helper-17 (Th17) cell plasticity towards a Thelper-2 (Th2) cell phenotype, controls suppression function of T regulatory (Treg) cells, and iNKT cell development (Albu et al., 2007; Albu et al., 2011; Califano et al., 2014; Li et al., 2010a; Li et al., 2010b; Uddin et al., 2014; Vanvalkenburgh et al., 2011; Zhang et al., 2010). Given that ILC2 development relies on the same regulators that are critical for T cell development, including Notch, TCF-1, Gata3 and Gfi1 (Hoyler et al., 2012; Spooner et al., 2013; Yang et al., 2013), and because Bcl11b transcripts are found highly expressed in ILC2s (Yang et al., 2013), we investigated its role in these cells. Though Bcl11b was not required for ILC2 development, and the numbers of mature ILC2s remained normal in the absence of Bcl11b, Bcl11b−/− ILC2s downregulated ST2 and the critical transcription factors for ILC2 lineage Gfi1, Gata3 and Rorα, and acquired expression of the ILC3 lineage transcription factors Rorγt and Ahr, as well as the receptor for IL-23. Bcl11b−/− ILC2s expanded similarly to wild type ILC2s in response to the protease allergen papain, but produced ILC3-type cytokines, resulting in reduced mucus production and increased airway infiltration of neutrophils instead of eosinophils, usually associated with the response to protease allergens. Our study thus demonstrates that Bcl11b maintains identity and function of mature ILC2s, changing the notion that Bcl11b is a T-cell specific transcription factor.
Results
Bcl11b is high in the peripheral ILC2 populations
Given that Bcl11b transcripts are expressed in ILC2s (Yang et al., 2013), we first evaluated Bcl11b protein in ILC2s, defined as Lin−CD90+CD127+ST2+ (Monticelli et al., 2011), as well as in ILC3s (Lin−CD90+CD127+Rorγt+) (Halim et al., 2012b; Monticelli et al., 2011). Whereas a large percentage of the lung and mesenteric lymph nodes (mLN) ILC2s showed high Bcl11b (Figure S1A and B), only a small percentage of the mLN ILC3s was positive for Bcl11b, and the amount was lower compared to ILC2s (Figure S1A–B). In the bone marrow (BM) ILC2 precursors (ILC2Ps) (Lin−CD127+Sca-1hi cKit−ST2+) the amount of Bcl11b was close to background, both in the Klrg1hi and Klrglo populations (Figure S1F). Given these results, we further focused our studies on the role of Bcl11b in mature ILC2s.
Bcl11b−/− ILC2s have reduced ST2
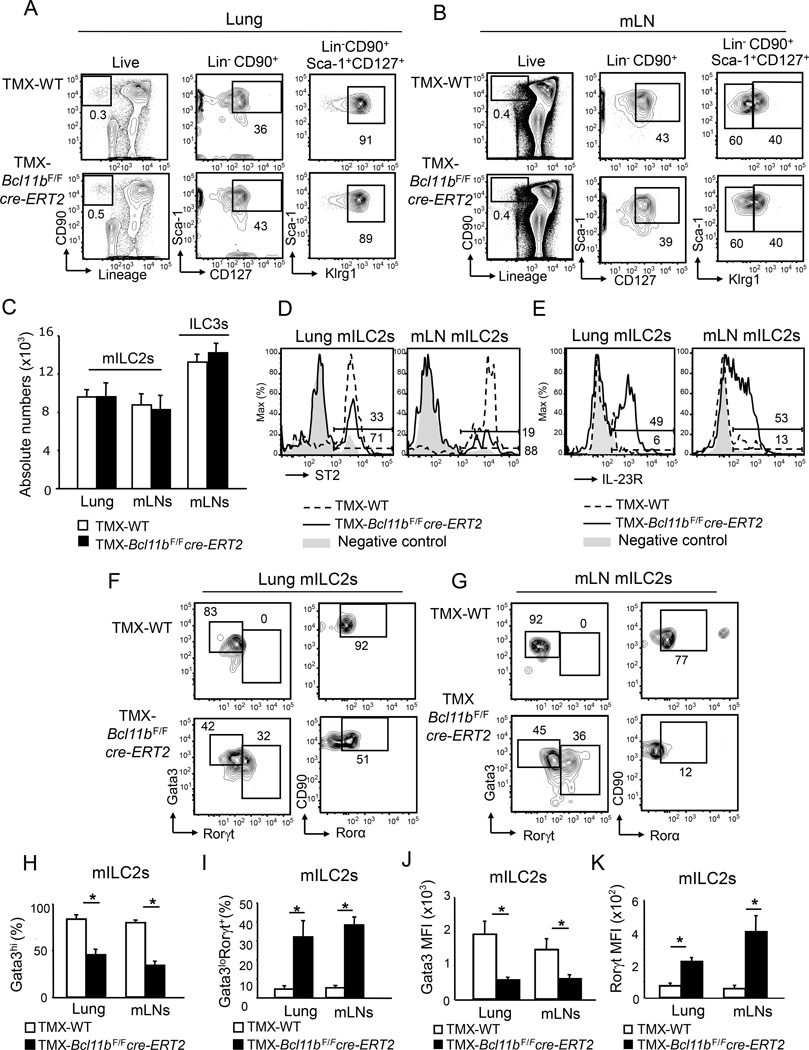
To remove Bcl11b in the ILC2s we utilized the inducible cre-estrogen receptor T2 (cre-ERT2), in which the estrogen receptor 1 gene (ESR1) has a mutation which makes it highly responsive to tamoxifen treatment (TMX) (Figure S1C). Given that Klrg1 was previously identified as a marker for mature ILC2s (Hoyler et al., 2012), we defined peripheral mature ILC2s (from here on mILC2s) as Lin−CD90+CD127+Sca-1+Klrg1hi (Figure 1A–C). Of note, the vast majority of Lin−CD90+CD127+Sca-1+Klrg1hi cells were ST2+Gata3+Rorγt− in wild type mice (Figure 1D, F and G), supporting the idea that these cells indeed represent the mature ILC2 population, while Lin−CD90+CD127+Sca-1+Klrg1lo cells were Rorγt+, hence ILC3s (Figure S2). Lung, mLNs and small intestine lamina propria (SILP) Bcl11b−/− mILC2s were similar in percentages and absolute numbers to wild type ILC2s (Figure 1A–C and Figure S3A), however Bcl11b−/− mILC2s exhibited a marked reduction in ST2 (Figure 1D), but not in IL-17Rβ, a subunit of the interleukin-25 receptor (IL-25R) (Figure S4A). Of note, Bcl11b was efficiently removed in the mILC2s of TMX-treated Bcl11bF/Fcre-ERT2 mice (Figure S1D), except in the subpopulation of cells that still maintained ST2 (Figure S1E). These results demonstrate that Bcl11b deficiency does not cause the loss of mILC2s, but instead results in reduction of ST2, but not of IL-17Rβ.
Figure 1.
Bcl11b's removal causes reduction of ST2, Gata3 and Rorα, and increase in Rorγt in mature ILC2s. A–B) Flow cytometry analysis of the Lin− CD90+ (left panel), Lin−CD90+Sca-1+CD127+ (central panel) and Lin−CD90+Sca-1+CD127+Klrg1hi and -Klrg1lo (right panel) populations in the lung (A) and mesenteric lymph nodes (m)LNs (B) of TMX-Bcl11bF/F cre-ERT2 and -wild type control mice. C) Absolute numbers of Lin−CD90+Sca-1+CD127+Klrg1hi mature (m)ILC2s and Lin−CD90+Sca-1+CD127+Klrg1lo (ILC3s) populations in the lung and mLNs of TMX-Bcl11bF/Fcre-ERT2 (black) and -wild type control (white) mice. Data (n=7), derived from four independent experiments, are presented as mean ± SEM. Significance was determined by Student's t test; * indicates p-value ≤ 0.05. D–E) Surface ST2 (D) and IL-23R (E) evaluated by flow cytometry in the mILC2s from the lung and mLNs of TMX-Bcl11bF/F cre-ERT2 (black) and -wild type control (dashed) mice. Gray shaded area represents negative control. F–G) Flow cytometry analysis of Gata3 and Rorγt (left panel), and CD90 and Rorα (right panel) in the mILC2s from the lung (F) and mLNs (G) of the indicated groups of mice. H–I) Average frequencies of Gata3+ (H) and Rorγt+Gata3lo (I) mILC2s in the lung and mLNs of the indicated groups of mice. J–K) MFI of Gata3 (J) and Rorγt (K) in mILC2s from lung and mLNs of the indicated groups of mice. (H–K) Data (n=6), from three independent experiments, are presented as mean ± SEM. Significance was determined by Student's t test; * indicates p-value ≤ 0.05. See also Supplemental Figures S1, S2, S3 and S5.
Bcl11b does not control development or maturation of ILC2 precursors (ILC2Ps) in the bone marrow (BM)
As shown above Bcl11b was close to background in the BM ILC2Ps (Figure S1F). Supporting this observation, there was no difference in the Lin−CD127+Sca-1hi cKit−ST2+ ILC2Ps or in the Klrg1lo and Klrg1hi populations in the BM of TMX-Bcl11bF/Fcre-ERT2 and -Bcl11bF/F mice (Figure S5A–C). We evaluated the response of BM ILC2Ps to IL-33 treatment, known to promote maturation of these progenitors (Spooner et al., 2013), and this treatment caused a similar increase in the numbers of BM ST2hiKlrg1hi ILC2s in both groups of mice (Figure S5D), demonstrating that Bcl11b deficiency did not restrict maturation of the ILC2 lineage.
Bcl11b−/− ILC2s have reduced Gata3 and Rorα and upregulate the ILC3-specific transcription factor Rorγt and the receptor for IL-23 (IL-23R)
Considering that ST2 expression is controlled by Gata3 in ILC2s (Hoyler et al., 2012), we further investigated Gata3. As expected the wild type mILC2 population showed high Gata3 and Rorα amounts in the lung, mesenteric lymph nodes (mLNs) and small intestine lamina propria (SILP) and did not express the ILC3 lineage transcription factor Rorγt (Figure 1F–G and S3B–C), which conversely was expressed in the ILC3 population (Figure S2). Different from wild type mice, TMX-Bcl11bF/Fcre-ERT2 mice had a major reduction in the Gata3hi and Rorα+ mILC2 population in the lung, mLN and SILP, in favor of a Gata3loRorγt+ population (Figure 1F–I and Figure S3B–C). Additionally, Bcl11b−/− mILC2s exhibited a significant reduction in Gata3 mean fluorescence intensity (MFI), while the Rorγt MFI was increased (Figure 1J–K) even in in the lung tissue, where ILC3s are restricted at steady state (Halim et al., 2012b). Not only was Rorγt increased in the Bcl11b−/− mILC2s, but also IL-23R (Figure 1E), normally expressed in ILC3s (Buonocore et al., 2010). Expression of Gata3 and Rorγt in the ILC3 population from mLNs of TMX-Bcl11bF/Fcre-ERT2 mice was equivalent to the wild type (Figure S2), indicating that the ILC3s keep their identity in the absence of Bcl11b. Gata3 was similar in the ILC2Ps from the BM of both TMX-Bcl11bF/Fcre-ERT2 mice and -Bcl11bF/F mice, while Rorγt was not expressed (Figure S5E), further indicating that Bcl11b does not control ILC2 identity in the BM. These results demonstrate that the absence of Bcl11b in the mILC2 population causes downregulation of the critical ILC2 lineage factors Gata3, Rorα and ST2, and upregulation of the ILC3 lineage factors Rorγt and IL-23R, resulting in loss of identity.
The shift in ILC2 identity in the absence of Bcl11b occurs in a T cell-independent manner
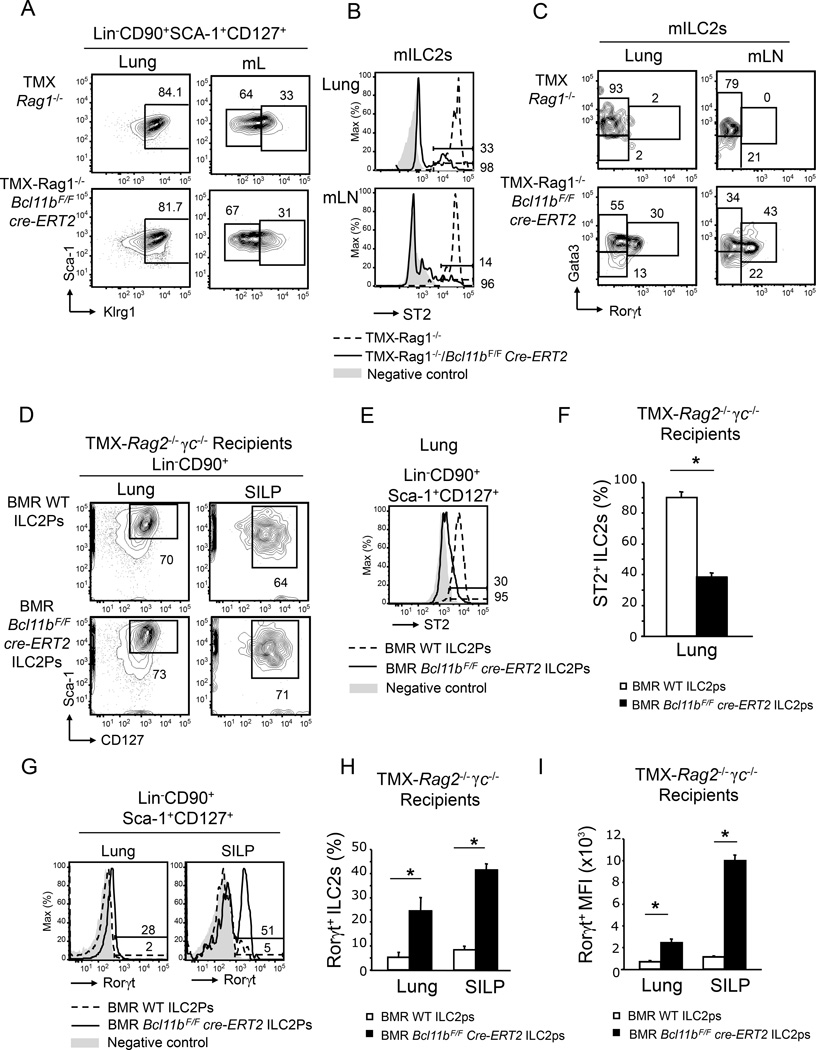
Bcl11b is also expressed in T cells, in which it plays critical roles. To exclude any implication of T cells in the observed phenotype, we generated Bcl11bF/F cre-ERT2 Rag1−/− mice, which lack T and B cells, but maintain the ILC populations (Halim et al., 2012a). Similar to the T cell sufficient mice, the percentages of mILC2s remained equivalent between TMX- Rag1−/−Bcl11bF/F cre-ERT2 and -Rag1−/−Bcl11bF/F mice (Figure 2A). Also similar to immunocompetent mice, ST2 was profoundly reduced in the absence of Bcl11b in the mILC2s (Figure 2B), and so were the percentages of the Gata3hi mILC2s, in favor of a Gata3loRorγt+ population (Figure 2C), indicative of the acquisition of an ILC3 phenotype. These results demonstrate that the loss of the ILC2 identity and acquisition of the ILC3 phenotype is caused by a T cell-independent mechanism.
Figure 2.
The phenotypic alterations of Bcl11b−/− ILC2s are T cell independent and intrinsic. A) Flow cytometry analysis of the mILC2s and ILC3s in lung and mLNs of TMX-Rag1−/− Bcl11bF/F cre-ERT2 and -Rag1−/− mice. B) Flow cytometry analysis of surface ST2 in the mILC2s from lung and mLNs of TMX-Rag1−/−Bcl11bF/Fcre-ERT2 (black) and -Rag1−/− (dashed) mice. Gray shaded area represents negative control. C) Gata3 and Rorγt evaluated by flow cytometry in the mILC2s from lung and mLNs of the indicated groups of mice. Data (n=6) is representative of three independent experiments. D) Flow cytometry analysis of Lin−CD90+Sca-1+CD127+ ILCs in the lung and small intestine lamina propria (SILP) of TMX-Rag2−/−γc−/− mice reconstituted with bone marrow (BM) ILC2Ps (Lin−CD127+Sca-1hiST2+) from Bcl11bF/Fcre-ERT2 or wild type mice. Of note, donor mice were not treated with TMX, which was only administered to the recipient mice two weeks after transfer, as described in Figure S6A. E–F) Surface ST2 (E) and average frequency of ST2+ cells (F) in the Lin−CD90+Sca-1+CD127+ population from lung of TMX-Rag2−/−γc−/− mice reconstituted as in (D), with BM ILC2Ps from the indicated groups of mice. Gray shaded area represents negative control. G–I) Rorγt (G), average frequency of Rorγt+-cells (H), and average MFI of Rorγt (I) in the Lin−CD90+Sca-1+CD127+ population in the lung and SILP of the indicated groups of mice. Data (n=5), derived from two independent experiments, are presented as mean ± SEM. Significance was determined by Student's t test, * indicates p-value ≤ 0.05. See also Supplemental Figure S6.
Identity loss and acquisition of the ILC3 phenotype in the absence of Bcl11b is cell intrinsic
To determine whether the observed phenotype is intrinsic to mILC2s, sub-lethally irradiated Rag2−/−γc−/− mice, which lack adaptive and innate lymphoid populations, were reconstituted with sorted BM Lin−CD127+Sca-1hi ST2+ ILC2 precursors from Bcl11bF/Fcre-ERT2 or Bcl11bF/F mice, not treated with TMX. Two and a half weeks post-transfer, the TMX treatment was initiated and the ILC populations were evaluated four weeks post-transfer (Figure S6A). While Lin−CD127+Sca-1hiST2+ ILC2 precursors from Bcl11bF/F donor mice generated ST2+ mILC2s, the same precursors from Bcl11bF/Fcre-ERT2 donor mice generated mILC2s with low ST2, which upregulated Rorγt (Figure 2D–I). To demonstrate that the non-hematopoietic populations do not participate in the observed phenotype, BM Lin−CD127+Sca-1hi ST2+ ILC2 precursors from Bcl11bF/Fcre-ERT2 or Bcl11bF/F mice, not treated with TMX, were transferred into lethally irradiated Bcl11bF/Fcre-ERT2 or Bcl11bF/F mice (Figure S6B), which were further treated as described above. Both Bcl11bF/Fcre-ERT2 and Bcl11bF/F mice reconstituted with ILC2Ps from Bcl11bF/F donor mice generated mILC2s with high ST2 and Gata3 (Figure S6C–D), while Bcl11bF/Fcre-ERT2 and Bcl11bF/F mice reconstituted with ILC2Ps from Bcl11bF/Fcre-ERT2 donor mice generated ST2loGata3loRorγt+IL-23R+ mILC2s (Figure S6C–D), demonstrating that the loss of ST2 and the shift in the mILC2 identity toward an ILC3 phenotype was not dependent on the non-hematopoietic compartment of TMX-Bcl11bF/Fcre-ERT2 mice. Taken together these data demonstrate that Bcl11b is required to maintain mILC2 identity and to block expression of the ILC3 transcription factor Rorγt in ILC2s in a cell intrinsic manner.
Bcl11b−/− ILC2s proliferate in response to IL-23 and IL-25, but not in response to IL-33
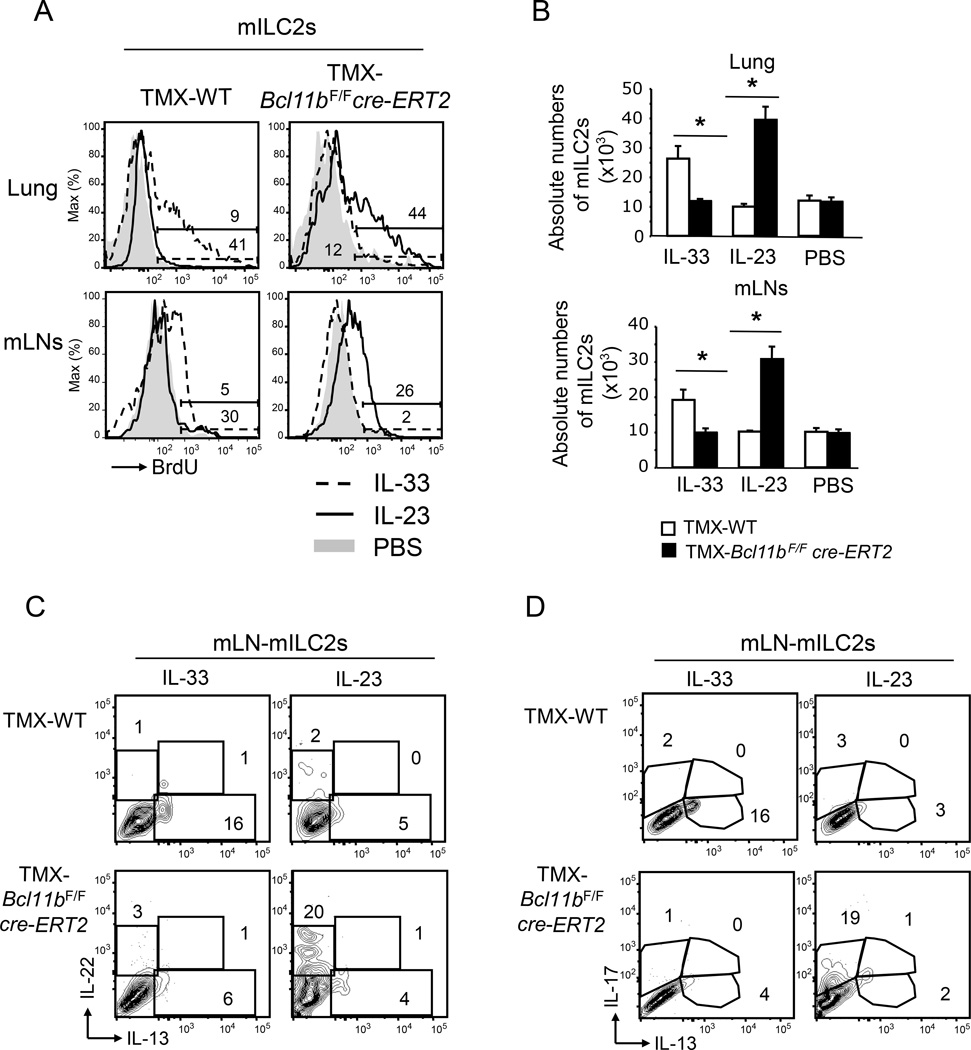
Bcl11b−/− mILC2s exhibited a reduction in ST2 concomitant with an increase in IL-23R (Figures 1D–E). We thus investigated how these cells respond to IL-33 vs. IL-23 in terms of proliferation. While wild type mILC2s exhibited enhanced BrdU incorporation and an increase in their absolute numbers in response to IL-33 injection (Figure 3A–B), Bcl11b−/− mILC2s exhibited minimal BrdU incorporation and the absolute numbers remained similar to PBS injection (Figure 3A–B). Conversely, IL-23 administration caused increased BrdU incorporation in the Bcl11b−/− mILC2s, as well as an increase in their absolute numbers, which did not occur in wild type cells (Figure 3A–B). Importantly, mILC2s from both TMX- Bcl11bF/Fcre-ERT2 and -Bcl11bF/F mice injected with IL-25 incorporated BrdU and underwent proliferative expansion equivalently (Figure S4B–C), in agreement with similar expression of IL-17Rβ in the mILC2s of both groups of mice (Figure S4A). These data taken together demonstrate that Bcl11b−/− mILC2s fail to expand in response to IL-33 and instead expand in respond to IL-23, while maintaining normal expansion in response to IL-25.
Figure 3.
Bcl11b−/− mILC2s respond to IL-23 instead of IL-33. A) BrdU incorporation in mILC2s from lung and mLNs of TMX-Bcl11bF/Fcre-ERT2 and -wild type control mice administered with either IL-23 (black), IL-33 (dashed) or PBS (gray shaded area). B) Absolute numbers of mILC2s from the lung and mLNs of the indicated groups of mice from the experiment in (A). Data (n=5), derived from two independent experiments, are presented as mean ± SEM. Significance was determined by Student's t test, * indicates p-value ≤ 0.05. C–D) Flow cytometry analysis of intracellular IL-22 vs. IL-13 (C), and IL-17 vs. IL-13 (D) in mILC2s from the mLNs of IL-33- or IL-23-treated TMX-Bcl11bF/Fcre-ERT2 and -wild type control mice. Data (n=4) derived from three independent experiments. See also Supplemental Figure S4.
Bcl11b−/− ILC2s produce ILC3 cytokines in response to IL-23, but not ILC2 cytokines in response to IL-33 or IL-25
We further investigated the cytokine production by Bcl11b−/− vs. wild type ILC2s in response to IL-33, IL-25 or IL-23 injections. Wild type ILC2s produced IL-13 in response to IL-33 or IL-25, and failed to produce IL-22 or IL-17 in response to IL-23 (Figure 3C–D and Figure S4D–E). Conversely, Bcl11b−/− ILC2s produced IL-22 and IL-17 in response to IL-23, but not IL-13 in response to IL-33 or IL-25 (Figure 3C–D and Figure S4D–E). These results demonstrate that Bcl11b−/− ILC2s respond by producing ILC3 cytokines to IL-23 stimulation, but not ILC2 cytokines in response to IL-33 or IL-25. Additionally, though Bcl11b−/− ILC2s have normal IL-17Rβ and proliferated in response to IL-25, they failed to produce IL-13 in response to this cytokine (Figure S4D–E), suggesting alterations in factors that control ILC2 cytokine production, though the expansion pathway remained likely intact.
Bcl11b−/− ILC2s respond by an ILC3-type of airway inflammation to the protease papain
ILC2s have been previously shown to play a critical role in the response to protease induced airway inflammation (Halim et al., 2012a; Wilhelm et al., 2011). As expected, intranasal administration of the protease allergen papain induced the production of the ILC2 cytokines IL-5 and IL-13 in the lung of wild type mice, while the production of these cytokines was reduced in TMX-Bcl11bF/Fcre-ERT2 mice, which instead produced the ILC3 cytokines IL-17 and IL-22 (Figure 4A–B). Importantly the Bcl11b−/− ILC2s expanded similarly to wild type ILC2s following administration of papain (Figure 4C). Related to the elevated production of IL-17 and decreased IL-5, increased neutrophil numbers were observed in the lung and bronchoalveolar lavage fluid (BALF) of TMX-Bcl11bF/Fcre-ERT2 mice, while the numbers of eosinophils were reduced, contrary to the response of the wild type mice, which presented increased eosinophils and less evident neutrophils (Figure 4D). We further evaluated mucus production in response to IL-13, and found that the lungs of TMX-Bcl11bF/Fcre-ERT2 mice treated with papain exhibited diminished mucus production compared to wild type mice (Figure 4E). These data taken together demonstrate that the lung Bcl11b−/− ILC2s respond to papain by expansion similar to wild type, however they produce the ILC3 cytokines IL-17 and IL-22, causing increased neutrophil numbers. In addition, Bcl11b−/− ILC2s exhibit reduced ability to produce ILC2 cytokines, which thus results in diminished mucus production and reduced eosinophils. These results further support the idea that Bcl11b−/− ILC2s are functionally shifted to an ILC3 phenotype.
Figure 4.
Bcl11b−/− ILC2s promote airway infiltration of neutrophils in response to papain, and confer increased protection to C. rodentium. A) Flow cytometry analysis of intracellular IL-13, IL-5, IL-17 and IL-22 in Lin−CD90+Sca-1+CD127+ ILC2s from the lungs of papain- or PBS-treated TMX- Bcl11bF/Fcre-ERT2 (red) and -wild type control mice (black). Gray shaded area represents PBS-treated mice. B) Average frequency of cytokine-producing Lin−CD90+Sca-1+CD127+ ILC2s from the lungs of papain-treated TMX-Bcl11bF/Fcre-ERT2 (black) and -wild type control (white) mice. Data (n=6), derived from two independent experiments, are presented as mean ± SEM. Significance determined by Student's t test, * indicates p-value ≤ 0.05. C) Absolute numbers of Lin−CD90+Sca-1+CD127+ ILC2s from the lungs of the indicated groups of mice. D) Absolute numbers of eosinophils (Lymph−CD11b+CD11cloLy6G−SiglecF+) and neutrophils (Lymph−CD11b+CD11cloLy6G+SiglecF−) in the lung and BALF of the indicated groups of mice, determined by flow cytometry analysis. Data (n=6), derived from three independent experiments, are presented as mean ± SEM. Significance determined by Student's t test, * indicates p-value ≤ 0.05. E) Mucus production evaluated by Periodic Acid Schiff (PAS) staining of lung tissues of papain-treated TMX- Bcl11bF/Fcre-ERT2 and -wild type control mice. Data is representative of three independent pairs. Scale bar - 200 µm. F) Bacterial colony forming units (CFU) from feces of Citrobacter-infected TMX-Rag1−/− Bcl11bF/Fcre-ERT2 (black) and -Rag1−/− mice (white) day 5 post-infection. Data (n=4), derived from two independent experiments, are presented as mean ± SEM. Significance determined by Student's t test; * indicates p-value ≤ 0.05. G–H) Flow cytometry analysis of intracellular IL-17 vs. IL-22 in mILC2s and ILC3s from the mLNs (G) and SILP (H) of Citrobacter-infected TMX-Rag1−/−Bcl11bF/Fcre-ERT2 and -Rag1−/− mice day 5 post-infection. Data (n=4) is derived from two independent experiments.
Small intestine Bcl11b−/− ILC2s produce ILC3 cytokines following infection with Citrobacter rodentium, conferring increased protection to infection
IL-22 and IL-17 produced by ILC3s play a critical role in the early response to infection with Citrobacter (Qiu et al., 2012). Given that Bcl11b−/− ILC2s behave as ILC3s during papain-induced asthma, we investigated their response following Citrobacter infection. Mice bearing Bcl11b−/− ILC2s had reduced bacterial burden on day 5 post-infection, associated with increased IL-22 and IL-17 production by small intestine and mLNs ILC2s (Figure 4F–H). The response of Bcl11b−/− ILC2s was similar to wild type mILC3s (Figure 4G–H). Thus Bcl11b−/− ILC2s efficiently participate in the protective response to Citrobacter together with ILC3s.
Bcl11b sustains expression of key ILC2 genes and blocks expression of critical ILC3 genes in ILC2s
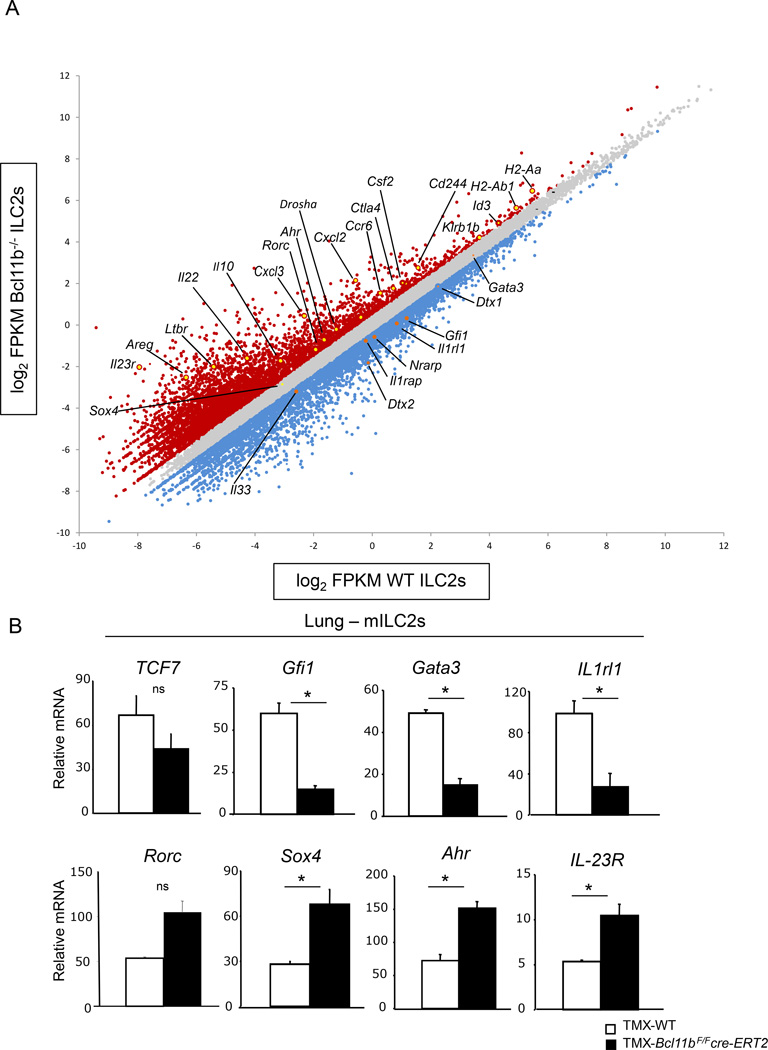
To establish the molecular functions of Bcl11b in mature ILC2 lineage, we performed RNA-sequencing (RNA-seq) analysis on sorted ILC2s (Lin−CD90+Klrg1+) from the mLNs of TMX-Bcl11bF/Fcre-ERT2 mice and -wild type controls at steady state. The analysis revealed that a broad ILC2 genetic program was downregulated, including the mRNAs for Il1rl1 (encoding ST2), as well as its coreceptor Il1rap, and importantly the transcription factor Gfi1 (Figure 5A), known to sustain the ILC2 genetic program and repress the ILC3 genetic program (Spooner et al., 2013). Additionally, mRNA for Gata3, known to be a downstream target of Gfi1, was also mildly downregulated in Bcl11b−/− mILC2s (Figure 5A), and when evaluated by qRT-PCR was significantly altered (Figure 5B). Tcf7 mRNA, encoding TCF-1, also known to regulate Gata3 (Spooner et al., 2013; Weber et al., 2011), had only a minor decrease, and not significant when evaluated by qRT-PCR (Figure 5A–B), suggesting that likely Bcl11b is positioned upstream of Gfi1 and Gata3, however not of Tcf7. Rorα mRNA remained unchanged (data not shown) even though Rorα protein was severely reduced (Figure 1F–G), suggesting that Rorα is regulated through an alternative mechanism. Related to this, Drosha mRNA was highly upregulated in Bcl11b−/− mILC2s (Figure 5A), suggesting the possibility of an increase in microRNAs, and thereby post-transcriptional regulation of ILC2 genes. The gene encoding amphiregulin (Areg), which is produced by ILC2s following influenza infection (Monticelli et al., 2011), was also upregulated in Bcl11b−/− mILC2s (Figure 5A), suggesting that Bcl11b may play an inhibitory role for this gene. In addition, a broad ILC3 genetic program (Cherrier et al., 2012; Mortha et al., 2014; Sonnenberg et al., 2011) was upregulated, including transcripts for Rorc, Ahr, IL-23R, IL-22, Cxcl3, Cxcl2L2, Ccr6, Ltbr and Csf2 (encoding GM-CSF) (Figure 5A). Furthermore, expression of Sox4 was marginally, but significantly upregulated (Figure 5A–B), similar to the profile found in Gfi1−/− ILC2s. Importantly, and different from Gfi1−/− ILC2s, Ahr mRNA was highly upregulated (Figure 5A). Of note, in Gfi1−/− ILC2s Ahr was downregulated. Furthermore, Bcl11b−/− mILC2s upregulated transcripts for MHC class II genes H2-Aa and H2-Ab1 (Figure 5A), expressed both in ILC3s and ILC2s (Hepworth et al., 2013; Oliphant et al., 2014). Consistent with lower expression of Notch signaling components in ILC3s (Cherrier et al., 2012), Nrarp, Dtx1 and Dtx2 were downregulated in Bcl11b−/− mILC2s, further supporting the idea that Bcl11b−/− mILC2s acquire an ILC3 phenotype. Additionally, Bcl11b−/− mILC2s exhibited a higher abundance of mRNAs for NK markers such as Cd244 and Klrb1b (Figure 5A), usually upregulated in the absence of Bcl11b from most T cells and progenitors (Avram and Califano, 2014). Bcl11b−/− mILC2s also upregulated several genes known to be important for Tregulatory cells, including Ctla4 and IL-10 (Figure 5A) (Asseman et al., 1999; Read et al., 2000), suggesting that these cells may acquire a suppressive phenotype. Taken together these results demonstrate that overall Bcl11b sustains a large fraction of the ILC2 genetic program and represses the ILC3 genetic program in mILC2s, further substantiating the claim that Bcl11b−/− mILC2s lose their genetic and functional identity.
Figure 5.
Bcl11b sustains expression of ILC2 program and restricts expression of the ILC3 program in mILC2s. A) RNA-seq analysis of mILC2s (Lin−CD90+Klrg1+) sorted from the mLNs of TMX-Bcl11bF/Fcre-ERT2 and -wild type control mice at steady state. Data is shown as a scatter plot of log2 of Fragments Per Kilobase of transcript per million mapped reads (FPKM) from Bcl11b−/− vs. wild type ILC2s. Blue dots represent downregulated genes and Red dots represent upregulated genes in Bcl11b−/− ILC2s. Genes of interest are indicated and marked in orange (downregulated) and yellow (upregulated). B) Quantitative RT-PCR analysis of the indicated mRNAs from mILC2s from the lung of TMX-Bcl11bF/Fcre-ERT2 and -wild type control mice. Data is representative for four independent experiments with three mice per group and is presented as mean ± SEM. Significance was determined by Student's t test; * indicates p-value ≤ 0.05. ns indicates non-significant: Rorc p = 0.06; TCF7 p = 0.24.
Bcl11b associates with conserved noncoding sequences (CNSs) within Gfi1 and Ahr loci, but not with Gata3 or Rorc in ILC2s
Given that Gfi1 was among the genes regulated by Bcl11b, and was recently found to control many of the genes changed in the absence of Bcl11b, we reasoned that Bcl11b may exert its control through direct regulation of Gfi1. We thus designed primers covering conserved noncoding sequences from approximately −800bp through the transcription start site (TSS) (Yucel et al., 2004), and using ChIP assays found that Bcl11b bound to a region located at −413bp through −295bp (Figure 6A–B), indicating that Bcl11b may regulate expression of the Gfi1 gene by direct association with its promoter. We further tested whether Bcl11b bound to the Gata3 proximal promoter, given that we previously found association of Bcl11b with sequences at this location in Th17 cells (Califano et al., 2014). In ILC2 cells Bcl11b did not associate with the Gata3 proximal promoter (Figure S7C), suggesting that the reduced Gata3 in Bcl11b−/− ILC2 cells is likely a consequence of lower Gfi1 amounts. We further tested the association of Bcl11b with CNSs at Ahr and Rorc loci. We found that while Bcl11b did not significantly associate with any CNSs at the Rorc locus (Figure S7D), it significantly associated with a CNS located at −5447bp through −5352bp at the Ahr locus (Figure 6C). These results thus suggest that while changes in Rorc may be a consequence of Gfi1 downregulation in Bcl11b−/− ILC2 cells, Bcl11b regulates Ahr directly. Taken together these results suggest that Bcl11b regulates the ILC2 lineage through positive regulation of Gfi1, which maintains the ILC2 lineage by sustaining Gata3, Il1rl1 and ILC2 cytokine expression, while also repressing expression of ILC3 genes through Rorc, also downstream of Gfi1. In addition, Bcl11b also represses the ILC3 program through direct repression of Ahr expression and consequently its downstream genes.
Figure 6.
Bcl11b associates with conserved noncoding sequences at the Gfi1 and Ahr loci. A) Chromatin immunoprecipitation (ChIP) by anti-Bcl11b antibodies (Blue) or IgG (Pink) of cross-linked nuclear extracts from sorted Lin−CD90+Klrg1+ ILC2s expanded in vitro as described in Supplementary Material and Methods. ChIP-ed DNA was amplified by qPCR with primers for the indicated conserved regions in the Gfi1 promoter (Yucel et al., 2004), and expressed as percent of the input. Data (n=6), derived from three independent experiments, are presented as mean ± SEM. Significance was determined by Student's t test; * indicates p-value ≤ 0.05. Sequence of the primers is provided in Figure S7E. B) Vista plot of the conserved noncoding sequences between mouse and human at the Ahr locus covering a region between −11000bp through the transcription start site (TSS) (red triangle). Five sets of primers were designed (1–5) for the conserved regions (pink peaks). C) ChIP-ed DNA was amplified by qPCR with the 5 sets of primers for the 5 indicated conserved regions (black triangles). Data (n=6), derived from three independent experiments, are presented as mean ± SEM. Significance was determined by Student's t test; * indicates p-value ≤ 0.05. Sequence of the primers is provided in Figure S7E. See also Supplemental Figure S7.
Enforced expression of Gfi1 in Bcl11b−/− ILC2s restores Gata3 and ST2 and reduces Rorγt and IL-23R
We further wish to substantiate the hypothesis that downregulation of Gata3 and ST2, and the repression of Rorc in Bcl11b−/− ILC2s are a consequence of Gfi1's regulation. We thus retrovirally transduced Bcl11b−/− ILC2s with Gfi1-NGFR or control NGFR viruses (Figure 7A). Gfi1 transduction of Bcl11b−/− ILC2s resulted in upregulation of Gata3 and ST2 and reduction of Rorγt and IL-23R (Figure 7B–C). However, Rorα was not upregulated as a result of Gfi1 transduction (Figure 7B–C), suggesting that its regulation is through a Gfi1-independent mechanism. These results demonstrate that Bcl11b exerts its impact on Gata3, ST2 and Rorγt, but not on Rorα, through Gfi1.
Figure 7.
Transduction with Gfi1 retroviruses reestablishes the ILC2 phenotype and restricts the ILC3 phenotype in Bcl11b−/− ILC2s. A) Flow cytometry analysis of the NGFR+ population in the Gfi1-NGFR- or NGFR-transduced lung Lin−CD90+Sca-1+CD127+ cells, isolated from TMX-Rag1−/−Bcl11bF/Fcre-ERT2 mice. B) Gata3, ST2, Rorγt, IL-23R and Rorα in NGFR+ Lin−CD90+Sca-1+CD127+ cells ((Gfi1-NGFR (black) and NGFR (dashed)). C) Average MFI of Gata3, ST2, Rorγt, IL-23R and Rorα in NGFR+ Lin−CD90+Sca-1+CD127+ cells (Gfi1-NGFR (black) or NGFR (white)). Data (n=4), derived from two independent experiments, are presented as mean ± SEM. Significance determined by Student's t test, * indicates p-value ≤ 0.05.
Discussion
In this study we have demonstrated that Bcl11b controls the lineage identity and function of mature ILC2s, but does not impact ILC2 development. Bcl11b has been suggested to be a T cell lineage specific transcription factor that maintains T cell lineage identity by repressing the genetic programs of innate cells (Li et al., 2010a; Li et al., 2010b). In contrast to this notion, here we report that Bcl11b is also highly expressed in mature ILC2s, and is essential in retaining mature ILC2 lineage fidelity by sustaining expression of essential ILC2 transcription factors and by suppressing key ILC3 transcription factors through an intrinsic mechanism. As a consequence of these alterations, Bcl11b−/− ILC2s downregulated critical ILC2 genes and derepressed essential ILC3 genes. Importantly, our study demonstrates that absence of Bcl11b alters not only the genetic program of mature ILC2s, but their functional program as well. Specifically, Bcl11b−/− ILC2s expanded and responded to the allergen papain with an ILC3 functional program, causing production of ILC3 cytokines and infiltration of neutrophils instead of eosinophils in the airways.
Our study positions Bcl11b in the hierarchy of transcription factors that control the mature ILC2 program upstream of Gfi1, Gata3 and Rorα, but not of TCF-1. Considering that Gfi1 has been demonstrated to regulate Gata3, and the fact that Bcl11b did not associate with the Gata3 promoter, but did bind the Gfi1 promoter, it is likely that reduced Gfi1 caused the decrease in Gata3 in the absence of Bcl11b. Additionally, reintroduction of Gfi1 in Bcl11b−/− mILC2s restored Gata3, however did not impact Rorα, suggesting that Bcl11b regulates Rorα independently of Gfi1. Previous evidence, which demonstrates that Gfi1 does not regulate Rorα, supports this idea (Spooner et al., 2013). Furthermore, we did not detect a reduction in Rorα mRNA following Bcl11b removal, even though protein amounts were decreased, suggesting that the regulation of Rorα occurs likely post-transcriptionally. The ST2 subunit of the IL-33 receptor was also reduced in the absence of Bcl11b, and this is likely related to diminished Gfi1 and Gata3, given that Il1rl1 expression is known to be regulated by Gata3 and Gfi1 (Hoyler et al., 2012; Spooner et al., 2013; Yagi et al., 2014; Yang et al., 2013). The reduction in ST2 likely made Bcl11b−/− ILC2s respond inefficiently to IL-33, however they expanded and produced ILC3 cytokines in response to IL-23. The ILC2 receptor IL-17Rβ, a subunit of IL-25R, remained unchanged, and Bcl11b−/− ILC2s expanded normally in response to IL-25. However they failed to produce IL-13, supporting the idea of alterations in factors controlling ILC2 cytokine production, such as Gata3, but intact proliferation abilities. Similar to what was observed Gata3−/− ILC2s (Yagi et al., 2014), Klrg1 was normal in Bcl11b−/− ILC2s. Therefore mature ILC2s were maintained, but they lost key features of ILC2 identity, as well as their function. In this regard, different from Gata3−/− ILC2s, but similar to Gfi1−/− ILC2s (Hoyler et al., 2012; Spooner et al., 2013; Yagi et al., 2014; Yang et al., 2013), Bcl11b−/− ILC2s, including those in the lung, expressed the transcription factor Rorγt together with IL-23R. However, different from Gfi1−/− ILC2s, which failed to respond to papain (Spooner et al., 2013), Bcl11b−/− ILC2s expanded, but rather than producing IL-5 and IL-13, produced IL-17 and IL-22, causing an ILC3-type of inflammation with increased airway neutrophils, and reduced mucus production. Gata3, ST2, Rorγt and IL-23R were all restored following transduction with Gfi1, suggesting that these genes are likely controlled by Bcl11b through Gfi1. Rorc was upregulated in Bcl11b−/− ILC2s close to significance, however we cannot exclude the possibility that, although Rorc may be partially regulated by Bcl11b through Gfi-Sox4, there may also be a Gfi1-independent, post-transcriptional mechanism in place, by which Rorγt is regulated in a manner dependent on Bcl11b. In addition to regulating Gfi1 expression directly, Bcl11b also associated with the Ahr locus and Ahr mRNA was highly upregulated in the absence of Bcl11b. Bcl11b−/− ILC2s produced IL-22, known to be controlled by Ahr in ILC3s (Qiu et al., 2012; Lee et al., 2012), which conferred enhanced resistance to Citrobacter infection. Different from Bcl11b−/− ILC2s, Gfi1−/− ILC2s showed a minor but significant decrease in Ahr expression (Spooner et al., 2013), suggesting that although Gfi1 is downstream of Bcl11b in mature ILC2s, there are some differences between their regulatory functions. This could be due to the fact that Gfi1−/− ILC2s exhibit developmental defects and therefore are not fully mature ILC2s, whereas Bcl11b−/− ILC2s mature normally from the ILC2Ps in the bone marrow. Different from the other transcription factors known to be critical for ILC2 lineage, including TCF-1, Gfi1, Gata3 and Rorα, Bcl11b is unique in the fact that it only controls mature ILC2s, being very modestly expressed in bone marrow ILC2Ps and thus not affecting their development and/or maturation. The ILC2 transcriptional program mirrors the network found in developing thymocytes. Early Notch signaling induces the expression of TCF-1, which promotes upregulation of Gata3 (Weber et al., 2011). Notch signaling, TCF-1 and Gata3 synergistically promote the induction of Bcl11b expression, which further restricts alternative lineage fates and solidifies T cell commitment (Garcia-Ojeda et al., 2013; Li et al., 2010a; Weber et al., 2011). In DN3 thymocytes, deficiency of Bcl11b causes reduction in Tcf7 and Gata3 transcripts, indicating that in later stages of development Bcl11b sustains the expression of these T cell determining factors (Li et al., 2010a; Li et al., 2010b). In ILC2s Bcl11b seems to be expressed following the induction of TCF-1, Gata3, and Gfi1 and ILC2P fate specification. However, once it is expressed, Bcl11b is required to sustain expression of Gfi1 in mature ILC2s, similar to how it sustains Gata3 and Tcf7 expression in DN3 thymocytes. Importantly and different from thymocytes, Bcl11b−/− ILC2s derepress the critical ILC3 transcription factors Rorγt and Ahr, and functionally respond similar to ILC3s. How expression of Bcl11b is induced in ILC2s remains to be established.
Our analysis of mRNAs in Bcl11b−/− ILC2s was conducted on steady state ILC2s, and not following culture with ILC2 stimulating cytokines, such as IL-25. This is especially important since recent reports suggest that IL-25 treatment can stimulate the proliferation of an ILC2-like cell which does not express ST2, possesses multipotency and can acquire IL-17 producing capabilities (Huang et al., 2015; Saenz et al., 2010). Whether Bcl11b regulates the phenotype of these ILC2-like cells remains to be determined. In addition to ILC3 functional genes, Bcl11b−/− ILC2s also upregulated several genes associated with the suppressive phenotype of Tregulatory cells, including Ctla4 and IL-10 (Asseman et al., 1999; Read et al., 2000). ILC3s are known to inhibit CD4+ T cell activation, through antigen presentation without costimulation. MHC class II genes involved in this process (Hepworth et al., 2013) were upregulated in Bcl11b−/− ILC2s. Whether Bcl11b−/− ILC2s contribute to the suppression of the T cell-mediated immune responses, resulting in protection from inflammation and autoimmunity, remains to be establish.
In conclusion, our studies extend Bcl11b's role beyond T lineage identity, demonstrating its ability to control mature type II innate lymphoid cell fate by maintaining expression of essential ILC2 transcription factors and guarding from expression of critical ILC3 transcription factors.
Experimental Procedures
Mice
Bcl11bF/F were previously described (Albu et al., 2007; Albu et al., 2011; Califano et al., 2014; Li et al., 2010b; Zhang et al., 2010) and bred with cre-ERT2 mice and further with Rag1−/− mice. cre-ERT2, Rag1−/− and Rag2−/−γc−/− mice were from the Jackson Laboratories and Taconic Biosciences, respectively. All mice were housed in pathogenfree conditions. All experiments were conducted on 7–12 week old mice.
Study Approval
All mouse procedures were approved by the Albany Medical Center Institutional Animal Care and Use Committee.
Tamoxifen treatment for cre-ERT2 activation
Mice were given intraperitoneal injections of 200µg of tamoxifen for five consecutive days. Seven-eight days following the last injection, mice were sacrificed and analyzed.
Papain-induced inflammation
Mice were intranasally administered 10 µg papain or PBS, on days 0–2 and further euthanized on day 3 and lungs and BALF were collected and processed as described in the supplementary material and methods, and populations were analyzed by flow cytometry. Additionally, lung tissue was fixed in 10% paraformaldehyde, paraffin embedded and sections were stained with Periodic Acid Schiff (PAS).
Citrobacter rodentium infection and bacterial burden
Mice were infected with C. rodentium via oral gavage with 1010 CFU in 200 ml PBS. Fecal pellets were collected, weighed and then homogenized in sterile PBS. Serially diluted homogenates were plated on MacConkey agar plates and incubated at 37°C. Colonies were counted after 24 hr.
Tissue digestion for release of cells
Lymphocytes were isolated from mesenteric lymph nodes, lung, small intestine and bone marrow. For lamina propria (LP) lymphocyte isolation, tissue was digested for 90 minutes at 37°C in 0.75ml complete media (CM) containing 100mg/ml collagenase D (Roche), 1mg/ml dispase II (Invitrogen), and 0.2 mg/ml DNAse (Roche). LP lymphocytes were further washed with CM and enriched by 33% Percoll gradient centrifugation. For lung lymphocyte isolation, the lung was perfused by injecting 10ml PBS into the right ventricle of the heart. Further, lung tissue was incubated at 37°C for one hour in CM containing 1mg/ml collagenase D (Roche) and 0.15mg/ml DNAse (Roche). Cells were washed, filtered through 40µm cell strainer and treated with red blood cell lysis buffer. To collect BAL fluid the mouse lungs were lavaged three times with 1 ml of PBS, and cells were centrifuged at 350g for 5 minutes and stained for flow cytometry.
mILC2 Isolation
Lin−CD90+CD127+Sca-1+Klrg1hi ILC2s were enriched from the lungs of Rag1−/− mice using Dynabeads FlowComp Mouse Pan T (CD90.2) kit (Invitrogen), followed by cell sorting on a FACSAria II (BD Biosciences).
mILC identification
The mILC populations were identified as previously described (Hoyler et al., 2012; Monticelli et al., 2011). Briefly, gating on CD90+ cells, negative for CD3, CD5, CD19, CD11c, CD11b, TCRβ, and NK1.1, and further as CD127+Sca-1+. The ILC2 vs. ILC3 cells were differentiated based on high or low expression of Klrg1, respectively.
Retroviral transduction of ILCs
Lung mILC2s were enriched from TMX-Rag1−/− Bcl11bF/Fcre-ERT2 mice using Dynabeads FlowComp Mouse Pan T (CD90.2) kit (Invitrogen) and cultured for two days in 20% FBS DMEM with 1U/ml IL-2, 20ng/ml IL-7 and 20ng/ml IL-33. Retroviruses were generated by transfecting Plat E cells with Gfi1-NGFR or NGFR retroviral vectors (Del Real and Rothenberg, 2013). Retroviral supernatants were collected two days after transfection and supplemented with 4µg/ml polybrene and ILC2s were infected by spinoculation, as previously described (Cismasiu et al., 2006). ILC2s were evaluated for NGFR, ST2, Gata3, Rorα, IL-23R and Rorγt by FACS analysis 5 days after transduction.
RNA-sequencing analysis
Lin−CD90+KLRG1+ mILC2s were sorted from the mLNs of TMX-Bcl11bF/Fcre-ERT2 and -wild type controls. RNA was extracted using RNeasy Plus Micro Kit (Qiagen) and RNA-seq experiments were performed. Sequenced reads were cleaned according to a rigorous pre-processing workflow (Trimmomatic-0.32) before mapping them to the mouse genome (mm10) with SHRiMP2.2.3. Gene expression levels were measured by FPKM (Fragments Per Kilobase of transcript per Million mapped reads). Cufflinks2.0.2 was used to perform differential expression analysis with an FDR cutoff of 0.05 (95% confidence interval).
Accession numbers
RNA-seq data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE67437.
Quantitative (q)RT-PCR
RNA was extracted from sorted lung mILC2s (Lin−CD90+CD127+Sca-1+KLRg1hi) using RNeasy Plus Micro Kit and Omniscript RT kit (Qiagen). qRT-PCR was conducted as previously described (Cismasiu et al., 2006). The primers for qRT-PCR were chosen such as to extend products under 200 bp, with no formation of primer dimmers, and cross introns. Sequence of the primers is available upon request. The relative abundance of each message was normalized to actin and calculated as: 2−(Ct gene−Ct actin), where Ct represents the threshold cycle for each transcript.
Statistical Analysis
Differences between TMX-Bcl11bF/Fcre-ERT2 mice and –wild type mice were determined by a two-tailed Student t test (unequal variance). p ≤ 0.05 was considered significant. All values were expressed as mean ± Standard error (SEM).
Supplementary Material
ACKNOWLEDGEMENTS
We greatly acknowledge Drs. Kate MacNamara, Jon Harton and Bill O'Connor (Albany Medical College) for valuable suggestions and exciting discussions. We further acknowledge Dr. Renjie Song (Wadsworth Center) for assistance with cell sorting. We greatly acknowledge Dr. Douglas Cohn, Ms. Victoria Boppert and Ms. Hattie Wang for care of the mice. Grant Support: NIH grants RO1AI067846 and R21AI112492 to Dorina Avram.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interest.
References
- Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu DI, VanValkenburgh J, Morin N, Califano D, Jenkins NA, Copeland NG, Liu P, Avram D. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci U S A. 2011;108:6211–6216. doi: 10.1073/pnas.1014304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 1999;190:995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Califano D. The Multifaceted Roles of Bcl11b in Thymic and Peripheral T Cells: Impact on Immune Diseases. J Immunol. 2014;193:2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartemes KR, Iijima K, Kobayashi T, Kephart GM, McKenzie AN, Kita H. IL-33-responsive lineage- CD25+ CD44(hi) lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J Immunol. 2012;188:1503–1513. doi: 10.4049/jimmunol.1102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano D, Sweeney KJ, Le H, VanValkenburgh J, Yager E, O'Connor W, Jr, Kennedy JS, Jones DM, Avram D. Diverting T helper cell trafficking through increased plasticity attenuates autoimmune encephalomyelitis. J Clin Invest. 2014;124:174–187. doi: 10.1172/JCI70103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, Dekruyff RH, Umetsu DT. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORgammat sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, Avram D. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–12019. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ojeda ME, Klein Wolterink RG, Lemaitre F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, Di Santo JP. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood. 2013;121:1749–1759. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012a;36:451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012b;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, Siebenlist U, Williamson PR, Urban JF, Jr, Paul WE. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential 'inflammatory' type 2 innate lymphoid cells. Nat Immunol. 2015;16:161–169. doi: 10.1038/ni.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010a;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010b;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, Hardwigsen J, Anguiano E, Banchereau J, Chaussabel D, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, Belz GT. TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol. 2013;191:4383–4391. doi: 10.4049/jimmunol.1301228. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant CJ, Hwang YY, Walker JA, Salimi M, Wong SH, Brewer JM, Englezakis A, Barlow JL, Hams E, Scanlon ST, et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 2014;41:283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, Golub R. Notch signaling is necessary for adult, but not fetal, development of RORgammat(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Siracusa MC, Perrigoue JG, Spencer SP, Urban JF, Jr, Tocker JE, Budelsky AL, Kleinschek MA, Kastelein RA, Kambayashi T, et al. IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature. 2010;464:1362–1366. doi: 10.1038/nature08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner CJ, Lesch J, Yan D, Khan AA, Abbas A, Ramirez-Carrozzi V, Zhou M, Soriano R, Eastham-Anderson J, Diehl L, et al. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat Immunol. 2013;14:1229–1236. doi: 10.1038/ni.2743. [DOI] [PubMed] [Google Scholar]
- Uddin MN, Zhang Y, Harton JA, MacNamara KC, Avram D. TNF-alpha-dependent hematopoiesis following Bcl11b deletion in T cells restricts metastatic melanoma. J Immunol. 2014;192:1946–1953. doi: 10.4049/jimmunol.1301976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanvalkenburgh J, Albu DI, Bapanpally C, Casanova S, Califano D, Jones DM, Ignatowicz L, Kawamoto S, Fagarasan S, Jenkins NA, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208:2069–2081. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C, Hirota K, Stieglitz B, Van Snick J, Tolaini M, Lahl K, Sparwasser T, Helmby H, Stockinger B. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat Immunol. 2011;12:1071–1077. doi: 10.1038/ni.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, et al. Transcription factor RORalpha is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel R, Kosan C, Heyd F, Moroy T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem. 2004;279:40906–40917. doi: 10.1074/jbc.M400808200. [DOI] [PubMed] [Google Scholar]
- Zhang S, Rozell M, Verma RK, Albu DI, Califano D, VanValkenburgh J, Merchant A, Rangel-Moreno J, Randall TD, Jenkins NA, et al. Antigen-specific clonal expansion and cytolytic effector function of CD8+ T lymphocytes depend on the transcription factor Bcl11b. J Exp Med. 2010;207:1687–1699. doi: 10.1084/jem.20092136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.