Abstract
Microbes produce a biofilm matrix consisting of proteins, extracellular DNA, and polysaccharides that is integral in the formation of bacterial communities. Historical studies of polysaccharides revealed that their overproduction often alters the colony morphology and can be diagnostic in identifying certain species. The polysaccharide component of the matrix can provide many diverse benefits to the cells in the biofilm, including adhesion, protection, and structure. Aggregative polysaccharides act as molecular glue, allowing the bacterial cells to adhere to each other as well as surfaces. Adhesion facilitates the colonization of both biotic and abiotic surfaces by allowing the bacteria to resist physical stresses imposed by fluid movement that could separate the cells from a nutrient source. Polysaccharides can also provide protection from a wide range of stresses, such as desiccation, immune effectors, and predators such as phagocytic cells and amoebae. Finally, polysaccharides can provide structure to biofilms, allowing stratification of the bacterial community and establishing gradients of nutrients and waste products. This can be advantageous for the bacteria by establishing a heterogeneous population that is prepared to endure stresses created by the rapidly changing environments that many bacteria encounter. The diverse range of polysaccharide structures, properties, and roles highlight the importance of this matrix constituent to the successful adaptation of bacteria to nearly every niche. Here, we present an overview of the current knowledge regarding the diversity and benefits that polysaccharide production provides to bacterial communities within biofilms.
INTRODUCTION
The ability to construct and maintain a structured multicellular bacterial community depends critically on the production of extracellular matrix components (1, 2). While the biofilm matrix may be composed of various molecules, the focus of this chapter is on the extracellular polysaccharides (PSs) important for biofilm formation.
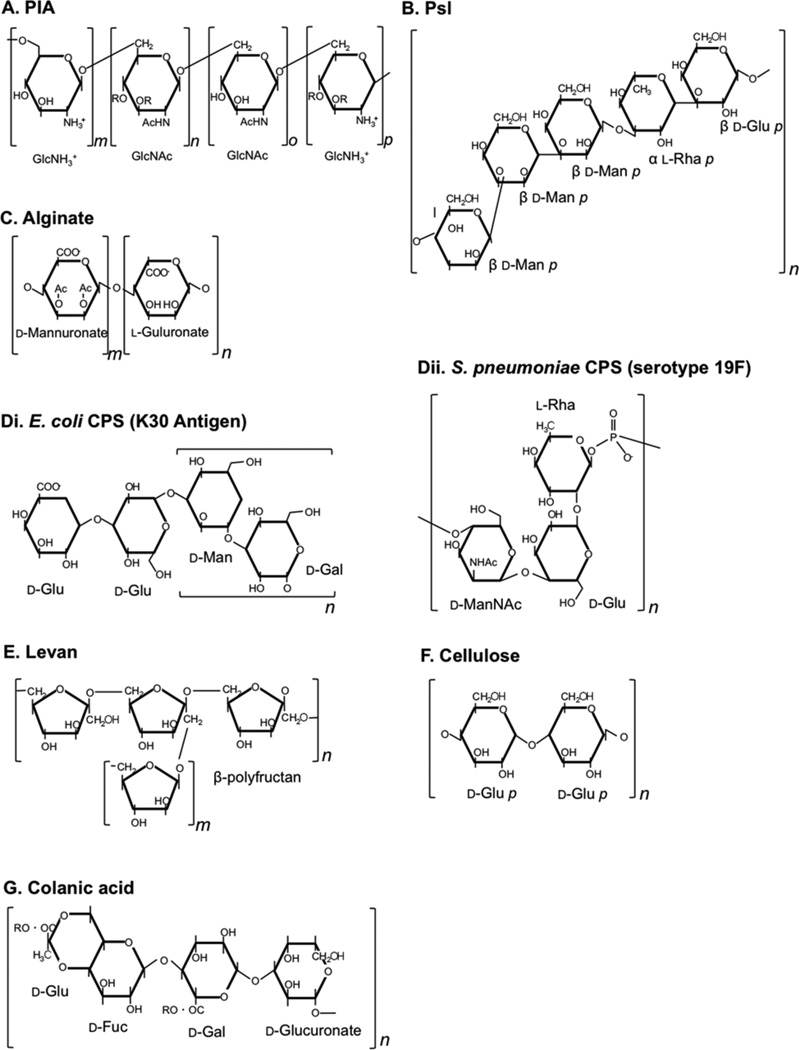
The PSs synthesized by microbial cells vary greatly in their composition and hence in their chemical and physical properties. Many are polyanionic, but others are neutral or polycationic (see Table 1 and Fig. 1) (2). In most natural and experimental environments, PSs are found in ordered compositions, with long, thin molecular chains, ranging in mass from 0.5 to 2.0 × 106 Da. PSs can be elaborated in a multitude of ways, influenced by the environment and association with other molecules such as lectins, proteins, lipids, and bacterial and host extracellular DNA (eDNA). Moreover, many biofilms are composed of multiple bacterial or even fungal species, whereby a range of PSs may interact to generate further permutations of unique architectures (3).
TABLE 1.
Summary of the cellular location, chemical composition, and functions of bacterial polysaccharides important for biofilm formation
| Functions |
|||||
|---|---|---|---|---|---|
| Localization | Charge | Aggregative | Protective | Architectural | |
| Pel | Secreted | NA | X | X | X |
| Psl | Secreted/cell associated | Neutral | X | X | X |
| PIA | Secreted | Polycationic | X | X | |
| Cellulose | Secreted | Neutral | X | X | |
| Alginate | Cell associated | Polyanionic | X | X | |
| CPS | Covalently attached | Polyanionic | X | ||
| Levan | Cell associated | Neutral | X | X | |
| Colanic acid | Cell associated | Polyanionic | X | ||
| VPS | Secreted | NA | X | X | X |
| Bacillus EPS | Secreted | Neutral | X | ||
FIGURE 1.
Adapted representative chemical structures of polysaccharides which participate in biofilm formation including (A) polysaccharide intercellular adhesin (PIA), (B) Psl, (C) alginate, capsular polysaccharide (CPS) from (Di) E. coli and (Dii) S. pneumoniae, (E) levan, (F) cellulose, and (G) colanic acid. Brackets depict repeating units. doi:10.1128/microbiolspec.MB-0011-2014.f1
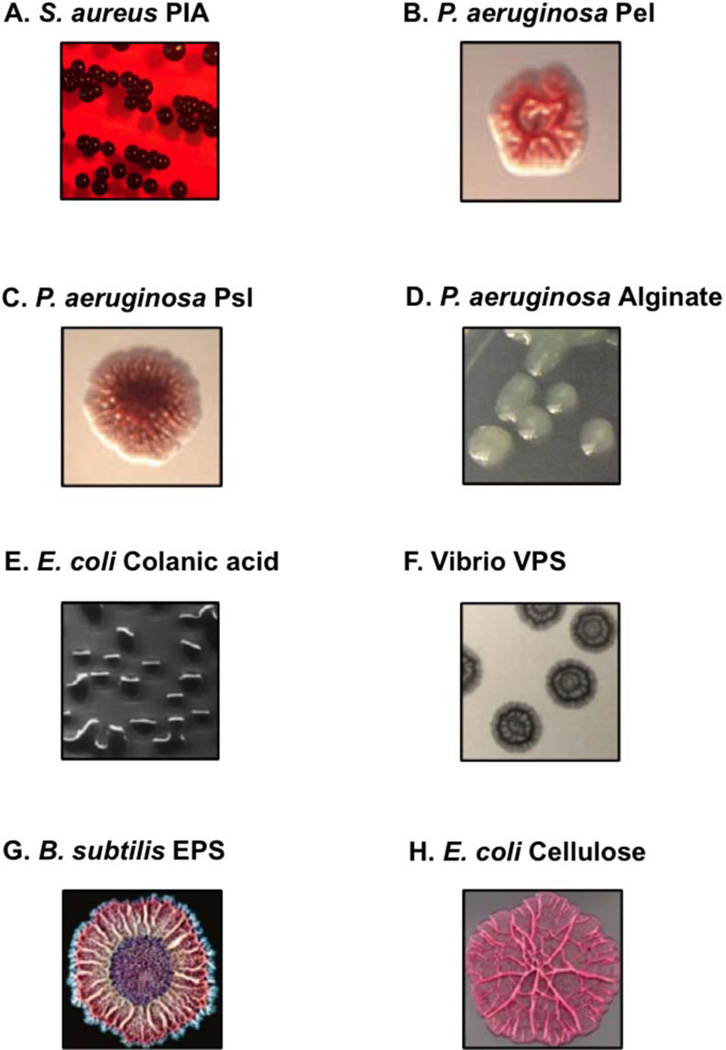
The diversity in PS structure also provides a range of functional roles for PSs in microbial biofilms. For many bacteria, structural and physical consequences of PS expression confer unique colony morphology phenotypes (Fig. 2). The PS in the biofilm matrix dictates a framework for the biofilm landscape. Inhabitants of the biofilm need to be protected from the environment (host cells, antimicrobials, desiccation, temperature, competing microbes, etc.) while maintaining access to nutrients and the ability to respond to changes in the environment. Bacteria generate multiple PSs to cope with these needs in a variety of different ways. PSs can help bacteria adhere to a multitude of different surfaces and host and bacterial cells, provide protection from the onslaught of antimicrobials in the environment, provide reservoirs for nutrient acquisition, and aid in the creation of distinct architectures, which further potentiate an environment suitable for microbes to persist. In this chapter, we will discuss PSs that are known to be important for microbial biofilm formation. For strictly organizational purposes, PSs are divided here into three functional categories to highlight their importance and diversity in biofilm biology. While these PSs are subjectively categorized into aggregative, protective, and architectural, these divisions are by no means exclusive. Several PSs have roles in each of these categories (see Table 1), which will also be discussed below.
FIGURE 2.
Colony phenotypes conferred upon expression or overexpression of PS by representative bacteria. (A) PS intercellular adhesion producing Staphylococcus aureus. Reprinted from World Journal of Microbiology and Biotechnology (214) with permission from the publisher. (B) Pel producing Pseudomonas aeruginosa (ΔwspFΔpsl). Reprinted from Molecular Microbiology (215) with permission from the publisher. (C) Psl producing P. aeruginosa (ΔwspFΔpel). Reprinted from Molecular Microbiology (215) with permission from the publisher. (D) Alginate overproducing P. aeruginosa (mucA22). Not previously published. Credit: Daniel Wozniak. (E) Colanic acid producing Escherichia coli. Reprinted from PLoS One (216) with permission from the publisher. (F) VPS producing rugose variant of Vibrio cholerae. Reprinted from The Proceedings of the National Academy of Sciences of the USA (66) with permission from the publisher. (G) EPS producing Bacillus subtilis. Reprinted from The Proceedings of the National Academy of Sciences of the USA (217) with permission from the publisher. (H) Cellulose producing E. coli (csgD::cm). Reprinted from The Journal of Medical Microbiology (218) with permission from the publisher. doi:10.1128/microbiolspec.MB-0011-2014.f2
AGGREGATIVE POLYSACCHARIDES
The formation of biofilms occurs in multiple stages: initial attachment, microcolony and macrocolony formation, and detachment or disassembly (4–6). Aggregative PSs play essential roles in each of these steps: aiding in adhesion to surfaces, formation of complex structures by promoting microbial interactions, and relief of these interactions promoting dissolution of the biofilm. Bacteria can elaborate multiple PSs, which are important in different strains and varying environmental conditions, including surface substrate, nutrition, and flow rate (7). The redundancy of aggregative PSs produced by many bacteria highlights the essentiality of bacteria remaining associated with the biofilm community. Moreover, the ability to modify PS production provides compensatory mechanisms to adapt to changing environments. The PSs described in this section highlight the importance of aggregation in the biofilm community lifestyle and demonstrate the range of functions of these PSs.
Polysaccharide Intercellular Adhesion
Significance, structure, and regulation
The PS intercellular adhesion (PIA) is the primary PS involved in biofilm formation of Staphylococcus aureus and Staphylococcus epidermidis, which contribute significantly to endocarditis, osteomyelitis, and infections associated with indwelling medical devices (4, 8). PIA was originally identified in S. epidermidis and named as such due its role in mediating cell-cell interactions during biofilm formation (9–11). Similar PSs were subsequently identified in S. aureus and referred to as polysaccharide/ adhesion (PS/A or PSA) (12), poly-N-acetylglucosamine (13) or S. aureus exopolysaccharide (14). It has since been verified that despite possible variations in the degree of N-acetylation, O-succinylation, and molecular weight, each PS represents the same homopolymer composed of β-1-6 linked 2-amino-2-deoxy-d-glucopyranosyl residues (Fig. 1) (11, 13, 15). PIA is synthesized by enzymes encoded by the icaADBC locus, which is negatively regulated by icaR located upstream of icaA (16–18). Importantly, icaADBC orthologous genes have also been identified in other human pathogens such as Escherichia coli (19), Yersinia pestis (20), Aggregatibacter actinomycetemcomitans, Actinobacillus pleuropneumoniae (21), Bordetella spp. (22), Klebsiella pneumoniae, Enterobacter cloacae, Stenotrophomonas maltophilia, the Burkholderia cepacia complex (BCC) (23), and Acinetobacter baumannii (24). The ica operon is tightly regulated in vitro and is induced by a number of environmental conditions. Although there is strain-to-strain variation, PIA can be induced in the presence of high NaCl, glucose, temperature and ethanol, anaerobiosis, and subinhibitory concentrations of some antibiotics (25–27). In S. epidermidis the quorum sensing LuxS system negatively regulates PIA expression, where a luxS mutant demonstrates increased biofilm formation and enhanced virulence in a rat model of biofilm-associated infection (28). σB and RsbU also positively regulate PIA. σB activates the ica operon by repressing transcription of icaR though an unknown intermediate (29). In S. aureus the Spx protein, which directly interacts with RNA polymerase to regulate transcription, negatively regulates the ica operon and biofilm formation (30, 31), while the global regulator SarA positively regulates PIA production and biofilms (32, 33). The importance of PIA during staphylococci infections has been extensively studied, and several roles have been attributed to PIA, such as promoting cellular aggregation and biofilm formation, increased virulence, and protection from host innate immune responses (34–36; reviewed in references 25, 37–40).
Role in biofilm biology
In S. aureus and S.epidermidis, initial attachment is mediated primarily by cell surface proteins that bind to mammalian extracellular matrix/plasma proteins (41). The micro- and macro- colony formation stage requires intercellular bacterial aggregation, and in staphylococci, this is mediated primarily by PIA. This has been illustrated by mutants in the ica operon retaining the ability to adhere to surfaces while being incapable of forming multilayered biofilms (16). In the detachment stage of the biofilm lifestyle, matrix components are often degraded; however, staphylococci do not seem to possess PIA hydrolytic enzymes (37). Instead, PIA is dispersed by elevating expression of detergent-like peptides, which disrupt noncovalent interactions between PIA and the bacterial cell surface (38). The importance of PIA for staphylococci biofilm formation has been demonstrated in vivo in several animal models where the ica operon is upregulated and required for biofilm formation during infection (42–46). However, staphylococcal strains demonstrate a wide range of biofilm phenotypes, and many strains maintain the ability to form biofilms in the absence of PIA (32, 47, 48), where several PIA-independent protein factors and/or eDNA seem to be more important (8, 49, 50). At least some strains are also able to switch between PIA-dependent and -independent phenotypes, which may aid in adaptation to changing environments (51). Collectively, these data demonstrate that biofilm formation and PIA production are integral to the staphylococcal lifestyle and its ability to cause a range of diseases.
Pseudomonas aeruginosa Pel
Significance, structure, and regulation
Pel is an aggregative PS produced by P. aeruginosa. It derives its name from the thick pellicle observed in strains overexpressing the pel operon. This operon encodes seven enzymes with homology to other PS synthesis proteins; however, several genes predicted to be essential for Pel production are missing from this operon (52–54). This indicates that there may be reliance on several other PS synthesis enzymes encoded elsewhere on the chromosome. The structure and composition of Pel remain unknown, but studies are underway to identify the sugars and linkages present in this PS. The common strain PA14 relies exclusively on Pel production for aggregation, because it is missing the pslABCD genes (see below) (52).
Pel production is increased by elevated amounts of the intracellular second messenger cyclic diguanylate (c-di-GMP) (55). Recent studies have described a role for the flagellum regulator FleQ in the regulation of the pel operon, where the cellular pool of c-di-GMP is sensed by FleQ, which causes FleQ to change from a repressor of the pel promoter to an activator (56, 57). In addition, c-di-GMP functions post-translationally in Pel synthesis by modulating activity of PelD (55).
Role in biofilm biology
The contribution of Pel to biofilm structure is most evident when observing biofilms that form at the air-liquid interface, called pellicles. The production of Pel is strongly correlated with increased pellicle formation, adherence to culture tubes, and formation of aggregates in broth culture (52, 58, 59). The biofilm properties of Pel and Psl (below) are closely linked, because there is evidence of functional overlap in these two PSs (59). It appears that the structural requirements of these two PSs for biofilm formation are strain specific, because there are examples of strains which are incapable of producing one or the other yet still can efficiently form biofilms (59).
Pseudomonas aeruginosa Psl
Significance, structure, and regulation
The polysaccharide synthesis locus produces another important P. aeruginosa PS, Psl (60). The psl operon consists of 15 genes (pslA-O), which are cotranscribed (61). Mutagenesis studies revealed that 11 of these genes (pslACDEFGHIJKL) are essential for Psl production and surface attachment (62). There are two forms of Psl, a high molecular weight cell-associated form and a low molecular weight form that appears to be released from cells (62). Released Psl consists of d-mannose, d-glucose, and l-rhamnose in a 3:1:1 ratio, respectively (Fig. 1) (62, 63). The structure of cell-associated Psl is unknown but is believed to be a polymer of mannose, glucose, and rhamnose and possibly galactose (63). Psl is produced primarily in nonmucoid strains, because expression of the psl operon is repressed in mucoid strains (64).
The effect of Psl is most easily studied in rugose small colony variant strains such as the ΔwspF mutant, where Psl is overproduced (65). The overproduction of Psl leads to a rough, wrinkled colony morphology and hyper-biofilm phenotype, which is consistent with overexpression of adhesive PSs in other bacteria (66, 67). As with Pel, high expression of Psl in this strain is due to elevated levels of the second messenger cyclic di-GMP (65, 68, 69). There are several pathways that lead to regulation of Psl production by altering the c-di-GMP level of the cell. The first of these regulatory pathways to be described was the regulation of the psl operon by the master flagellum regulator, FleQ (56, 57). In this instance, c-di-GMP relieves the repressive effect of FleQ at the psl operon promoter, leading to expression of the psl operon (56, 65).
Another important observation is that patients who survive systemic infections with P. aeruginosa often produce neutralizing antibodies against Psl (70). Passive immunization with these antibodies is protective against P. aeruginosa infection in several murine models (70). Psl also increases adherence to epithelial cell layers and evasion of phagocytosis by neutrophils, emphasizing that Psl plays important roles during the initiation and persistence of chronic infections (71, 72).
Role in biofilm biology
PAO1 Δpsl mutants form thin, diffuse biofilms lacking structure compared to the wild type PAO1, while the psl overexpressing strains form thicker biofilms with significantly more biomass and microcolony height (73). While this suggests that Psl is essential for biofilm formation in P. aeruginosa strain PAO1, it is important to note that this requirement of Psl for biofilm formation varies from strain to strain. For example, the common strain PA14 does not synthesize Psl yet retains the ability to form biofilms (74). This discovery led to an investigation of the roles of the adhesive PSs, Psl, and Pel in various P. aeruginosa isolates. It was concluded that there are strain-specific requirements for PS production in biofilms. In some strains, Psl is exclusively required for biofilm formation, while in other strains, Psl and Pel are functionally redundant (59). Importantly, one of these two adhesive PSs is required in all strains tested to form a mature biofilm in vitro (59). Recent work indicates that Psl can also function as a signaling molecule that stimulates biofilm formation (75). Psl is also protective against many classes of antibiotics, serving yet another role in the biofilm matrix (76). Psl-producing cells were more resistant to a wide range of antibiotics than cells unable to produce Psl. This protective effect was transferable to cells unable to produce Psl when grown in coculture, including other species of bacteria such as E. coli and S. aureus (76). Collectively, these observations indicate that Psl is important for initial attachment, resistance, and biofilm structure.
PROTECTIVE POLYSACCHARIDES
Microbial protection from the onslaught of host and environmental factors is one of the most quintessential and frequently described attributes of the biofilm mode of growth. In fact, a current criteria used to define infectious biofilms is increased recalcitrance to antimicrobials and the host immune response (77, 78). While several factors including metabolic heterogeneity, altered chemical microenvironments, and persister cell populations contribute to biofilm antimicrobial resistance, an important role for PSs is clearly evident (79–81). It had been initially proposed that PSs in the matrix pose a diffusion barrier, which protects biofilm-grown cells from antibiotic penetration (82). However, studies have demonstrated that several antibiotics can readily penetrate the biofilm (83, 84). While diffusion may not be prevented, it may be delayed enough to induce expression of genes that mediate tolerance (85) or be neutralized by enzymes within the matrix before reaching the bacterial cells (86, 87).
Another prevalent and important role for PSs is protection from opsonic and nonopsonic phagocytosis. Several studies demonstrate that antibodies or inflammatory cells cannot efficiently penetrate biofilms embedded in a PS matrix (8, 49, 88–91). Protection from the immune response results in poor detection, promoting maintenance of stable chronic infections and decreased virulence. Importantly, PS-producing bacteria may confer resistance to nonproducing bacteria within the biofilm (76). PSs are also important for providing a hydrated biofilm environment, which can provide protection from desiccation and promote biofilm fluidity (2, 92). The PSs described in this section provide evidence for a diversity of protective functions toward biofilm bacteria.
Alginate
Significance, structure, and regulation
The first P. aeruginosa PS described was alginate due to its prevalence in cystic fibrosis (CF) pulmonary infections, whereby up to 90% of P. aeruginosa clinical isolates overproduce alginate (referred to as mucoid) (60, 93, 94). CF patients are initially colonized by nonmucoid P. aeruginosa (Psl and/or Pel producing) but convert to the mucoid phenotype during the course of infection. Mucoid conversion confers a selective advantage to P. aeruginosa in the CF lung (discussed further below) (95). Mucoid conversion is directly correlated with an increase in morbidity and mortality of CF patients and remains a significant clinical challenge for eradicating P. aeruginosa infections (96). While the majority of alginate research has focused on P. aeruginosa, alginate production has also been reported in other Pseudomonads and Azotobacter vinelandii (54, 97). Alginate is a random linear polymer of variably acetylated 1,4-linked β-d-mannuronic acid and its C5 epimer α-l-guluronic acid, originally identified in brown seaweeds (Fig. 1) (98, 99). This acetylated polymer structure yields a highly hygroscopic PS promoting biofilm fluidity and resistance to desiccation (2, 92).
Alginate synthesis is tightly regulated due to the high metabolic cost of production (alginate regulation has been extensively reviewed in references 95, 100–102). In brief, expression of the alginate biosynthetic operon (algD-A) is dependent on the alternative sigma factor AlgT (also called AlgUAσE/o22/RpoE). The activity of AlgT is antagonized by the anti–sigma factor MucA, which sequesters AlgT to the cell membrane, preventing interaction with AlgT-dependent promoters (100). Alginate is constitutively overproduced (as seen in mucoid CF isolates) upon acquisition of mutation(s) in the mucA gene, which results in a truncated, inactive protein. The mutant MucA is unable to inactivate AlgT, allowing expression of the AlgT regulon, including the alginate operon (95, 100, 103). Additional regulators of alginate production include AlgR, IHF, AlgB, and AmrZ (algD transcriptional regulators) and MucB to E and P, AlgW, KinB, and ClpX proteases (regulation of MucA stability) (100, 101, 104–106).
Role in biofilm biology
Early microscopy evidence demonstrated that mucoid P. aeruginosa forms microcolonies embedded in an extracellular matrix in the alveoli of CF patients (88, 107). Alginate was initially thought to be essential for P. aeruginosa adherence and biofilm formation, (108–111). However, this paradigm has been challenged by multiple groups who, by comparing isogenic mucoid and nonmucoid strains, demonstrated that alginate is not required for biofilm formation (112–15). These studies, corroborated by more recent work, demonstrate that the Psl and Pel PSs provide attachment and structural support for nonmucoid and mucoid biofilms (116, 117), while alginate contributes to generation of a unique architecture (112, 114, 115). Alginate acetylation further influences mucoid biofilm architecture by promoting cell-cell adhesion (112, 114, 115, 118).
The biofilm architecture constructed by mucoid P. aeruginosa, combined with the intrinsic properties of alginate, provide P. aeruginosa significant protection from antimicrobials in the microenvironment. Mucoid P. aeruginosa biofilms are more resistant to antibiotic treatment in vitro (115, 119, 120), and even the most aggressive antibiotic treatments are unable to eradicate mucoid P. aeruginosa infections in CF patients (96). Alginate also provides protection from the innate immune response by inhibiting both opsonic and nonopsonic phagocytosis, further influenced by the levels of alginate acetylation (121–125). Protection is additionally afforded by the ability of alginate to scavenge reactive oxygen intermediates (126, 127) and inhibit killing by cationic antimicrobial peptides (128–130). Although antibodies to alginate are found in the sera of chronically infected CF patients, these antibodies fail to mediate opsonic killing of P. aeruginosa in vitro (131). Additionally, the anionic nature of alginate promotes cation chelation (Ca2+ preferentially) (132). Calcium chelation by alginate biofilms induces type III secretion, whose effectors may provide further protection from host immune responses (133). Contemporary studies further emphasize the protective nature of P. aeruginosa alginate, demonstrating that mucoid biofilms contain more viable cells than nonmucoid biofilms and are more resistant to DNase treatment (116, 117). Moreover, compared to nonmucoid isolates, mucoid P. aeruginosa maintains in vitro biofilm formation capacity and gene expression profiles during chronic lung infection of CF patients (134). This suggests that protection from the CF microenvironment afforded by alginate results in a more stable population over time and may help to explain how clonal mucoid strains can persist for decades in CF patients.
Capsular Polysaccharide
Significance, structure, and regulation
Bacterial capsular polysaccharides (CPSs) are found on the cell surface of a broad range of species. The CPS is tightly associated with the bacterial cell surface via covalent attachments to either phospholipid or lipid-A molecules. CPSs are highly hydrated molecules composed of repeating monosaccharides joined by glycosidic linkages (135). CPSs are extremely diverse, not only in the monosaccharide constituents, but also in glycosidic linkages, branching, and substitutions with noncarbohydrate residues, leading to a nearly unlimited range of structures (two examples in Fig. 1) (136). For example, over 80 CPSs (K antigens) have been identified in E. coli, and 93 CPSs (serotypes) in Streptococcus pneumoniae (137, 138). Despite the variety of CPS structures, many of the details of polymer synthesis and regulation are shared among Gram-positive and Gram-negative bacteria. Capsule synthesis requires nucleotide diphosphosugar precursors available in the cytoplasm and concludes with assembly of the polymer at the periplasmic face of the plasma membrane (136). The three primary bio-synthetic pathways are termed Wyz dependent, synthase dependent, and ATP-binding cassette transporter dependent (139). Many CPS-producing bacteria respond to environmental conditions by varying capsule production between a low CPS-producing phase and a high CPS-producing phase, whereby stable phenotypic variants may also be selected for under specific environmental conditions (138, 140, 141). Moreover, CPS plays pivotal roles in bacterial survival in the environment and infection in the host, providing protection from desiccation, opsonophagocytosis, and complement-mediated and cationic antimicrobial peptide-mediated killing (135).
Role in biofilm biology
The complexity of biofilm formation in varying environments and stages of infection, the ubiquity and diversity of CPSs among bacteria, and the propensity of bacteria to synthesize multiple PSs have created a complex and inconclusive view of the role of CPSs in biofilm biology. In general, CPS production is thought to inhibit bacterial adherence and biofilm formation. CPS-deficient strains in S. pneumoniae, Neisseria meningitidis, S. aureus, and Vibrio vulnificans demonstrate increased adherence to epithelial cells and surfaces and more robust biofilm formation (142–146). CPS is also often downregulated during biofilm formation and upon contact with epithelial cells (143, 147, 148). It is hypothesized that decreased CPS further enhances the quiescent nature often associated with biofilm formation: potentiating immune evasion, decreased virulence, and persistence (142, 143, 149). This is supported by observations that low-CPS-producing strains are more frequently isolated from chronic infections, and high-CPS strains, from acute infections (143, 146).
However, this simplistic, binary view may not illustrate the complete role of CPSs in biofilm formation. Studies have demonstrated that CPSs may be important in mature biofilms, aiding in the maintenance of biofilm size and dispersal. In V. vulnificans, CPS was found to be expressed in later stages of biofilm formation and was hypothesized to be synthesized and secreted after biofilms mature and reach a threshold of cell density (150). In support of this, CPS expression in mature biofilms and maintenance of biofilm size is regulated by quorum sensing molecules (151). Similarly, in S. pneumoniae, higher CPS expression has been demonstrated in biofilm towers, compared to cells at the biofilm surface (148). Further evidence supporting a role of CPS in biofilm maturation is seen during formation of intracellular biofilm-like communities within bladder epithelial cells during urinary tract infections by uropathogenic E. coli (149). Collectively, these data point toward an overall view for multiple pathogens that CPS plays an important, yet incompletely defined role in biofilm biology. Several lines of evidence support an inhibitory role during initial attachment and biofilm formation; however, the ability to then enhance CPS synthesis in later stages may prove to be critical during infection. This clearly requires further study.
Levan
Significance, structure, and regulation
Levans are high molecular mass, neutral homopolymers composed of β-d-fructans with extensive and irregular branching (Fig. 1) (152). Levan production confers a mucoid phenotype to bacteria expressing this and has been described in several plant pathogens (Pseudomonas syringae, Erwinia amylovora, and Bacillus subtilis) and Streptococcus mutans oral biofilms (153–155). Levan is produced exclusively from extracellular sucrose catalyzed by excreted levansucrase (153, 156, 157). Levansucrase is activated in the presence of sucrose and is frequently regulated by two-component sensor kinases (LadS in P. syringae; SacX/SacY and DegS/DegU in B. subtilis; RscC/RscB, GacS/GacR (GrrS/GrrA), and EnvZ/OmpR in E. amylovora; and CovS/CovR [VicK/ VicR] in S. mutans [152, 158, 159]). In B. subtilis sacB and sacC are involved in levansucrase production and modification. Levansucrase is secreted from the cell by the SecA pathway (160, 161). In P. syringae and E. amylovora the levansucrase biosynthetic operon is composed of three genes: iscA-C (153, 162). Proposed functions for levan include protection from desiccation and bacteriophages, nutrient storage, and virulence (157, 163, 164).
Role in biofilm biology
The role of levan production in biofilms has predominately been described in S. mutans dental biofilms. Upon exposure to sucrose, levan accumulates on dental plaques and is catabolized to acid when the environmental sugar is depleted. Prolonged exposure promotes dental carries (165). While levan is not required for S. mutans biofilm formation, supplementation of sucrose to biofilms in vitro yields thicker structures, which contain more levan and are more resistant to shear stress (92, 166, 167). Interestingly, levan is a water-soluble PS, and levan produced by B. subtilis possesses a particularly low viscosity. At typical concentrations where other PSs display gel behavior, levan remains fluid, suggesting that it may not form an adhesive framework (168). In P. syringae, levan is not required for biofilm formation and is not responsible for maintenance of biofilm structure. Instead, levan may function as a storage molecule, utilized during periods of starvation (164). Levan was identified in the voids and blebs of P. syringae biofilms, whose formation required supplementation of sucrose in the media (164). This theory of nutrient retention by levan was previously suggested for S. mutans in the oral cavity (163). Collectively, these data suggest that levan contributes to the physio-chemical properties of bacterial biofilms but is not required for its formation.
ARCHITECTURAL POLYSACCHARIDES
Some of the first biofilm-related PSs identified were studied due to the role that they play in biofilm structure. These PSs have provided extensive insight into the regulation of biofilm formation and structure, primarily due to the readily observable phenotypes associated with mutants or overproducers of these products. For example, through studying the rugose phenotype associated with PS produced by Vibrio cholerae (Vibrio polysaccharide [VPS]), important studies in gene regulation, second messenger signaling, and biofilm matrix assembly have been reported (169–171).
Colanic Acid
Significance, structure, and regulation
Colanic acid (CA), or M antigen, is a branched PS composed of glucose, galactose, and glucuronic acid (Fig. 1) (172). The 19-gene wca cluster is responsible for producing CA in several species of Enterobacteriaceae (172–174). This cluster was formerly called the cps cluster, but it was renamed when it was discovered that these genes encode enzymes that produce CA (172). As with most PSs, several of the precursors of CA, such as UDP-d-glucose and UDP-d-galactose are produced by enzymes that are located in other loci, while many of the enzymes dedicated to CA synthesis are included in the wca cluster (172). For many years, the study of CA was focused on its effect on colony morphology, but advances in molecular techniques allowed a genetic approach to the regulation, structure, and role of CA in the biofilm matrix. The wca gene cluster is regulated by the well-characterized Rcs phosphorelay system. In this variation of a two-component system, RcsC transfers a phosphate to RcsD, which subsequently transfers it to RcsB. RcsB is a LuxR family helix-turn-helix response regulator, which binds the promoter of the wca operon and activates transcription (175–178). The highly unstable accessory protein RcsA can enhance this interaction. Collectively, this signaling cascade leads to upregulation of the wca cluster and production of CA, which allows the bacterium to endure environmental stresses such as desiccation (179). CA is primarily produced at lower temperatures, indicating that it is most beneficial to the bacterium in the environmental phase of its life cycle (175).
Role in biofilm biology
CA was one of the first PSs studied with respect to its role in biofilm formation. Early in vitro work demonstrated that an E. coli mutant incapable of producing CA had two interesting phenotypes. First, the CA mutant demonstrated attachment capabilities similar to the wild type strain. This was interesting to the biofilm field, because previous work on the role of PSs in biofilms indicated that the PS was involved in the initial attachment to surfaces (12, 180). Second, though the CA mutant was able to attach at wild type levels, it formed biofilms with less three-dimensional structure than the biofilms formed by the wild type strain (181). These biofilms were described as “collapsed,” with cells densely packed against the substrate. It was concluded that CA was critical for the development of mature, three-dimensional biofilms by E. coli (181). This work was integral to the biofilm field, because it was one of the first observations that implicated PSs as structural members of the biofilm matrix.
In E. coli, CA is produced at low temperatures and therefore is more important for environmental survival than during infection (182). In contrast, the role of CA produced by the pathogen Salmonella enterica serovar Typbimurium is dispensable for binding abiotic surfaces (183). Further characterization of the role of CA in S. enterica biofilms indicated that the CA mutants were unable to build a three-dimensional biofilm on either HEp-2 cells or chicken intestinal epithelium, suggesting that CA is involved in pathogenesis (183). These divergent roles for CA in two relatively closely related bacteria demonstrate how PSs can enhance the diversity of bacterial colonization niches and biofilm phenotypes.
Vibrio Polysaccharide
Significance, structure, and regulation
V. cholerae is a Gram-negative aquatic bacterium that is the causative agent of the diarrheal disease cholera. This bacterium must endure many environmental stresses as it transits from an aquatic environment to the human gastrointestinal system. One of the mechanisms that are important in maintaining this infectious cycle is the production of the VPS. High expression of this PS is the cause of the rugose variant of an El Tor V. cholerae strain (66, 184). VPS was demonstrated to be essential for biofilm formation and resistance to chlorine. The NtrC family response regulator VpsR activates transcription of the vps operon, allowing for production of the PS (185).
The VPS is encoded by two operons, vpsA through vpsK, and vpsL through vpsQ. These operons are positively regulated by the response regulators VpsR and VpsT (185, 186). Expression analysis indicates that VpsR is epistatic to the positive regulator VpsT (187). Additionally, both VpsT and VpsR induce expression of each other, presenting a rapid positive feedback loop resulting in production of VPS (186). The negative regulator HapR represses VpsR, VpsT, and the vps operons. Collectively, these three regulators affect the expression of VPS and therefore biofilm phenotypes and colony rugosity. Deletion of hapR in a smooth strain relieves the negative regulation of vps expression, resulting in the transition to a rugose colony. Conversely, deletion of vpsR or vpsT in a rugose strain produces a smooth strain (187).
Several studies have indicated that vps genes are expressed during intestinal colonization and infection. VPS could aid in the formation of hyper-infectious aggregates of cells, as well as attachment and colonization of the intestine. In support of this, vps operon mutants demonstrated a significant defect in colonization of infant mouse intestines, supporting VPS being involved in host colonization (188).
Recent work indicates that the expression of the vps gene clusters is dependent on elevated intracellular levels of the second messenger c-di-GMP. VpsT binds c-di-GMP, resulting in oligomerization and upregulation of the vps operons (169, 170). Several diguanylate cyclases and phosphodiesterases affect VPS production by altering the cellular pool of c-di-GMP. Specifically, the phosphodiesterase VieA reduces vps expression by degrading c-di-GMP (189), while the diguanylate cyclases CdgA, H, K, L, and M all contribute to the activation of VpsT by producing elevated levels of c-di-GMP (170).
VPS consists mainly of glucose (52.6%) and galactose (37.0%), with small amounts of N-acetylglucosamine, mannose, and xylose (5.1%, 3.8%, and 1.5%, respectively). The primary linkages in this PS are 4-linked glucose and 4-linked galactose, suggesting that these sugars form the linear backbone. Branching of the PS is suggested by the detection of small amounts of 3,4- and 4,6-linked galactose and glucose, as well as 2,4-linked galactose (66).
Role in biofilm biology
Much of what is known regarding the role of VPS in biofilm biology stems from the observation of V. cholerae rugose variants. These colonies have a highly structured, wrinkled morphology due to the overproduction of VPS. When the VPS genes are deleted from a rugose strain, the colony reverts to a smooth morphology. In addition to the rough colony morphology, rugose strains demonstrate enhanced attachment and biofilm formation phenotypes (66, 185). Both vps operons are essential for VPS production and, therefore, biofilm development (188). Super-resolution confocal microscopy revealed that VPS promotes the retention of daughter cells in the biofilm, as well as the accumulation of the biofilm matrix proteins RbmA, Bap1, and RbmC (171). In the absence of VPS, biofilms were restricted to the monolayer stage. During biofilm growth, VPS was extruded from the cell and formed spherical foci on the cell surface. It was proposed that the VPS, along with the proteins Bap1 and RbmC, forms the biofilm matrix envelope around cell clusters. Collectively, VPS is regarded as an essential part of the V. cholerae biofilm matrix, which is important in both the environmental and host phases of the infectious cycle.
B. subtilis Polysaccharide
Significance, structure, and regulation
B. subtilis produces several PSs that affect biofilm structure and function, but the prevalent architectural PS is EPS (157, 190, 191). During a study of the formation of fruiting bodies, it was observed that ΔyveQ (epsG) and ΔyveR (epsH) mutants were unable to form structured pellicles or produce multicellular fruiting bodies (190). Sixteen genes encoding EPS are in an operon, and most are homologous to those involved in PS production and modification in other bacteria (190). A subsequent report updated the operon annotation to 15 genes and renamed it epsA-O (67). Further characterization of the genes involved in biofilm formation indicated that an unlinked gene, ybxB, is essential for biofilm formation (191). The ΔybxB mutant forms pellicles that are similar to the EPS-deficient ΔyveR mutant. yhxB has sequence similarity to both phosphoglucomutase and phospho-mannomutase sugar-modifying enzymes that are involved in PS synthesis in other bacteria. It was concluded that yhxB along with the epsA-O operon are essential for the production of EPS and biofilms (191). Though this PS has been extensively studied, the structure is unknown (157).
The epsA-O operon is repressed by SinR (67), which binds to an operator upstream of the operon (67, 192). PS repression is relieved by SinI, which forms a complex with SinR and antagonizes its activity (67). As a result, SinR is deemed the master biofilm regulator of B. subtilis (67).
Role in biofilm biology
EPS is essential for the formation of pellicles and surface structures by B. subtilis (191). A ΔepsH mutant forms weak pellicles and smooth colonies, in contrast to the robust pellicle and rugose colonies formed by the wild type strain (67). Increased EPS expression in a ΔsinR mutant exhibits enhanced cell aggregation, pellicle formation, colony rugosity, and impaired swarming motility (67). The importance of PS in biofilm formation and cell aggregation is highlighted with the observation that addition of norspermidine can induce biofilm dissolution by targeting PS and interfering with its ability to aggregate and provide biofilm structure (193), though recent work has contradicted some of the claims about the effect of norspermidine (194), indicating that further research is required on this topic. Collectively, these data demonstrate the importance of the structural role of EPS in biofilm formation and stability.
Cellulose
Significance, structure, and regulation
Cellulose is one of the most abundant PSs in nature. The structure of cellulose fibrils is conserved throughout evolution, with bacterial cellulose indistinguishable from cellulose produced by higher-order algae, fungi, and plants. This polymer consists of repeating chains of β-1,4 linked d-glucose that form fibrils, which resemble cables (Fig. 1). Fibrils range from 1 to 25 nm in width and 1 to 9 µm in length and are assembled in the extracellular space immediately adjacent to the cell. These cables can align to form sheets, introducing the structural rigidity that plant cellulose provides due to its insoluble and inelastic nature. Cellulose fibrils have a tensile strength similar to steel (195). This structure is most evident in wood and paper products, which are largely composed of cellulose.
The role of cellulose in bacteria was first described in the Gram-negative bacterium Gluconacetobacter xylinus (formerly Acetobacter xylinum) (196–198). This bacterium produces large quantities of highly pure cellulose that shares many properties with cellulose produced by higher-order organisms, which aided in research on the role of plant and algal cellulose. Static cultures of G. xylinus produce a robust pellicle that is rich in cellulose, facilitating the isolation of the polymer from the matrix. Subsequently, many bacteria have been found to produce cellulose, including commensal organisms such as E. coli, pathogens such as S. enterica, and environmental organisms such as many Pseudomonas spp. (199–201). The diversity in the niches of the organisms that produce cellulose highlights the variety of functions that it can play in the biofilm matrix.
The cellulose production genes have several names, depending on the species. These include acs (Acetobacter cellulose synthesis), bcs (bacterial cellulose synthesis), and cel (cellulose) (200). In general, the operon consists of two conserved genes and several accessory genes. The first conserved gene is bcsA (acsA or celA), which encodes the cellulose synthase enzyme. This is followed by bcsB (acsB or celB), which encodes a c-di-GMP binding protein (200). Additional genes in the operon can vary but generally include genes including a cellulase (bcsZ or celC) (200). The most thorough characterization of the cellulose biosynthetic machinery has been conducted in G. xylinus. In this bacterium, the cellulose synthase enzyme complexes are linked to the cytoplasmic membrane by 8 to 10 transmembrane domains. Approximately 50 synthase complexes form in a row along the long axis of the bacterium, excreting cellulose fibers that rapidly aggregate with nearby fibers to form a ribbon (200, 202).
Regulation of cellulose production is linked to c-di-GMP levels. In fact, the initial characterization of c-di-GMP and the diguanylate cyclase and phosphodiesterase enzymes responsible for this molecule were made in G. xylinus as a result of studies investigating factors that promote cellulose production (203, 204). c-di-GMP binds to BcsB and promotes the production of cellulose. In addition, cellulose production in E. coli and S. enterica is dependent on AdrA, which contains a GGDEF domain (199, 205). The cellulose synthase is expressed constitutively but is not active without the c-di-GMP inducer. It has been proposed that AdrA is the diguanylate cyclase that provides the c-di-GMP required for cellulose production to the cellulose synthase. This is supported by the evidence that expression of adrA from a plasmid is sufficient to induce cellulose production (199). This system provides rapid production of cellulose, since the biosynthetic machinery is present, requiring only production of AdrA to provide the allosteric inducer c-di-GMP.
Role in biofilm biology
Cellulose provides both structure and protection to bacterial cells in biofilms. The first reports of the contribution of cellulose to G. xylinus pellicles indicated that cellulose was responsible for cell aggregation in the pellicle, allowing the biofilm to float to the surface of the culture where oxygen was readily available to the bacteria (198). In addition to this role, cellulose provides protection from the mutagenic effects of ultraviolet light (195, 206).
The role of cellulose in biofilm formation has expanded across many species. One report describes the production of cellulose in many Enterobacteriaceae, including E. coli, Salmonella enteritidis, Salmonella typbimurium, and K. pneumoniae (199). In these species, cellulose often interacts with the curli fimbriae to develop the biofilm matrix that is responsible for the rdar (rough, dry, and red) colony phenotype (199, 200). Other groups have observed that cellulose production results in pellicle formation in many Pseudomonas spp. (201). Cellulose production enhances binding of epithelial cells and reduces immune response to the bacteria in the probiotic strain of E. coli Nissle 1917 (207). These functions are important for establishing a commensal relationship with the host epithelium. Cellulose is produced by many bacteria to utilize the properties of this simple polymer for a wide variety of functions, providing a good example of the functional diversity of PSs.
FUTURE PERSPECTIVES
A wealth of knowledge has been gained regarding the composition and functions of PS components of the biofilm matrix by utilizing methods such as traditional genetic techniques combined with biochemistry and immunochemistry, lectin/carbohydrate stains, and microscopy (208). While necessary for initial observations, this reductionist point of view leaves significant gaps in our understanding of biofilm biology. PS isolation is difficult to achieve, especially from environmental or host sources, which contain a diverse range of components. Moreover, the paucity of animal models, particularly for chronic infections, presents a significant challenge for correlating in vitro findings to infection. Many bacteria produce multiple PSs, and in vivo infections are rarely comprised of a single microbe. Our current understanding of how PSs interact in polymicrobial communities in the environment or the host is extremely limited. Bridging this gap will require the development of more extensive in situ techniques to visualize, measure, and fully define PSs in real-world situations. Further development and utilization of microscopy techniques such as super-resolution and Raman scattering microscopy, combined with fully hydrated living models, will be essential to provide a complete interpretation of the complexity of microbial biofilms and the contribution of PS production (209).
While PSs are a predominant and well-studied biofilm component, the matrix is clearly composed of a range of macromolecules, including eDNA, proteins, and lipids. It stands to reason and is frequently speculated that these molecules interact in the matrix to form specific and dynamic interactions to fine-tune the biofilm community to varying environments. However, information regarding the biological significance, regulation, and direct visualization of these interactions are currently lacking. For example, since the seminal discovery by Whitchurch and colleagues in 2002 that eDNA is an important component of P. aeruginosa biofilms (210), only a few studies have defined how eDNA interacts with PS in the matrix. It has been suggested that in Myxococcus xanthus biofilms, eDNA directly interacts with PS, enhancing the physical strength and resistance to biological stress (211). Accordingly, a mathematical model of biofilm stress relaxation reveals evidence that filamentous eDNA may interact with PS to produce well-defined agglomerate structures with unique physioelastic properties (212). For protein-EPS interactions, an additional observation in M. xanthus illustrated the first direct interaction of EPS and type IV pili (213). However, the biological significance and the nature of these interactions remain unclear, representing a significant gap in our understanding of the biofilm matrix.
In addition to interacting with other components of the biofilm matrix, it is likely that various PSs, produced by either the same bacteria or by other species in the environment, interact with each other to produce unique and/or compensatory functions. Some PSs may be exclusively expressed at specific stages of biofilm formation or under certain environmental conditions. Interrogating the spatial and temporal production of PSs and how they interact with other molecules in the environment will significantly enhance our understanding of how biofilms develop and how they may be modulated.
ACKNOWLEDGMENTS
Dominique H. Limoli and Christopher J. Jones contributed equally to this work.
Footnotes
Conflicts of interest: We disclose no conflicts.
REFERENCES
- 1.Branda SS, Vik Å, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland IW. The biofilm matrix: an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 3.Jenkinson HF, Lamont R. Streptococcal adhesion and colonization. Crit Rev Oral Biol Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 4.Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010;5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harbor Perspect Med. 2013;3:a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JKM, Ragunath C, Kaplan JB, Mack D. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 9.Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun. 1992;60:2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenney D, Hübner J, Muller E, Wang Y, Goldmann DA, Pier GB. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun. 1998;66:4711–4720. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maira-Litran T, Kropec A, Abeygunawardana C, Joyce J, Mark G, III, Goldmann DA, Pier GB. Immunochemical properties of the Staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce JG, Abeygunawardana C, Xu Q, Cook JC, Hepler R, Przysiecki CT, Grimm KM, Roper K, Ip CCY, Cope L, Montgomery D, Chang M, Campie S, Brown M, McNeely TB, Zorman J, Maira-Litrán T, Pier GB, Keller PM, Jansen KU, Mark GE., III Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res. 2003;338:903–922. doi: 10.1016/s0008-6215(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 15.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun. 2005;73:3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 18.Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184:4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Preston JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JKM, Ramasubbu N. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parise G, Mishra M, Itoh Y, Romeo T, Deora R. Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol. 2007;189:750–760. doi: 10.1128/JB.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skurnik D, Davis MR, Benedetti D, Moravec KL, Cywes-Bentley C, Roux D, Traficante DC, Walsh RL, Maira-Litran T, Cassidy SK, Hermos CR, Martin TR, Thakkallapalli EL, Vargas SO, McAdam AJ, Lieberman TD, Kishony R, LiPuma JJ, Pier GB, Goldberg JB, Priebe GP. Targeting pan-resistant bacteria with antibodies to a broadly conserved surface polysaccharide expressed during infection. J Infect Dis. 2012;205:1709–1718. doi: 10.1093/infdis/jis254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litran T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cue D, Lei MG, Lee CY. Genetic regulation of the intercellular adhesion locus in Staphylococci. Front Cell Infect Microbiol. 2012;2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Anti-microb Agents Chemother. 2000;44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuryastuti T, Krom BP, Aman AT, Busscher HJ, van der Mei HC. Ica-expression and gentamicin susceptibility of Staphylococcus epidermidis biofilm on orthopedic implant biomaterials. J Biomed Mater Res. 2010;96A:365–371. doi: 10.1002/jbm.a.32984. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2005;74:488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knobloch JKM, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frees D, Chastanet A, Qazi S, Sørensen K, Hill P, Msadek T, Ingmer H. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 31.Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol. 2006;188:4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valle J, Toledo-Arana A, Berasain C, Ghigo J-M, Amorena B, Penadés JR, Lasa I. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 34.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 35.Kristian SA, Birkenstock TA, Sauder U, Mack D, Götz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis. 2008;197:1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 36.França A, Vilanova M, Cerca N, Pier GB. Monoclonal antibody raised against PNAG has variable effects on static S. epidermidis biofilm accumulation in vitro. Int J Biol Sci. 9:518–520. doi: 10.7150/ijbs.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto M. Staphylococcus epidermidis: the “accidental” pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong K-F, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 39.Mack D, Becker P, Chatterjee I, Dobinsky S, Knobloch JKM, Peters G, Rohde H, Herrmann M. Mechanisms of biofilm formation in Staphylococcus epidermidis and Staphylococcus aureus: functional molecules, regulatory circuits, and adaptive responses. Int J Med Microbiol. 2004;294:203–212. doi: 10.1016/j.ijmm.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Rohde H, Frankenberger S, Zähringer U, Mack D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of bio-material-associated infections. Eur J Cell Biol. 2010;89:103–111. doi: 10.1016/j.ejcb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Patti JM, Allen BL, McGavin MJ, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 42.Rupp ME, Fey PD, Heilmann C, Götz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 43.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/ hemagglutinin of Staphylococcus epidermidis in the pathogenesis of bio-material-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/ hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999;67:2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenney D, Pouliot KL, Wang Y, Murthy V, Ulrich M, Doring G, Lee JC, Goldmann DA, Pier GB. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 46.Fluckiger U, Ulrich M, Steinhuber A, Doring G, Mack D, Landmann R, Goerke C, Wolz C. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by Staphylococci in a device-related infection model. Infect Immun. 2005;73:1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hennig S, Nyunt Wai S, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297:117–122. doi: 10.1016/j.ijmm.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. The Pel and Psl poly-saccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysac-charide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak DJ. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353–8356. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J Bacteriol. 2013;195:1637–1644. doi: 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysac-charide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 68.Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiGiandomenico A, Warrener P, Hamilton M, Guillard S, Ravn P, Minter R, Camara MM, Venkatraman V, MacGill RS, Lin J, Wang Q, Keller AE, Bonnell JC, Tomich M, Jermutus L, McCarthy MP, Melnick DA, Suzich JA, Stover CK. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med. 2012;209:1273–1287. doi: 10.1084/jem.20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byrd MS, Pang B, Mishra M, Swords WE, Wozniak DJ. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappaB activation in A549 cells. MBio. 2010;1:e00140–e00110. doi: 10.1128/mBio.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. Pseudomonas aeruginosa Psl poly-saccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012;14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 75.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013;9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 78.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 79.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 80.Brown MR, Allison DG, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- 81.Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 82.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 83.Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Beer D, Stoodley P, Lewandowski Z. Measurement of local diffusion coefficients in biofilms by microinjection and confocal microscopy. Biotechnol Bioeng. 1997;53:151–158. doi: 10.1002/(SICI)1097-0290(19970120)53:2<151::AID-BIT4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 85.Jefferson KK, Goldmann DA, Pier GB. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2005;49:2467–2473. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dibdin GH, Assinder SJ, Nichols WW, Lambert PA. Mathematical model of beta-lactam penetration into a biofilm of Pseudomonas aeruginosa while undergoing simultaneous inactivation by released beta-lactamases. J Antimicrob Chemother. 1996;38:757–769. doi: 10.1093/jac/38.5.757. [DOI] [PubMed] [Google Scholar]
- 87.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 88.Lam J, Chan R, Lam K, Costerton JW. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leid JG, Shirtliff ME, Costerton JW, Stoodley AP. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol. 2003;171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 92.Sutherland I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 93.Doggett RG, Harrison GM, Stillwell RN, Wallis ES. An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J Pediatr. 1966;68:215–221. [Google Scholar]
- 94.Elston HR, Hoffman KC. Increasing incidence of encapsulated Pseudomonas aeruginosa strains. Am J Clin Pathol. 1967;48:519–523. doi: 10.1093/ajcp/48.5_ts.519. [DOI] [PubMed] [Google Scholar]
- 95.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaspar MC, Couet W, Olivier JC, Pais AACC, Sousa JJS. Pseudomonas aeruginosa infection in cystic fibrosis lung disease and new perspectives of treatment: a review. Eur J Clin Microbiol Infect Dis. 2013;32:1231–1252. doi: 10.1007/s10096-013-1876-y. [DOI] [PubMed] [Google Scholar]
- 97.Clementi F. Alginate production by Azotobacter vinelandii. Crit Rev Biotechnol. 1997;17:327–361. doi: 10.3109/07388559709146618. [DOI] [PubMed] [Google Scholar]
- 98.Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franklin MJ, Ohman DE. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993;175:5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 101.Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol. 2012;84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hay ID, Wang Y, Moradali M, Rehman ZU, Rehm BHA. Genetics and regulation of bacterial alginate production. Environ Microbiol. 2014 doi: 10.1111/1462-2920.12389. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 103.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wood LF, Ohman DE. Identification of genes in the σ22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. MBio. 2012;3:e00094–e00012. doi: 10.1128/mBio.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, Goldberg JB, Schurr MJ, Yu HD. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol. 2011;194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol. 2011;193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 108.Ramphal R, Pier GB. Role of Pseudomonas aeruginosa mucoid exopolysaccharide in adherence to tracheal cells. Infect Immun. 1985;47:1–4. doi: 10.1128/iai.47.1.1-4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davies DG, Geesey GG. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61:860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoyle BD, Williams LJ, Costerton JW. Production of mucoid exopolysaccharide during development of Pseudomonas aeruginosa biofilms. Infect Immun. 1993;61:777–780. doi: 10.1128/iai.61.2.777-780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stapper AP, Narasimhan G, Ohman DE, Barakat J, Hentzer M, Molin S, Kharazmi A, Høiby N, Mathee K. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J Med Microbiol. 2004;53:679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 115.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ghafoor A, Hay ID, Rehm BHA. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang L, Hengzhuang W, Wu H, Damkiær S, Jochumsen N, Song Z, Givskov M, Høiby N, Molin S. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2012;65:366–376. doi: 10.1111/j.1574-695X.2012.00936.x. [DOI] [PubMed] [Google Scholar]
- 118.Tielen P, Strathmann M, Jaeger K-E, Flemming H-C, Wingender J. Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol Res. 2005;160:165–176. doi: 10.1016/j.micres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 119.Hengzhuang W, Wu H, Ciofu O, Song Z, Hoiby N. Phar-macokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Peudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2011;55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alkawash MA, Soothill JS, Schiller NL. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS. 2006;114:131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- 121.Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 123.Schwarzmann S, Boring JR. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect Immun. 1971;3:762–767. doi: 10.1128/iai.3.6.762-767.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Simpson JA, Smith SE, Dean RT. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J Gen Microbiol. 1988;134:29–36. doi: 10.1099/00221287-134-1-29. [DOI] [PubMed] [Google Scholar]
- 125.Mai GT, Seow WK, Pier GB, McCormack JG, Thong YH. Suppression of lymphocyte and neutrophil functions by Pseudomonas aeruginosa mucoid exopolysaccharide (alginate): reversal by physicochemical, alginase, and specific monoclonal antibody treatments. Infect Immun. 1993;61:559–564. doi: 10.1128/iai.61.2.559-564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Simpson JA, Smith SE, Dean RT. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med. 1989;6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 127.Learn DB, Brestel EP, Seetharama S. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect Immun. 1987;55:1813–1818. doi: 10.1128/iai.55.8.1813-1818.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chan C, Burrows LL, Deber CM. Helix induction in antimicrobial peptides by alginate in biofilms. J Biol Chem. 2004;279:38749–38754. doi: 10.1074/jbc.M406044200. [DOI] [PubMed] [Google Scholar]
- 129.Chan C, Burrows LL, Deber CM. Alginate as an auxiliary bacterial membrane: binding of membrane-active peptides by polysaccharides. J Pept Res. 2005;65:343–351. doi: 10.1111/j.1399-3011.2005.00217.x. [DOI] [PubMed] [Google Scholar]
- 130.Limoli DH, Rockel AB, Host KM, Jha A, Kopp BT, Hollis T, Wozniak DJ. Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog. 2014;10:e1004083. doi: 10.1371/journal.ppat.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pier GB, Boyer D, Preston M, Coleman FT, Llosa N, Mueschenborn-Koglin S, Theilacker C, Goldenberg H, Uchin J, Priebe GP, Grout M, Posner M, Cavacini L. Human monoclonal antibodies to Pseudomonas aeruginosa alginate that protect against infection by both mucoid and nonmucoid strains. J Immunol. 2004;173:5671–5678. doi: 10.4049/jimmunol.173.9.5671. [DOI] [PubMed] [Google Scholar]
- 132.Lattner D, Flemming H-C, Mayer C. 13C–NMR study of the interaction of bacterial alginate with bivalent cations. Int J Biol Macromol. 2003;33:81–88. doi: 10.1016/s0141-8130(03)00070-9. [DOI] [PubMed] [Google Scholar]
- 133.Horsman SR, Moore RA, Lewenza S. Calcium chelation by alginate activates the type III secretion system in mucoid Pseudomonas aeruginosa biofilms. PLoS One. 2012;7:e46826. doi: 10.1371/journal.pone.0046826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee B, Schjerling CK, Kirkby N, Hoffmann N, Borup R, Molin S, Høiby N, Ciofu O. Mucoid Pseudomonas aeruginosa isolates maintain the biofilm formation capacity and the gene expression profiles during the chronic lung infection of CF patients. APMIS. 2011;119:263–274. doi: 10.1111/j.1600-0463.2011.02726.x. [DOI] [PubMed] [Google Scholar]
- 135.Roberts IS. The biochemistry and genetics of capsular poly-saccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 136.Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 137.Bratcher PE, Kim KH, Kang JH, Hong JY, Nahm MH. Identification of natural Pneumococcal isolates expressing serotype 6D by genetic, biochemical and serological characterization. Microbiology. 2010;156:555–560. doi: 10.1099/mic.0.034116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol. 2011;14:205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 139.Yother J. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol. 2011;65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 140.Lukáčová M, Barák I, Kazár J. Role of structural variations of polysaccharide antigens in the pathogenicity of Gram-negative bacteria. Clin Microbiol Infect. 2008;14:200–206. doi: 10.1111/j.1469-0691.2007.01876.x. [DOI] [PubMed] [Google Scholar]
- 141.Kim JO, Weiser JN. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis. 1998;177:368–377. doi: 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- 142.Qin L, Kida Y, Imamura Y, Kuwano K, Watanabe H. Impaired capsular polysaccharide is relevant to enhanced biofilm formation and lower virulence in Streptococcus pneumoniae. J Infect Chemother. 2013;19:261–271. doi: 10.1007/s10156-012-0495-3. [DOI] [PubMed] [Google Scholar]
- 143.Sanchez CJ, Kumar N, Lizcano A, Shivshankar P, Dunning Hotopp JC, Jorgensen JH, Tettelin H, Orihuela CJ. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS One. 2011;6:e28738. doi: 10.1371/journal.pone.0028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Joseph LA, Wright AC. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J Bacteriol. 2004;186:889–893. doi: 10.1128/JB.186.3.889-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yi K, Rasmussen AW, Gudlavalleti SK, Stephens DS, Stojiljkovic I. Biofilm formation by Neisseria meningitidis. Infect Immun. 2004;72:6132–6138. doi: 10.1128/IAI.72.10.6132-6138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tuchscherr LPN, Buzzola FR, Alvarez LP, Caccuri RL, Lee JC, Sordelli DO. Capsule-negative Staphylococcus aureus induces chronic experimental mastitis in mice. Infect Immun. 2005;73:7932–7937. doi: 10.1128/IAI.73.12.7932-7937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deghmane A-E, Giorgini D, Larribe M, Alonso J-M, Taha M-K. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol Microbiol. 2002;43:1555–1564. doi: 10.1046/j.1365-2958.2002.02838.x. [DOI] [PubMed] [Google Scholar]
- 148.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, Johnson C, Hu FZ, Stoodley P, Ehrlich GD, Post JC. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goller CC, Seed PC. Revisiting the Escherichia coli poly-saccharide capsule as a virulence factor during urinary tract infection: contribution to intracellular biofilm development. Virulence. 2010;1:333–337. doi: 10.4161/viru.1.4.12388. [DOI] [PubMed] [Google Scholar]
- 150.Kim H-S, Park S-J, Lee K-H. Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol. 2009;74:436–453. doi: 10.1111/j.1365-2958.2009.06875.x. [DOI] [PubMed] [Google Scholar]
- 151.Lee K-J, Kim J-A, Hwang W, Park S-J, Lee K-H. Role of capsular polysaccharide (CPS) in biofilm formation and regulation of CPS production by quorum-sensing in Vibrio vulnificus. Mol Microbiol. 2013;90:841–857. doi: 10.1111/mmi.12401. [DOI] [PubMed] [Google Scholar]
- 152.Velázquez-Hernández ML, Baizabal-Aguirre VM, Bravo-Patiño A, Cajero-Juárez M, Chávez-Moctezuma MP, Valdez-Alarcón JJ. Microbial fructosyltransferases and the role of fructans. J Appl Microbiol. 2009;106:1763–1778. doi: 10.1111/j.1365-2672.2008.04120.x. [DOI] [PubMed] [Google Scholar]
- 153.Osman SF, Fett WF, Fishman ML. Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J Bacteriol. 1986;166:66–71. doi: 10.1128/jb.166.1.66-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bereswill S, Geider K. Characterization of the rcsB gene from Erwinia amylovora and its influence on exoploysaccharide synthesis and virulence of the fire blight pathogen. J Bacteriol. 1997;179:1354–1361. doi: 10.1128/jb.179.4.1354-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dedonder R, Peaud-Lenoel C. Studies on the levansucrase of Bacillus subtilis. I. Production of levans and levansucrase (levansuccharotransfructosidase) by cultures of Bacillus subtilis. Bull Soc Chim Biol (Paris) 1957;39:483–497. [PubMed] [Google Scholar]
- 156.Li H, Ullrich MS. Characterization and mutational analysis of three allelic isc genes encoding levansucrase in Pseudomonas syringae. J Bacteriol. 2001;183:3282–3292. doi: 10.1128/JB.183.11.3282-3292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marvasi M, Visscher PT, Casillas Martinez L. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010;313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 158.Records AR, Gross DC. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol. 2010;192:3584–3596. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li J, Kim IH. Effects of levan-type fructan supplementation on growth performance, digestibility, blood profile, fecal microbiota, and immune responses after lipopolysaccharide challenge in growing pigs. J Anim Sci. 2013;91:5336–5343. doi: 10.2527/jas.2013-6665. [DOI] [PubMed] [Google Scholar]
- 160.Shida T, Mukaijo K, Ishikawa S, Yamamoto H, Sekiguchi J. Production of long-chain levan by a sacC insertional mutant from Bacillus subtilis 327UH. Biosci Biotechnol Biochem. 2002;66:1555–1558. doi: 10.1271/bbb.66.1555. [DOI] [PubMed] [Google Scholar]
- 161.Leloup L, Driessen AJ, Freudl R, Chambert R, Petit-Glatron MF. Differential dependence of levansucrase and alpha-amylase secretion on SecA (Div) during the exponential phase of growth of Bacillus subtilis. J Bacteriol. 1999;181:1820–1826. doi: 10.1128/jb.181.6.1820-1826.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hettwer U, Jaeckel FR, Boch J, Meyer M, Rudolph K, Ullrich MS. Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola. Appl Environ Microbiol. 1998;64:3180–3187. doi: 10.1128/aem.64.9.3180-3187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Burne RA, Chen YYM, Wexler DL, Kuramitsu H, Bowen WH. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J Dent Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- 164.Laue H, Schenk A, Li H, Lambertsen L, Neu TR, Molin S, Ullrich MS. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology. 2006;152:2909–2918. doi: 10.1099/mic.0.28875-0. [DOI] [PubMed] [Google Scholar]
- 165.Kiska DL, Macrina FL. Genetic analysis of fructan-hyperproducing strains of Streptococcus mutans. Infect Immun. 1994;62:2679–2686. doi: 10.1128/iai.62.7.2679-2686.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dogsa I, Brloznik M, Stopar D, Mandic-Mulec I. Exopolymer diversity and the role of levan in Bacillus subtilis biofilms. PLoS One. 2013;8: e62044. doi: 10.1371/journal.pone.0062044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arvidson SA, Rinehart BT, Gadala-Maria F. Concentration regimes of solutions of levan polysaccharide from Bacillus sp. Carbohydr Polym. 2006;65:144–149. [Google Scholar]
- 169.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shikuma NJ, Fong JCN, Yildiz FH. Cellular levels and binding of c-di-GMP control subcellular localization and activity of the Vibrio cholerae transcriptional regulator VpsT. PLoS Pathog. 2012;8:e1002719. doi: 10.1371/journal.ppat.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Anderson ES, Rogers AH. Slime polysaccharides of the Enterobacteriaceae. Nature. 1963;198:714–715. [Google Scholar]
- 174.Grant WD, Sutherland IW, Wilkinson JF. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J Bacteriol. 1969;100:1187–1193. doi: 10.1128/jb.100.3.1187-1193.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Brill JA, Quinlan-Walshe C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1988;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stout V, Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 179.Ophir T, Gutnick DL. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl Environ Microbiol. 1994;60:740–745. doi: 10.1128/aem.60.2.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Navasa N, Rodríguez-Aparicio L, Martínez-Blanco H, Arcos M, Ferrero MÁ. Temperature has reciprocal effects on colanic acid and polysialic acid biosynthesis in E. coli K92. Appl Microbiol Biotechnol. 2009;82:721–729. doi: 10.1007/s00253-008-1840-4. [DOI] [PubMed] [Google Scholar]
- 183.Ledeboer NA, Jones BD. Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar typhimurium on HEp-2 cells and chicken intestinal epithelium. J Bacteriol. 2005;187:3214–3226. doi: 10.1128/JB.187.9.3214-3226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ali A, Mahmud ZH, Morris JG, Sozhamannan S, Johnson JA. Sequence analysis of TnphoA insertion sites in Vibrio cholerae mutants defective in rugose polysaccharide production. Infect Immun. 2000;68:6857–6864. doi: 10.1128/iai.68.12.6857-6864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fong JCN, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 193.Kolodkin-Gal I, Cao S, Chai L, Böttcher T, Kolter R, Clardy J, Losick R. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell. 2012;149:684–692. doi: 10.1016/j.cell.2012.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 194.Hobley L, Kim SH, Maezato Y, Wyllie S, Fairlamb AH, Stanley-Wall NR, Michael AJ. Norspermidine is not a self-produced trigger for biofilm disassembly. Cell. 2014;156:844–854. doi: 10.1016/j.cell.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brown AJ. XLIII.—On an acetic ferment which forms cellulose. J Chem Soc. 1886;49:432–439. [Google Scholar]
- 197.Hestrin S, Schramm M. Synthesis of cellulose by Acetobacter xylinum. Biochem J. 1954;58:345–352. doi: 10.1042/bj0580345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cook KE, Colvin JR. Evidence for a beneficial influence of cellulose production on growth of Acetobacter xylinum in liquid medium. Curr Microbiol. 1980;3:203–205. doi: 10.1007/BF02602449. [DOI] [PubMed] [Google Scholar]
- 199.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 200.Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 201.Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol. 2006;8:1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x. [DOI] [PubMed] [Google Scholar]
- 202.Kimura S, Chen HP, Saxena IM, Brown RM, Itoh T. Localization of c-di-GMP-binding protein with the linear terminal complexes of Acetobacter xylinum. J Bacteriol. 2001;183:5668–5674. doi: 10.1128/JB.183.19.5668-5674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 204.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Römling U. Genetic and phenotypic analysis of multicellular behavior in Salmonella typhimurium. Methods Enzymol. 2001;336:48–59. doi: 10.1016/s0076-6879(01)36577-1. [DOI] [PubMed] [Google Scholar]
- 206.Williams WS, Cannon RE. Alternative environmental roles for cellulose produced by Acetobacter xylinum. Appl Environ Microbiol. 1989;55:2448–2452. doi: 10.1128/aem.55.10.2448-2452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Monteiro C, Saxena I, Wang X, Kader A, Bokranz W, Simm R, Nobles D, Chromek M, Brauner A, Brown RM, Römling U. Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ Microbiol. 2009;11:1105–1116. doi: 10.1111/j.1462-2920.2008.01840.x. [DOI] [PubMed] [Google Scholar]
- 208.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 209.Roche Y, Cao-Hoang L, Perrier-Cornet J-M, Waché Y. Advanced fluorescence technologies help to resolve long-standing questions about microbial vitality. Biotechnol J. 2012;7:608–619. doi: 10.1002/biot.201100344. [DOI] [PubMed] [Google Scholar]
- 210.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 211.Hu W, Li L, Sharma S, Wang J, McHardy I, Lux R, Yang Z, He X, Gimzewski JK, Li Y, Shi W. DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS One. 2012;7:e51905. doi: 10.1371/journal.pone.0051905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Peterson BW, van der Mei HC, Sjollema J, Busscher HJ, Sharma PK. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. MBio. 2013;4:e00497–e00413. doi: 10.1128/mBio.00497-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hu W, Yang Z, Lux R, Zhao M, Wang J, He X, Shi W. Direct visualization of the interaction between pilin and exopolysaccharides of Myxococcus xanthus with eGFP-fused PilA protein. FEMS Microbiol Lett. 2011;326:23–30. doi: 10.1111/j.1574-6968.2011.02430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zmantar T, Bettaieb F, Chaieb K, Ezzili B, Mora-Ponsonnet L, Othmane A, Jaffrézic N, Bakhrouf A. Atomic force microscopy and hydrodynamic characterization of the adhesion of Staphylococcus aureus to hydrophilic and hydrophobic substrata at different pH values. J Microbiol Biotechnol. 2011;27:887–896. [Google Scholar]
- 215.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lacour S, Bechet E, Cozzone AJ, Mijakovic I, Grangeasse C. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS One. 2008;3:e3053. doi: 10.1371/journal.pone.0003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Nat Acad Sci USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bokranz W, Wang X, Tschäpe H, Römling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Micrbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]