Abstract
Prokaryotes often reside in groups where a high degree of relatedness has allowed the evolution of cooperative behaviors. However, very few bacteria or archaea have made the successful transition from unicellular to obligate multicellular life. A notable exception is the myxobacteria, in which cells cooperate to perform group functions highlighted by fruiting body development; an obligate multicellular function. Like all multicellular organisms, myxobacteria face challenges in how to organize and maintain multicellularity. These challenges include maintaining population homeostasis, carrying out tissue repair and regulating the behavior of non-cooperators. Here we describe the major cooperative behaviors that myxobacteria use: motility, predation and development. In addition, this review emphasizes recent discoveries in the social behavior of outer membrane exchange (OME), wherein kin share OM contents. Finally, we review evidence that OME may be involved in regulating population homeostasis, thus serving as a social tool for myxobacteria to make the cyclic transitions from unicellular to multicellular states.
Keywords: Cooperation, outer membrane exchange, cell-cell communication, Myxococcus xanthus
Graphical Abstract
INTRODUCTION
Transitions from unicellular to multicellular life are classified as major evolutionary events [1]. Within the three kingdoms of life, it has been estimated that unicellular organisms have made successful multicellular transitions at least 25 times, with eukaryotes constituting the most complex and spectacular examples [2]. In contrast, for reasons that are unclear, bacteria and archaea have made only limited forays into multicellularity. However, among these latter kingdoms, the myxobacteria have arguably made the most sophisticated transition into multicellularity. In so doing, the myxobacteria have also retained a unicellular life stage. Strikingly, the myxobacteria life cycle is functionally analogous to that of social eukaryotic slime molds (amoebae), in particular the model organism Dictyostelium discoideum. Both of these organisms can exist as single cells or small groups of cells during vegetative growth that transition into obligate multicellular fruiting bodies in response to starvation. It is also striking that these are the only known groups of organisms to share this strategy of building multicellular structures by gathering cells from the environment, which in both cases results in fruiting body formation. Given their evolutionary success [3], the ability to transition from unicellular to multicellular life (fruiting bodies) on a temporal basis offers important fitness advantages for these organisms.
For multicellular organisms to succeed, the individual cells within the collective must cooperate. Such cells need to communicate and coordinate their behaviors to create a functional unit — a tissue. Challenges in this evolutionary transition include the development of (i) biochemical mechanisms for self-recognition; (ii) a means to communicate, organize and synchronize cell behaviors and (iii) a means for the population to reach homeostasis. These ‘bioengineering’ steps represent significant evolutionary hurdles and likely account, at least in part, for why there have been relatively few successful transitions toward multicellularity in the tree of life. In addition to the bioengineering challenges, there are counter-productive Darwinian forces at play [1]. The multicellular environment, in which cells share their resources or ‘public goods,’ provides a breeding ground in which cells can mutate and exploit their clonal environment with selfish and detrimental outcomes. In animals, these Darwinian forces manifest in the relentless development of cancerous cells. To counter this pervasive threat, animals have developed complex immune systems to recognize and remove detrimental cancerous cells, as well as foreign cells, from the body. Multicellular or cooperative microbial communities are also threatened by such exploiter cells. If the population has no mechanism to counteract exploiters, then the demise of the population will likely ensue by a ‘tragedy of the commons’ mechanism [4]. In this scenario, the population loses its cooperative fitness advantage. As described below, cooperative cells have evolved mechanisms to regulate selfish behaviors.
The transition to multicellularity requires cooperation among individual cells. In the last quarter century there has been a greater appreciation for bacterial cooperation within and even across species. Within the realm of biofilms, diverse mechanisms of cell-cell adhesion, public commodity sharing and communication have shown how groups of bacteria can work together [5]. This is not surprising when one considers the advantages inherent in cooperation over individuality [6]. It is plausible that communication and cooperation among related bacteria is the rule rather than the exception. However, very few bacteria are ‘obligate cooperators’ in which cell autonomy has been lost in a commitment toward multicellularity. In contrast to biofilms or colonies, obligate cooperators function exclusively as a multi-celled unit, as found in plants and animal species. In bacteria, a rare example is filamentous cyanobacteria. Here vegetative and heterocyst cells must function together as a unit, since the growth of one cell type is dependent on the fixed carbon or nitrogen provided by the other type of cell [7].
Myxobacteria have been studied for over half a century for their ability to coordinate cells as a unit during the processes of social motility and predation and the formation of elaborate developmental structures (Fig. 1). As individual cells, myxobacteria are thought to struggle to survive [8], but as a collective they are very successful and even a dominant species [3], as evident by their great abundance in a wide range of soil and water habitats [8; 9; 10]. Their two gliding motility engines and molecular arsenal of exoenzymes and secondary metabolites make them mobile predators. When nutrients are exhausted, a myxobacterial swarm will develop into fruiting bodies that contain environmentally resistant spores (Fig. 1) [11]. Curiously, many cells lyse during development, while others help form the fruiting body structure, leaving only a fraction of the original population as viable spores [12]. Whether this cell lysis is a product of programmed cell death and altruism or intra-swarm competition, or both, is still inconclusive [13]. Fruiting body formation provides the benefit of survival and dispersal at the expense of the majority of the cells that initiated the process.
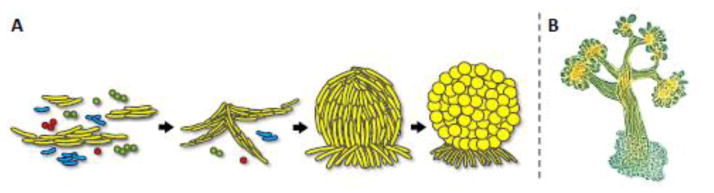
Fig. 1.
Cell recognition and fruiting body development in myxobacteria. A) A schematic model of fruiting body formation by M. xanthus cells (yellow). The natural soil environment of myxobacteria contains diverse microbial communities (depicted by cells of different shapes and colors, left). Upon starvation, M. xanthus cells recognize kin and migrate into aggregates. The aggregates increase in size and form a haystack-shaped fruiting body, within which cells sporulate (depicted by circles). B) Micrograph of Chondromyces crocatus fruiting body [20] (courtesy of Hans Reichenbach).
As outlined by Hamilton’s theory of kin selection (inclusive fitness) [14; 15], the evolution and maintenance of cooperation are possible only in groups of closely related individuals. This process requires (i) a mechanism to discriminate kin from non-kin and (ii) the means to keep those kin in close proximity as a viscous or dense population. Myxobacteria use both strategies to ensure that cooperative behaviors will most likely benefit cells that are highly related. Herein we will review these strategies.
The success of myxobacteria as a functional collective is governed by cell-cell communication, coordination and cooperation among individual cells. A fundamental challenge then is how to create a large, functionally homogeneous collective from a diverse group of cells. That is, in spite of mechanisms to maintain genetic relatedness within a group, cell diversity nevertheless exists within myxobacterial populations [16]. Moreover, within some microenvironments genetic diversity can vary and be extremely high [3; 17], as spores are stable to environmental stresses and readily disperse by wind and water, creating a global microbial melting pot of species and sub-species. In other cases, heterogeneous nutrient availability in soil limits clonal expansion, and thus islands of separate populations can develop and change, and groups that are compatible may eventually merge (Fig. 1)[17]. Of course, mutations within populations will constantly be introduced, leading to mutants which might be defective cooperators. In other cases cells might be genetically identical but physiologically distinct due to aging, starvation or cellular wounds. Therefore the collective functions of a swarm can be threatened by a multitude of factors including non-cooperators, cheaters, incompatible strains, and damaged cells. The ability to maintain coordinated behavior in spite of these challenges is likely crucial to the success of an obligate cooperator.
Much research into bacterial cooperation has focused on the use of public goods. For instance, quorum sensing is a communication strategy in which chemical signals are produced by a community. When the density of the signal reaches the minimum threshold concentration, it elicits a response from surrounding cells, which coordinate their gene expression in response to the local concentration of the signal [reviewed in [18]]. In another example, siderophores are secreted to sequester iron for the producer and its kin [19]. In myxobacteria, one cooperative behavior involves membrane fusion and the exchange of large amounts of outer membrane (OM) components between cells [reviewed in [20]]. Therefore, in this system, OM components including lipids, lipoproteins and lipopolysaccharide (LPS) can be viewed as public goods. However, unlike the conventional sense of a public good, this resource is guarded from non-kin cells—the homophilic cell surface receptor TraA both identifies kin and catalyzes the membrane fusion event (Fig. 2) [21]. OM exchange (OME) therefore is a unique platform for the sharing of cell contents that we propose contributes to the cooperative behavior of myxobacteria. In the following sections, we discuss these behaviors and expand on the mentioned concepts.
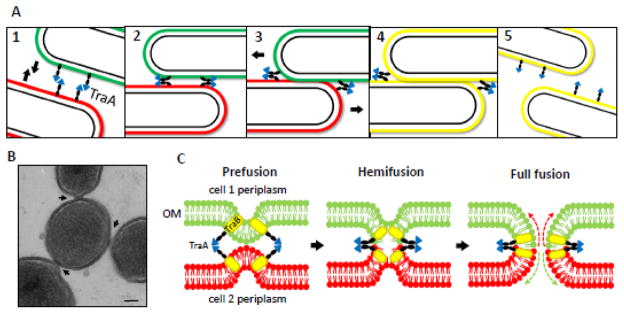
Fig. 2.
Model for outer membrane exchange (OME) in myxobacteria. A) 1) TraA-TraA recognition occurs between neighboring cells. Green and red symbolize different fluorescent markers in the OM. 2) Cells are brought into close proximity upon recognition and motility. 3) ‘Prefusion junctions’ form and cells continue to move. 4) Motility helps trigger fusion to facilitate OM exchange. 5) Cells separate after OME. B) TraAB overexpression leads to cell-cell ‘prefusion junctions’ (arrows) as seen by cryo-electron microscopy. Cross-section shows center cell interacting with three neighboring cells. Bar, 100 nm [85]. C) Schematic model of OM fusion dynamics (based on eukaryotic models) during OME. Upon TraA-TraA interaction, the OMs of two aligned cells come into close contact and form a prefusion junction. The outer leaflets of the two OMs then fuse creating a hemifusion complex. Subsequently, full fusion of the OM (both outer and inner leaflets) is presumed to occur, though soluble periplasmic proteins have not been found to transfer. Dashed arrows indicate lateral diffusion of OM components between cells.
Cooperative behaviors used by myxobacteria
Social movement
The intricate myxobacterial lifestyle involves a number of multicellular behaviors, one of which is coordinated group movement. M. xanthus cells typically travel within dynamic multicellular structures called swarms. Although single cells moving at the edges of M. xanthus swarms are frequently observed, the majority of cells on an agar plate are found within the swarm, where they use cooperative social (S) motility to facilitate swarm expansion [22]. S-motility requires type IV pili (TFP) and exopolysaccharides (EPSs). Briefly, TFP are assembled at the leading cell pole and bind to EPSs present on neighboring cells or on the gliding surface. The retraction of TFP generates a force to propel the cell forward [23; 24; 25]. Moreover, S-motility is a cell contact–dependent behavior that requires a large number of cells in close proximity, and a maximum swarm expansion rate is achieved at high cell density [22; 26]. One advantage for individuals living within the high-density swarms might be that those cells are able to share the pool of secreted EPSs (public good) contributed by the group, and therefore the burden of EPS production on each individual is reduced. A recent study showed that M. xanthus EPSs not only help mediate surface adhesion but also exhibit lubricating properties to facilitate cooperative movement [27]. This finding provides an explanation for the ‘stick-slip’ movement observed in S-motility, where cells move in bursts followed by pauses. Additionally, ‘evolved cooperative motility’ has been described in myxobacteria [28]. Here, in the absence of functional TFP, mutants are selected that develop a de novo cooperative swarming phenotype. Mechanistically, this occurs by enhanced production of an extracellular fibril matrix that co-opts the M. xanthus adventurous (A) motility system to function socially by bundling cells together [28]. Despite the cost of bril over-production, the resulting cell-cell attachment provides the benefit of achieving mobility on a specific medium (semi-solid agar) and results in a new pathway to achieve multicellular behavior.
M. xanthus cells constantly interact with each other to coordinate their movements in a swarm. Interestingly, cells within a swarm often pause and reverse directions [29; 30], and this periodic reversal of gliding direction helps to coordinate swarm expansion [31]. Reversals are controlled by the Frz chemosensory pathway, which drives a small G-protein switch (MglA) to reverse the gliding direction [32; 33; 34; 35; 36]. Mutations in frz genes can cause non-optimal reversal frequencies, which lead to defects in swarming, lipid chemotaxis and developmental aggregation [29; 31; 32; 35; 37]. A relatively recent study suggested that FrzCD, a methyl-accepting chemotaxis protein, might play a role in cell-cell interactions [38]. FrzCD protein clusters transiently realign between adjacent cells upon side-by-side contact to trigger cell reversals [38]. These interactions between side-by-side cells may help the population to coordinate and synchronize cell movements [39]. Thus, M. xanthus uses a variety of cell-cell interactions to organize the macro-scale movement of the swarm unit.
Predation
When prey or other nutrients are present in the environment, M. xanthus swarms will feed upon them collectively. Unlike other predation strategies (e.g., phagocytosis [40], prey cell invasion [41], far-ranging diffusible antibiotic secretion [42]), myxobacteria can penetrate prey colonies and secrete extracellular enzymes and antibiotics to kill and consume prey cells [reviewed in [43]]. During predation, populations of M. xanthus cells ripple, a phenomenon where cells form self-organized and rhythmically travelling high-density waves [44]. Rippling is a cooperative behavior involving cell-cell alignment and periodic reversals [35; 45]. The opposite-moving waves of cells appear to collide and subsequently reverse direction after contact, so that each wave appears to oscillate back and forth [46]. By allowing more-frequent prey-predator contacts, this judicious oscillation behavior is thought to help enhance predation efficiency [43; 46]. Rippling has also been observed during starvation and fruiting body formation [44; 47]. Initial studies regarding this behavior primarily focused on developmental rippling, whereas predatory rippling was rarely mentioned [46]. However, it was recently suggested that rippling in M. xanthus is solely a predatory behavior and that developmental rippling results from nutrient scavenging of lysed cells [46]. As for the physiological role of rippling behavior, both computational simulations and experimental data suggest that ripples cause faster penetration of predators into prey colonies via a signaling mechanism that is dependent on side-by-side contact between M. xanthus cells [38; 48]. Also, rippling helps the predators remain on the prey region for a longer time, which is likely due to organized cell-cell alignments and periodic cell reversals that prevent cells from drifting randomly [48].
In addition to rippling, predation efficiency is also enhanced by a high cell density [43]. M. xanthus predation is often compared to a ‘wolf-pack’ model [43; 49; 50], suggesting that cells living in a high density work cooperatively as an efficient predatory unit [43]. Generally, the high-density predatory swarms secrete increased concentrations of antibiotics and hydrolytic enzymes (public goods) to their local environment [43; 49]. Individual predators thus benefit from this group effort by sharing the pool of hydrolyzed products from the prey cells to promote growth. Recent studies also suggested that M. xanthus produce OM vesicles, containing multiple antibacterial molecules and enzymes, to assist with predation [51; 52]. The packaging of multiple secondary metabolites and enzymes within one vesicle might be delivered as a lethal cocktail to help ensure that predation occurs efficiently [52].
Development
Perhaps the most striking example of cooperation in myxobacteria is fruiting body development (Fig. 1). Upon starvation, M. xanthus enters a developmental program and forms multicellular fruiting bodies, within which the cells differentiate into spores [53]. Fruiting body morphology ranges from relatively simple mounds found in M. xanthus to elaborate tree-like structures of Chondromyces crocatus (Fig. 1) [54]. Following starvation, individual cells of M. xanthus arrange themselves into simple aggregates that develop into dome-shaped mounds containing ~100,000 cooperating cells [55]. In addition to mound formation, rippling behaviors can also be observed in some areas during the early developmental stage (referred to as the aggregation phase) [44; 56]. It has been suggested that these travelling waves carry piles of cells that collide with each other and form ‘traffic jams’ [56; 57]. These masses of cells enter the mound from different directions and are thought to get caught within the jams, which leads to aggregate formation [56; 57]. After the aggregation phase, sporulation proceeds and a small fraction of the cells differentiate into spherical dormant spores [12; 55]. The remaining cells in the population differentiate into peripheral rods (persister-like cells) or lyse [12; 58; 59]. Development is also a cell density–dependent behavior in M. xanthus [60]. Cells sporulate less efficiently, or not at all, if their density is below a threshold concentration [61; 62].
Coordinated cell aggregation into multicellular fruiting bodies is regulated by cell-cell signaling. During the initiation of development, M. xanthus cells accumulate elevated levels of (p)ppGpp in response to starvation and produce A-signal, an extracellular quorum-like signal that is thought to monitor local cell density [63; 64]. Once the extracellular A-signal exceeds the minimum threshold, ensuring that a minimum population (a quorum) of starving cells exists, the expression of development-specific genes regulated by A-signal is initiated [63; 65]. Cooperative aggregation ensues and further morphogenesis is dependent on another signal, the C-signal, which is a morphogen that requires cell-cell contact for its activity [11; 66; 67]. The nature of the C-signal is not clear, although it has been suggested that the CsgA protein may act as a morphogen itself or as an enzyme to generate the C-signal [68; 69]. The transmission of the C-signal seems to rely on end-to-end contacts between cells [70; 71], and the frequent cell-cell interactions that occur within high-density aggregates increase the level of C-signaling [66]. One model suggests that C-signal transmission provides a positional clue to regulate directional cell movement [69]. Here, during the aggregation stage, C-signaling helps recruit cells into chains via pole-to-pole interactions [69]. According to this model, cells from disordered fields are organized into streams and move toward the aggregation centers with a higher speed and a lower reversal frequency, compared with cells that are not recruited [66]. The increased cell density within the aggregates in turn provides positive feedback that helps raise the level of C-signaling until it reaches the threshold required for spore differentiation [72; 73]. However, a more recent study proposed a different model, suggesting that the reduction in cell velocity, rather than the suppression of cell reversal, is responsible for aggregate formation [74]. Here, cells were found to slow down and orient themselves in parallel within high-density aggregates. It has also been suggested that different experimental conditions might account for some of the different observations found during development [35; 74].
Outer membrane exchange (OME)
Previously, we described a novel process whereby OM proteins and lipids from M. xanthus are exchanged between individual cells [75; 76]. Studies relating to OME can be traced back to 1977 when extracellular complementation (stimulation) was first discovered for motility mutants in M. xanthus [77]. In those studies, a small subset of nonmotile mutants was found to be rescued after transient contact with mutants from a different complementation group. Later studies revealed the mechanism of stimulation, in which the functional motility proteins are transferred from a donor strain to restore the motility defect of the mutant recipient [78; 79]. Stimulation involves only transient phenotypic changes upon cell-cell contact, and the ‘rescued’ mutant becomes nonmotile over time if it is physically separated from the donor cells, suggesting that only proteins, instead of DNA, are transferred [20; 21; 63]. By this genetic approach six OM proteins (CglB/C/D/E/F and Tgl) have been identified as substrates for transfer by OME [20; 80]. One important feature of these stimulatable proteins is that they contain either type I or type II signal sequence (SS) peptides [20; 81; 82; 83]. Based on these findings, a heterologous reporter (SSOM-mCherry) was constructed and shown to transfer in live cells. Here the type II SS fusion directs the mCherry reporter to the OM, where it then becomes a competent substrate for cell-cell transfer [76]. Lipids were also found to be transferred via OME by tracking transfer among live cells labeled with lipophilic fluorescent dyes [75]. These observations, combined with the fact that an inner membrane–localized mCherry reporter (SSIM-mCherry) is not transferred [76], suggested that OME involves the transient fusion of the OMs between cells. Strikingly, both endogenous and heterologous cargo transfer experiments found that the donor and recipient cells equally share the amount of exchangeable OM materials within a relatively short period of time, indicating that transfer is an efficient process [75; 76; 78].
OME is a mutual or cooperative behavior, as it requires two or more cells to make direct contact and because transfer is bidirectional (Fig. 2). Physically the cells need to be on a hard surface, as exchange does not occur in liquid medium or on semi-solid agar [76; 84]. Motility (either A- or S-motility), which helps to align and facilitate cell contacts, also plays a critical role in OME [76; 84]. Among nonmotile cells, transfer is barely detected; instead, for efficient transfer, at least one partner (either donor or recipient), or even a third-party cell, needs to be motile [20]. To date, OM lipoproteins, proteins, lipids [75], LPS [85] and a putative signal(s) have been shown to be transferred [86].
Two host genetic determinants, called traA and traB, are required for OME [75]. OME requires that the TraA and TraB proteins be present in both engaged cells, as disruption of either of the two proteins blocks OME [75]. Additional analyses found that TraA functions as a cell surface receptor, governing the cell-cell interactions involved in OME [21; 75]. The TraA protein contains a distant PA14-like domain, Cys-rich tandem repeats, and a putative protein sorting tag called MYXO-CTERM, thought to facilitate the localization of TraA to the cell surface [75].
Interestingly, TraA has a domain architecture and sequence that are similar to those of the FLO1 cell surface receptor, which is required for flocculation in Saccharomyces cerevisiae [87]. The FLO1 sequence varies across environmental isolates, and it plays a key role in self-recognition and has been described as a ‘greenbeard’ gene that directs cooperative interactions toward other cells that bear the same allele [87]. We have reported that TraA has an analogous role to FLO1 in myxobacteria. That is, TraA acts as a molecular specificity determinant and exhibits allele-specific interactions during OME between different environmental isolates [21]. In this case, OME is a cooperative behavior that involves the sharing of public goods among individuals carrying identical or very similar traA alleles. Thus the TraA receptor represents a perceptible trait that is used to distinguish kin from non-kin and governs OME among related individuals. In this regard, TraA can be classified as a rare greenbeard gene and, to our knowledge, is the first greenbeard gene in bacteria known to be involved in beneficial or altruistic-like behaviors [21].
A further requirement for a greenbeard classification is that the gene must be polymorphic to allow specificity during recognition; that is, the greenbeard function allows select individuals in a diverse population to benefit from interactions, whereas others are excluded. Importantly, TraA is polymorphic within the PA14-like domain, and sequence conservation within this domain correlates with cell-cell recognition [21]. Based on these and other experiments, we propose that the polymorphic domain serves as the molecular recognition determinant through homotypic interactions between TraA receptors. In support of this model, we have shown that cell-cell recognition can be ‘reprogrammed’ by swapping traA alleles from different strains [21]. The C-terminal portion of TraA contains Cys-rich repeats that likely form disulfide bonds, and we hypothesize that this region functions as a rigid stalk to display the polymorphic binding domain on the cell surface, which appears to also function in membrane fusion (Fig. 2).
The role of TraB in OME is not well understood. It contains an OmpA-like domain at its C-terminus that is predicted to bind the cell wall [75]. Sequence analysis of traB alleles suggests the N-terminal region contains a β-barrel domain. Therefore, as a putative βbarrel protein in the OM, one possible function of TraB would be to facilitate TraA transport to the cell surface. TraB might also interact with TraA to form a functional complex for OM fusion.
The OME mechanism is proposed to involve the fusion of the OM (Fig. 2) [63]. Although OM-enclosed tubes (OMTs) are produced in large quantities by myxobacteria [88], we do not think that OMTs are required for OME [89]. Under certain conditions, however, OMTs might be produced as a byproduct of OME and motility [90]. According to our current model, M. xanthus cells physically interact with each other, and TraA-TraA interactions then force the opposing membranes to make contact, effectively displacing water between them, to catalyze OM fusion (Fig. 2B) [91; 92]. Fusion is followed by lateral diffusion and exchange of OM contents between aligned cells (Fig. 2A). Supporting the concept of OM fusion, we discovered that TraA/B functions as a cell-cell adhesin. Specifically, TraA/B overexpression leads to the formation of tight cell-cell adhesion zones, which based on cryo-electron microscopy suggest they form ‘prefusion junctions’ [85] (Fig. 2B). However, for OM fusion to occur, cells need to be placed on a hard surface, and, we presume, the prefusion junctions are stressed through cell movements, which in turn helps to destabilize the OM and trigger fusion as outlined in Figure 2.
How OME benefits the individual and group
TraA is highly conserved across most Myxococcales, and its function has apparently been maintained despite significant phylogenetic divergence within the order. In addition there are no TraA/B orthologs found in sequenced genomes outside of this order. Given these findings, what then is the driving force to maintain conservation of TraA/B and why is it unique to the myxobacteria? In other words, what is the function of OME and how is it important to the myxobacterial lifestyle? Though some information has recently come to light [85], the answer may not be straightforward. Here we discuss what is known about the utility of OME, including how this social behavior can benefit the individual cell and the group, and the other ways it may contribute to the behavior of myxobacteria.
The ‘Allee effect’ [93], or the idea that fitness is positively correlated with population density, figures prominently in the literature on myxobacteria [61]. As described above, the importance of population density has been observed during growth [49], social motility [22], feeding [49] and development [62; 65] and may be important for competitive interactions between isolates. However, population homogeneity is not a given. For instance, the prevalence of myxobacteria in small-scale soil patches indicates that encounters among motile populations may occur frequently [94; 95]. Even when stochastic encounters occur among compatible environmental strains, they may not be genotypically identical or may have adapted phenotypic differences because of environmental circumstances. Given these plausible scenarios, how can a heterogeneous collection of cells become a coherent functional unit?
One answer to the above dilemma may lie with OME. Recently we used a genetic model of membrane damage in which affected cells have deleterious or even lethal changes to their LPS and showed that damaged cells were repaired by OME with healthy LPS donors [85]. More specifically, LPS mutants with impaired motility and abolished sporulation were rescued when mixed with healthy traA+ cells, but not with ΔtraA cells. Furthermore, mutants unable to synthesize lipid A by a conditionally lethal mutation were maintained in a population of healthy cells when OME could occur but perished in the absence of OME. Therefore, during the merging of healthy cells and cells with a damaged OM in nature, OME can enable membrane homeostasis to be reached, which can also rejuvenate or heal wounds on ailing cells (Fig. 3).
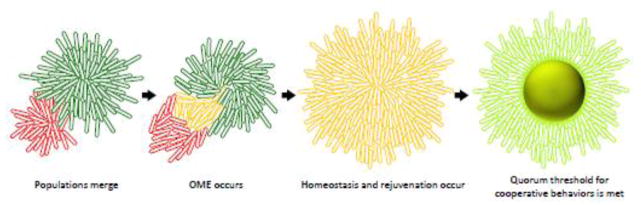
Fig. 3.
A model for the utility of OME in a bacterial community. Two distinct but compatible populations meet (green and red, left), OME catalyzes OM mixing (yellow cells contain components from red and green cells). The mixed population then transitions towards membrane homeostasis (middle right), in which membrane content from both populations is equally distributed among cells. This integrated population is now larger, more homogenous and better equipped for cooperative behaviors (fruiting body, right). Damage that has occurred to some cells can be repaired by dilution followed by active repair (yellow to light green transition).
When we simulated the merging of membrane-damaged (in this case sporulation deficient) and healthy wild-type (WT) populations, we found that an increase in cell density could also lead to a significant increase in sporulation outcomes during development as compared with healthy cells incubated alone [85]. This increase far surpassed what can be explained by the restoration of sporulation to the damaged cell population, indicating that an increase in cell density above a threshold level along with cell rejuvenation had synergistically increased sporulation efficacies. However, when the healthy population was incapable of OME (ΔtraA), the increase in cell density did not improve the sporulation outcome. Put simply, whereas the ΔtraA strain has no obvious developmental phenotype in a pure culture [75], it has a clear developmental phenotype in heterogeneous populations. Therefore, during the merging of damaged and healthy colonies, an increase in cell density (above a threshold level) is beneficial only when the damaged cells can be healed and contribute to group behaviors. In this manner, there is a benefit for healthy cells to temporarily sacrifice their fitness by merging with damaged cells. In this scenario, both the donor and recipient cells benefit; this can explain how OME is evolutionarily maintained by mutual benefit to donor and recipient cells that bear compatible TraA alleles.
In total, motility, development, antibiotic resistance and even lethal biosynthetic defects can be phenotypically complemented by OME in myxobacteria [85]. From an evolutionary standpoint this finding is curious because it suggests that deleterious mutations to some OM proteins or OM biosynthesis proteins could be maintained in the population by phenotypic complementation. Could this mean that all cells that contribute to a population are important regardless of the functional properties of their OMs? And if so, how long will these defective cells be maintained? During development, the majority of cells die, apparently to support the sporulation of the minority population [96]. Perhaps this strategy promotes intra-swarm competition to eliminate less fit cells from the spore or ‘germ’ pool [13]. Alternatively, perhaps the maintenance of diverse OM phenotypes within the spore pool contributes to the phenotypic diversity of strains found within and between populations [3; 16; 97; 98]. Last, it cannot be forgotten that myxobacteria transition between single and multicellular life stages and, consequently, selection against deleterious mutations in a haploid genome is strong when solitary cells cannot be complemented by OME. Future research is required to resolve these possibilities.
Although the benefit of OME to heterogeneous populations is reasonably clear, there may be other benefits to the ‘shared membrane concept.’ As cell-to-cell interactions are guarded from exploitation by non-kin cells, OME could provide a platform to communicate intra-swarm signals. Indeed, we have observed that some motility mutants inhibit the motility and development of their WT kin in a Tra-dependent manner (see below) [75; 86]. In addition, OME allows phenotypic plasticity, in which the OM phenotype of a single cell is not completely dictated by its genotype. For instance, the LPS of some myxobacteria are modified during development [99] and perhaps those modified molecules are transferable. In addition, it has long been known that bacteria can modify their OM in response to antimicrobial compounds or stress [100]. Because myxobacteria transfer lipids, proteins and LPS, it is likely that phenotypic changes can be relayed to cells that have not yet encountered the forthcoming stimuli in their environment. In this and other scenarios, scout cells might relay information in the form of an adapted OM proteome and lipidome to the central group to quickly respond to stimuli at the periphery of the swarm.
Finally, the TraA cell surface adhesin in its own right could contribute to cooperation in myxobacteria. Population viscosity, which involves limited dispersal of related individuals, is one strategy for maintaining a high degree of relatedness between nearby cells, which is critical for cooperation to evolve [1]. As an adhesin that discriminates kin from non-kin [21], TraA could contribute to population viscosity in myxobacteria. Indeed, as mentioned above, when traAB is overexpressed cells specifically adhere to one another (Fig. 2). Perhaps when surrounded by both kin and non-kin, TraA can help parse the populations.
Cheating
Cooperation benefits individual members of a population and also increases the fitness of the population as a whole [101]. Cooperation can, however, be exploited. Although the production of public goods benefits the population at a sustainable cost to its actor [102], some individuals can exploit this situation by using public goods without contributing toward their production. Such individuals have a fitness advantage, as they do not devote their own resources to the behavior yet they enjoy the benefits. This leads to an increase in the number of these social cheaters in the population. As exploiters, their population outcompetes cells that cooperate, and eventually the entire social structure will collapse as the critical public good will not be available at sufficient levels. This phenomenon is referred to as the ‘tragedy of the commons’, in which the exploitative nature of a few individuals can lead to the destruction of a cooperative population [103]. For example, increased virulence of a few phage in a population of phage with low virulence can lead to the abolishment of the host and, consequently, the destruction of the phage population [104]. In another example, the mixing of myxobacteria cheaters with cooperators results in a scenario where the whole population is destroyed by the exploitative nature of the cheaters [105].
Policing
Cooperation is subject to exploitation. One strategy to help prevent exploitation involves kin recognition: cooperative behavior is directed toward related individuals and excluded from non-kin. In fact, such behavior is found in D. discoideum and M. xanthus natural fruiting bodies, in which individuals are highly related to each other [16; 106]. The evolution of ‘policing’ functions is another way to help protect the integrity of cooperating communities. The concept of punishment as a form of policing is found in eukaryotes [107; 108; 109]. Thus cooperation and policing are both important in maintaining a healthy population [110]. For instance, cell competition and culling occur to allow eukaryotic tissues to become more homogenous and fit [111]. Additionally, higher eukaryotes have immune systems that provide surveillance and protection against foreign and cancerous cells across tissues. In soil, there is a large diversity of myxobacteria, based on the 16S rRNA sequence analysis [97], and thus cell-cell surveillance and culling may be an important step to build functional and healthy communities from diverse populations. Indeed, in the laboratory myxobacteria can evolve policing capabilities to control cheating associated with a csgA mutant [112]. Here, continuous exploitation by cheaters of multicellular development and sporulation leads to the development of an evolved strain from a cooperator strain that can police or control the csgA cheaters. In this case, the mechanism of policing is unknown but could involve the production of an inhibitory agent. Recent evidence has shed mechanistic light on policing in a different bacterium. Here, Pseudomonas aeruginosa, can police cheaters through quorum sensing, in which cooperators use a regulatory cascade to catalyze cyanide synthesis and cheaters are unable to synthesize the enzyme that degrades cyanide, thus becoming susceptible to killing [113].
As noted above, OME involves kin recognition and is highly selective. The polymorphic domain in TraA acts as a molecular identity card to recognize individuals that bear identical or very similar TraA receptors [21]. Because OME involves the bulk transfer of proteins and lipids, and does so efficiently, cells would be at a major disadvantage to exchange with non-cooperators. Thus, a priori, one might predict there is a mechanism(s) to prevent exploitation. Indeed this appears to be the case. The fact that both partnering cells must express compatible TraA proteins helps to ensure that public goods are guarded: OME occurs among kin and transfer is bidirectional, meaning one cell cannot unilaterally siphon goods. An advantage of this system is that the goods are exchanged only after the involved parties are verified in a cell contact–dependent manner. This system is in contrast to secreted public goods that are poorly controlled once they leave the cell boundary. As outlined below, we are investing whether there is another level of policing function involved in OME.
Swarm inhibition
OME can regulate the behavior of mixed cell populations. In this phenomenon, motile M. xanthus cells are inhibited from swarming when mixed with nonmotile cells [75]. Inhibition is Tra dependent, as a tra mutation in either the motile or nonmotile strain relieves swarm inhibition. Using swarm inhibition as an assay, we devised a forward genetic screen to investigate OME in more detail [86]. The two questions we sought to address were (i) whether other proteins in addition to TraA/B are directly involved in OME, and (ii) what downstream pathway(s) controls swarm inhibition. Importantly, from the screen no other players, besides TraA/B, were found by this criteria to be directly involved in OME, as all mutants isolated (>50) mapped to the traAB locus [86]. We did, however, isolate a new mutant class that is not defective in OME yet can be relieved from swarm inhibition. The mutant was named OmrA for outer membrane exchange response. OmrA contains a single domain that is homologous to the lipid flippase domain found in the Staphylococcus aureus protein MprF [86]. In S. aureus, MprF controls cytoplasmic membrane homeostasis by flipping modified phospholipid components from the inner leaflet to the outer leaflet [114]. MprF is a bifunctional enzyme in which a second domain catalyzes the synthesis of lysyl-phosphatidylglycerol from lysyl-tRNA and phosphatidylglycerol precursors. Thus once the aminoacylated phosphatidylglycerol is synthesized, MprF flips it to the outer leaflet [114].
In S. aureus, gain-of-function mutations in mprF confer resistance to cationic antibiotics, such as daptomycin, by modifying the surface charge on the cytoplasmic membrane [114]. We found a second gene with homology to the synthase domain of MprF in the M. xanthus genome and constructed a mutant and tested it for a swarm relief phenotype. As predicted, this mutant, omrB, exhibits a swarm relief phenotype (partial) in M. xanthus [86]. From these results we hypothesized that OmrA/B are involved in modifying and regulating inner membrane homeostasis and that inactivating these genes perturbs the inner membrane in such a way that the motile responder cells are no longer regulated by ‘signals’ transferred from nonmotile cells [86].
To understand what causes swarm inhibition and to gain insight into OME, we labeled motile and nonmotile strains with distinguishable fluorescent proteins to track their fates in a mixed colony. Interestingly, the motile cells disappear by 72 hours when mixed with nonmotile cells, and their disappearance is Tra dependent (Dey et al). Additionally, the motile cells become filamentous after a 24-hour incubation in co-culture. From these results we concluded that the nonmotile cells are killing the motile cells by a Tra-dependent mechanism. One possible mechanism for this is that the nonmotile cells exclusively express a toxin/antitoxin pair. OME is then presumed to deliver the toxin to the susceptible motile strain. Consistent with our above findings, when the motile strain contains an omrA mutation, killing is blocked (Dey et al). From these results we concluded that among certain closely related strains there is a sibling killing mechanism. Although it remains unclear why there is killing between related myxobacteria strains, we suspect that it is a form of social surveillance or a policing behavior. Thus in contrast to cooperative and beneficial functions described for OME, there are also adversarial functions.
CONCLUSION
Surviving in hostile natural environments is a constant struggle. Like many eukaryotes, myxobacteria have evolved multicellularity, whereby unity of numbers provides advantages over individuality [6]. These multicellular-related functions require communication, coordination and cooperation among related cells in a diverse population. Because of these properties myxobacteria represent ideal organisms for the study of complex multicellular behaviors that are experimentally tractable. We hypothesize that OME provides a mechanism that helps individual cells come together to form a multicellular unit that has tissue-like qualities, involving resource sharing and maintaining homeostasis. Myxobacteria may also use OME to police social interactions and our future work will explore these possibilities.
HIghlights.
Cooperation plays a pivotal role in the multicellular behaviors of myxobacteria
Myxobacteria build multicellular communities from cells in their environment
Fruiting body development, swarming and predation all benefit from cooperative cell interactions
Outer membrane exchange provides a mechanism for kin recognition and social behaviors
Acknowledgments
This work was supported by NIH grant GM101449 to D.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bourke AF. Principles of social evolution. Oxford University Press; Oxford: 2011. [Google Scholar]
- 2.Grosberg RK, Strathmann RR. The evolution of multicellularity: A minor major transition. Annu Rev Ecol. 2007;38:621–654. [Google Scholar]
- 3.Zhou XW, Li SG, Li W, Jiang DM, Han K, Wu ZH, Li YZ. Myxobacterial community is a predominant and highly diverse bacterial group in soil niches. Environ Microbiol Rep. 2014;6:45–56. doi: 10.1111/1758-2229.12107. [DOI] [PubMed] [Google Scholar]
- 4.Hardin G. The tragedy of the commons. The population problem has no technical solution; it requires a fundamental extension in morality. Science. 1968;162:1243–8. [PubMed] [Google Scholar]
- 5.Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–67. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 6.Lyons NA, Kolter R. On the evolution of bacterial multicellularity. Curr Opin Microbiol. 2015;24:21–8. doi: 10.1016/j.mib.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wingreen NS, Levin SA. Cooperation among microorganisms. PLoS Biol. 2006;4:e299. doi: 10.1371/journal.pbio.0040299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichenbach H. The ecology of the myxobacteria. Environ Microbiol. 1999;1:15–21. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 9.Singh B. Myxobacteria in soils and composts; their distribution, number and lytic action on bacteria. Microbiology. 1947;1:1–10. doi: 10.1099/00221287-1-1-1. [DOI] [PubMed] [Google Scholar]
- 10.Li SG, Zhou XW, Li PF, Han K, Li W, Li ZF, Wu ZH, Li YZ. The existence and diversity of myxobacteria in lake mud - a previously unexplored myxobacteria habitat. Environ Microbiol Rep. 2012;4:587–95. doi: 10.1111/j.1758-2229.2012.00373.x. [DOI] [PubMed] [Google Scholar]
- 11.Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–49. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 12.Lee B, Holkenbrink C, Treuner-Lange A, Higgs PI. Myxococcus xanthus developmental cell fate production: heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J Bacteriol. 2012;194:3058–68. doi: 10.1128/JB.06756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khare A, Shaulsky G. First among equals: competition between genetically identical cells. Nat Rev Genet. 2006;7:577–83. doi: 10.1038/nrg1875. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton WD. The genetical evolution of social behaviour. I J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton WD. The genetical evolution of social behaviour. II J Theor Biol. 1964;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- 16.Kraemer SA, Velicer GJ. Endemic social diversity within natural kin groups of a cooperative bacterium. Proc Natl Acad Sci U S A. 2011;108:10823–10830. doi: 10.1073/pnas.1100307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vos M, Velicer GJ. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus. Curr Biol. 2009;19:1763–7. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 19.West SA, Buckling A. Cooperation, virulence and siderophore production in bacterial parasites. Proc Biol Sci. 2003;270:37–44. doi: 10.1098/rspb.2002.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall D. Molecular recognition in myxobacterial outer membrane exchange: functional, social and evolutionary implications. Mol Microbiol. 2014;91:209–20. doi: 10.1111/mmi.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pathak DT, Wei X, Dey A, Wall D. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet. 2013;9:e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser D, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–245. [Google Scholar]
- 23.Li Y, Lux R, Pelling AE, Gimzewski JK, Shi W. Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili. Microbiology. 2005;151:353–360. doi: 10.1099/mic.0.27614-0. [DOI] [PubMed] [Google Scholar]
- 24.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:01–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 25.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berleman JE, Vicente JJ, Davis AE, Jiang SY, Seo Y-E, Zusman DR. FrzS regulates social motility in Myxococcus xanthus by controlling exopolysaccharide production. PLoS ONE. 2011;6:e23920. doi: 10.1371/journal.pone.0023920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibiansky ML, Hu W, Dahmen KA, Shi W, Wong GCL. Earthquake-like dynamics in Myxococcus xanthus social motility. Proc Natl Acad Sci U S A. 2013;110:2330–2335. doi: 10.1073/pnas.1215089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velicer GJ, Yu Y-tN. Evolution of novel cooperative swarming in the bacterium Myxococcus xanthus. Nature. 2003;425:75–78. doi: 10.1038/nature01908. [DOI] [PubMed] [Google Scholar]
- 29.Blackhart BD, Zusman DR. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhlwein HRH. Swarming and morphogenesis in Myxobacteria. Film C893/1965 Institut fur den Wissensch Film; Gottingen, Germany: 1968. [Google Scholar]
- 31.Wu Y, Kaiser AD, Jiang Y, Alber MS. Periodic reversal of direction allows myxobacteria to swarm. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0811662106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser D, Warrick H. Myxococcus xanthus swarms are driven by growth and regulated by a pacemaker. J Bacteriol. 2011;193:5898–5904. doi: 10.1128/JB.00168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mauriello EMF, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spormann AM, Kaiser D. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J Bacteriol. 1999;181:2593–2601. doi: 10.1128/jb.181.8.2593-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauriello EMF, Mignot T, Yang Z, Zusman DR. Gliding motility revisited: How do the myxobacteria move without flagella? Microbiol Mol Biol Rev. 2010;74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartzell P, Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J Bacteriol. 1991;173:7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearns DB, Shimkets LJ. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol. 9:126–129. doi: 10.1016/s0966-842x(01)01948-5. [DOI] [PubMed] [Google Scholar]
- 38.Mauriello EMF, Astling DP, Sliusarenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc Natl Acad Sci U S A. 2009;106:4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser D, Warrick H. Transmission of a signal that synchronizes cell movements in swarms of Myxococcus xanthus. Proc Natl Acad Sci U S A. 2014;111:13105–13110. doi: 10.1073/pnas.1411925111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke M, Maddera L. Phagocyte meets prey: Uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur J Cell Biol. 2006;85:1001–1010. doi: 10.1016/j.ejcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Jurkevitch E, Minz D, Ramati B, Barel G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol. 2000;66:2365–2371. doi: 10.1128/aem.66.6.2365-2371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H, Ochi K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl Environ Microbiol. 2001;67:1885–1892. doi: 10.1128/AEM.67.4.1885-1892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimkets LJ, Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch R, Kaiser D. Cell behavior in traveling wave patterns of myxobacteria. Proc Natl Acad Sci U S A. 2001;98:14907–14912. doi: 10.1073/pnas.261574598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berleman JE, Chumley T, Cheung P, Kirby JR. Rippling is a predatory behavior in Myxococcus xanthus. J Bacteriol. 2006;188:5888–5895. doi: 10.1128/JB.00559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gronewold TMA, Kaiser D. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol Microbiol. 2001;40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Vaksman Z, Litwin DB, Shi P, Kaplan HB, Igoshin OA. The mechanistic basis of Myxococcus xanthus rippling behavior and its physiological role during predation. PLoS Comput Biol. 2012;8:e1002715. doi: 10.1371/journal.pcbi.1002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenberg E, Keller KH, Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Y, Wei X, Ebright R, Wall D. Antibiotic production by myxobacteria plays a role in predation. J Bacteriol. 2011;193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 52.Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, Cunha J, Hadi MZ, Zusman DR, Northen TR, Witkowska HE, Auer M. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol. 2014;5:474. doi: 10.3389/fmicb.2014.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimkets LJ. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Connor KA, Zusman DR. Patterns of cellular interactions during fruiting-body formation in Myxococcus xanthus. J Bacteriol. 1989;171:6013–6024. doi: 10.1128/jb.171.11.6013-6024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser D. Coupling cell movement to multicellular development in myxobacteria. Nat Rev Micro. 2003;1:45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- 57.Kuner JM, Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harvey CW, Du H, Xu Z, Kaiser D, Aranson I, Alber M. Interconnected cavernous structure of bacterial fruiting bodies. PLoS Comput Biol. 2012;8:e1002850. doi: 10.1371/journal.pcbi.1002850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connor KA, Zusman DR. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J Bacteriol. 1991;173:3318–33. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaiser D. Bacteria also vote. Science. 1996;272:1598–9. doi: 10.1126/science.272.5268.1598. [DOI] [PubMed] [Google Scholar]
- 61.Kadam SV, Velicer GJ. Variable patterns of density-dependent survival in social bacteria. Behav Ecol. 2006;17:833–838. [Google Scholar]
- 62.Kaplan HB, Plamann L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett. 1996;139:89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x. [DOI] [PubMed] [Google Scholar]
- 63.Pathak DT, Wei X, Wall D. Myxobacterial tools for social interactions. Res Microbiol. 2012;163:579–591. doi: 10.1016/j.resmic.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris BZ, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuspa A, Plamann L, Kaiser D. A-signalling and the cell density requirement for Myxococcus xanthus development. J Bacteriol. 1992;174:7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jelsbak L, Søgaard-Andersen L. Pattern formation: fruiting body morphogenesis in Myxococcus xanthus. Curr Opin Microbiol. 2000;3:637–642. doi: 10.1016/s1369-5274(00)00153-3. [DOI] [PubMed] [Google Scholar]
- 67.Kim SK, Kaiser D. C-factor: A cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 68.Avadhani M, Geyer R, White DC, Shimkets LJ. Lysophosphatidylethanolamine is a substrate for the short-chain alcohol dehydrogenase SocA from Myxococcus xanthus. J Bacteriol. 2006;188:8543–50. doi: 10.1128/JB.01047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sogaard-Andersen L, Kaiser D. C factor, a cell-surface-associated intercellular signaling protein, stimulates the cytoplasmic Frz signal transduction system in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1996;93:2675–9. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kruse T, Lobedanz S, Berthelsen NMS, Søgaard-Andersen L. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 71.Kim SK, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990;249:926–8. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 72.Kim SK, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S, Lee BU, Shimkets LJ. csgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–10. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 74.Sliusarenko O, Zusman DR, Oster G. Aggregation during fruiting body formation in Myxococcus xanthus is driven by reducing cell movement. J Bacteriol. 2007;189:611–619. doi: 10.1128/JB.01206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81:315–26. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 77.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 79.Wall D, Wu SS, Kaiser D. Contact Stimulation of Tgl and Type IV Pili inMyxococcus xanthus. J Bacteriol. 1998;180:759–761. doi: 10.1128/jb.180.3.759-761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 81.Rodriguez-Soto JP, Kaiser D. Identification and localization of the Tgl protein, which is required for Myxococcus xanthus social motility. J Bacteriol. 1997;179:4372–4381. doi: 10.1128/jb.179.13.4372-4381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriguez AM, Spormann AM. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J Bacteriol. 1999;181:4381–4390. doi: 10.1128/jb.181.14.4381-4390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pathak DT, Wall D. Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J Bacteriol. 2012;194:1940–1949. doi: 10.1128/JB.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci U S A. 1998;95:3054–8. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, Wall D. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dey A, Wall D. A genetic screen in Myxococcus xanthus identifies mutants that uncouple outer membrane exchange from a downstream cellular response. J Bacteriol. 2014;196:4324–4332. doi: 10.1128/JB.02217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé J-P, Fink GR, Foster KR, Verstrepen KJ. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S, Witkowska HE, Costerton JW, Berleman JE, Auer M. Bacterial social networks: Structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol. 2014;16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wei X, Vassallo CN, Pathak DT, Wall D. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J Bacteriol. 2014;196:1807–1814. doi: 10.1128/JB.00850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ducret A, Fleuchot B, Bergam P, Mignot T. Direct live imaging of cell–cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. ELife. 2013;2:e00868. doi: 10.7554/eLife.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–56. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 92.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–83. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Allee WC, Park O, Emerson AE, Park T, Schmidt KP. Principles of animal ecology. WB Saundere Co. Ltd; 1949. [Google Scholar]
- 94.Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu Rev Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 95.Vos M, Velicer GJ. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl Environ Microbiol. 2006;72:3615–3625. doi: 10.1128/AEM.72.5.3615-3625.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wireman JW, Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1977;129:798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu ZH, Jiang DM, Li P, Li YZ. Exploring the diversity of myxobacteria in a soil niche by myxobacteria-specific primers and probes. Environ Microbiol. 2005;7:1602–1610. doi: 10.1111/j.1462-2920.2005.00852.x. [DOI] [PubMed] [Google Scholar]
- 98.Velicer GJ, Mendes-Soares H, Wielgoss S. Whence comes social diversity? Ecological and evolutionary analysis of the myxobacteria. In: Zhaomin Yang PIH, editor. Myxobacteria: Genomics, Cellular and Molecular Biology. Caister Academic Press; pp. 1–28. [Google Scholar]
- 99.Panasenko SM, Jann B, Jann K. Novel change in the carbohydrate portion of Myxococcus xanthus lipopolysaccharide during development. J Bacteriol. 1989;171:1835–40. doi: 10.1128/jb.171.4.1835-1840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- 101.West SA, Griffin AS, Gardner A. Evolutionary explanations for cooperation. Curr Biol. 2007;17:R661–72. doi: 10.1016/j.cub.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 102.Gordo FDaI. The tragedy of the commons, the public goods dilemma, and the meaning of rivalry and excludability in evolutionary biology. Evol Ecol Res. 2006;8:321–332. [Google Scholar]
- 103.Rankin DJ, Bargum K, Kokko H. The tragedy of the commons in evolutionary biology. Trends Ecol Evol. 2007;22:643–51. doi: 10.1016/j.tree.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 104.Kerr B, Neuhauser C, Bohannan BJ, Dean AM. Local migration promotes competitive restraint in a host-pathogen ‘tragedy of the commons’. Nature. 2006;442:75–8. doi: 10.1038/nature04864. [DOI] [PubMed] [Google Scholar]
- 105.Fiegna F, Velicer GJ. Competitive fates of bacterial social parasites: persistence and self-induced extinction of Myxococcus xanthus cheaters. Proc Biol Sci. 2003;270:1527–34. doi: 10.1098/rspb.2003.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gilbert OM, Foster KR, Mehdiabadi NJ, Strassmann JE, Queller DC. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc Natl Acad Sci U S A. 2007;104:8913–7. doi: 10.1073/pnas.0702723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oldroyd BP. Social evolution: policing without genetic conflict. Curr Biol. 2013;23:R208–10. doi: 10.1016/j.cub.2013.01.051. [DOI] [PubMed] [Google Scholar]
- 108.Hartmann A, Wantia J, Torres JA, Heinze J. Worker policing without genetic conflicts in a clonal ant. Proc Natl Acad Sci U S A. 2003;100:12836–40. doi: 10.1073/pnas.2132993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Foster KR, Ratnieks FL. Convergent evolution of worker policing by egg eating in the honeybee and common wasp. Proc Biol Sci. 2001;268:169–74. doi: 10.1098/rspb.2000.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Strassmann JE, Queller DC. Evolution of cooperation and control of cheating in a social microbe. Proc Natl Acad Sci U S A. 2011;108 (Suppl 2):10855–62. doi: 10.1073/pnas.1102451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morata G, Ballesteros-Arias L. Developmental Biology. Death to the losers Science. 2014;346:1181–2. doi: 10.1126/science.aaa2345. [DOI] [PubMed] [Google Scholar]
- 112.Manhes P, Velicer GJ. Experimental evolution of selfish policing in social bacteria. Proc Natl Acad Sci U S A. 2011;108:8357–62. doi: 10.1073/pnas.1014695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci U S A. 2015;112:2187–91. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ernst CM, Peschel A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol. 2011;80:290–9. doi: 10.1111/j.1365-2958.2011.07576.x. [DOI] [PubMed] [Google Scholar]