Abstract
Contact-dependent growth inhibition (CDI) is a widespread mechanism of inter-bacterial competition mediated by the CdiB/CdiA family of two-partner secretion proteins. CdiA effectors carry diverse C-terminal toxin domains (CdiA-CT), which are delivered into neighboring target cells to inhibit growth. CDI+ bacteria also produce CdiI immunity proteins that bind specifically to cognate CdiA-CT toxins and protect the cell from auto-inhibition. Here, we compare the structures of homologous CdiA-CT/CdiI complexes from Escherichia coli EC869 and Yersinia pseudotuberculosis YPIII to explore the evolution of CDI toxin/immunity protein interactions. Both complexes share an unusual β-augmentation interaction, in which the toxin domain extends a β-hairpin into the immunity protein to complete a six-stranded anti-parallel sheet. However, the specific contacts differ substantially between the two complexes. The EC869 β-hairpin interacts mainly through direct H-bond and ion-pair interactions, whereas the YPIII β-hairpin pocket contains more hydrophobic contacts and a network of bridging water molecules. In accord with these differences, we find that each CdiI protein only protects target bacteria from its cognate CdiA-CT toxin. The compact β-hairpin binding pocket within the immunity protein represents a tractable system for the rationale design of small molecules to block CdiA-CT/ CdiI complex formation. We synthesized a macrocyclic peptide mimic of the β-hairpin from EC869 toxin and solved its structure in complex with cognate immunity protein. These latter studies suggest that small molecules could potentially be used to disrupt CDI toxin/immunity complexes.
Keywords: bacterial competition, interspecies growth inhibition, DNase, toxin/immunity proteins
Introduction
Bacteria possess many strategies to compete and cooperate with other microorganisms in the environment. Contact-dependent growth inhibition (CDI) is one competitive mechanism used by some Gram-negative species to inhibit the growth of neighboring bacteria [1,2]. CDI+ cells express CdiB/CdiA two-partner secretion systems, which deliver protein toxins into target bacteria through a receptor-mediated process. CdiB is an Omp85 outer-membrane protein that exports and assembles toxic CdiA effectors onto the surface of CDI+ cells. CdiA proteins range from 180 to 630 kDa depending on bacterial species and form β-helical filaments that are predicted to extend several hundred angstroms (Å) from the inhibitor-cell surface. CdiA binds to specific receptors on susceptible bacteria and subsequently delivers a toxin domain derived from its C-terminus (CdiA-CT) into the target cell [3–5]. CDI+ bacteria deploy a variety of CdiA-CT toxins with distinct activities. The CdiA-CTEC93 from Escherichia coli EC93 dissipates ion gradients by forming membrane pores [6], but most other characterized CDI toxins have specific nuclease activities. CDI toxins from E. coli EC869 and Dickeya dadantii 3937 are potent DNases capable of degrading target-cell chromosomes [5,7], and CdiA-CTECL from Enterobacter cloacae ATCC 13047 cleaves 16S rRNA to block protein synthesis [8]. CDI+ bacteria protect themselves from auto-inhibition by producing small CdiI immunity proteins that bind to the CdiA-CT and block its toxin activity. Because CDI toxins are diverse, CdiA-CT/CdiI protein interactions are necessarily specific between cognate pairs. Therefore, CdiI immunity proteins neutralize their cognate CdiA-CT but provide no protection against the toxins deployed by other bacteria [7,9]. This diverse network of toxin/ immunity pairs suggests that CDI plays an important role in inter-cellular competition and self/non-self recognition.
We recently surveyed the UniProt database and identified at least 120 distinct CdiA-CT toxin families. Only 26 of these toxins have Pfam designations [10] and the remaining domains are uncharacterized. We initiated structural studies of these protein pairs to discover new toxin activities and toxin/immunity binding interactions. The first CDI toxin/immunity protein complex structures to be determined were from Burkholderia pseudomallei 1026b and enterohemorrhagic E. coli strain EC869 [7]. The CdiA-CT toxin sequences from these bacteria are not related, yet the three-dimensional structures of the domains superimpose with an rmsd of 3.9 Å. Structural homology searches revealed significant similarity to type IIS restriction endonucleases, suggesting that both toxins are DNases. Indeed, the C-terminal domain of CdiA-CTo11 EC869 has potent Zn2 +dependent DNase activity in vitro and in vivo [7]. However, CdiA-CTII Bp1026b has no detectable activity on DNA, and instead this toxin preferentially cleaves near the 3′-end of tRNAAla molecules [11]. Thus, the same toxin fold is used to target different nucleic acid substrates. Though CdiA-CTo11EC869 and CdiA-CTII Bp1026b are similar in structure, other CDI toxins do not share the type IIS restriction endonuclease fold. The crystal structure of CdiA-CT-ECL from E. cloacae ATCC 13047 reveals similarity to the C-terminal nuclease domain of colicin E3 [8,12,13], and sequence homology and activity studies strongly suggest that CdiA-CTK96243 from B. pseudomallei K96243 is related to the C-terminal nuclease domain of colicin E5 [2,11]. Moreover, Aravind and colleagues have predicted that CDI systems deploy two classes of RNA deaminase (Pfam: PF14424 and PF14437), as well as homologues of the EndoU poly(U)-specific endonuclease that processes eukaryotic snoRNAs (Pfam: PF14436) [10,14,15]. Thus, CDI represents a versatile platform to deliver structurally diverse toxins into Gram-negative bacteria.
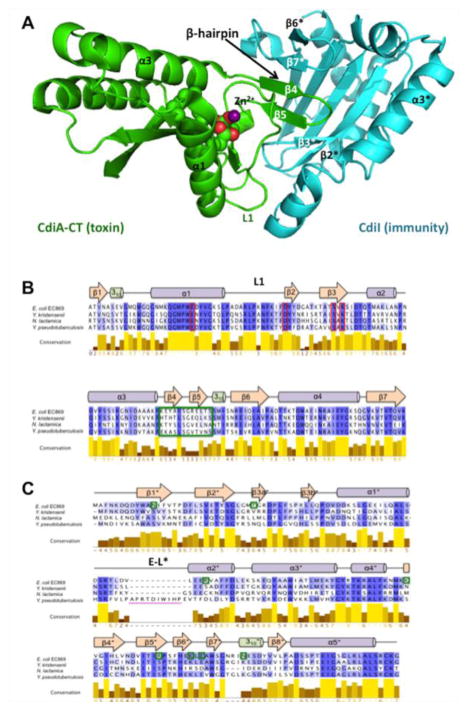
Although toxin/immunity pairs within a given family are homologous, there is often considerable sequence diversity between members, suggesting that families continue to diverge and evolve. When viewed in the context of available crystal structures, it is apparent that residues at the interface of the toxin/ immunity protein complexes are diversifying most rapidly. This phenomenon is exemplified by toxin/ immunity proteins that are homologous to the orphan-11 (o11) CdiA-CT/CdiI pair from E. coli EC869 [7,9]. CdiA-CTo11EC869 interacts with CdiIo11EC869 through β-augmentation, in which the toxin domain extends a β-hairpin to complete a six-stranded anti-parallel β-sheet within the immunity protein (Fig. 1a) [7]. The sequences encoding the β-hairpin (corresponding to β4 and β5) are the most variable between members of the CdiA-CTo11EC869 nuclease family (Fig. 1b). Moreover, CdiIo11EC869 residues that interact with the toxin are not conserved between related immunity proteins (Fig. 1c), suggesting that each immunity protein is specific for its cognate toxin. Here, we use structure–function analyses to examine the β-augmentation interactions of two homologous CdiA-CT/CdiI complexes to study the diversification of CDI toxin/immunity protein interfaces. We find that the CdiA-CT/CdiIYPIII complex from Yersinia pseudotuberculosis YPIII also features a β-augmentation interaction; however, the precise intermolecular contacts differ substantially between the complexes. In accord with these differences, each CdiI immunity protein only protects against its cognate toxin, demonstrating that each pair is a distinct cognate toxin/immunity pair. Finally, we synthesized a macrocyclic peptide mimic of the β-hairpin from CdiA-CTo11 EC869 and solved its crystal structure in complex with the CdiIo11 EC869 immunity protein. This structure forms the basis to refine the β-hairpin mimic to increase affinity with the goal of producing compounds that activate DNase toxins through sequestration of immunity proteins.
Fig. 1.
Structure of the CdiA-CT/CdiIo11EC869 complex and alignments of toxin and immunity homologues. (a) Cartoon representation of the CdiA-CT/CdiIo11EC869 complex structure (PDB ID: 4G6U) with the toxin and immunity colored green and cyan, respectively. Toxin active-site residues are rendered in space-filling model and the Zn2+ ion is represented by a purple sphere. (b) Protein sequence alignment of the CdiA-CTo11EC869 nuclease domain and its homologues. Active-site residues are outlined in red boxes, and the β4/β5-hairpin is outlined in a green box. (c) Protein sequence alignment of the CdiIo11EC869 immunity protein and its homologues. Residues that form H-bond or ion-pair interactions with CdiA-CTo11EC869 are marked with green boxes. The location of CdiIYPIII elongated loop (E-L*) is indicated with a magenta bar. For (b) and (c), alignments were prepared using Jalview, with progressively darker shades of purple indicating greater residue conservation. The secondary-structure elements shown are from the CdiA-CT/CdiIo11EC869 complex structure.
Results
Structure of the CdiA-CT/CdiIYPIII complex reveals conservation of the β-augmentation interaction
Alignment of CdiA-CTo11EC869 toxin homologues indicates that most secondary-structure elements have high level of sequence conservation with the exception of strands β4 and β5, which mediate the β-augmentation interaction with CdiIo11EC869 (Fig. 1a and b). To determine whether β-augmentation occurs in other homologous toxin/immunity pairs, we performed structural and functional analyses of the CdiA-CT/CdiIYPIII complex encoded by the YPK_0575/YPK_0576 genes of Y. pseudotuberculosis YPIII. The C-terminal nuclease domain of CdiA-CTYPIIIis 70.4% identical with CdiA-CTo11EC869 and shares all of the predicted active-site residues (Fig. 1b). Similarly, the CdiIYPIII and CdiIo11EC869 immunity proteins share 49.1% identity, though CdiIYPIII contains a 10-residue insertion between α1 and α2 that is predicted to produce an elongated loop (Fig. 1c). We solved the crystal structure of the CdiA-CT/CdiIYPIII complex to 2.1 Å resolution by molecular replacement using the structure of the CdiA-CT/CdiIo11EC869 complex (PDB code: 4G6U) as a search model (Fig. 2a). As with other CdiA-CT domains [7,8], the N-terminal region (residues Val1–Gly173) was not resolved in the structure. The final model included CdiA-CTYPIII residues 174–298 and 148 water molecules resulting in an Rwork/Rfree (%) of 20.5/25.6 (Table 1). The CdiA-CT/CdiIo11EC869 and CdiA-CT/CdiIYPIII complexes have very similar structures. The toxin domains superimpose with rmsd of 0.84 Å over 101 of 123 α-carbons, and the immunity proteins superimpose with rmsd of 1.01 Å over 133 of 173 α-carbons (Fig. 2b). The CdiA-CT/ CdiIYPIII complex also contains a β-augmentation interaction in which the toxin extends its β4/β5-hairpin into binding pocket within the immunity protein (Fig. 2a and b). However, in contrast to CdiA-CTo11EC869, which contains an ordered Zn2+ ion in the active site [7], no zinc was detected by metal K-edge absorption analysis of multiple CdiA-CT/CdiIYPIII crystals and the electron density spheres within the active-site vicinity of CdiA-CTYPIII were not within zinc coordinating distances with the catalytic residues or would form a zinc tetracoordination or hexacoordination sphere and thus were modeled as water molecules (Fig. S2).
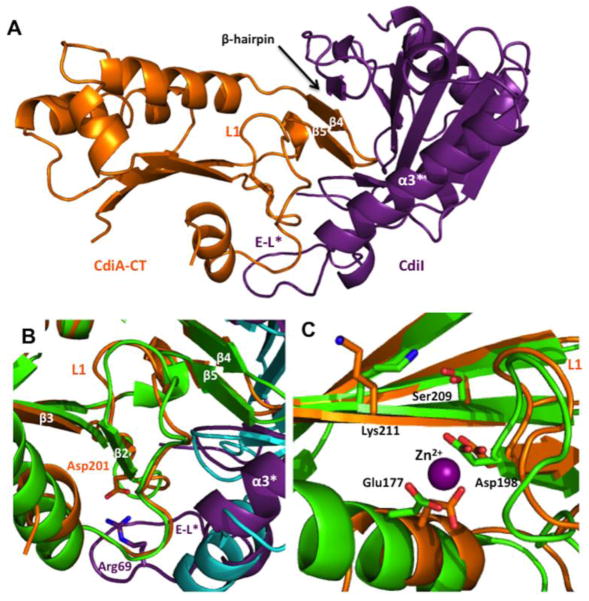
Fig. 2.
The structure of the CdiA-CT/CdiIYPIII complex. (a) Ribbon representation of the CdiA-CT/CdiIYPIII complex with toxin and immunity proteins colored orange and purple, respectively. Location of the CdiA-CTYPIII β4/β5-hairpin is indicated. (b) Structural superimposition of the β-hairpin binding regions of CdiA-CT/CdiIo11EC869 and CdiA-CT/CdiIYPIII. CdiA-CTo11EC869 and CdiIo11EC869 are colored green and cyan, respectively. CdiA-CTYPIII residues that form a salt-bridge via loop L1 are depicted as sticks. (c) Predicted active-site residues of CdiA-CTo11EC869 (carbons in green) and CdiA-CTYPIII (carbons in orange). Residue labels correspond to both toxins. The Zn2+ ion was observed in the CdiA-CTo11EC869 structure and is shown as a purple sphere. Extended loop (E-L*) of CdiIYPIII is labeled in (a) and (b).
Table 1.
X-ray diffraction data and atomic refinement.
| CdiA-CT/CdiIYPIII | CdiIYkris | MAC/CdiIo11 EC869 | |
|---|---|---|---|
| Space group | C2 | P31 | P21 |
| Unit cell dimensions | |||
| a, b, c (Å) | 65.5, 65.5, 71.5 | 54.4, 54.4, 54.4 | 34.8, 128.2, 45.0 |
| β (°) | 92.18 | 112.73 | |
| pH of crystallization condition | 7.0 | 7.4 | 6.0 |
| Protein concentration (mg/mL) | 10 | 12.5 | 7.5 |
| Dataset | |||
| Wavelength (Å) | 1.0 | 1.0 | 1.0 |
| Resolution range | 46.49–2.1 | 50.0–1.8 | 50.0–2.0 |
| Unique reflections (total) | 18,152 | 16,618 | 23,561 |
| Completeness (%)a | 99.4 (98.3) | 99.7 (100) | 96.5 (92.1) |
| Redundancya | 3.5 (3.5) | 5.4 (5.5) | 3.1 (3.1) |
| Rmerge a,b | 0.059 (0.460) | 0.161 (0.607) | 0.076 (0.485) |
| I/σ(I)a | 10.0 (1.6) | 9.48 (3.1) | 11.1 (3.1) |
| NCS copies | 1 | 1 | 2 |
| Other ions | 2 Cl− | — | 2 Cl− |
| Model refinement | |||
| Resolution range (Å) | 46.49–2.09 | 23.58–1.80 | 34.81–2.00 |
| No. of reflections (working/free) | 18,149/1851 | 16,580 (1673) | 23,526 (2004) |
| No. of protein atoms | 2308 | 1283 | 2596 |
| No. of water molecules | 148 | 130 | 132 |
| No. of CT-MAC atoms | — | — | 222 |
| Missing residues | 1–173, 298 (CdiA) 1–2 (CdiI) |
None | 1, 168, 169 |
| Rwork/Rfreec (%) | 20.5/25.6 | 18.1/22.1 | 18.4/23.1 |
| rmsd | |||
| Bond lengths (Å) | 0.003 | 0.007 | 0.009 |
| Bond angles (°) | 0.694 | 0.998 | 1.222 |
| Ramachandran plot | |||
| Most favorable region (%) | 95.25 | 98.77 | 97.13 |
| Additional allowed region (%) | 4.75 | 1.23 | 2.87 |
| Disallowed region | 0 | 0 | 0 |
| PDB code | 4ZQU | 4ZQV | 4ZQW |
Rfree was computed identically except where all reflections belong to a test set of 10% randomly selected data.
Statistics for the highest-resolution shell are given in brackets.
Rmerge = Σ|I − 〈I〉|/Σ 〈I〉.
Rwork = Σ|Fo − Fc|/ΣFo.
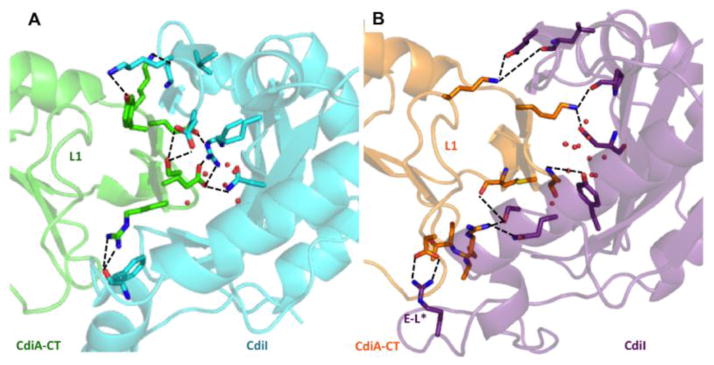
CdiA-CTYPIII and CdiIYPIII bind one another through a network of 14 direct H-bonds and ion pairs (Table S1) combined with several hydrophobic interactions. Only two CdiA-CTYPIII β-hairpin side chains (Glu242 and Lys243) interact directly with CdiIYPIII, compared to the six direct side-chain interactions in the CdiA-CT/CdiIo11EC869 complex (Table S1 and Fig. 3). The CdiA-CT/CdiIYPIII β-hairpin pocket also contains a network of bridging water molecules and several more hydrophobic interactions than the CdiA-CT/CdiIo11 EC869 complex (Fig. 3). In addition, extensive interactions outside of the β-augmentation region contribute to CdiA-CT/CdiIYPIII complex stability. Loop L1 of CdiA-CTYPIII forms several hydrophobic contacts with the elongated loop of CdiIYPIII. Residue Lys195 within L1 forms a salt-bridge with CdiIYPIII residue Glu137 within β6. The loop connecting strands β2 and β3 also has several H-bond interactions and Asp201 forms a prominent salt-bridge with Arg69 in the elongated loop of CdiIYPIII (Table S1 and Fig. 2 b). In contrast, the CdiA-CT/CdiIo11EC869 complex has a less extensive interaction network outside of the β-augmentation region. Loop L1 of CdiA-CTo11 EC869 has fewer hydrophobic contacts and one ionic interaction between Asp183 and Arg71 of CdiIo11 EC869 (Table S1). Together, these structures reveal overall conservation between the toxin/immunity protein pairs but reveal important differences in the network of bonds that stabilize each complex (Fig. S3).
Fig. 3.
Comparison of the β-augmentation interactions. (a) Ribbon representation of the CdiA-CT/CdiIo11EC869 complex with toxin and immunity proteins are colored green and cyan, respectively. Residues at the complex interface involved in direct ion pair or H-bond interactions are shown in stick representation, with carbon atoms colored as stated for above: oxygen and nitrogen atoms are colored red and blue, respectively. Water molecules at the interface are represented as red spheres. (b) Ribbon representation of the CdiA-CT/CdiIYPIII complex with toxin and immunity proteins colored orange and purple, respectively. Residues and water molecules represented and colored as in (a).
CdiIo11 EC869 and CdiIYPIII immunity proteins are specific for their cognate toxins
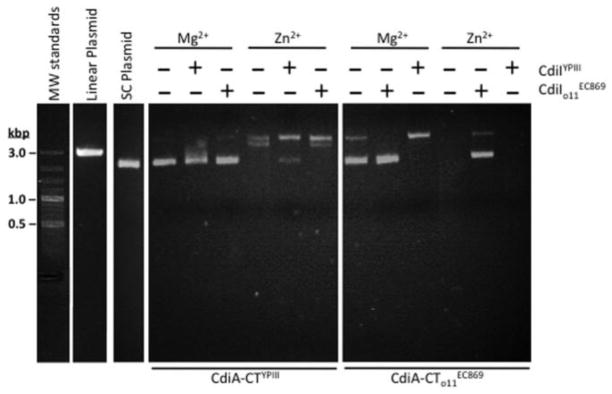
The conservation of nuclease active-site residues strongly suggests that CdiA-CTYPIII has DNase activity similar to that described for CdiA-CTo11 EC869 [7]. We isolated the CdiA-CTYPIII domain from its immunity protein and tested for DNase activity in vitro using supercoiled plasmid as a substrate. CdiA-CTYPIII had no detectable nuclease activity in the presence of Mg2+ ions, but it converted the supercoiled plasmid into open-circular form when the reactions were supplemented with Zn2+ (Fig. 4). CdiA-CTYPIII appears to be less active than the CdiA-CTo11EC869 toxin, which had detectable DNase activity with Mg2+ and completely degraded the plasmid in the presence of Zn2+ ions (Fig. 4). We next tested the CdiIYPIII and CdiIo11EC869 immunity proteins for the ability to neutralize DNase activity in vitro. Each immunity protein was able to partially block the activity of its cognate toxin but had no effect on non-cognate toxin activity (Fig. 4). These results strongly suggest that each immunity protein only binds to its cognate toxin. We measured the dissociation constants (Kd) for cognate and non-cognate complexes using biolayer interferometry. CdiA-CTYPIII and CdiIYPIII form a high-affinity complex with Kd = 16 ± 1 nM, which is similar to the value (18 ± 7 nM) previously reported for the complex [7]. In contrast, CdiIYPIII has ~ 1000-fold lower affinity for non-cognate CdiA-CTo11 EC869 with a Kd of 13 ± 2 μM. This highly reduced affinity between CdiA-CTo11EC869 and CdiIYPIII compared to cognate protein pairs is perhaps due, in part, to the shape and electrostatic incompatibility of the β-hairpin with the binding pocket of CdiIYPIII immunity protein (Fig. S4).
Fig. 4.
CdiA-CTYPIII has DNase activity in vitro. Supercoiled plasmid DNA was incubated with purified CdiA-CTo11EC869 or CdiA-CTYPIII in the presence of either Mg2+ or Zn2+ and then analyzed by agarose gel electrophoresis and ethidium bromide staining. Reactions were supplemented with purified CdiIo11EC869 or CdiIYPIII immunity proteins where indicated. Untreated supercoiled plasmid substrate and linearized plasmid were included as controls for the migration of undigested DNA. The migration positions of linear molecular weight (MW) DNA standards are indicated in kilobase pairs (kb).
Immunity proteins only provide protection against their cognate toxins during CDI
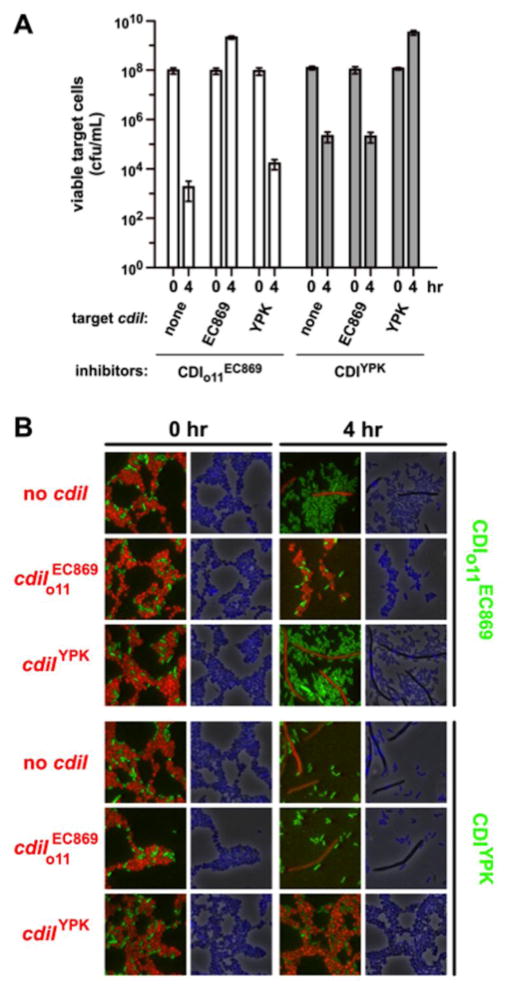
The CdiA-CTYPIII toxin is encoded within a defective cdi locus that has been inactivated by complex gene rearrangements and deletions. To ascertain whether the toxin is functional in cell-mediated CDI, we fused the CdiA-CTYPIII nuclease domain to the C-terminus of CdiAEC93 and tested the resulting chimera for inhibition activity against E. coli target cells. Inhibitor cells that express the CdiAEC93-CTYPIII chimera reduced viable target-cell counts more than 500-fold after 4 h of co-culture, but target cells that express the cognate CdiIYPIII immunity protein were completely protected from inhibition (Fig. 5a). In contrast, target cells that express immunity protein were inhibited to the same extent as cells that lack any immunity gene (Fig. 5a). Similarly, the CdiIYPIII immunity protein was unable to protect target cells from inhibitor cells that deploy the toxin (Fig. 5a). We also examined competition co-cultures by fluorescence microscopy to detect DNase activity in target bacteria. We labeled inhibitor cells with yellow fluorescent protein (YFP) and target cells with mKate2 to differentiate the two populations and also stained the cells with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nucleoids. Immediately after cell mixing, both inhibitor-cell and target-cell populations had similar morphologies and nucleoid staining was uniform (Fig. 5b). After 4 h of co-culture, target cells that lack the appropriate immunity protein became filamentous and lost DAPI staining (Fig. 5b and Fig. S5), indicating significant damage to the chromosome. In contrast, target cells that express cognate CdiI immunity proteins retained normal morphology and DAPI staining (Fig. 5b and Fig. S5). Together, these data demonstrate that these two toxin/immunity systems have diverged into distinct non-overlapping immunity groups.
Fig. 5.
CdiIo11EC869 and CdiIYPIII confer specific immunity to CDI. (a) Competition co-cultures. Inhibitors cells that deploy CdiA-CTo11EC869 (from pCH9305) or CdiA-CTYPIII (from pCH2409) were incubated at a 1:1 ratio with target cells that express CdiIo11EC869 (from pCH9315), CdiI-YPIII (from pCH848) or no immunity at all (none, pTrc99a vector). Viable target cells were quantified as colony-forming units (c.f.u.) per milliliter at the beginning of the co-culture and after 4 h. Data represent the average ± standard error of the mean for three independent experiments. (b) Fluorescence microscopy of competition co-cultures. YFP-labeled inhibitor cells were co-cultured with mKate2-labeled target strains that carry the indicated immunity genes. Cells were stained with DAPI to visualize genomic DNA at 0 and 4 h of co-culture.
β-Augmentation is required for toxin/immunity protein complex formation
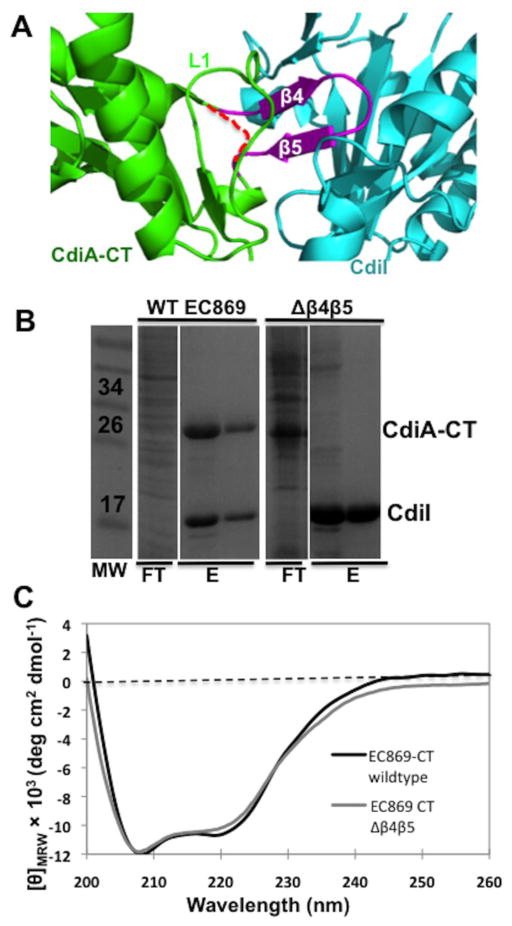
The β-augmentation interactions observed in the two toxin/immunity protein complexes suggest that the proteins bind using a lock-and-key mechanism. Therefore, we sought to crystallize and solve the structures of isolated toxins and immunity proteins to determine if the proteins undergo any conformational changes upon complex formation. Neither of the isolated toxin domains formed crystals, but we were able to crystallize and solve the 1.8-Å structure of an isolated immunity protein (CdiIYkris) encoded by the Ykris_10740 locus of Yersinia kristensenii ATCC 33638 (Table 1). CdiIYkris shares 68.9% and 51.5% sequence identity with and CdiIYPIII (respectively) (Fig. 1c), and its structure superimposes onto and CdiIYPIII with rmsd of 0.626 and 0.984 Å over all α-carbons (respectively) (Fig. S6). In addition, structural homology searches identified yet another immunity protein homologue (CdiINMB) encoded by the NMB0488 locus in Neisseria meningitidis MC58 (PDB code: 2GKP). CdiINMB superimposes onto each of the other immunity proteins with rmsd values <0.7 Å over all α-carbons (Fig. S6). Collectively, these structures indicate that homologues retain the same architecture of the β-hairpin binding pocket in the absence of bound toxin. This finding suggests that β-hairpins are modular, raising the possibility that interaction specificity could be altered by exchanging β4/β5 (β-hairpin) sequences between toxins.
To test whether β-augmentation is required for stable complex formation, we replaced the β-hairpin (residues Lys242–Thr252) with a Gly-Ser-Gly peptide linker to generate (Fig. 6a). Wild-type binds to its cognate immunity protein with nanomolar affinity and co-purifies with His6-tagged during Ni2+-affinity chromatography (Fig. 6b). In contrast, did not co-purify with His6-tagged (Fig. 6b), suggesting that the mutant domain has lower affinity for immunity protein. We purified to homogeneity by anion-exchange chromatography and measured its affinity for using biolayer interferometry. However, no binding interaction was detected, indicating that the dissociation constant is >300 μM. To test whether deletion of the β-hairpin disrupts toxin structure, we examined the domain using circular dichroism (CD) spectroscopy. This analysis revealed that has essentially the same secondary-structure content as the wild-type domain (Fig. 6c). Taken together, these result demonstrate that the β-hairpin is critical for complex formation. Moreover, despite the very low affinity of CdiI to , a high level of expression of was observed (Fig. 6b) together with healthy cell growth, suggesting that the β-hairpin is also required for toxic DNase activity. This was confirmed by testing in vitro DNase activity in the presence of super-coiled plasmid DNA and Zn2+, which showed that the domain had no observable DNase activity (data not shown).
Fig. 6.
Deletion of the β-hairpin disrupts complex formation. (a) Ribbon representation of the CdiA-CT/Cdilo11EC869 complex interface. CdiA-CTo11EC869, Cdilo11EC869 and the β-hairpin colored green, cyan and olive, respectively. The Gly - Ser - Gly linker in is depicted as a red broken line. (b) His6-tagged was co-expressed with or , followed by purification via Ni2+-affinity chromatography and then analysis by SDS/PAGE gel. Relevant molecular weight standards are labeled. The flow-through (FT), wash (W) and elution fractions (E) are indicated. (c) CD spectra of purified and .
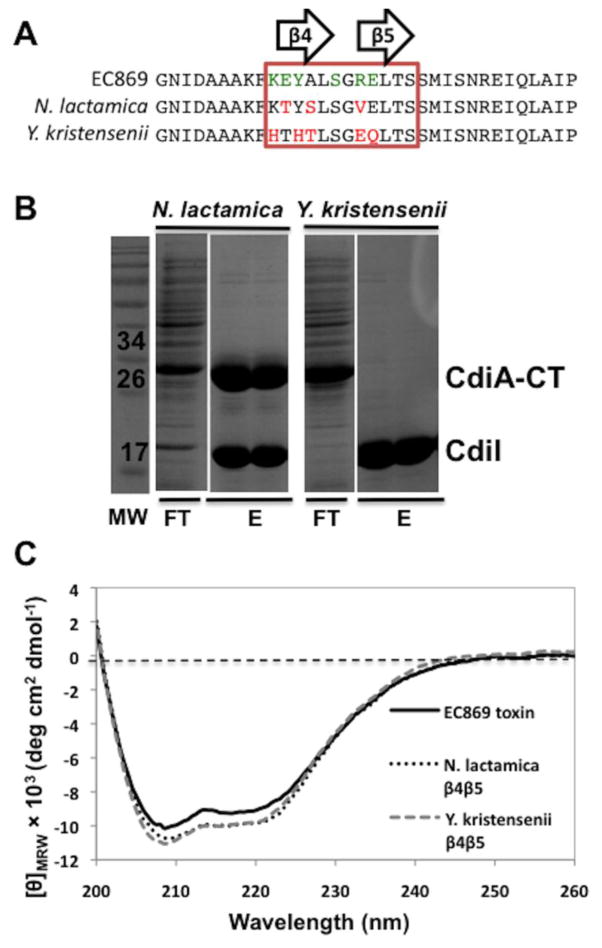
We next asked whether CdiI binding specificity can be altered by grafting heterologous β-hairpins onto the toxin. Using a catalytically inactive version of that contains the Asp198Ala mutation, we replaced residues Lys242–Glu250 with the corresponding sequences from homologous toxins from Y. kristensenii ATCC 33638 (CdiA-CTYkris encoded by ykris0001_10730) and Neisseria lactamica ATCC 23970 (CdiA-CTNlact encoded by NEILACOT_05635) (Fig. 7a). We co-expressed and together with His6-tagged and purified the tagged immunity protein by Ni2+-affinity chromatography. Chimeric toxin co-purified with His6-tagged , but eluted in the void volume of the column (Fig. 7b). These results suggest that binds with relatively high affinity to , whereas the toxin does not. We first confirmed that each chimeric toxin was folded properly using CD spectroscopy (Fig. 7c), then measured binding affinities for using biolayer interferometry. and bound to with dissociation constants of 180 ± 100 nM and 46 ± 36 μM, respectively (Table 2), consistent with the co-purification data. The difference in affinities of the N. lactamica and Y. kristensenii chimeric toxins for the immunity protein is in part due to the differences in electrostatic and shape complementarity (Fig. S7). The N. lactamica and EC869 β-hairpins share similar electrostatics and shape (Fig. S7a and b), allowing to retain nanomolar affinity for (Table 2). In contrast, the Y. kristensenii β-hairpin has a different shape and altered electrostatics compared with , which results in low micromolar affinity of chimeric toxin for (Fig. S7a and c). We then tested whether the grafted β-hairpins confer higher affinity for CdiINlact and CdiIYkris immunity proteins. The chimera bound to CdiIYkris with about the same affinity as , but somewhat surprisingly, this domain bound to CdiINlact with ~10-fold higher affinity (Table 2). The domain bound to CdiINlact with essentially the same affinity as for and interacted with CdiIYkris with approximately 10-fold lower affinity (Table 2).
Fig. 7.
Toxin β-hairpin sequence contributes to complex binding affinity. (a) Protein sequence alignment of the β-hairpin region (boxed) of CdiA-CTo11EC869, CdiA-CTNlact and CdiA-CTYkris. CdiA-CTo11EC869 residues that interact with CdiIo11EC869 are shown in green. Red residues indicate sequence differences with respect to CdiA-CTo11EC869. (b) CdiIo11 EC869-His6 was co-expressed with CdiA-CTo11EC869 containing the β-hairpins from either N. lactamica or Y. kristensenii then purified by Ni2+-affinity chromatography and analyzed by SDS-PAGE. The molecular mass standards are indicated in kilodaltons (kDa). The flow-though (FT) and wash fractions (W1 and W2) are indicated followed by elution with imidazole gradient. (c) CD spectra of purifiedCdiA-CTo11EC869/Nlact and CdiA-CTo11 EC869/Ykris show similar secondary-structure content compared to wild-type CdiA-CTo11EC869.
Table 2.
Dissociation constants (μM) of CdiA-CT and CdiI interactions determined using biolayer interferometry.
|
|
|
|
||||
|---|---|---|---|---|---|---|
|
|
0.018 ± 0.007 | 46 ± 36 | 0.18 ± 0.10 | |||
| CdiIYkris | 5.0 ± 0.4 | 24 ± 14 | 5.8 ± 3 | |||
| CdiINlact | 82 ± 18 | 3.2 ± 2 | 0.17 ± 0.09 |
Structure of the complex
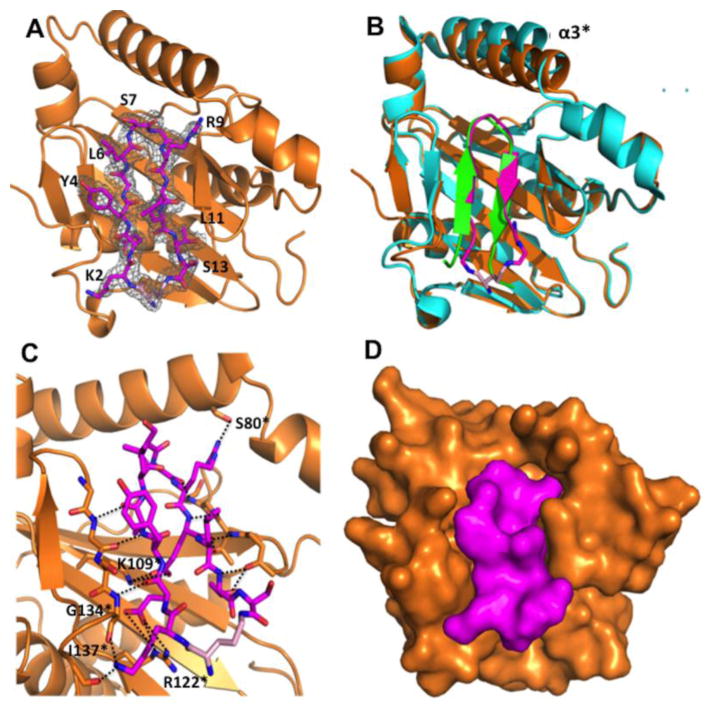
In principle, molecules that disrupt CdiA-CT/CdiI complexes should liberate the toxin domain and induce CDI+ bacteria to undergo auto-inhibition. Nowick and coworkers have previously developed macrocyclic peptides containing δ-linked ornithine turn units that adopt a β-hairpin conformation and should be suitable to disrupting the CdiA-CT/CdiI complex [16,17]. To test the feasibility of this strategy, we designed a macrocyclic peptide (MAC) that mimics the β-hairpin of . The MAC peptide contains residues corresponding to Lys242–Ser253 of , which were connected through a δ-linked ornithine residue (Fig. S1). Biolayer interferometry experiments failed to detect a binding interaction between MAC and ; the MAC peptide was unable to promote DNase activity when added in ~103-fold molar excess to the complex in vitro (data not shown). Despite its low affinity for , we were able to crystallize the MAC peptide in complex with the immunity protein and solve the structure to 2.0 Å resolution by molecular replacement (Fig. 8a). As anticipated, the MAC peptide forms a two-stranded β-sheet, though there are only four cross-strand H-bonds compared to the five in the complex. The ornithine turn creates a bulge that prevents formation of the fifth H-bond (Fig. 8b). The structure superimposes well with (rmsd of 0.437 Å over all α-carbons), though helix α3* is displaced 3.0 Å to create a slightly altered β-hairpin binding pocket (Fig. 8b). Five MAC peptide side chains form H-bonds or ion-pair interactions with the immunity protein, in contrast to the six direct interactions observed in the complex. In the complex, β-hairpin residue Ser247 interacts with the immunity protein. However, the corresponding Ser7 residue within the MAC peptide does not interact with (Fig. 8c). Additionally, the side-chain conformation of MAC residue Arg10 is altered compared to that of Arg249 in the structure, resulting in a H-bond interaction with the hydroxyl of Ser80 rather than the backbone carbonyl of Phe75 in the toxin/immunity structure. The structure establishes that structure-based designed macrocyclic peptides can bind in the β-hairpin binding pocket (Fig. 8d), forming contacts that mimic those found in the CdiA-CT/CdiI complexes.
Fig. 8.
Structure of the complex. (a) Crystal structure with depicted as orange ribbons and MAC displayed as sticks, carbon, nitrogen, oxygen and ornithine carbons colored magenta, blue, red and pink, respectively. The MAC 2Fo - Fc electron density map is shown in a gray mesh and contoured at 1.0 σ. (b) Structural superimposition of MAC [colored same as (a)] and the complex. Only residues Lys242–Ser253 of the β-hairpin are shown (green). is colored teal and helix α3* is labeled. (c) MAC interacts with through a network of H-bonds and ion pairs. Interacting bonds are shown as black dotted lines. β-strands that H-bond with MAC (β3* and β7*) are shown as sticks. (d) Surface representation of the CdiICT-MAC structure, oriented as in (a) and (b), depicting the complementarity of MAC and .
Discussion
CDI toxin/immunity protein pairs are diverse and comprise more than 100 distinct families. Even within a given family, there is considerable sequence variability suggesting that toxin/immunity protein families continue to evolve. This phenomenon is well illustrated by homologues of the toxin/immunity protein pair. Alignment of 26 closely related toxin domains from this family reveals that nearly all of the secondary-structure elements are highly conserved (Fig. S3a). The obvious exception is the β4/β5-hairpin, which mediates the β-augmentation interaction and is the least conserved element in the family. Loop L1 of the toxin domain is responsible for all other contacts with the immunity protein; however, in contrast to the β-hairpin, this region is well conserved with five invariant residues (Arg189, Leu190, Pro19, Phe194 and Asp198). Although loop L1 is highly conserved, it interacts with immunity proteins using distinct contacts in the and CdiA-CT/CdiIYPIII complexes. Loop L1 of engages almost exclusively in hydrophobic and van der Waals interactions, whereas the C-terminal portion of the CdiA-CTYPIII loop is dominated by direct H-bond and ion-pair interactions involving residues Asp201, Ala203 and Thr204. The differences are striking because these three residues are also present in yet do not form the same interactions. Similar phenomena are observed for the immunity proteins. CdiI strand β3* anneals with the toxin’s variable β5 strand during β-augmentation; accordingly, β3* varies between immunity proteins in the family (Fig. S3b). However, CdiI strands β7*, β8* and β9* and the intervening loops are highly conserved, yet this region interacts with cognate toxins using distinct molecular contacts. In several instances, highly conserved residues engage in direct H-bonds in one complex but fail to make any intermolecular contact in another closely related complex. Therefore, even conserved sequence elements can be exploited to discriminate against near-cognate partners. The idiosyncratic nature of these interactions most likely explains why immunity binding specificity cannot be switched through a simple exchange of β4/β5-hairpins between homologous toxins.
The divergence of toxin/immunity protein interactions was first recognized and characterized in a subset of E-class colicins. Colicins are diffusible protein toxins released by some strains of E. coli to kill other competing bacteria [18]. Though colicins and CdiA proteins are not related, there are several features common to both competition systems. One striking parallel is the variability of C-terminal toxin domains. The eight characterized E-class colicins share nearly identical N-terminal domains, but their C-terminal nuclease domains are distinct with DNase (E2, E7, E8 and E9), ribosomal RNase (E3, E4 and E6) or tRNase (E5) activities [18]. Like CdiA proteins, colicins are always encoded in tandem with a specific immunity protein that binds the nuclease domain and blocks its activity. Colicins E2, E7, E8 and E9 carry homologous DNase domains, yet their respective immunity proteins do not protect against near-cognate toxins [19,20]. Structure–function analyses show that E-class immunity proteins bind to a contiguous stretch of ~30 residues that are highly variable between the different nuclease domains [21–23]. Similarly, the interaction surfaces on the immunity proteins are also variable but contain a conserved core interaction comprised of Tyr54 and Tyr55 (ImE9 numbering) [21,24]. This invariant core provides significant binding energy and near-cognate colicin toxin/immunity protein interactions often have dissociation constants of 10−8 M [25], which are similar in affinity to the cognate CdiA-CT/CdiI complexes studied here. An analogous core interaction centered at the tip of the β-hairpin is found in the toxin/immunity family. Leu246 of the toxin engages in a hydrophobic interaction with an aliphatic residue in the immunity protein (Ala131 in and Val141 in CdiIYPIII). Similarly, toxin residue Ser247 interacts with a Tyr residue (Tyr84 in and Tyr94 in CdiIYPIII) that is invariant in the immunity protein family (Fig. S3b). However, these core interactions do not provide significant binding affinity for near-cognate toxin/immunity pairs. A final important parallel between the colicin E-class and DNases is that both the immunity proteins bind to exosites, leaving the nuclease active site exposed in the toxin/immunity complex [7,18]. The spatial segregation of substrate and immunity binding sites presumably provides the flexibility to evolve unique protein–protein interactions while retaining catalytic activity. The fact that two unrelated DNase toxin/immunity pairs appear to be diverging rapidly suggests that this is a general and perhaps universal feature of toxin/immunity systems.
Protein–protein interactions presumably evolve through mutational drift followed by reciprocal changes in the binding partner to maintain overall affinity while the underlying molecular contacts change. Riley and colleagues have proposed a diversification–selection model to explain the observed diversity in E-class colicin/immunity protein pairs. According to their model, some mutations expand immunity function and allow the newly evolved immunity protein to protect not only against its cognate toxin but also against the colicins released by other strains [26,27]. Such mutations would appear to be rare but have been identified and characterized experimentally [28,29]. One striking example that supports this model is the Asp33Leu mutation in ImE2 immunity protein, which increases affinity for non-cognate colicin E9 more than a 3000-fold [29]. The advantage conferred by the new immunity gene would provide the selective pressure to retain the allele and allow it to become fixed in the population. This in turn allows for subsequent mutations in the linked colicin gene. Further mutations in the colicin are predicted to produce “super-killer” toxins, to which the ancestral bacteria are not immune [27]. Thus, the evolved colicin/immunity pair kills ancestral cells, thereby allowing fixation of the new pair in the population. Multiple iterations of this process are predicted to eventually produce a family of divergent toxin/ immunity pairs. Of course, mutations that disrupt the toxin/immunity protein complex should be lethal to the cell; thus, the pressure to retain high-affinity interactions is presumably a significant barrier to diversification. However, colicin/immunity protein complexes have some of the highest known binding affinities, with cognate pairs characterized by femto-molar dissociation constants [22,25,29,30]. Therefore, even if a mutation results in a 1000-fold decrease in affinity, the complex will still have a sub-nanomolar dissociation constant, which is sufficient to provide complete protection against toxicity [25,30]. Thus, the extraordinarily high affinity of cognate colicin/immunity protein complexes provides a buffer against the potentially lethal effects of mutations that disrupt the toxin/immunity protein interface. In contrast, the CDI toxin/immunity proteins studied here have much lower binding affinities with dissociation constants of about 20 nM for cognate pairs. Therefore, CDI toxin/immunity systems must exploit other biophysical mechanisms to avoid self-intoxication during evolution. One possible mechanism involves the over-expression of immunity proteins relative to the toxins. The majority (21 of 25) of homologues presented in Fig. S3a are encoded by truncated cdiA gene fragments that lack the N-terminal coding sequences required for secretion. These pseudogene pairs are termed “orphan” modules because they resemble cdiA-CT/cdiI coding sequences that have been displaced from full-length cdiA genes [9]. Orphan cdiA-CT reading frames usually lack translation initiation signals, whereas the linked orphan cdiI genes have canonical ribosome binding sites upstream of the initiating Met codon. These observations suggest that the toxins are expressed at very low levels, but the immunity proteins are highly expressed. Under these conditions, the selective pressure to retain immunity function would be relieved and allow the immunity gene to undergo drift without lethal consequences. This hypothetical scenario is supported by the observation that non-cognate/mutated immunity proteins can fully protect cells when over-expressed [25,31]. Therefore, we propose that the organization of cdiA-CT/cdiI gene pairs into orphan modules serves to accelerate toxin/immunity evolution by attenuating toxin expression. We note that this could be a general strategy to generate diversity in inter-bacterial competition systems because similar clusters of orphan gene pairs are associated with rhs genes in type VI secretion systems [9,32] and the mafB genes of Neisseria species [33].
CDI systems are widespread throughout proteo-bacteria and are most commonly found in pathogenic species, such as Yersinia pestis, N. meningitidis and B. pseudomallei [2,34]. Because CDI+ bacteria exchange CdiA-CT toxins with one another, it may be possible to induce bacterial suicide with small molecules that specifically disrupt CDI toxin/immunity protein binding interactions. The β-hairpin binding pocket within and homologous immunity proteins is an attractive target to test this antimicrobial strategy. Small cyclic peptides that fold into β-hairpins have been used to study protein–protein and protein–DNA interactions and, in some instances, have been used to specifically disrupt protein complexes [35,36]. As illustrated by the structure, cyclic β-hairpin mimics can be designed to bind CdiI immunity proteins. Our design could be improved to enhance binding affinity and possibly be utilized as a protein–protein complex inhibitor by increasing the number of residues or designing additional contacts. Although the current MAC contains pentapeptide strands, we have previously reported cyclic β-hairpin mimics containing heptapeptide and nonapeptide β-strands [37,38]. Homologous MACs containing larger β-hairpin mimics and designed to achieve more contacts may allow rational design of a higher affinity macrocyclic peptide that specifically may disrupt toxin/immunity complexes within bacterial pathogens, setting the stage for the development of a new class of antibacterials.
Materials and Methods
Bacterial strains and plasmid constructs
All bacterial strains and plasmids used in this study are presented in Table S2. All primers used in this study are presented in Table S3. YFP-labeled E. coli EPI100 cells were generated by integrating the yfp coding sequence at the gal locus. First, a genomic integration construct was made by amplifying the kanamycin-resistance cassette from plasmid pKAN [39] with primers Kan-1/Kan-2, followed by blunt-end ligation to SmaI-digested plasmid pBluescript. One plasmid clone was identified with the kanamycin-resistance cassette in the opposite orientation as pKAN, and this plasmid was termed pNAK. A fragment of galM was then amplified using primers CH3789/ CH3790, and the product was ligated to SacI/BamHI-digested plasmid pNAK to produce pCH2500. A yfp-galT fragment was amplified from E. coli DA28100 (a gift from Sanna Koskiniemi, Uppsala University) using primers CH3787/CH3788, digested with KpnI/EcoRI and then ligated into pCH2500 to yield plasmid pCH2503. The large KpnI/SacI fragment from pCH2503 was recombined into E. coli EPI100 cells that harbor plasmid pSIM6 as described previously [40,41]. mKate2-labeled target bacteria were generated by integrating the coding sequence of mKate2 at the phage HK022 attP site using plasmids pDE1013 and pAH69 as described previously [42].
The coding sequence for CdiA-CT/CdiIYPIII was amplified from Y. pseudotuberculosis YPIII genomic DNA with primers YPK0575-Kpn-for/YPK0576-Xho-rev. The resulting product was digested with KpnI/XhoI and ligated pET21S to generate plasmid pCH10413. The expression construct was generated by replacing the β4/β5-hairpin coding sequence with a Gly-Ser linker. The 5′-end of the construct was amplified with primers β-deletion-for1/β-deletion-rev1 and the 3′-end with primers β-deletion-for2/β-deletion-rev2. The two PCR fragments were ligated at the BamHI site, and the joined fragments re-amplified with β-deletion-for1/ β-deletion-rev2. The resulting product was ligated to pET21d using NcoI and XhoI restriction sites to generate pCH10369. Catalytically inactive domains carrying the Asp198Ala mutation and heterologous β-hairpin sequences were generated by PCR. Plasmid pCH10164 was amplified with primers EC869-CT-Nco/ EC869-Nlact(beta)-rev and EC869-Nlact(beta)-for/ EC869-cdiI-Spe, and the two products combined by overlap extension PCR (OE-PCR) [43] using primers EC869-CT-Nco/EC869-cdiI-Spe. The final product was digested with NcoI/SpeI and ligated to pET21S to generate plasmid pCH10365. The same procedure was used to introduce the Y. kristensenii β-hairpin by PCR with primers EC869-CT-Nco/EC869-Ykris(beta)-rev and EC869-Ykris (beta)-for/EC869-cdiI-Spe. The two products were combined by OE-PCR and ligated to pET21S to generate plasmid pCH10175. The coding sequences for CdiIYkris (ykris0001_10740) and CdiINlact (NEILACOT_05636) were chemically synthesized (GenScript, Inc.) with flanking restriction sites and ligated to plasmid pUC57. The ykris0001_10740 sequence was subcloned into pTrc99KX to generate plasmid pCH10103, which was then used as a template for PCR with primers pTrc-seq2/Ykris-cdiI-Spe-rev. The resulting product was digested with KpnI/SpeI and ligated to pET21K to generate plasmid pCH10170. The NEILACOT_05636 sequence was subcloned into pCH450 to generate plasmid pCH10101, which was then used as a template for PCR with primers pCH450-for/Nlact-cdiI-Spe-rev. The resulting product was digested with NcoI/ SpeI and ligated to pET21S to generate plasmid pCH10172.
The chimeric CDI system that deploys CdiA-CTYPIII toxin was generated by replacing the DNase domain with the corresponding region of CdiA-CT-YPIII. Regions upstream and downstream of the sequence were amplified from plasmid pCH9305 using primers DL1527/EC869o11-G173-rev (upstream) and EC93-YPIII-down-for/DL2368 (downstream). The cdiA-CT/cdiIYPIII sequence was amplified from Y. pseudotuberculosis YPIII genomic DNA using primers EC869o11-G173-for/EC93-YPIII-chim-rev. The three PCR products were combined by OE-PCR using primers DL1527/ DL2368. The final product was electroporated together with plasmid pCH10163 into E. coli strain DY378 as described previously [7,8]. Recombinants were selected on yeast extract/glucose-agar supplemented with 33 μg/mL chloram-phenicol and 10 μM D/L-o-chlorophenylalanine. All plasmid constructs were verified by DNA sequence analysis.
Protein purification
All proteins were over-produced from pET21-derived plasmid using either E. coli CH2016 or E. coli BL21-Gold(DE3). Cells were grown aerobically at 37 °C in LB medium containing 150 μg/mL ampicillin. CdiA-CT/CdiI-YPIII expression was induced by the addition of 1 mM isopropyl-β-D-thiogalactoside at OD600 ~ 0.8 and grown for a further 3–4 h before harvesting. Cells were collected by centrifugation at 5500g for 25 min and then washed with resuspension buffer [20 mM sodium phosphate (pH 7.0) and 150 mM NaCl]. Cells were resuspended and disrupted by sonication on ice in resuspension buffer containing 10 mg/mL lysozyme and 1 mM phenylmethyl-sulfonyl fluoride. Cell debris was removed by centrifugation at 18,000g for 30 min followed by filtration through a 1.0-μm filter. Clarified lysates were loaded onto a Ni2+-charged HiTrap column (5 mL; GE Healthcare) or Ni2 +-nitrilotriacetic acid (Ni2 +-NTA) agarose resin (MCLAB) and washed with resuspension buffer supplemented with 10 mM imidazole. Proteins were eluted with a linear gradient of imidazole (10–500 mM) in resuspension buffer. Fractions were collected, combined and concentrated to a volume of ~500 μL using a 10-kDa centrifugal concentrator (Centricon; Millipore). Proteins were further purified by gel filtration on a Superdex 200 column for the CdiA-CT/CdiIYPIII complex or on a Superdex 75 for individual immunity proteins (GE Healthcare). Gel-filtration columns were equilibrated with 20 mM sodium phosphate (pH 7.0) and 150 mM NaCl using an AKTA FPLC. Purification of CdiIYkris and followed the same protocol, except that all buffers contained 20 mM Tris–HCl (pH 7.4) instead of sodium phosphate. CdiA-CT/CdiIYPIII, CdiIYkris and were concentrated to 10, 12.5 and 7.5 mg/mL (respectively) for crystallization trials.
The individual His6-tagged CdiI proteins were over-produced from plasmid pET21d constructs and purified as described above for . CdiA-CT proteins were isolated from co-expressed His6-tagged CdiI proteins by two methods, depending on whether the two proteins co-eluted following Ni2 +-affinity chromatography. CdiA-CT/CdiI-His6 complexes were denatured overnight in 6 M urea and then subjected to Ni2+-affinity chromatography in buffers containing 6 M urea. Denatured CdiA-CT toxins were collected from the void volume, refolded by dialysis into 20 mM Tris–HCl (pH 8.0) and 10 mM NaCl and then concentrated on a HiTrap Q anion-exchange column and eluted with a salt gradient, yielding 95% pure CdiA-CT protein. Purified toxins were then exchanged into 20 mM Tris–HCl (pH 7.4) and 150 mM NaCl by gel filtration on a S75 column.
Crystallization and structure determination
Protein crystals were grown by hanging-drop vapor diffusion, with drops containing a 1:1 ratio (vol/vol) of protein solution to reservoir liquor. Crystals were mounted and collected under cryo-conditions with the addition of 40% glycerol as cryoprotectant to the reservoir solution. Datasets were collected at 70 K at a wavelength of 1.0 Å and images were indexed, integrated and reduced using either iMOSFLM (CdiA-CT/CdiIYPIII complex) [44] or the HKL2000 suite (CdiIYkris and ) [45]. Initial phases were determined by molecular replacement by autoMR in PHENIX using the structure (PDB code: 4G6U) as a search model. Initial model building was performed by Autobuild in PHENIX. The final models were built through iterative manual building in Coot and refined with phenix.refine. Data collection and refinement statistics are presented in Table 1. All molecular graphics were prepared with PyMOL [46].
CdiA-CT/CdiIYPIII crystals were grown from a solution (10 mg/mL) and a reservoir containing 50 mM Hepes (pH 7.0), 20% polyethylene glycol (PEG) 3350 and 1% tryptone. The complex crystallized in space group C2 with unit cell dimensions 65.51 Å × 65.51 Å × 71.49 Å and one complex per asymmetric unit. The model contains residues Met174–Lys297 (numbered from Val1 of the VENN motif) of CdiA-CTYPIII and residues Asp3–Lys176 of CdiIYPIII. CdiA-CTYPIII residues Lys182, Lys220, Lys240 and Lys297 were modeled as Ala due to lack of observable side-chain density. Similarly, CdiIYPIII residues Asp3, Lys108, Lys118, Lys148 and Lys176 were modeled as Ala residues. The final CdiA-CT/CdiIYPIII model includes 148 water molecules resulting in an Rwork/Rfree (%) of 20.5/ 25.6 (Table 1). CdiIYkris immunity protein crystals were grown from a solution (12.5 mg/mL) over a reservoir containing 0.2 M ammonium fluoride and 20% PEG 3350. The crystal space group was P31 with unit cell dimensions 54.448 Å × 54.448 Å × 54.472 Å and one molecule per asymmetric unit. The final model contains CdiIYkris residues Met1–Gly165 and 130 water molecules resulting in an Rwork/Rfree (%) of 18.1/22.1. CdiIYkris residues Lys4, Glu67, Lys96, Lys126 and Lys136 were modeled as Ala due to lack of observable side-chain density. In addition, the CdiIYkris-His6 expression construct contained an Ala84Thr mutation.
The macrocyclic peptide (MAC) that mimics the β-hairpin (Fig. S1) was prepared according to previously described procedures [16,47,48]. MAC peptide (2 mg) was added to 200 μL of 7.5 mg/mL to yield a solution at a ~10:1 peptide-to-protein ratio. co-crystals were grown over 2 days in 0.2 M sodium acetate (pH 5.6), 0.1 M 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol propane (pH 6.9) and 20% (wt/vol) PEG 3350, using the protein/peptide mixture previously described. Initial crystals were of poor quality and resulted in highly mosaic diffraction data. Crystal quality was improved by microseeding [49]. Briefly, crystals were harvested into 80 μL of crystallization solution and a seed stock was generated using a seed bead (Hampton). Following optimization, suitable diffraction-quality crystals were generated using a hanging drop containing 1 μL of seed stock and 1 μL of the protein/peptide mixture following a 3-fold dilution. The complex crystallized in space group P21 with unit cell dimensions 34.776 Å × 128.166 Å × 44.953 Å (Table 1). Each asymmetric unit contained two complexes. The final model contains two molecules of residues Ala2–Gly167, two macrocyclic peptides and 132 water molecules, resulting in Rwork/Rfree (%) of 18.4/23.1. Residues Lys5, Gln43, Glu78, Lys85, Glu93 of one molecule (chain D only), Asp117 and Glu139 of both molecules were modeled as Ala due to lack of observable side-chain density.
Protein analyses
The secondary structure of purified toxins (0.1 mg/mL in 20 mM Tris–HCl, pH 7.4) was analyzed by CD spectroscopy on a Jasco J-720 spectropolarimeter using a 0.1-cm pathlength. Spectra were collected at 20 nm/min with a 2-nm bandwidth and a 4-s response time. Three consecutive scans were collected and averaged for each analysis. CdiA-CT/CdiI binding affinities were determined by biolayer interferometry as described previously [7]. Binding reactions were performed at 25 °C in 20 mM Tris–HCl (pH 7.4) and 150 mM NaCl. CdiI-His6 immunity proteins were immobilized onto Ni2+-NTA biosensors and exposed to cognate or heterologous CdiA-CT toxins at 0.5–300 μM. A reference was subtracted from all binding curves before curve fitting. Curve fitting and data processing were performed using BLitz Pro software (ForteBio, Inc.).
In vitro analysis of nuclease activities
The activities of purified and CdiA-CTYPIII were assayed in vitro using supercoiled plasmid pUC18 as a substrate. CdiA-CT (at 1 μM final concentration) was incubated with 250 ng of plasmid DNA in 20 mM Tris–HCl (pH 7.5), 100 mM NaCl and 0.1 mg/mL bovine serum albumin supplemented with 2 mM MgCl2 or ZnCl2 for 1 h at 37 °C. Where indicated, purified CdiI-His6 proteins were included at 2 μM final concentration and allowed to bind CdiA-CT for 30 min at room temperature prior to adding substrate DNA. Reactions were quenched with 10 mM ethylenediaminetetraacetic acid followed by the addition of 300 μL of denaturing solution (4 M guanidine HCl and 33% 2-propanol). The reactions were purified over silica membrane spin columns (Epoch Life Sciences). Columns were then washed with 70% ethanol and 10 mM Tris–HCl (pH 8.0) followed by elution with 10 mM Tris–HCl (pH 8.0). Purified DNA from reactions was run on 1% agarose gels containing ethidium bromide and visualized using Bio-Rad Gel Doc 2000.
Competition co-cultures and fluorescence microscopy
Inhibitor cells (E. coli EPI100 carrying plasmid pCH9305, pCH2409 or pDAL878) and target cells (CH8251 carrying plasmid pTrc99a, pCH9315 or pCH848) were grown individually in LB media with 33 μg/μL Cm for inhibitors and 150 μg/μL Amp for targets. The overnight cultures were diluted into fresh LB medium without antibiotics and grown in baffled flasks at 37 °C. At mid-log phase, the inhibitor and target strains were mixed together at a 1:1 ratio in baffled flasks and a sample was withdrawn to score viable target cells as colony-forming units per milliliter on LB agar supplemented with 200 μ/mL rifampicin (Rif). After 4 h of co-culture, another sample was taken and viable target cells were enumerated on Rif-supplemented LB agar. Viable target-cell counts are the mean colony-forming units per milliliter ± the standard error of the mean for three independent experiments. Competitions with fluorescent inhibitor and target bacteria were conducted as described above, except that YFP-labeled inhibitor CH2550 cells and mKate2-labeled targets cells were used. Cells were diluted into fresh LB medium and grown to late-log phase at 30 °C in the dark to maximize fluorescence. Inhibitor and target cells were mixed at a 1:1 ratio in baffled flasks and incubated at 37 °C with shaking in the dark for the duration of the experiment. Samples (equivalent to OD600 = 0.2) were removed at the indicated times and cells were collected by centrifugation. Cells were briefly resuspended in freshly prepared 4% formaldehyde in 1× phosphate-buffered saline, and the fixation reaction was quenched with 125 mM glycine. Fixed cells were washed with 1× phosphate-buffered saline and spotted onto a slide coated with poly-D-lysine (Gold Seal Fluorescent Antibody Rite-On Slides from Fisher prepared by coating with a 1% poly-D-lysine solution prior to addition of cells). Unbound cells were removed gently with Nanopure water, and the slides were treated with Fluorogel II with DAPI mounting medium (Fisher Scientific/EMS) and a coverslip was overlaid prior to imaging. Images were acquired on an Olympus fluorescent microscope with a 100× oil objective using an Optronics MacroFire digital microscope camera. Lightfield images were captured with a 12-ms exposure (gain 2) and DAPI images were acquired in grayscale with a 48-ms exposure (gain 2). Fluorescent images were captured in grayscale using a 502-ms exposure/gain 5 (for YFP) or a 1-s exposure/gain 5 (for mKate2). Images were overlaid and false-colored using Fiji [50], and stacked images were cropped to 400 × 400 pixels using GIMP. The same microscope images used to display fluorescence were used to obtain cell length measurements. Cells were manually measured using the line tool in Fiji, and between 175 and 328 cells from three microscopy fields were measured for each co-culture competition. Each object plotted represents a single cell length measurement. P values were obtained using two-tailed unpaired t tests.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (GM102318 to C.S.H. and C.W.G.) and the National Science Foundation (CHE-1058825 to J.S.N. and DGE-1144085 to J.L.E.W.). Structure determination was, in part, supported by the Advanced Light Source (U.S. Department of Energy under Contract No. DE-AC02-05CH11231) at Berkeley National Laboratories and the Stanford Synchrotron Radiation Lightsource (supported in part by National Institutes of Health P41 GM103393 and U.S. Department of Energy DE-AC02-76SF00515). Funding for open access charge is from National Institutes of Health. We would like to tank the staff at Advanced Light Source and Stanford Synchrotron Radiation Light-source for their invaluable help in data collection. We would also like to thank Elias Gerrick and Sonya Donato for technical support. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Abbreviations used
- CDI
contact-dependent growth inhibition
- DAPI
4′,6-diamidino-2-phenylindole
- YFP
yellow fluorescent protein
- OE-PCR
overlap extension PCR
- PEG
polyethylene glycol
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2015.09.020.
Footnotes
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 4ZQU, 4ZQV and 4ZQW.
References
- 1.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 2.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, Low DA. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio. 2013;4:e00480–13. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb JS, Nikolakakis KC, Willett JL, Aoki SK, Hayes CS, Low DA. Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS ONE. 2013;8:e57609. doi: 10.1371/journal.pone.0057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol. 2009;191:1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A. 2012;109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, Hayes CS. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure. 2014;22:707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t’Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, Tuanyok A, Keim PS, Peacock S, Hayes CS, Low DA. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr S, Walker D, James R, Kleanthous C, Hemmings AM. Inhibition of a ribosome-inactivating ribonuclease: The crystal structure of the cytotoxic domain of colicin E3 in complex with its immunity protein. Structure. 2000;8:949–960. doi: 10.1016/s0969-2126(00)00186-6. [DOI] [PubMed] [Google Scholar]
- 13.Soelaiman S, Jakes K, Wu N, Li C, Shoham M. Crystal structure of colicin E3: Implications for cell entry and ribosome inactivation. Mol Cell. 2001;8:1053–1062. doi: 10.1016/s1097-2765(01)00396-3. [DOI] [PubMed] [Google Scholar]
- 14.Gioia U, Laneve P, Dlakic M, Arceci M, Bozzoni I, Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem. 2005;280:18996–19002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- 15.Renzi F, Caffarelli E, Laneve P, Bozzoni I, Brunori M, Vallone B. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc Natl Acad Sci U S A. 2006;103:12365–12370. doi: 10.1073/pnas.0602426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woods RJ, Brower JO, Castellanos E, Hashemzadeh M, Khakshoor O, Russu WA, Nowick JS. Cyclic modular beta-sheets. J Am Chem Soc. 2007;129:2548–2558. doi: 10.1021/ja0667965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowick JS, Brower JO. A new turn structure for the formation of beta-hairpins in peptides. J Am Chem Soc. 2003;125:876–877. doi: 10.1021/ja028938a. [DOI] [PubMed] [Google Scholar]
- 18.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper PC, James R. Two new E colicins, E8 and E9, produced by a strain of Escherichia coli. J Gen Microbiol. 1984;130:209–215. doi: 10.1099/00221287-130-1-209. [DOI] [PubMed] [Google Scholar]
- 20.Watson R, Rowsome W, Tsao J, Visentin LP. Identification and characterization of Col plasmids from classical colicin E-producing strains. J Bacteriol. 1981;147:569–577. doi: 10.1128/jb.147.2.569-577.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 22.Wojdyla JA, Fleishman SJ, Baker D, Kleanthous C. Structure of the ultra-high-affinity colicin E2 DNase–Im2 complex. J Mol Biol. 2012;417:79–94. doi: 10.1016/j.jmb.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7:91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 24.Kleanthous C, Walker D. Immunity proteins: Enzyme inhibitors that avoid the active site. Trends Biochem Sci. 2001;26:624–631. doi: 10.1016/s0968-0004(01)01941-7. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Keeble AH, Giffard C, James R, Moore GR, Kleanthous C. Highly discriminating protein–protein interaction specificities in the context of a conserved binding energy hotspot. J Mol Biol. 2004;337:743–759. doi: 10.1016/j.jmb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Riley MA. Positive selection for colicin diversity in bacteria. Mol Biol Evol. 1993;10:1048–1059. doi: 10.1093/oxfordjournals.molbev.a040054. [DOI] [PubMed] [Google Scholar]
- 27.Tan Y, Riley MA. Positive selection and recombination: Major molecular mechanisms in colicin diversification. Trends Ecol Evol. 1997;12:348–351. doi: 10.1016/s0169-5347(97)01127-0. [DOI] [PubMed] [Google Scholar]
- 28.Masaki H, Akutsu A, Uozumi T, Ohta T. Identification of a unique specificity determinant of the colicin E3 immunity protein. Gene. 1991;107:133–138. doi: 10.1016/0378-1119(91)90306-v. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Hamill SJ, Hemmings AM, Moore GR, James R, Kleanthous C. Dual recognition and the role of specificity-determining residues in colicin E9 DNase-immunity protein interactions. Biochemistry. 1998;37:11771–11779. doi: 10.1021/bi9808621. [DOI] [PubMed] [Google Scholar]
- 30.Wallis R, Leung KY, Pommer AJ, Videler H, Moore GR, James R, Kleanthous C. Protein–protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry. 1995;34:13751–13759. doi: 10.1021/bi00042a005. [DOI] [PubMed] [Google Scholar]
- 31.Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS. Following evolutionary paths to protein–protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009;16:1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- 32.Koskiniemi S, Garza-Sanchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, Andersson DI, Hayes CS, Low DA. Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar typhimurium. PLoS Genet. 2014;10:e1004255. doi: 10.1371/journal.pgen.1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamet A, Jousset AB, Euphrasie D, Mukorako P, Boucharlat A, Ducousso A, Charbit A, Nassif X. A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog. 2015;11:e1004592. doi: 10.1371/journal.ppat.1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughlin WA, Tyndall JD, Glenn MP, Fairlie DP. Beta-strand mimetics. Chem Rev. 2004;104:6085–6117. doi: 10.1021/cr040648k. [DOI] [PubMed] [Google Scholar]
- 36.Fasan R, Dias RL, Moehle K, Zerbe O, Vrijbloed JW, Obrecht D, Robinson JA. Using a beta-hairpin to mimic an alpha-helix: Cyclic peptidomimetic inhibitors of the p53-HDM2 protein–protein interaction. Angew Chem Int Ed Engl. 2004;43:2109–2112. doi: 10.1002/anie.200353242. [DOI] [PubMed] [Google Scholar]
- 37.Cheng PN, Liu C, Zhao M, Eisenberg D, Nowick JS. Amyloid beta-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity. Nat Chem. 2012;4:927–933. doi: 10.1038/nchem.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham JD, Chim N, Goulding CW, Nowick JS. Structures of oligomers of a peptide from beta-amyloid. J Am Chem Soc. 2013;135:12460–12467. doi: 10.1021/ja4068854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 40.Datta S, Costantino N, Court DL. A set of recombineering plasmids for Gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 41.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. Recombineering: Genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb0116s78. http://dx.doi.org/10.1002/0471142727.mb0116s78. [DOI] [PubMed]
- 42.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012;109:8884–8889. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aiyar A, Xiang Y, Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol. 1996;57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
- 44.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System, 1.3 ed. Schro€dinger, LLC; 2010. [Google Scholar]
- 47.Cheng PN, Nowick JS. Giant macrolactams based on beta-sheet peptides. J Org Chem. 2011;76:3166–3173. doi: 10.1021/jo102598n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer RK, Li H, Nowick JS. X-ray crystallographic structures of trimers and higher-order oligomeric assemblies of a peptide derived from Abeta(17–36) J Am Chem Soc. 2014;136:5595–5598. doi: 10.1021/ja5017409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergfors T. Seeds to crystals. J Struct Biol. 2003;142:66–76. doi: 10.1016/s1047-8477(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 50.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.