Abstract
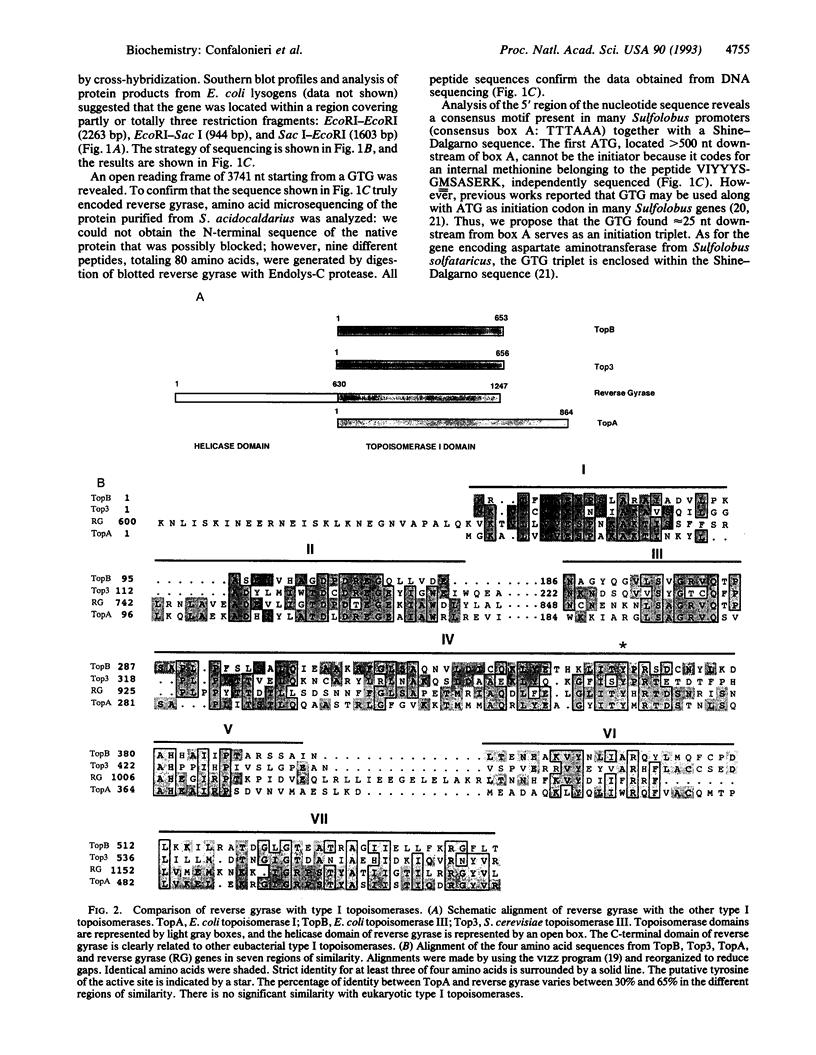
Reverse gyrase is a type I DNA topoisomerase able to positively supercoil DNA and is found in thermophilic archaebacteria and eubacteria. The gene coding for this protein was cloned from Sulfolobus acidocaldarius DSM 639. Analysis of the 1247-amino acid sequence and comparison of it with available sequence data suggest that reverse gyrase is constituted of two distinct domains: (i) a C-terminal domain of approximately 630 amino acids clearly related to eubacterial topoisomerase I (Escherichia coli topA and topB gene products) and to Saccharomyces cerevisiae top3; (ii) an N-terminal domain without any similarity to other known topoisomerases but containing several helicase motifs, including an ATP-binding site. These results are consistent with those from our previous mechanistic studies of reverse gyrase and suggest a model in which positive supercoiling is driven by the concerted action of helicase and topoisomerase in the same polypeptide: this constitutes an example of a composite gene formed by a helicase domain and a topoisomerase domain.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Bokranz M., Klein A. Nucleotide sequence of the methyl coenzyme M reductase gene cluster from Methanosarcina barkeri. Nucleic Acids Res. 1987 May 26;15(10):4350–4351. doi: 10.1093/nar/15.10.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C., Portemer C., Huber R., Forterre P., Duguet M. Reverse gyrase in thermophilic eubacteria. J Bacteriol. 1991 Jun;173(12):3921–3923. doi: 10.1128/jb.173.12.3921-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C., Portemer C., Nadal M., Stetter K. O., Forterre P., Duguet M. Reverse gyrase, a hallmark of the hyperthermophilic archaebacteria. J Bacteriol. 1990 Dec;172(12):6803–6808. doi: 10.1128/jb.172.12.6803-6808.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubellis M. V., Rozzo C., Nitti G., Arnone M. I., Marino G., Sannia G. Cloning and sequencing of the gene coding for aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. Eur J Biochem. 1989 Dec 8;186(1-2):375–381. doi: 10.1111/j.1432-1033.1989.tb15219.x. [DOI] [PubMed] [Google Scholar]
- Depew R. E., Liu L. F., Wang J. C. Interaction between DNA and Escherichia coli protein omega. Formation of a complex between single-stranded DNA and omega protein. J Biol Chem. 1978 Jan 25;253(2):511–518. [PubMed] [Google Scholar]
- Drlica K. Bacterial topoisomerases and the control of DNA supercoiling. Trends Genet. 1990 Dec;6(12):433–437. doi: 10.1016/0168-9525(90)90306-q. [DOI] [PubMed] [Google Scholar]
- Forterre P., Mirambeau G., Jaxel C., Nadal M., Duguet M. High positive supercoiling in vitro catalyzed by an ATP and polyethylene glycol-stimulated topoisomerase from Sulfolobus acidocaldarius. EMBO J. 1985 Aug;4(8):2123–2128. doi: 10.1002/j.1460-2075.1985.tb03902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Gallagher M. P., Mimmack M. L., Pearce S. R. A family of closely related ATP-binding subunits from prokaryotic and eukaryotic cells. Bioessays. 1988 Apr;8(4):111–116. doi: 10.1002/bies.950080406. [DOI] [PubMed] [Google Scholar]
- Jaxel C., Nadal M., Mirambeau G., Forterre P., Takahashi M., Duguet M. Reverse gyrase binding to DNA alters the double helix structure and produces single-strand cleavage in the absence of ATP. EMBO J. 1989 Oct;8(10):3135–3139. doi: 10.1002/j.1460-2075.1989.tb08466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A., Asai K. Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984 Jun 21;309(5970):677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Lau K., Wu H. Y., Liu L. F. Identification of a DNA supercoiling activity in Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Oct 11;20(19):5067–5072. doi: 10.1093/nar/20.19.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee E. H., Masai H., Allen G. C., Jr, Kornberg A. The priA gene encoding the primosomal replicative n' protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4620–4624. doi: 10.1073/pnas.87.12.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal M., Jaxel C., Portemer C., Forterre P., Mirambeau G., Duguet M. Reverse gyrase of Sulfolobus: purification to homogeneity and characterization. Biochemistry. 1988 Dec 27;27(26):9102–9108. doi: 10.1021/bi00426a006. [DOI] [PubMed] [Google Scholar]
- Nielsen P. J., Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988 Jul;7(7):2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., DiGate R. J., Zavitz K. H., Marians K. J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pühler G., Lottspeich F., Zillig W. Organization and nucleotide sequence of the genes encoding the large subunits A, B and C of the DNA-dependent RNA polymerase of the archaebacterium Sulfolobus acidocaldarius. Nucleic Acids Res. 1989 Jun 26;17(12):4517–4534. doi: 10.1093/nar/17.12.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26(3-4):335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin W., Marliere P. Comparaison de plusieurs séquences protéiques par reconnaissance de blocs conservés. C R Acad Sci III. 1986;303(13):541–546. [PubMed] [Google Scholar]
- Schmid S. R., Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992 Feb;6(3):283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Shibata T., Nakasu S., Yasui K., Kikuchi A. Intrinsic DNA-dependent ATPase activity of reverse gyrase. J Biol Chem. 1987 Aug 5;262(22):10419–10421. [PubMed] [Google Scholar]
- Slesarev A. I., Kozyavkin S. A. DNA substrate specificity of reverse gyrase from extremely thermophilic archaebacteria. J Biomol Struct Dyn. 1990 Feb;7(4):935–942. doi: 10.1080/07391102.1990.10508533. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Caron P. R., Kim R. A. The role of DNA topoisomerases in recombination and genome stability: a double-edged sword? Cell. 1990 Aug 10;62(3):403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Recent studies of DNA topoisomerases. Biochim Biophys Acta. 1987 Jun 6;909(1):1–9. doi: 10.1016/0167-4781(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Zhang H., Jessee C. B., Liu L. F. A protein factor from Xenopus oocytes with simian virus 40 large tumor antigen-like DNA supercoiling activity. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9078–9082. doi: 10.1073/pnas.87.23.9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Stetter K. O., Janeković D. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur J Biochem. 1979 Jun 1;96(3):597–604. doi: 10.1111/j.1432-1033.1979.tb13074.x. [DOI] [PubMed] [Google Scholar]