ABSTRACT
Many symbiotic gut bacteria possess the ability to degrade multiple polysaccharides, thereby providing nutritional advantages to their hosts. Like microorganisms adapted to other complex nutrient environments, gut symbionts give different metabolic priorities to substrates present in mixtures. We investigated the responses of Bacteroides thetaiotaomicron, a common human intestinal bacterium that metabolizes more than a dozen different polysaccharides, including the O-linked glycans that are abundant in secreted mucin. Experiments in which mucin glycans were presented simultaneously with other carbohydrates show that degradation of these host carbohydrates is consistently repressed in the presence of alternative substrates, even by B. thetaiotaomicron previously acclimated to growth in pure mucin glycans. Experiments with media containing systematically varied carbohydrate cues and genetic mutants reveal that transcriptional repression of genes involved in mucin glycan metabolism is imposed by simple sugars and, in one example that was tested, is mediated through a small intergenic region in a transcript-autonomous fashion. Repression of mucin glycan-responsive gene clusters in two other human gut bacteria, Bacteroides massiliensis and Bacteroides fragilis, exhibited variable and sometimes reciprocal responses compared to those of B. thetaiotaomicron, revealing that these symbionts vary in their preference for mucin glycans and that these differences occur at the level of controlling individual gene clusters. Our results reveal that sensing and metabolic triaging of glycans are complex processes that vary among species, underscoring the idea that these phenomena are likely to be hidden drivers of microbiota community dynamics and may dictate which microorganisms preferentially commit to various niches in a constantly changing nutritional environment.
IMPORTANCE
Human intestinal microorganisms impact many aspects of health and disease, including digestion and the propensity to develop disorders such as inflammation and colon cancer. Complex carbohydrates are a major component of the intestinal habitat, and numerous species have evolved and refined strategies to compete for these coveted nutrients. Our findings reveal that individual bacteria exhibit different preferences for carbohydrates emanating from host diet and mucosal secretions and that some of these prioritization strategies are opposite to one another. Thus, we reveal new aspects of how individual bacteria, some with otherwise similar metabolic potential, partition to “preferred niches” in the complex gut ecosystem, which has important and immediate implications for understanding and predicting the behavioral dynamics of this community.
INTRODUCTION
Bacteria in the human colon have developed several strategies to forage the complex carbohydrates (polysaccharides or glycans) present in dietary fiber. With the exception of starch, members of this diverse group of nutrients transit the proximal intestine undigested due to a lack of appropriate human enzymes. Residual dietary glycans therefore require the activity of symbiotic bacteria in the distal gut first for hydrolysis (1) and then for fermentation into host-absorbable short-chain fatty acids, which provide a significant contribution to our dietary energy harvest (2). Human intestinal cells also secrete endogenous glycans, largely as mucin glycoproteins and epithelial glycocalyx, which contribute to a protective barrier for host tissue. Most glycans associated with intestinal mucins are O-linked glycans (herein referred to as O-glycans) to the side chains of serine and threonine residues in mucin polypeptides. Encompassing ~102 different linear and branched structures, the O-glycans that are attached to mucins are remarkably diverse and also distinct from dietary fiber glycans (3, 4). Thus, to metabolize O-glycans, gut bacteria require highly diversified sensory systems coupled to expression of dozens of appropriate enzymes (5, 6).
A number of species from the dominant human gut bacterial phyla (Bacteroidetes, Firmicutes, Actinobacteria, and Verrucomicrobia) have been associated with degradation of mucus and/or its attached O-glycans, while other species in these phyla lack this behavior (7–11). Several experiments with simplified or transplanted gut microbial communities assembled in gnotobiotic mice have implied that the ability to forage host-derived glycans provides a metabolic alternative when dietary glycans wane or are absent or when microorganisms are forced into competition by the addition of additional microbial species (12–17).
Detrimental host effects associated with disrupted mucin glycosylation and bacterial degradation of host glycan structures are beginning to emerge as well. Studies using mouse models that lack normal mucin expression or glycosylation have observed spontaneous colonic inflammation or increased susceptibility to inflammatory agents, including individual bacteria (18–21), raising the possibility that microbe-catalyzed alterations to mucus could also be harmful. This notion is supported by findings in humans with ulcerative colitis, which show that O-glycosylation patterns are simpler in people with active disease versus the same patients during remission (22). Symbionts such as Bacteroides thetaiotaomicron can liberate sialic acid from host glycoconjugates, providing a sugar that it cannot use itself to pathogens such as Salmonella enterica and Clostridium difficile (23). Finally, a recent study involving mice that are genetically susceptible to acute colonic inflammation that can be induced by B. thetaiotaomicron or other commensal Bacteroidetes demonstrated that genetic disruption of bacterial sulfatase production blocks the progression of disease (24). The precise, disease-causing target(s) of B. thetaiotaomicron sulfatases has not yet been identified. However, the observation that most of the 28 different predicted sulfatase-encoding genes in the colitogenic B. thetaiotaomicron type strain (25) are contained in gene clusters that are responsive to host mucin O-glycans or glycosaminoglycans (6, 24) suggests that bacterial interactions with the sulfated carbohydrates abundant in colonic mucus or the glycocalyx are a critical step.
B. thetaiotaomicron and several other mucin-degrading Bacteroides species possess the ability to catabolize other carbohydrates in addition to O-glycans. To date, essentially all of these metabolic abilities that have been described are mediated by tightly regulated expression of corresponding polysaccharide utilization loci (PULs) that encode specific degradative machinery for the particular substrate(s) present (7). We have previously shown that when B. thetaiotaomicron is grown in a mixture containing a dozen different glycans, it represses expression of genes involved in O-glycan utilization until the latest stages of growth when most or all of the other substrates have been depleted (26). Here, we investigate the ability of individual glycans to modulate O-glycan metabolism in B. thetaiotaomicron and other gut species and provide molecular genetic insight into the basis of this phenomenon. We identify two species, Bacteroides massiliensis and Bacteroides fragilis that exhibit different, often reciprocal, O-glycan preferences compared to those of B. thetaiotaomicron. Given the likelihood that such metabolic variations extend to other nutrients, the phenomena described here represent an underappreciated and perhaps pivotal driver of dynamics in the diet-microbiota-host relationship.
RESULTS
O-linked glycans are given low metabolic priority by B. thetaiotaomicron.
The metabolic preferences and corresponding regulatory mechanisms exhibited by symbiotic bacteria in the human colon remain largely unexplored. We previously found that B. thetaiotaomicron preferentially metabolizes glycosaminoglycans prior to O-glycans when they are present together in a mixture (6). To test the hypothesis that other glycans repress O-glycan use, we grew B. thetaiotaomicron in pairwise mixtures of porcine mucin O-glycans (PMOG) and several other carbohydrates (Fig. 1). If either substrate was prioritized, we anticipated “diauxic” growth patterns, indicating metabolic shifts between the most- and least-preferred nutrients. In addition, we harvested bacterial RNA at “early” and “late” points in growth and compared glycan-specific PUL expression by quantitative PCR (qPCR) on reverse-transcribed RNA.
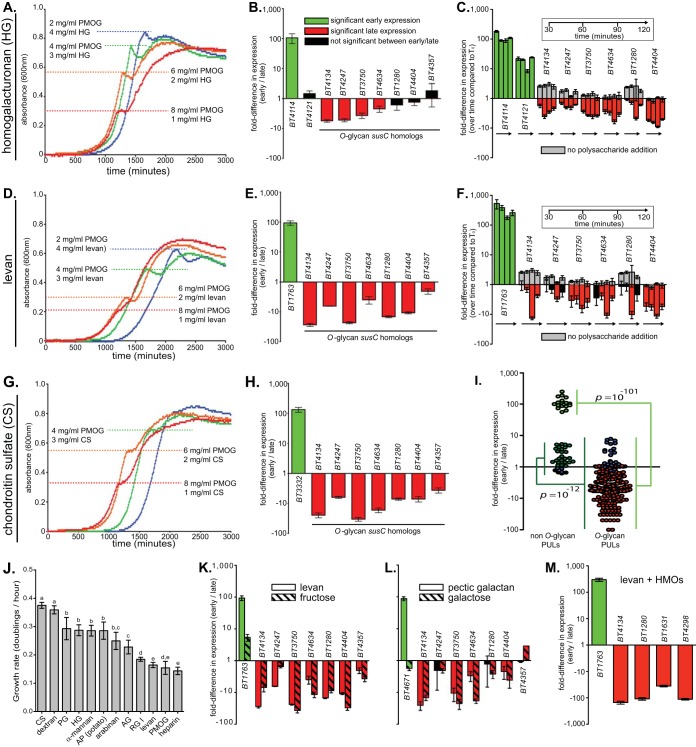
FIG 1 .
B. thetaiotaomicron deprioritizes O-glycan metabolism in the presence of competing glycans and monosaccharides. (A) B. thetaiotaomicron growth curves in mixtures of homogalacturonan (HG) and PMOG. The dashed horizontal lines indicate the pause in exponential growth that occurs as a function of changing glycan mix concentrations. (B) Relative expression differences between early and late growth phases for B. thetaiotaomicron grown in a mixture of HG and PMOG (1.5 mg/ml and 7 mg/ml, respectively). Early and late samples were harvested before and after the pause, respectively. A total of seven previously identified O-glycan-responsive PULs (6) were monitored for changes in expression based on the criteria that they were induced more than 8.5-fold in pure PMOG and are not part of recombinational shufflons that might obfuscate temporal expression. PULs with statistically significant (P ≤ 0.05 by one-tailed Student’s t test) increases in expression during early growth, PULs with statistically significant increases in later growth, and PULs with no significant changes in early versus late growth are shown. (C) A “spike-in” experiment in which B. thetaiotaomicron was grown to mid-exponential phase on 10 mg/ml PMOG alone and then HG (final concentration of 2.5 mg/ml) was abruptly introduced. Bacterial samples were harvested at 30-, 60-, 90-, and 120-min intervals after the introduction of HG. A corresponding negative control (minimal medium with no carbon) was used to monitor changes in O-glycan PUL expression (gray bars). Expression changes were determined relative to the culture grown in PMOG only (time zero [T0]). (D to H) Experiments similar to those described above, but with levan or chondroitin sulfate (CS) as the competing glycan. Additional experiments for pairwise combinations are shown in Fig. S1 in the supplemental material. (I) Aggregate expression differences of all PULs involved in metabolizing O-glycans and the non-O-glycans tested. Each symbol shows the value for one separate biological replicate of three conducted for each combination. (J) Exponential growth rates of B. thetaiotaomicron on all individual glycans tested. PG, pectic galactan; AP, amylopectin; AG, arabinogalactan; RG I, rhamnogalacturonan I. (Letters over each bar indicate data groupings in which the t test P values were <0.001). (K and L) Additional early/late growth phase experiments comparing the responses to a competing polysaccharide (levan and pectic galactan) and their corresponding monosaccharides (fructose and galactose). (M) HMO-responsive PUL (11) expression during early and late growth in a mixture of levan and human milk oligosaccharides (HMOs). A total of three biological replicates were conducted for each experimental condition, including bacterial growth curves. All values are means ± standard deviations (SD) (error bars), except in panels C and F, where they are means ± standard errors (SE) (error bars).
Data for a plant cell wall pectin (homogalacturonan), a bacterial β2,6-linked fructan capsule (levan), and an animal tissue glycosaminoglycan (chondroitin sulfate) are shown in Fig. 1A to H. Consistent with our hypothesis, we observed diauxic growth in which the early growth phase was prolonged by increasing homogalacturonan, levan, or chondroitin sulfate (Fig. 1A, D, and G). Gene expression comparisons further support repression of O-glycan metabolism with high expression of sentinel susC-like genes from the homogalacturonan, levan, and chondroitin sulfate PULs early and reciprocal patterns for nearly all of the seven different O-glycan-responsive PULs that were tested (Fig. 1B, E, and H; see Fig. S1I in the supplemental material). In addition, B. thetaiotaomicron cells that were pregrown in PMOG abruptly repressed O-glycan PULs when homogalacturonan, levan, or a mixture of all glycans used was introduced (Fig. 1C and F; Fig. S2A).
Results similar to those with homogalacturonan, levan, and chondroitin sulfate were observed with nearly all other glycans tested (see Fig. S1A to S1H in the supplemental material). In aggregate, gene expression trends for glycans tested against PMOG revealed significant overall repression of O-glycan genes, underscoring that utilization of this host-derived nutrient is low in B. thetaiotaomicron’s metabolic hierarchy (Fig. 1I). Although B. thetaiotaomicron growth rates on PMOG are among the lowest compared to other glycans (Fig. 1J; see Fig. S3 for schematics of all glycans used), strong repression of O-glycan PULs in mixtures with levan or heparin, which both had similarly low growth rates, argues that growth rate is insufficient to account for this trend. Only dextran failed to exhibit significant expression changes between early and late growth (blue data points in Fig. 1I), although it still elicited a diauxic growth profile (Fig. S1C).
To determine whether the signals that induce repression of O-glycan PULs originate from the transcriptional sensor/regulators that activate expression of their cognate PULs in response to the alternative glycans that were presented with PMOG, as was reported in one previous study (27), or their component simple sugars, we performed experiments with the monosaccharides fructose and galactose. Fructose is the sole component of levan and is sufficient to trigger expression of the single levan PUL (28). Galactose is abundant in both pectic galactan and PMOG (6). The presence of either monosaccharide was sufficient to repress expression of O-glycan PULs (Fig. 1K and L). The observation that galactose decreases expression of O-glycan PULs reveals that a monosaccharide, in sufficient quantity, is capable of suppressing utilization of another polymer in which it is also contained. Moreover, activation of the repressing glycan’s cognate PUL, via its associated regulator, is not required because galactose does not induce expression of the pectic galactan PUL (Fig. 1L). Additional monosaccharide experiments further support the idea that simple sugars mediate repression, albeit with notable variation with respect to sugar identity and timing (see Fig. S2B to S2L in the supplemental material).
Finally, we grew B. thetaiotaomicron on mixtures containing human milk oligosaccharides (HMOs), glycans with structural similarity to O-glycans, plus either homogalacturonan or levan to determine whether O-glycan/HMO-responsive PULs are repressed. Both of these substrate pairs elicited clear diauxic growth (see Fig. S4A and S4B in the supplemental material) and repression of known HMO utilization genes when mixed with levan (11) (Fig. 1M).
Expression of some O-glycan utilization functions in other Bacteroides species is opposite that in B. thetaiotaomicron.
In seven other B. thetaiotaomicron strains with ≥98% 16S rRNA genes identity to the type strain VPI 5482, we observed preferred use of fructans, either levan or inulin (chosen based on which linkage type each strain optimally used), and homogalacturonan when individually mixed with PMOG (see Fig. S4C and S4D in the supplemental material). This commonality among B. thetaiotaomicron strains raises the question of whether more distantly related gut symbionts also behave similarly and assign low metabolic priority to O-glycans when they are presented with alternative polysaccharides. Thus, we investigated two other O-glycan-degrading species, Bacteroides massiliensis and Bacteroides fragilis, with various phylogenetic distances relative to B. thetaiotaomicron (Fig. S5A). A survey of the glycans that these two species can use as the sole carbon source revealed that, among the substrates tested, they are restricted to PMOG and starch (B. massiliensis), and PMOG, starch, and inulin (B. fragilis), respectively (see Table S1 in the supplemental material). Transcriptome sequencing (RNAseq)-based whole-genome transcriptional profiling identified 6 of 33 PULs in B. massiliensis and 8/52 PULs in B. fragilis that are active on PMOG (Tables S2 and S3). A genome survey revealed that B. massiliensis has a PUL with identical organization to the B. thetaiotaomicron starch utilization system (sus) and is activated on starch (Fig. S5B and S5C). Likewise, B. fragilis has two previously described PULs that mediate starch and inulin utilization (28, 29).
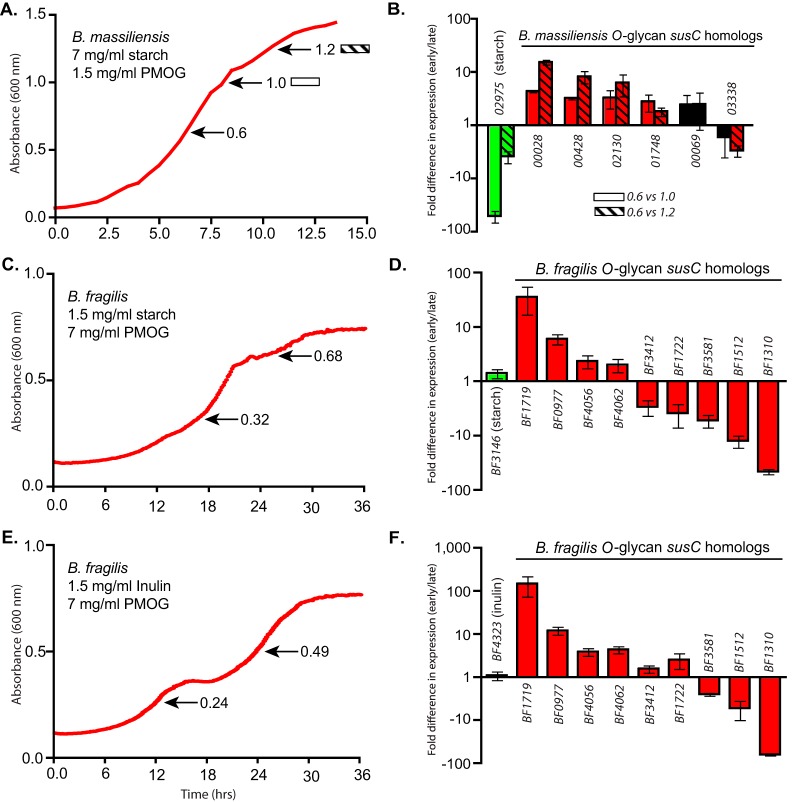
Growth of B. massiliensis in a mixture of PMOG and starch failed to reveal a distinct diauxic pattern as clear as that observed for B. thetaiotaomicron. However, two short pauses in late growth suggested a possible metabolic shift (Fig. 2A) that was supported by individual growth levels on PMOG and starch when tested alone (see Fig. S5F in the supplemental material). Early versus late expression measurements revealed that B. massiliensis represses starch utilization until later growth and exhibits neutral or preferred expression of five of the six O-glycan-responsive PULs tested (Fig. 2B). Despite B. fragilis exhibiting diauxic growth patterns that appeared to favor either starch or inulin over PMOG (Fig. S5G and S5H), the PULs for these two glycans did not show preferential expression at the growth points analyzed (Fig. 2C and E). In contrast, some O-glycan-responsive PULs were prioritized in early growth, while others remained neutral or were activated later (Fig. 2D and F). The latter finding highlights that independent gene clusters, which otherwise are activated by related glycans from the same source (here, those present in gastric mucin) can exhibit opposing behavior. This observation suggests that these PULs are individually regulated by opposing signals that vary in their influence on overall expression. These signals likely emanate from both positive and negative cues: recognition of the cognate glycan or repression in the presence of other glycans/sugars, respectively. The contributions of these opposing forces are considered further below.
FIG 2 .
(A) B. massiliensis growth curve in a potato amylopectin (starch)/PMOG mix. (B) Relative expression of B. massiliensis O-glycan-responsive PULs between early and late growth. Two later growth point transcripts were analyzed (absorbance at 600 nm values of 1.0 and 1.2) and compared separately back to the early growth point (A600 = 0.6). (C) B. fragilis growth curve in a potato amylopectin (starch)/PMOG mix. (D) Relative expression of B. fragilis O-glycan-responsive PULs between early and late growth in an APpot (potato amylopectin) (starch)/PMOG mix. (E) B. fragilis growth curve in an inulin/PMOG mix. (F) Relative expression of B. fragilis O-glycan-responsive PULs between early and late growth in an inulin/PMOG mix. Note the reciprocal amounts of starch and PMOG required for B. massiliensis analysis compared to similar experiments with B. thetaiotaomicron. All expression values are significant (P ≤ 0.05 by one-tailed Student’s t test), except for those values labeled in black. All values are mean ± SD.
Differential substrate recognition in complex mixtures contributes to prioritized PUL expression.
The above results suggest that simple sugars mediate substantial repression of O-glycan metabolism (Fig. 1K and L), similar to 3′-5′ cyclic AMP (cAMP) catabolite repression in Escherichia coli and Bacillus subtilis (30). However, no evidence for cAMP-responsive pathways has been reported in Bacteroides species (31), and we also failed to observe decreased repression of O-glycan-targeting PULs with the addition of cAMP (see Fig. S6A in the supplemental material). We concluded that the presence of polysaccharide surface capsule, which is both phase variable and a known barrier to incoming glycan nutrients (32), is not required for repression of O-glycan PULs under the conditions tested (Fig. S6B). Finally, experiments with a mutant that is unable to utilize levan (ΩBT1754) revealed that the mere presence of this preferred glycan in the extracellular environment of a strain that cannot degrade it is insufficient to trigger repression of genes involved in O-glycan use (Fig. S6C and S6D). Collectively, these results lead us to conclude that a cAMP-independent repression mechanism exists in Bacteroides that is not passively governed by the presence of bacterial or environmental polysaccharides at the cell surface.
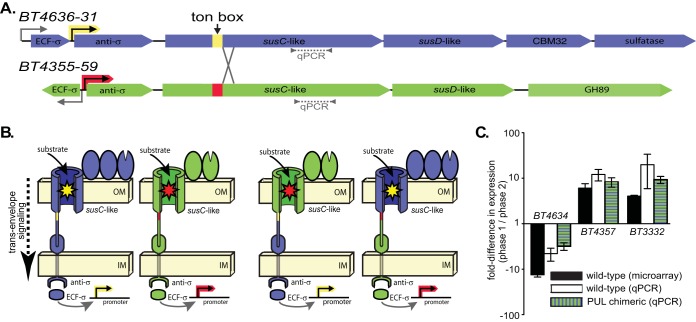
To begin to understand the mechanism(s) underlying B. thetaiotaomicron’s prioritization of different glycans, we chose to separate the glycan-sensing capacity of PULs with different responses to O-glycans from their corresponding regulatory elements. To do this, we created two chimeric PULs that were recombined just downstream of their predicted Ton box sequences (Fig. 3A and B; see Fig. S6E in the supplemental material). One of the O-glycan PULs that was chosen for this recombination (genetic locus from BT4355 to BT4359 [BT4355-59]) was most active during early growth in previous experiments with a partially defined mixture of O-linked glycans and glycosaminoglycans that is termed the “100 mM fraction” (6) based on how it is purified (see Fig. S7A to S7C and the corresponding legend in the supplemental material for details). This same PUL was also weakly and inconsistently repressed in the pairwise glycan mixtures shown in Fig. 1 and Fig. S1. In contrast, the other PUL (BT4636-31) showed reciprocal expression during multiphasic growth in the 100 mM fraction (Fig. 3C) and also tended to be more strongly repressed in the pairwise mixtures.
FIG 3 .
(A) A schematic of the O-glycan-responsive PULs, BT4636-31 and BT4355-59, showing the presumed locations, based on previous microarray data (6), of noninducible promoters (bent arrows), ECF-σ-inducible promoters (colored bent arrows), predicted Ton boxes, and chimeric recombination sites. (B) Cellular organization of chimeric Sus-like systems encoded by the above PULs separating glycan sensing at outer membrane TonB-dependent transporters (yellow and red stars) and genetic regulation via inducible promoters and genes encoding ECF-σ and anti-σ factors. See Fig. S6E in the supplemental material for a detailed illustration of the genetic recombination scheme used. OM, outer membrane; IM, inner membrane. (C) Growth phase-specific expression of BT4634 and BT4357 in an undefined mixture containing O-glycans and glycosaminoglycans (GAGs), termed the “100 mM fraction” from PMOG purification (6). Previous microarray (black bars) and current qPCR (white bars and green striped bars) measurements are shown for comparison.
The extracytoplasmic function sigma factor (ECF-σ)/anti-σ factor pairs that regulate these PULs work with outer membrane TonB-dependent transporters via a mechanism termed trans-envelope signaling (33). In this sensory strategy, outer membrane transporters recognize glycans during passage across the outer membrane (illustrated as stars in Fig. 3B), and this information is relayed to the corresponding ECF-σ-dependent promoters via protein contacts between the N-terminal domain of the outer membrane transporter, inner membrane anti-σ factor, and cytoplasmic ECF-σ factor (5). Since all of these protein domains are encoded upstream of the Ton box recombination sites, our chimeric PULs were designed to separate the regulatory elements from the downstream regions of the susC-like genes that encode glycan-sensing transporter domains (Fig. 3B). Despite their discordant responses in the 100 mM fraction, these PULs are both activated on PMOG and N-acetyllactosamine (LacNAc), a major disaccharide building block of O-glycans, as the sole carbon source, suggesting that they respond to similar ligands that may share overlapping structural features (see Fig. S7D in the supplemental material).
Both chimeric PULs were activated during growth on O-glycans as the sole carbon source, suggesting that chimeric recombination did not eliminate their function (see Fig. S7E in the supplemental material). We then grew wild-type B. thetaiotaomicron and a strain that simultaneously contained both chimeric PULs in place of the wild-type loci in the 100 mM fraction that produces discordant phase-specific PUL expression (Fig. 3C). Interestingly, the susC-like genes in the chimeric PULs, which were uncoupled from their native regulatory regions, exhibited discordant growth between early and late growth phases that matched the pattern observed for wild-type B. thetaiotaomicron. A gene from a control PUL (BT3332), which mediates high-priority chondroitin sulfate catabolism (6), did not show altered phase-specific expression in the chimeric strain. This observation indicates that the regulatory components of these two PULs do not mediate expression differences in this particular case, instead the differential sensing of nutrients via the outer membrane TonB-dependent transporter causes this behavior. Despite the fact that these two PULs show responses to similar O-glycan saccharides, such as LacNAc, this behavior could be explained by the sequential release of recognizable saccharides from more-complex structures present in the 100 mM mixture that drive the selective and sequential activation of the two PULs monitored during growth in this mixture. In contrast, if promoter level regulatory events, such as those mediated by cAMP receptor protein (CRP)-cAMP complexes during catabolite repression in other bacteria, predominate in this system, we would have expected growth phase-dependent expression changes to track with each PUL’s respective promoter and regulator-encoding genes instead of with susC-like sensor/transport regions.
A transcribed intergenic region mediates conditional repression of one O-glycan PUL.
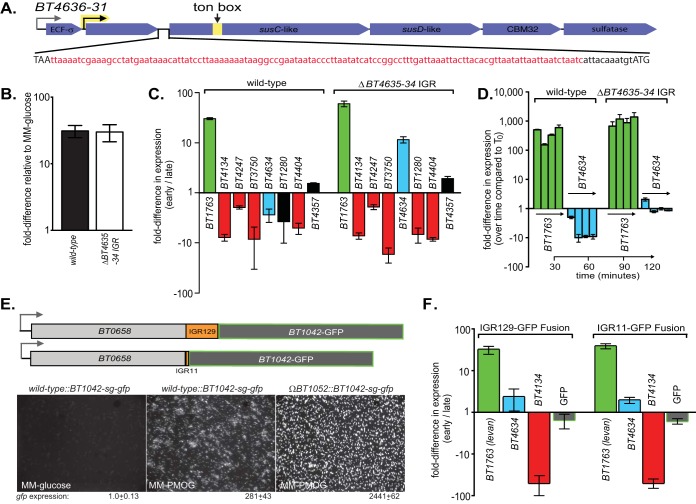
In considering additional transcriptional mechanisms that could mediate repression of O-glycan utilization, we noted that six of seven O-glycan PULs that were repressed in Fig. 1 are controlled by ECF-σ factor/anti-σ protein pairs (5), and the genes encoding anti-σ factors and downstream TonB-dependent (SusC-like) receptors are separated by sometimes large intergenic regions (IGRs) ranging from 50 to 197 bp and with no obvious nucleotide homology to each other (see Fig. S1I in the supplemental material for PUL schematics). A previous study in which polar insertions were made in anti-σ coding genes suggested that these genes are cotranscribed (5), and we confirmed this for one PUL (BT4636-31) where the genes flanking a 129-bp region could be detected within a single transcript by reverse transcription-PCR (RT-PCR) (Fig. S7F). On the basis of these observations, we hypothesized that these IGRs play a role in O-glycan repression. To test this, we deleted 118 bp (leaving an 11-bp ribosome binding sequence [RBS]) at the 5′ end of the cotranscribed IGR in BT4636-31 PUL that is consistently repressed by alternative glycans (Fig. 4A). Deletion of this region in a mutant strain (ΔBT4635-34 IGR mutant) had no effect on downstream susC-like gene expression when PMOG was the only carbon source, indicating that it does not harbor a required promoter or other sequence required for activation (Fig. 4B). However, when this mutant was grown in either a levan and PMOG mix (Fig. 4C) or pregrown to mid-exponential phase on PMOG and then introduced to levan (Fig. 4D), expression of the downstream BT4634 transcript escaped normal repression. In fact, it was significantly upregulated during early growth phase in batch culture. Deletion of this particular IGR had no effect on the expression patterns of the six other PMOG PULs (Fig. 4C), indicating that its effect is largely autonomous and restricted to the cis-encoded genes that flank it. Despite this discordant PUL expression, both wild-type and ΔBT4635-34 IGR strains had similar growth profiles on PMOG alone (Fig. S7G). Taken together, these data suggest that the BT4636-31 O-glycan PUL contains its own cis-encoded sequence through which repression is mediated at either the DNA or mRNA level, a phenomenon that may extend to PULs specific for other polysaccharides and in other species.
FIG 4 .
(A) A schematic of the O-glycan-responsive BT4636-31 PUL showing the IGR deletion sequence. The full IGR sequence is shown in lowercase type with the deleted region shown in red and the retained ribosome binding site (RBS) shown in black. The respective stop (BT4635) and start (BT4634) codons are in uppercase type. (B) qPCR-based expression analysis of the wild-type and ΔBT4635-34 IGR B. thetaiotaomicron strains grown in PMOG as the only carbon source showing full PUL expression in the IGR deletion mutant. (C) Growth phase-specific expression profiles between wild-type and IGR deletion strains in a mixture of levan/PMOG. All other PMOG-responsive PULs previously tested were not altered during IGR deletion. All expression values are significant (P ≤ 0.05 by one-tailed Student’s t test) except those labeled in black. (D) Expression analysis from an experiment in which levan was introduced into cells actively growing on PMOG with the same strains shown in panel C and normalized to a no-carbon control as shown in Fig. 1C and F. (E) Fusion of a Bacteroides-adapted super-glo gfp (sg-gfp) reporter gene downstream of the BT4635-34 IGR sequence and its 118-bp deletion variant and a constitutively expressed gene (BT0658) in the ΔBT4635-34 IGR strain as a genetic background. The micrograph images at the bottom validate expression of this reporter. The values below the micrographs are the 16S rRNA gene-normalized expression values by qPCR in MM-glucose and MM-PMOG. The sg-gfp gene was adapted for expression in Bacteroides by making a translational fusion to the first eight codons of a host glycan-responsive susC-like gene, BT1042. This gene is expressed in response to PMOG and is also derepressed in a mutant lacking expression of a gene encoding an associated anti-σ factor, BT1052. Note that combined loss of BT1052 and growth in PMOG result in increased gfp expression as measured by both qPCR and cell fluorescence. Cells were exposed to oxygen on ice for 30 min prior to imaging to allow the green fluorescent protein (GFP) fluorophore to fold. (F) Relative expression differences between early and late growth phases for the sg-gfp reporter strains (with or without the full 129-bp IGR) grown in a mixture of levan/PMOG. Parallel measurements of the levan-responsive BT1763 gene and the O-glycan-responsive genes, BT4134 and BT4634 (here deprioritized in the ΔIGR background) serve as controls.
To determine whether the 129-bp BT4635-34 IGR was sufficient to cause repression of genes with which it is genomically and transcriptionally associated, we used the ΔBT4635-34 IGR strain that lacks this sequence in its native PUL as a background and made two genomic constructs in which a Bacteroides-adapted gfp reporter gene (Fig. 4E) was fused downstream of a constitutively expressed, candidate nonessential (34) B. thetaiotaomicron gene, BT0658. In previous microarray experiments (6, 7, 12), this gene was expressed, across multiple in vitro and in vivo conditions, to levels similar to those of genes in the BT4635-31 transcript during growth on PMOG as the sole carbon source. The two different reporter strains varied only with respect to inclusion of the full IGR between BT0658 and the downstream gfp allele, with one strain containing only the 11-bp RBS sequence that was left behind in the ΔBT4635-34 strain (Fig. 4E). Using this approach, we were able to determine whether the ΔBT4635-34 IGR contained all of the sequence required for orchestrating repression of its native O-glycan-responsive PUL. Results from a batch culture experiment with levan and PMOG as the combined carbon sources showed similar gfp transcript levels between strains, regardless of whether they contained the full IGR sequence (Fig. 4F). As internal controls for the anticipated repressive environment, expression of PUL genes involved in degrading levan (BT1763, prioritized) and O-glycans (BT4634 and BT4134) was measured and behaved as expected (note that the background strain in this experiment lacks the 118-bp IGR upstream of BT4634, which is why this gene does not show normal repression). Thus, this observation implies that although the 129-bp IGR that lies upstream of BT4634 is necessary for PUL repression when higher priority nutrients are present, this sequence is not sufficient for this function. It is therefore likely that sequences contained in the flanking anti-σ and/or susC-like genes are required, and future experiments will focus on defining these sequences. Toward this point, it is worth noting that, while we observe conditional repression of the susC-like gene (BT4634) and other downstream open reading frames (ORFs) in this and other O-glycan PULs, the upstream anti-σ gene (BT4635) was not subject to the same growth phase-specific repression (see Fig. S7H in the supplemental material). Provided the insight reported here regarding the role of the intervening IGR in PUL repression, it is probable that this region is the point at which PUL transcript is destabilized by an unknown mechanism and that this event does not equally influence the upstream anti-σ gene.
DISCUSSION
Many bacteria that thrive in complex nutrient environments are capable of utilizing several different substrates. Such microorganisms are often programmed to give preference to certain compounds either because they represent higher energetic value or because they are more easily or consistently accessed. We have previously reported that B. thetaiotaomicron exhibits preferential metabolism of some of the >12 different glycans that it can degrade and places O-glycans low in this hierarchy when all are present together (26). Data presented here extend this finding to nearly all single glycan sources accessible to B. thetaiotaomicron as well as simple sugars. A possible explanation for this behavior is the cost of degrading the ~102 different glycan structures present on mucins. B. thetaiotaomicron activates expression of 86 genes ≥10-fold during growth in PMOG (6). In contrast, activation of only five genes is sufficient (7) to catabolize the simpler and highly prioritized dietary glycan, pectic galactan (Fig. 1L). Indeed, one reason why animals create such diverse O-glycan structures attached to mucins may be to complicate the degradation process of these barrier molecules by microorganisms in order to retain their protective capacity. Given this scenario, it is possible that some bacteria have adapted as specialists for mucin degradation, perhaps by optimizing enzymatic strategies for O-glycan deconstruction or focusing on the dominant, and therefore most economical to harvest, sugars and linkages that are present. B. massiliensis and B. fragilis appear to be exhibiting a tendency toward specialization for mucin O-glycan degradation, since they utilize only one glycan or two additional glycans, respectively. More strikingly, B. massiliensis has even restricted the repertoire of simple sugars that it can metabolize to those found in O-glycans. B. massiliensis shows mostly reciprocal preference for O-glycans relative to B. thetaiotaomicron when grown on starch, suggesting that it has further optimized its preference for host mucin glycans over dietary substrates.
Collectively, our results provide several new lines of insight into the phenomenon and mechanism of PUL repression in Bacteroides. In addition to showing for the first time that individual species can prioritize nutrient mixtures differently, we also delineate part of the pathway through which alternative carbohydrate signals modulate individual PULs and explain variation in PUL expression during growth in mixed nutrients (see Fig. 5 for a working model). On the basis of our experiments with chimeric O-glycan PULs that respond to similar O-glycan cues (Fig. 3), we conclude that substrate sensing is a paramount first step that can vary between PULs in the context of the particular suite of nutrients that are present. This is especially important with mucin O-glycans that contain many dozen related but variable structures, some of which may harbor critical cues, such as LacNAc disaccharide, within larger chains and are potentially occluded by other surrounding linkages.
FIG 5 .
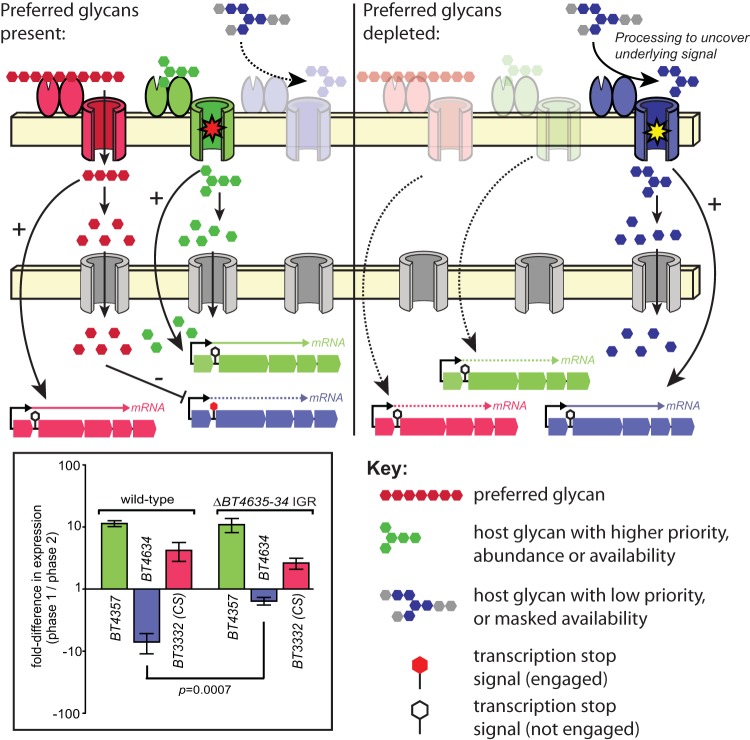
A model to explain the results reported in this study and based particularly on results of expression with the substrate at 100 mM. (Left) In the presence of preferred glycans, such as the CS and heparin glycosaminoglycans, PULs involved in catabolizing high-priority nutrients (red) are activated and liberate simple sugars that are ultimately transported into the cytoplasm, where they mediate repression of PULs that target lower-priority substrates. Since the PULs involved in degradation of some substrates that repress utilization of O-glycans can also be repressed in more-complex nutrient environments (26), it is possible that these gene clusters also harbor elements that mediate transcript repression via either the IGR-mediated mechanism described here or other unknown mechanisms. In the case of the nutrients present in the 100 mM fraction, the inducing substrate for the BT4355-59 PUL (green) is presumably present in either higher abundance and/or is more accessible, leading to deployment of this system early in growth. Alternatively, systems like BT4355-59 may be less susceptible to repression by monosaccharides present in certain high-priority substrates, which is consistent with the data shown in Fig. 1 and Fig. S1 in the supplemental material. (Right) After the preferred glycans are depleted and the corresponding repressive sugars are no longer present in the cytoplasm, expression of PULs that target low-priority glycans is released, but may require the action of other enzymes/Sus-like systems to uncover the glycan structure that is recognized by some systems such as that encoded by BT4636-31 (blue). (Left, inset) Thus, when the BT4636-31 IGR is absent during growth in the 100 mM substrate, repression is partially relieved (compare blue bars; P = 0.0007 by one-tailed Student’s t test). Since the BT4636-31 target ligand is sensed later in growth (Fig. 3), possibly because it is only enzymatically uncovered after the action of other systems, this system is not fully deployed early in growth even when its associated IGR is missing.
When substrate is successfully recognized and degraded, the released monosaccharides, which vary in identity based on the particular glycans present, flow into the cell where they can be sensed by an unknown mechanism, and this information is relayed to individual PULs to modulate their expression. Experiments with one IGR (Fig. 4) imply that this regulatory mechanism requires this region at either the DNA or mRNA level to reduce the abundance of low-priority PUL transcripts. While the complete molecular mechanism through which PUL repression occurs is not elucidated here, it is clear that individual PULs are capable of exhibiting variable responses to different carbohydrates, both with respect to the identity of the alternative glycan (Fig. 1; see Fig. S1 in the supplemental material) or the identity of monosaccharides and timing of the response (Fig. S2). For example, genes within the BT4636-31 PUL are repressed in the presence of several chemically distinct glycans (levan, chondroitin sulfate, amylopectin, etc.), but not in the presence of heparin, which imposes repression of other PULs in the same bacterial population sampled (Fig. S1A). In contrast, the PUL containing BT4247 is repressed under the same conditions listed for BT4636-31, including heparin, but escapes repression during growth in arabinan, a glycan that imposes strong repression on BT4634 (Fig. S1H). Deciphering how multiple, chemically distinct signals are integrated into regulation of each individual PUL and also distinguished from one another to create discordant regulatory effects at different PULs will illuminate a key area of global gene regulation in this diverse phylum of important gut symbionts.
Consistent with substrate sensing (i.e., transcriptional activation via glycan cues that may be present but masked by other linkages) being dominant relative to transcription repression in mediating sequential expression of PULs, an additional experiment using the 100 mM fraction as a growth substrate and wild-type and ΔBT4635-34 IGR B. thetaiotaomicron strains revealed that growth phase-specific repression of BT4634 was only partially, yet still significantly (P = 0.0007), alleviated with IGR deletion (Fig. 5, inset). Growth phase-specific deployment of the BT4634 PUL is not governed by the identity of its promoter and regulatory elements, or even its IGR, but instead tracks with the portion of the gene encoding its SusC-like transporter and downstream enzymatic functions involved in recognizing an unknown host glycan structure (Fig. 3C). Thus, we conclude that eliminating IGR-mediated repression of this PUL in this particular substrate mixture results in modest loss of repression because its inducing substrate may still not be revealed until later in growth. Prerequisite processing steps (shown as gray sugars attached to the blue inducing saccharide in Fig. 5) may therefore be required to fully activate this system, even in the absence of IGR-mediated repression, and these steps could be conferred by the action of the several additional O-glycan-responsive PULs in B. thetaiotaomicron.
Our finding that many different simple sugars exert repression of B. thetaiotaomicron O-glycan PULs adds to a previous conclusion by Lynch and Sonnenburg (27), where despite reporting that one monosaccharide (fructose) represses expression of a single PUL involved in arabinan utilization, they concluded that activation of the transcriptional regulator associated with arabinan-dependent gene expression is responsible for repression of three genomically unlinked O-glycan PULs (two of which are also monitored here and shown to be repressed). In this case, the natural ligand for the causative transcriptional regulator is known to be arabinan oligosaccharide and not arabinose (7), suggesting that this oligosaccharide cue signals repression of PULs involved in utilizing O-glycans and other substrates. A potential confounder in interpreting these data is that fructose was used via exposure to a chimeric hybrid two-component system regulator that sensed this monosaccharide and activated genes involved in arabinan utilization. While this hybrid regulator was expressed in a B. thetaiotaomicron mutant that was impaired for fructose uptake and metabolism, it was still capable of substantial growth on fructose (28), making it unclear whether the monosaccharide could have also had a direct effect. Nevertheless, these data collectively indicate that multiple mechanisms may be at work to fine-tune global expression of B. thetaiotaomicron glycan utilization functions in complex nutrient environments.
Understanding the complex metabolic relationships between dietary and mucosal glycans in the behavior of symbiotic gut bacteria will undoubtedly illuminate new aspects of how the gut microbiota contributes to health and disease. For example, several PULs shown here and in previous work (6) to be repressed by alternative nutrients harbor sulfatase-encoding genes. We have recently shown that sulfatases are required by B. thetaiotaomicron to cause inflammation in susceptible mice (24). If knowledge of diet or prebiotic nutrient conditions can be leveraged to suppress expression of these harmful pathways, it may be possible to reduce or eliminate their contribution to disease states.
Given the vast number of microbial lineages that compose the human gut microbiota and the often large number of different carbohydrate and other nutrient utilization traits encoded in individual species, it is likely that many additional metabolic relationships exist like those described in this report. Indeed, if additional opposing preferences for carbohydrate utilization exist among other gut microorganisms, these behaviors could be a major and unappreciated determinant of how different microbes execute their metabolic potential in vivo (35). Understanding these behaviors and their underlying mechanisms will be critical to predicting the behavior and assembly of human gut microbial communities during health and disease and designing therapeutic interventions.
MATERIALS AND METHODS
Growth and genetic manipulation of Bacteroides.
The type strains Bacteroides thetaiotaomicron ATCC 29148 (VPI 5482), Bacteroides massiliensis B84634 (DSM17679), and Bacteroides fragilis ATCC 25285 (NCTC9343) were used for all experiments except those in Fig. S4 in the supplemental material. Growth profiles and nearly all early/late gene expression experiments were performed using the same conditions and carbons sources as previously described (7), except when noted. B. thetaiotaomicron was pregrown in TYG broth overnight at 37°C in an anaerobic chamber (Coy Lab Products) under an 85% N/10% H2/5% CO2 atmosphere for all experiments. B. fragilis and B. massiliensis were pregrown in a custom chopped-meat (CM) broth under the same conditions (36). Prior to any growth or transcript expression experiments, all B. thetaiotaomicron and B. fragilis strains, including B. thetaiotaomicron strains that are not the type strain in Fig. S4, were transferred to minimal medium with glucose (MM-G) for overnight growth. When initially growing B. massiliensis in minimal medium, we discovered that it is somewhat fastidious. First, it was unable to grow on glucose alone so we used N-acetylglucosamine (GlcNAc) as a carbon source. Second, the addition of the supplements enhanced growth on minimal medium. Thus, we grew this strain in minimal medium containing Balch’s vitamins, trace minerals, amino acids, purine/pyrimidines, and beef extract (final concentration of 0.5 mg/ml) also found in our custom CM broth. We then tested for growth on these additional medium components using water as a negative control and GlcNAc as a positive control. We determined that B. massiliensis does not have the ability to grow on these substances alone. All strains were then washed in 2× concentration minimal medium with no carbon (MM-NC) prior to conducting any experiment with one source of carbon, glycan mixes, or alternative glycan introductions (spike-in experiments).
All growth curves, except where noted below, were performed in 200-µl cultures in 96-well microtiter plates and a robotic plate stacking and handling device (BioTek) (7). All growth curves were performed at 37°C except that B. thetaiotaomicron titration curves from Fig. S1 in the supplemental material were performed at 30°C to help accent the diauxic shift and help determine where harvest points for early and late time points were based on the substrate (three replicate cultures for each mixture ratio; run on separate 96-well plates). However, at 37°C, B. thetaiotaomicron behaved similarly when samples were taken for expression profiles. We determined that B. fragilis and B. massiliensis have limited glycan-degrading capabilities, under the conditions tested, by growing them on a custom glycan array in the same 200-µl culture robotic plate stacking and handling device. All spike-in, RNA-Seq, and 100 mM fraction cell harvests as well as B. massiliensis starch/porcine mucin O-glycans (PMOG) growth curves were performed in glass tubes using a UV-visible (UV-Vis) spectrophotometer (Thermo) to monitor absorbance at 600 nm (A600). B. thetaiotaomicron mutants and chimeric PUL recombinants were prepared by allelic exchange as previously described (28).
Transcript measurements by qPCR and whole-genome transcriptional profiling by RNAseq.
For each glycan mixture tested at early and late time points, three replicate culture sets were assayed. For B. thetaiotaomicron and B. fragilis, all glycan mixes tested were 1.5 mg/ml non-PMOG and 7 mg/ml PMOG (or human milk oligosaccharides [HMOs]) except for rhamnogalacturonan I (RG I) that was used at 3 mg/ml combined with 7 mg/ml PMOG. For B. massiliensis, a mixture of 7 mg/ml amylopectin from potato (APpot) and 1.5 mg/ml PMOG was used for cell harvests. To provide sufficient RNA for analysis, a total of 12 replicate 200-ml cultures were combined into a single 2.4-ml sample representing a series of slightly different harvest points. Thus, the sampling points shown in Fig. S1 in the supplemental material are shown as a distribution of 12 different A600 values and their corresponding average. For B. massiliensis early and two late glycan-specific PUL transcripts, cells were grown in glass tubes to keep consistent with initial glycan mix growth curves.
To measure spike-in glycan-specific PUL transcripts, B. thetaiotaomicron was grown in triplicate under 37°C conditions as described above. Cells were grown on 10 mg/ml minimal medium with PMOG (MM-PMOG) in glass tubes to mid-exponential phase, and the cells were harvested at time zero (T0). Then, either MM-levan, MM-HG (minimal medium with homogalacturonan), MM-polysaccharide mix (2.5 mg/ml final), or MM-NC was introduced to the MM-PMOG culture. The culture was then allowed to incubate for 2 h while harvesting cells every 30 min. All harvested cells were treated with RNAprotect bacterial reagent (Qiagen) and stored at −80°C until processed. Total RNA was extracted from cells using an RNeasy kit (Qiagen), treated with Turbo DNase I (Ambion), and reverse transcribed using Superscript III (Invitrogen). The 16S rRNA gene-normalized PUL gene abundance was assayed on an Eppendorf Realplex thermocycler and using SYBR FAST premixed qPCR master mix (Kapa Biosystems). All cultures assayed were actively growing on one or more carbon sources, and the use of 16S rRNA gene normalization is rationalized based on extensive comparisons of qPCR measurements with microarray or RNAseq data that were normalized using other algorithms that were not based on 16S rRNA genes (for example, Fig. 3C) (6, 7).
For RNAseq, the total RNA from B. fragilis and B. massiliensis cells grown on PMOG with glucose and GlcNAc, respectively, used as reference comparisons, was extracted using an RNeasy kit (Qiagen) and treated with Turbo DNase I (Ambion), and finally, rRNA was depleted using the Bacteria Ribo-Zero rRNA removal kit (Epicentre). Residual mRNA was converted to sequencing libraries using TruSeq barcoded adaptors (Illumina) and sequenced at the University of Michigan Sequencing Core in an Illumina HiSeq instrument with 24 samples multiplexed per lane. Barcoded data were demultiplexed and analyzed using the Arraystar software package with Qseq (DNAstar).
Illumina RNA sequencing data.
Sequencing data were deposited in NCBI under SRA accession numbers SRP035212 and SRP063781.
SUPPLEMENTAL MATERIAL
(A to H) Growth- and phase-specific gene expression analysis on pairwise glycan combinations as described for levan, HG, and CS in Fig. 1 of the main text. (I) Gene schematics of the seven B. thetaiotaomicron PMOG PULs analyzed in this study. Functional annotations are indicated for genes predicted to encode hybrid two-component system (HTCS) regulators (pink), ECF-σ (green), anti-σ (red), susC-like (purple), susD-like (orange), glycoside hydrolase (GH) (blue) (with family membership indicated). Additional annotations are listed for some other cotranscribed genes shown in gray: carbohydrate binding module (CBM, with family indicated), M60-like protease, and sulfatase. Download
(A) Time course experiment in which a polysaccharide mixture, containing 2.5 mg/ml total of 11 different glycans (0.25 mg/ml each) that B. thetaiotaomicron can degrade, was added to actively growing PMOG cultures. A negative control in which no polysaccharides were added was performed as described in the legend to Fig. 1. (B to L) Spike in expression results from an experiment in which B. thetaiotaomicron was grown to mid-exponential phase on PMOG (T0) and then an equal amount of prereduced minimal medium containing one of the individual monosaccharides indicated was added to measure its ability to initiate PUL repression. Samples were taken 30 min after the addition for 2 h. Only one replicate was performed, and values represent the means of two quantitative reverse transcription-PCR (qRT-PCR) technical replicates. Download
Schematic representations of the various glycan structures presented to Bacteroides species in this study. The full names of the abbreviated polysaccharides are provided in the legends to Fig. 1 and Fig. S1 in the supplemental material. A bracketed end(s) of a glycan chain indicates that the glycan structure can be longer than that which is shown. Definitions of symbols used are provided in the key at the top right. Note that although the structures drawn here represent predominant linkages and monosaccharides in each of the glycans, there may be some differences based on extraction method and species or tissue source. Download
(A) Biphasic growth on different proportions of levan and human milk oligosaccharides (HMOs) showing that HMOs are given lower priority in this mixed growth condition. (B) Biphasic growth on different proportions of homogalacturonan and HMOs showing that HMOs are given lower priority in this mixed growth condition. (C) Biphasic growth of different B. thetaiotaomicron (Bt) strains on combinations of PMOG and one of two different fructans, levan or inulin, depending on which substrate the indicated strain showed optimal growth rate on. (D) Biphasic growth of different B. thetaiotaomicron strains on combinations of PMOG and homogalacturonan. For strains in both panels C and D, percent 16S rRNA gene identity to the type strain VPI 5482 is indicated for each strain along with the host source. Download
(A) Phylogeny of several Bacteroidetes, including B. thetaiotaomicron, as well as Bacteroides fragilis and Bacteroides massiliensis, used in this study. (B) Cellular schematic of B. thetaiotaomicron’s starch utilization system (sus) and homology with B. massiliensis’s identically organized starch PUL. (C) B. massiliensis’s putative starch-responsive susC is activated in the presence of starch compared to N-acetylglucosamine (GlcNAc) (B. massiliensis did not grow on glucose as the sole carbon source, despite growth on starch). (D) Growth curves of B. thetaiotaomicron when grown on PMOG (red), starch (green), and GlcNAc (gray). (E) Growth curves of B. massiliensis grown on PMOG (red), starch (green), and GlcNAc (gray) as the sole carbon source. (F) B. massiliensis grown on PMOG alone (red), starch alone (green), and an APpot/PMOG mix (3.5 mg/ml and 3 mg/ml, respectively; which is the same mix used in Fig. 2) to help determine possible metabolic shifts. (G) Growth profiles of B. fragilis grown on pairwise mixes of starch/PMOG. (H) Growth profiles of B. fragilis grown on pairwise mixes of inulin/PMOG. Download
(A) Analysis of 16S rRNA gene-normalized qPCR threshold cycle (Ct) values during the first growth phase in a mixture of PMOG and levan to determine whether 3′-5′ cyclic AMP (cAMP) is a signal (at 10 mM) for catabolite repression in B. thetaiotaomicron. There is no effect of cAMP under the conditions tested to suggest that this molecule causes derepression of O-glycan-responsive PULs assayed, as the only two significant changes result in increased repression. Values are the means ± SD of three replicates. Values that are significantly different by t test are indicated by asterisks as follows: *, P < 0.05; **, P < 0.001. Values that are not significantly different (n.s.) by t test are also indicated. (B) Expression profiles for cells grown on a mix of levan and PMOG using a strain lacking all eight capsular polysaccharide loci (B. thetaiotaomicron Δcps). We determined that the various capsular polysaccharides that B. thetaiotaomicron employs in an in vitro mixed cell population does not affect glycan prioritization. Values are the means ± SD of three replicates. Statistical significance by t test is indicated as follows: *, P < 0.05; **, P < 0.001; n.s., not significant. (C and D) Comparison of expression values between wild-type B. thetaiotaomicron and a mutant containing a disruption in the BT1754 gene, which encodes an essential regulator for levan utilization, when pregrown on PMOG and levan is introduced. Cells were harvested as described above with T0 and 30-min harvest for 2 h. Only one replicate was performed, and values represent the means of two qRT-PCR technical replicates. (E) Genetic manipulation scheme for generating the chimeric strain for BT4636-31 and 4355-59 PULs. Download
We isolated the 100 mM NaCl-eluted fraction from porcine mucosal glycans (PMG, referred to herein as the “100 mM fraction”) similar to a previous study (6). Although most neutral porcine mucin O-glycans (PMOG) flow through an anion exchange column, the resulting fraction of 100 mM NaCl-eluted retained PGM contains residual MOG in addition to glycosaminoglycans (GAGs) and some N-linked glycans. The O-glycans contain more neutral sugars, while GAGs, including the chondroitin sulfate that is highest on the metabolic hierarchy of B. thetaiotaomicron (Bt), contain higher negative charge due to uronic acid, sulfate, and sialic acid content. Thus, growing B. thetaiotaomicron on this 100 mM fraction elicits a number of responses, including the chondroitin sulfate PUL, BT3328-3334, as well as O-glycan PULs. (A) Activation of B. thetaiotaomicron genes from the two O-glycan PULs used for chimera construction, BT4355-59 (red) and BT4636-31 (blue), when the bacteria were grown on PMOG as the sole carbon source. Values are the means ± SD of three replicates. (B) Growth on the 100 mM fraction exhibits an early (red) phase and a series of late (blue) growth phases. (C) Discordant transcript profiles from two B. thetaiotaomicron O-glycan-responsive PULs where one susC-like gene BT4357 is upregulated early in growth on the 100 mM fraction and another, BT4634, is repressed until later. This suggests that even individual PULs that target similar substrates can respond differently, possibly due to substrate structural/linkage diversity or sequential cleaving of substrates making other sugars or linkages available. Values are the means ± SD of two replicates determined from previous microarray data (6). (D) The two susC-like genes from the PULs described above with discordant expression grown on 100 mM NaCl fractionated PMG are both activated by the disaccharide, N-acetyllactosamine (LacNAc), that is a major component of the more-elaborate O-glycan chains contained in PMOG. (E) When the regulatory regions are swapped in the chimeric PUL B. thetaiotaomicron strain, activation still occurs, indicating that the PULs retain functionality. (F) Agarose gel image showing transcription of a region of the BT4635-34 PUL with an intergenic region (IGR) deleted. Notice the 118-bp difference between wild-type (wt) B. thetaiotaomicron and the ΔBT4635-34 IGR strain in reverse-transcribed cDNA and genomic DNA. Also, no amplifiable DNA for IGR-flanking genes was detected in only the extracted RNA. (G) Growth profiles for wild-type B. thetaiotaomicron and the ΔBT4635-34 IGR strain on PMOG alone (10/mg/ml total PMOG). (H) Growth phase-specific expression differences for the anti-σ (BT4635) and susC-like (BT4634) genes in either the 100 mM fraction or levan/PMOG, showing that these two adjacent genes, which flank the deleted IGR sequence, behave discordantly with the upstream anti-σ gene not showing similar repression. Download
In vitro growth ability of Bacteroides thetaiotaomicron VPI 5482, Bacteroides massiliensis DSM17679, and Bacteroides fragilis NCTC 9343 on various glycans and simple sugars. Boxes in gray highlight substrates on which growth was not observed, while boxes in green represent substrates that support detectable growth.
RNAseq-based gene expression changes in the whole-genome transcriptional profile of Bacteroides massiliensis DSM17679 grown on PMOG as the sole carbon source compared to an N-acetylglucosamine reference
RNAseq-based gene expression changes in the whole-genome transcriptional profile of Bacteroides fragilis NCTC 9343 grown on PMOG as a sole carbon source compared to glucose reference
ACKNOWLEDGMENTS
We thank Christopher Alteri for insightful discussions regarding molecular mechanisms.
This work was supported by awards from the NIH (DK084214 and GM099513), the Global Probiotics Council Young Investigator Grant for Probiotics Research, and by funds provided by the University of Michigan Biological Sciences Scholars Program.
Footnotes
Citation Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6(6):e01282-15. doi:10.1128/mBio.01282-15.
REFERENCES
- 1.El Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 2.McNeil NI. 1984. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr 39:338–342. [DOI] [PubMed] [Google Scholar]
- 3.Larsson JM, Karlsson H, Sjovall H, Hansson GC. 2009. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19:756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 4.Johansson MEV, Sjövall H, Hansson GC. 2013. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martens EC, Roth R, Heuser JE, Gordon JI. 2009. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem 284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. 2010. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 9.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 10.Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, Niedermeyer G. 1985. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 75:944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL.. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 13.Bjursell MK, Martens EC, Gordon JI. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 14.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. 2010. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem 285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. 2013. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol 11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. 2007. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med 204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu J, Wei B, Wen T, Johansson MEV, Liu X, Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, McDaniel JM, Sferra TJ, Turner JR, Chen H, Hansson GC, Braun J, Xia L. 2011. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest 121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Muñoz ME, Bergstrom K, Peng V, Schmaltz R, Jimenez-Cardona R, Marsteller N, McGee S, Clavel T, Ley R, Fu J, Xia L, Peterson DA. 2014. Discordance between changes in the gut microbiota and pathogenicity in a mouse model of spontaneous colitis. Gut Microbes 5:286–295. doi: 10.4161/gmic.28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson JMH, Karlsson H, Crespo JG, Johansson MEV, Eklund L, Sjövall H, Hansson GC. 2011. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis 17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 23.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, Glowacki RW, Hansson GC, Allen PM, Martens EC, Stappenbeck TS. 2015. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe 17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjdia A, Martens EC, Gordon JI, Berteau O. 2011. Sulfatases and a radical S-adenosyl-l-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem 286:25973–25982. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers TE, Pudlo NA, Koropatkin NM, Bell JSK, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch JB, Sonnenburg JL. 2012. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol 85:478–491. doi: 10.1111/j.1365-2958.2012.08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spence C, Wells WG, Smith CJ. 2006. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: regulation by carbon source and oxygen. J Bacteriol 188:4663–4672. doi: 10.1128/JB.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 31.Siegel LS, Hylemon PB, Phibbs PV Jr. 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J Bacteriol 129:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM, Martens EC. 2014. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio 5:e01441-14. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koebnik R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol 13:343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. 2012. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci U S A 109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A to H) Growth- and phase-specific gene expression analysis on pairwise glycan combinations as described for levan, HG, and CS in Fig. 1 of the main text. (I) Gene schematics of the seven B. thetaiotaomicron PMOG PULs analyzed in this study. Functional annotations are indicated for genes predicted to encode hybrid two-component system (HTCS) regulators (pink), ECF-σ (green), anti-σ (red), susC-like (purple), susD-like (orange), glycoside hydrolase (GH) (blue) (with family membership indicated). Additional annotations are listed for some other cotranscribed genes shown in gray: carbohydrate binding module (CBM, with family indicated), M60-like protease, and sulfatase. Download
(A) Time course experiment in which a polysaccharide mixture, containing 2.5 mg/ml total of 11 different glycans (0.25 mg/ml each) that B. thetaiotaomicron can degrade, was added to actively growing PMOG cultures. A negative control in which no polysaccharides were added was performed as described in the legend to Fig. 1. (B to L) Spike in expression results from an experiment in which B. thetaiotaomicron was grown to mid-exponential phase on PMOG (T0) and then an equal amount of prereduced minimal medium containing one of the individual monosaccharides indicated was added to measure its ability to initiate PUL repression. Samples were taken 30 min after the addition for 2 h. Only one replicate was performed, and values represent the means of two quantitative reverse transcription-PCR (qRT-PCR) technical replicates. Download
Schematic representations of the various glycan structures presented to Bacteroides species in this study. The full names of the abbreviated polysaccharides are provided in the legends to Fig. 1 and Fig. S1 in the supplemental material. A bracketed end(s) of a glycan chain indicates that the glycan structure can be longer than that which is shown. Definitions of symbols used are provided in the key at the top right. Note that although the structures drawn here represent predominant linkages and monosaccharides in each of the glycans, there may be some differences based on extraction method and species or tissue source. Download
(A) Biphasic growth on different proportions of levan and human milk oligosaccharides (HMOs) showing that HMOs are given lower priority in this mixed growth condition. (B) Biphasic growth on different proportions of homogalacturonan and HMOs showing that HMOs are given lower priority in this mixed growth condition. (C) Biphasic growth of different B. thetaiotaomicron (Bt) strains on combinations of PMOG and one of two different fructans, levan or inulin, depending on which substrate the indicated strain showed optimal growth rate on. (D) Biphasic growth of different B. thetaiotaomicron strains on combinations of PMOG and homogalacturonan. For strains in both panels C and D, percent 16S rRNA gene identity to the type strain VPI 5482 is indicated for each strain along with the host source. Download
(A) Phylogeny of several Bacteroidetes, including B. thetaiotaomicron, as well as Bacteroides fragilis and Bacteroides massiliensis, used in this study. (B) Cellular schematic of B. thetaiotaomicron’s starch utilization system (sus) and homology with B. massiliensis’s identically organized starch PUL. (C) B. massiliensis’s putative starch-responsive susC is activated in the presence of starch compared to N-acetylglucosamine (GlcNAc) (B. massiliensis did not grow on glucose as the sole carbon source, despite growth on starch). (D) Growth curves of B. thetaiotaomicron when grown on PMOG (red), starch (green), and GlcNAc (gray). (E) Growth curves of B. massiliensis grown on PMOG (red), starch (green), and GlcNAc (gray) as the sole carbon source. (F) B. massiliensis grown on PMOG alone (red), starch alone (green), and an APpot/PMOG mix (3.5 mg/ml and 3 mg/ml, respectively; which is the same mix used in Fig. 2) to help determine possible metabolic shifts. (G) Growth profiles of B. fragilis grown on pairwise mixes of starch/PMOG. (H) Growth profiles of B. fragilis grown on pairwise mixes of inulin/PMOG. Download
(A) Analysis of 16S rRNA gene-normalized qPCR threshold cycle (Ct) values during the first growth phase in a mixture of PMOG and levan to determine whether 3′-5′ cyclic AMP (cAMP) is a signal (at 10 mM) for catabolite repression in B. thetaiotaomicron. There is no effect of cAMP under the conditions tested to suggest that this molecule causes derepression of O-glycan-responsive PULs assayed, as the only two significant changes result in increased repression. Values are the means ± SD of three replicates. Values that are significantly different by t test are indicated by asterisks as follows: *, P < 0.05; **, P < 0.001. Values that are not significantly different (n.s.) by t test are also indicated. (B) Expression profiles for cells grown on a mix of levan and PMOG using a strain lacking all eight capsular polysaccharide loci (B. thetaiotaomicron Δcps). We determined that the various capsular polysaccharides that B. thetaiotaomicron employs in an in vitro mixed cell population does not affect glycan prioritization. Values are the means ± SD of three replicates. Statistical significance by t test is indicated as follows: *, P < 0.05; **, P < 0.001; n.s., not significant. (C and D) Comparison of expression values between wild-type B. thetaiotaomicron and a mutant containing a disruption in the BT1754 gene, which encodes an essential regulator for levan utilization, when pregrown on PMOG and levan is introduced. Cells were harvested as described above with T0 and 30-min harvest for 2 h. Only one replicate was performed, and values represent the means of two qRT-PCR technical replicates. (E) Genetic manipulation scheme for generating the chimeric strain for BT4636-31 and 4355-59 PULs. Download
We isolated the 100 mM NaCl-eluted fraction from porcine mucosal glycans (PMG, referred to herein as the “100 mM fraction”) similar to a previous study (6). Although most neutral porcine mucin O-glycans (PMOG) flow through an anion exchange column, the resulting fraction of 100 mM NaCl-eluted retained PGM contains residual MOG in addition to glycosaminoglycans (GAGs) and some N-linked glycans. The O-glycans contain more neutral sugars, while GAGs, including the chondroitin sulfate that is highest on the metabolic hierarchy of B. thetaiotaomicron (Bt), contain higher negative charge due to uronic acid, sulfate, and sialic acid content. Thus, growing B. thetaiotaomicron on this 100 mM fraction elicits a number of responses, including the chondroitin sulfate PUL, BT3328-3334, as well as O-glycan PULs. (A) Activation of B. thetaiotaomicron genes from the two O-glycan PULs used for chimera construction, BT4355-59 (red) and BT4636-31 (blue), when the bacteria were grown on PMOG as the sole carbon source. Values are the means ± SD of three replicates. (B) Growth on the 100 mM fraction exhibits an early (red) phase and a series of late (blue) growth phases. (C) Discordant transcript profiles from two B. thetaiotaomicron O-glycan-responsive PULs where one susC-like gene BT4357 is upregulated early in growth on the 100 mM fraction and another, BT4634, is repressed until later. This suggests that even individual PULs that target similar substrates can respond differently, possibly due to substrate structural/linkage diversity or sequential cleaving of substrates making other sugars or linkages available. Values are the means ± SD of two replicates determined from previous microarray data (6). (D) The two susC-like genes from the PULs described above with discordant expression grown on 100 mM NaCl fractionated PMG are both activated by the disaccharide, N-acetyllactosamine (LacNAc), that is a major component of the more-elaborate O-glycan chains contained in PMOG. (E) When the regulatory regions are swapped in the chimeric PUL B. thetaiotaomicron strain, activation still occurs, indicating that the PULs retain functionality. (F) Agarose gel image showing transcription of a region of the BT4635-34 PUL with an intergenic region (IGR) deleted. Notice the 118-bp difference between wild-type (wt) B. thetaiotaomicron and the ΔBT4635-34 IGR strain in reverse-transcribed cDNA and genomic DNA. Also, no amplifiable DNA for IGR-flanking genes was detected in only the extracted RNA. (G) Growth profiles for wild-type B. thetaiotaomicron and the ΔBT4635-34 IGR strain on PMOG alone (10/mg/ml total PMOG). (H) Growth phase-specific expression differences for the anti-σ (BT4635) and susC-like (BT4634) genes in either the 100 mM fraction or levan/PMOG, showing that these two adjacent genes, which flank the deleted IGR sequence, behave discordantly with the upstream anti-σ gene not showing similar repression. Download
In vitro growth ability of Bacteroides thetaiotaomicron VPI 5482, Bacteroides massiliensis DSM17679, and Bacteroides fragilis NCTC 9343 on various glycans and simple sugars. Boxes in gray highlight substrates on which growth was not observed, while boxes in green represent substrates that support detectable growth.
RNAseq-based gene expression changes in the whole-genome transcriptional profile of Bacteroides massiliensis DSM17679 grown on PMOG as the sole carbon source compared to an N-acetylglucosamine reference
RNAseq-based gene expression changes in the whole-genome transcriptional profile of Bacteroides fragilis NCTC 9343 grown on PMOG as a sole carbon source compared to glucose reference