Abstract
Hybridization thermodynamics on solid supports are compared with those in solution for two types of hybridization probe, DNA and uncharged morpholino oligonucleotides of identical sequences. Trends in hybridization affinity are discussed with respect to ionic strength, temperature, and surface behavior.
Solid phase hybridization, in which nucleic acids from solution bind to immobilized complementary “probe” sequences, is widely used in life science research and, increasingly, in clinical diagnostics.1 Surface hybridization also finds frequent use in materials chemistry.2 Despite its wide applications, surface hybridization has not reached the predictive understanding of its more thoroughly investigated solution counterpart, although existence of both kinetic and thermodynamic differences between solution and surface hybridization is widely acknowledged. In the case of thermodynamics, behavior of surface hybridization could be predicted from that in solution if the excess state functions to account for nonidealities stemming from surface specific effects were known. The present study considers origins of these offsets for six different sequences, for DNA and for an uncharged DNA mimic called morpholino (MO), as the surface-immobilized probe.
We are interested in addressing how DNA and MO probes differ in their surface vs solution hybridization behavior as a function of ionic strength, temperature, and surface-derived effects. Morpholinos are synthetic DNA mimics with an uncharged backbone consisting of morpholine rings connected by phosphorodiamidate groups;3 because morpholinos are uncharged, their comparison to DNA probes serves to highlight the role of electrostatics. Their charge neutrality also makes morpholinos similar to peptide nucleic acids (PNAs).4 Compared to PNAs, morpholinos offer flexibility with regard to oligo length and base composition, have an approximately 100-fold higher aqueous solubility than PNAs, and exhibit more moderate hybridization affinity that should reduce background signals when long probes are used for assaying sequence concentrations in complex mixtures,3 as in gene expression analysis.
DNA-DNA and MO-DNA melting curves were used to analyze surface and corresponding solution hybridizations of six 25mer DNA targets with complementary 25mer DNA and MO probe sequences (Table S1, Electronic Supplementary Information (ESI)). Surface melting curves were obtained in a total-internal-reflection-fluorescence geometry in which fluorescently-labeled Cy5-targets bound to probes on aldehyde-functionalized slides. The probe coverage was estimated to be 2.3 ± 0.3 × 1012 cm−2 for MO probes and 2.9 ± 0.8 × 1012 cm−2 for DNA probes. These coverages correspond to ~ 6 nm average distance between probe sites. Although 25mer probes can readily come into contact over such distances, these probe densities remain well below those (~ 5 × 1012 cm−2) at which steric crowding becomes a significant barrier to hybridization.5 Solution melting transitions were determined with UV absorbance. Full experimental details are provided in the ESI.
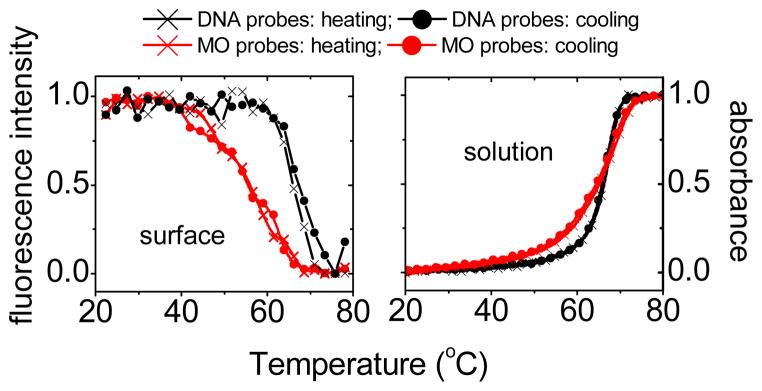
Figure 1 shows examples of melting transitions on surfaces and in solution. As temperature increases, surface transitions (left panel in Figure 1) manifest in decreasing signals due to dehybridization of fluorescent target from the slides. Solution transitions (right panel in Figure 1) lead to an increase in absorbance due to higher extinction coefficients of the single-stranded species. Experiments were performed in 0.012, 0.021, 0.037, and 0.11 mol L−1 phosphate buffers. For immobilized DNA probes melting transitions were increasingly difficult to observe as ionic strength decreased, with none of the six DNA probes yielding transitions in 0.012 mol L−1. When present, DNA probes invariably exhibited sharper transitions than MO probes, in solution as well as on surfaces (cf. Figure 1). The more gradual transitions of MO-DNA hybridization may reflect dispersion in thermodynamics due to stereochemical variations at the chiral P centers on the MO backbone, that arise during synthesis.5
Fig. 1.
Melting transition data. Left panel: Surface melting transitions for MO and DNA probes, measured at 0.3 °C min−1 scan rate. Right panel: Corresponding solution melting transitions, measured at 0.2 °C min−1. Black curves: DNA-DNA hybridization; red curves MO-DNA hybridization. Buffer: 0.11 mol L−1 sodium phosphate buffer, pH 7.0, no other salt. Target concentration for surface measurements: 0.1 μmol L−1. Sequence: sequence #1 (Table S1, ESI).
Thermodynamic analysis rests on equilibrium data. Equilibrium can be confirmed from superposition of heating and cooling cycles, as in Figure 1. Moreover, to minimize irreversibility that can arise during melting6 due to the high activation barrier posed by separation of the strands in a duplex, analysis was instead performed on cooling half-cycles since the activation barrier for hybridization is close to zero.7
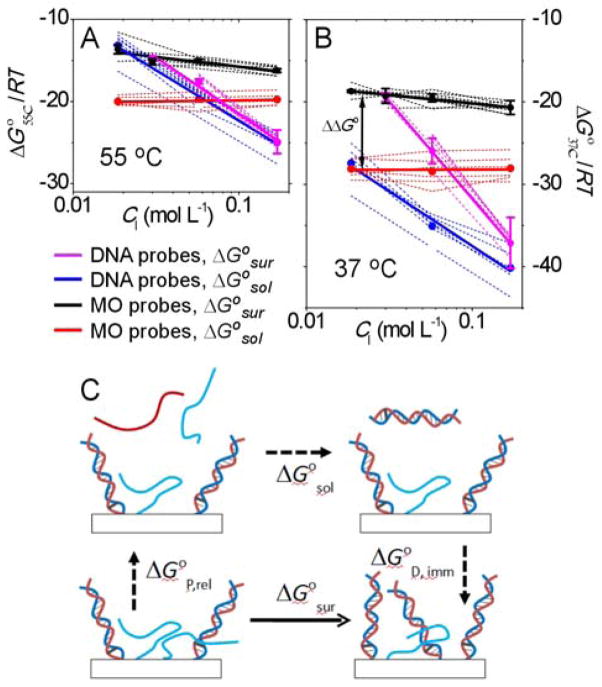
The enthalpy ΔH° and entropy ΔS° were obtained from a two-state model8 with both treated as independent of temperature.9 This simplest model considers each probe site to be either in an unhybridized or a hybridized state, with the physical nature of these states assumed uniform for all sites as well as constant in time. As such, a two-state treatment does not explicitly model dispersion in hybridization behavior that may arise from heterogeneity in probe coverage, synthetic uncertainties, chemistry of the solid support, or some other source. Two-state analysis also does not explicitly model changes in hybridization thermodynamics that arise as hybridization progresses; e.g., due to readjustments in the charge density or steric constraints in the probe layer.6b, 10 Despite their simplicity, two-state fits satisfactorily captured the character of observed melting transitions (Figure S3, ESI) to provide ΔG° = ΔH° – TΔS° values (Tables S2 and S3, ESI) that allow convenient comparison of the two probe types without the need to invoke structural models for both types of films. The derived hybridization free energies are plotted in Figures 2A and 2B. Figure 2A is for a temperature of 55 °C, which is close to most of the observed melting temperatures. Figure 2B shows free energies at 37 °C to illustrate the predicted effect of a temperature change.
Fig. 2.
Comparison of free energies of hybridization ΔG° on solid supports and in solution. (A) At 55 °C. (B) At 37 °C. Both (A) and (B) show data for individual probe sequences (dashed curves), sequence-averaged values (points), and linear fits to the averaged values (thick solid lines). CI is the concentration of Na+ cations, equal to 1.43 times the phosphate concentration. None of the DNA probes yielded clear surface hybridization transitions in the lowest ionic strength buffer (0.012 mol L−1). ΔΔG° = ΔG°sur − ΔG°sol is the difference between surface and solution hybridization free energies. (C) Consideration of equivalent paths shows that ΔΔG° can be expressed as the sum of the free energy of release of unhybridized probes, ΔG°P, rel, and that of immobilization of probe-target duplexes, ΔG°D, imm.
A number of conclusions can be summarized from Figure 2. For both probe types, surface hybridization tended to be less favorable than in solution (ΔG°sur > ΔG°sol). Thus, immobilization imposed a hybridization penalty, in agreement with prior studies.10b, 11 Second, the surface penalty to hybridization was higher for MO probes, as evidenced in a greater offset ΔΔG° = ΔG°sur − ΔG°sol between the surface ΔG°sur and solution ΔG°sol free energies. The observation that adaptation of MO probes to a surface format elicited a greater penalty than for DNA probes will be discussed further below. Third, lower temperatures made all reactions more favorable, as signified by more negative free energies in Figure 2B compared to 2A. This is expected for complexation reactions characterized by a negative ΔS°. Fourth, due to sharper transitions in solution, solution hybridization yielded more negative ΔH° and ΔS° values (Tables S2 and S3, ESI). A consequence of this is that lowering of temperature favored solution more than surface hybridization (approximating ∂ΔG°/∂T = − ΔS°), causing the offset ΔΔG° to increase in favor of the solution reaction as temperature decreased. A practical implication of this trend is that lower temperatures make it more difficult for surface hybridization to compete with double-stranded or folded structures present in a solution analyte. Fifth, for DNA probes, a decrease in ionic strength was accompanied by a strong increase (i.e. destabilization) in ΔG°, both at the surface and in solution. Destabilization at low ionic strength is expected due to poorer screening of charge-charge repulsions between a DNA probe and a DNA target. The data in addition indicate that ΔΔG° tended to increase somewhat as ionic strength decreased (cf. purple and blue curves in Figure 2), so that lower ionic strength favored solution over surface hybridization. Compared to DNA-DNA hybridization, MO-DNA hybridization was much more weakly affected by ionic strength. In solution all six MO probe sequences exhibited ΔG° values that did not change appreciably with buffer concentration (red curves in Figure 2). This is attributed to the neutral character of MO probes.5 On the solid support (black curves in Figure 2) ΔG° acquired a modest dependence on buffer concentration such that hybridization became less favorable at lower ionic strengths. This dependence caused the surface penalty ΔΔG° to increase with a decrease in ionic strength, leading to increased preference for solution hybridization, qualitatively similar to the trend for DNA probes.
Further discussion of the results is facilitated by Figure 2C, which compares solution and surface hybridization in terms of equivalent paths. From Figure 2C it follows that the surface penalty ΔΔG° can be expressed as
where ΔG°P,rel is the free energy of releasing a probe from the surface into solution, and ΔG°D, imm is the free energy of immobilizing a duplex from solution. ΔG°P,rel includes disruption of interactions of an unhybridized probe with other probes, with neighboring duplexes, or with the solid support, as well as associated changes in ionic and solvent distributions that would accompany transfer of a probe from the surface into solution. ΔG°D,imm represents analogous effects from duplex immobilization, including interactions with other duplexes, with unhybridized probes, and with the surface. Since duplexes sequester their bases in their interior, they are primarily expected to experience steric and electrostatic interactions via exposed duplex surfaces. Unhybridized probes are in addition capable of base-mediated interactions, leading to intra- or inter-probe base pairing and stacking, adsorption of bases to the support, or other effects. Such interactions are expected to affect probe folding as well as hybridization activity.12
Given the many avenues available to unhybridized probes for interacting with their surroundings, it is relevant to consider how MO and DNA probes differ in this regard. One important difference is that lack of charge renders MO probes less soluble in water, with solubilities in the 1 to 100 mmol L−1 range at the 25mer lengths used.3 At concentrations typical of immobilized films, this lowered solubility has been implicated to cause MO probes to aggregate and thus exist in a desolvated state on solid supports.13 Another study noted that MO probes adsorbed to the type of aldehyde slides used.14 Such physisorption as well as probe-probe aggregation would manifest in ΔG°P,rel, and thus in ΔΔG°, as penalties that contribute to the difference between surface and solution hybridization in Figure 2.
The importance of probe-surface interactions to ΔΔG° was tested by considering whether surface chemical treatments affected hybridization. These treatments included (1) modifying aldehyde slides with tris (hydroxymethyl) aminomethane to render the surface more hydrophilic and (2) immobilizing probes to p-phenylene diisothiocyanate (PDITC) modified slides (see ESI). It was reasoned that nucleotide bases may interact with the phenyl group in PDITC so as to hinder hybridization to target molecules. Measurements were performed using sequence #4 (Table S1, ESI) in 0.037 mol L−1 buffer.
Although PDITC surfaces did lead to slightly less favorable ΔG°sur for DNA probes by about 10 %, or ~1 kcal mol−1, surprisingly, for MO probes, changes in surface chemistry did not significantly affect ΔΔG° (Figure S4.C, ESI). This indicates that MO probes either interact similarly with the various surfaces or that the surface penalty ΔΔG° was primarily a reflection of probe aggregation at the surface (due to MO solubility limits), rather than probe-surface interactions. This conclusion can be compared to surface hybridization of peptide nucleic acids.15 Jensen et al 15b and Park et al 15c compared surface and solution thermodynamics of PNA probes hybridizing to DNA targets using probes immobilized via streptavidin-biotin chemistry. For a 15mer PNA probe, Jensen et al found a 51 % decrease in ΔG° of hybridization due to immobilization, while Park et al reported a 43 % and a 51 % decrease for another 15mer PNA probe, at two different salt concentrations. In comparison, for MO probes immobilization caused ΔG° to decrease about 30 % (Figure 2). The higher offsets for PNA probes are in line with their lower aqueous solubility, and thus presumably higher tendency to aggregate or adsorb to the support.
For uncharged probes like morpholinos, ΔG°P,rel should not depend on ionic strength. Moreover, measurements showed that ΔG°sol was also salt independent (Figure 2A, red curve). Figure 2C then implies that dependence of MO-DNA surface hybridization on ionic strength (Figure 2A, black curve) must be attributed to ΔG°D,imm. This dependence is attributed to charge interactions between MO-DNA duplexes and possibly between duplexes and repulsive (negative) charges on the solid support, such as from aldehyde oxidation16 or dissociated silanols. Amplification of these surface-specific electrostatic penalties at lower ionic strengths would lead to less favorable surface hybridization, as seen in Figure 2.
For DNA-DNA duplexes, theory predicts a stronger dependence on ionic strength at the surface than in solution because, in addition to a solution-like salt-dependence of duplex formation, surface hybridization changes the charge in the probe layer the cost of which also depends on ionic strength.10a, 10c Our results are not inconsistent with this prediction (cf. purple and blue lines in Figure 2). However, other studies have found weaker or comparable sensitivity.10b, 17 A weaker dependence was attributed to base-pairing between unhybridized probes that decreased the net gain in base pairs from target hybridization.10b The salt dependence of surface hybridization is thus expected to reflect sequence-specific effects, such as partial hybridization or self-folding among the probes. The six sequences of the present study were selected to minimize strong probe-probe and intramolecular associations.18
Although at the higher temperature of Figure 2A ΔG°sur was close to ΔG°sol for DNA probes, it is relevant to note that the surface and solution processes were not equivalent. The surface transitions were typically broader, especially at lower ionic strengths (Figure S3.B), as also reflected in their smaller enthalpic and entropic changes (Table S3). Various mechanisms can contribute to transition broadening such as dispersion in probe affinity due to local variations in steric and electrostatic factors, shift in hybridization energetics with extent of hybridization, and formation of only partly zipped duplexes (e.g. due to greater fraying at duplex ends19 at lower ionic strengths). As noted above, the outcome of broadened transitions, for both DNA and MO probes, is a weaker dependence of surface ΔG° on temperature that leads to an increasing surface penalty ΔΔG° as temperature decreases.
The balance between solution and surface thermodynamics is relevant in diagnostic applications where it is desirable to maximize selectivity for hybridization on the solid support in competition with folding or inter-strand base pairing in solution analyte. For DNA probes, a scenario in which surface hybridization is preferred, ΔG°sur < ΔG°sol, does not appear practicable based on Figure 2 results. On the other hand, the data indicate a crossover from solution DNA-DNA hybridization (blue line, Figure 2A) to surface MO-DNA hybridization (black line) as the preferred form of base pairing at CI below 0.015 mol L−1; below this concentration, the most attractive binding partner for a DNA target will be an immobilized MO probe even if there are fully complementary DNA sequences in solution. At lower temperatures, the crossover shifts to lower ionic strengths. Figure 3 considers how combinations of salt and temperature affect the balance between ΔG° of solution DNA-DNA and surface MO-DNA hybridization. The surface reaction wins (more negative ΔG°) for conditions in the lower right, providing a guideline for selecting settings to keep analyte in a partly denatured state while still allowing target-probe hybridization.
Fig. 3.
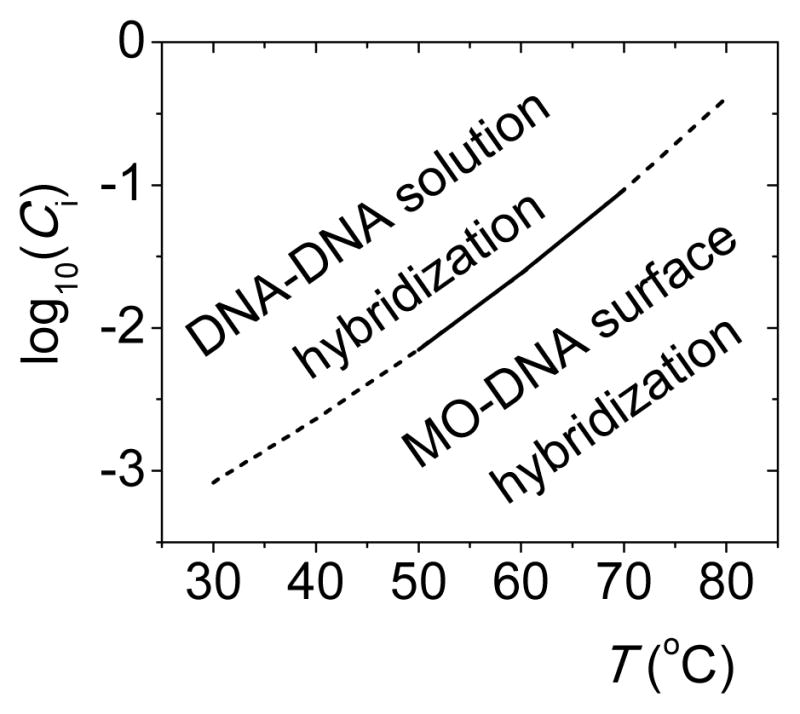
Separation in CI -T space between conditions favoring DNA-DNA solution hybridization and MO-DNA surface hybridization, based on sequence-averaged results from Tables S2 and S3 (ESI). The solid portion is interpolated from measurements, while dashed portions are extrapolated.
Surface hybridization is encountered in genomics technologies including DNA microarrays and biosensors, as well as finds numerous applications in fabrication of structures. By comparing hybridization thermodynamics of morpholino and DNA probes, on surfaces and in solution, the present report advances fundamental understanding of morpholino properties of direct relevance to such applications. It also motivates development of protocols for controlling the balance between surface and solution interactions, not only through selection of optimal probe type based on experimental need, but through control over hybridization thermodynamics. Such control could be pursued, for example, through application of electric fields,20 a strategy that should be especially effective under the low salt conditions optimally compatible with MO probes.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NHGRI R01HG004512), the National Science Foundation (DMR 12-06754), and by New York University.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental methods; ΔH° and ΔS° tables; results of surface chemistry studies.
Notes and references
- 1.(a) Harrison A, Binder H, Buhot A, Burden CJ, Carlon E, Gibas C, Gamble LJ, Halperin A, Hooyberghs J, Kreil DP, Levicky R, Noble PA, Ott A, Pettitt BM, Tautz D, Pozhitkov AE. Nucleic Acids Res. 2013;41:2779. doi: 10.1093/nar/gks1358. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rao AN, Grainger DW. Biomaterials Science. 2014;2:436. doi: 10.1039/C3BM60181A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Macfarlane RJ, Lee B, Jones MR, Harris N, Schatz GC, Mirkin CA. Science. 2011;334:204. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]; (b) Baron R, Willner B, Willner I. Chem Commun. 2007;323 doi: 10.1039/b610721b. [DOI] [PubMed] [Google Scholar]
- 3.Summerton JE. Lett Pept Sci. 2004;10:215. [Google Scholar]
- 4.Egholm M, Buchardt O, Christensen L, Behrens C, Freler SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 5.Gong P, Wang K, Liu Y, Shepard K, Levicky R. J Am Chem Soc. 2010;132:9663. doi: 10.1021/ja100881a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Belozerova I, Levicky R. J Am Chem Soc. 2012;134:18667. doi: 10.1021/ja3066368. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fuchs J, Fiche JB, Buhot A, Calemczuk R, Livache T. Biophys J. 2010;99:1886. doi: 10.1016/j.bpj.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloomfield VA, Crothers DM, Tinoco JI. Nucleic Acids - Structures, Properties, and Functions. University Science Books; Sausalito: 2000. [Google Scholar]
- 8.Puglisi JD, Tinoco I., Jr Methods Enzymol. 1989;180:304. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 9.(a) Fiche JB, Buhot A, Calemczuk R, Livache T. Biophys J. 2007;92:935. doi: 10.1529/biophysj.106.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ge D, Wang X, Williams K, Levicky R. Langmuir. 2012;28:8446. doi: 10.1021/la301165a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Halperin A, Buhot A, Zhulina EB. Biophys J. 2004;86:718. doi: 10.1016/S0006-3495(04)74150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Irving D, Gong P, Levicky R. J Phys Chem B. 2010;114:7631. doi: 10.1021/jp100860z. [DOI] [PubMed] [Google Scholar]; (c) Vainrub A, Pettitt BM. Phys Rev E. 2002;66:art. 041905. doi: 10.1103/PhysRevE.66.041905. [DOI] [PubMed] [Google Scholar]
- 11.Peterson AW, Wolf LK, Georgiadis RM. J Am Chem Soc. 2002;124:14601. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 12.Kastantin M, Schwartz DK. Small. 2013;9:933. doi: 10.1002/smll.201202335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Irving D, Qiao W, Ge D, Levicky R. J Am Chem Soc. 2011;133:11588. doi: 10.1021/ja202631b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao W, Kalachikov S, Liu Y, Levicky R. Anal Biochem. 2013;434:207. doi: 10.1016/j.ab.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Ghosh S, Mishra S, Banerjee T, Mukhopadhyay R. Langmuir. 2013;29:3370. doi: 10.1021/la400125x. [DOI] [PubMed] [Google Scholar]; (b) Jensen KK, Orum H, Nielsen PE, Norden B. Biochemistry. 1997;36:5072. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]; (c) Park H, Germini A, Sforza S, Corradini R, Marchelli R, Knoll W. Biointerphases. 2007;2:80. doi: 10.1116/1.2746871. [DOI] [PubMed] [Google Scholar]; (d) Sato Y, Fujimoto K, Kawaguchi H. Colloids Surf B - Biointerfaces. 2003;27:23. [Google Scholar]
- 16.McNesby JR, Heller CA., Jr Chem Rev. 1954;54:325. [Google Scholar]
- 17.(a) Azam MS, Gibbs-Davis JM. Anal Chem. 2013;85:8031. doi: 10.1021/ac401009u. [DOI] [PubMed] [Google Scholar]; (b) Peterlinz KA, Georgiadis RM, Herne TM, Tarlov MJ. J Am Chem Soc. 1997;119:3401. [Google Scholar]; (c) Watterson JH, Piunno PAE, Wust CC, Krull UJ. Langmuir. 2000;16:4984. [Google Scholar]
- 18.The probe sequences of the present study avoid intra- or inter-probe complementarity higher than a run of four GC base pairs.
- 19.Jose D, Datta K, Johnson NP, Von Hippel PH. Proc Natl Acad Sci USA. 2009;106:4231. doi: 10.1073/pnas.0900803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Heaton RJ, Peterson AW, Georgiadis RM. Proc Natl Acad Sci USA. 2001;98:3701. doi: 10.1073/pnas.071623998. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Johnson RP, Richardson JA, Brown T, Bartlett PN. Journal of the American Chemical Society. 2012;134:14099. doi: 10.1021/ja304663t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.