Abstract
Defining mechanisms that generate intratumour heterogeneity and branched evolution may inspire novel therapeutic approaches to limit tumour diversity and adaptation. SETD2 (Su(var), Enhancer of zeste, Trithorax-domain containing 2) trimethylates histone-3 lysine-36 (H3K36me3) at sites of active transcription and is mutated in diverse tumour types, including clear cell renal carcinomas (ccRCCs). Distinct SETD2 mutations have been identified in spatially separated regions in ccRCC, indicative of intratumour heterogeneity. In this study, we have addressed the consequences of SETD2 loss-of-function through an integrated bioinformatics and functional genomics approach. We find that bi-allelic SETD2 aberrations are not associated with microsatellite instability in ccRCC. SETD2 depletion in ccRCC cells revealed aberrant and reduced nucleosome compaction and chromatin association of the key replication proteins minichromosome maintenance complex component (MCM7) and DNA polymerase δ hindering replication fork progression, and failure to load lens epithelium-derived growth factor and the Rad51 homologous recombination repair factor at DNA breaks. Consistent with these data, we observe chromosomal breakpoint locations are biased away from H3K36me3 sites in SETD2 wild-type ccRCCs relative to tumours with bi-allelic SETD2 aberrations and that H3K36me3-negative ccRCCs display elevated DNA damage in vivo. These data suggest a role for SETD2 in maintaining genome integrity through nucleosome stabilization, suppression of replication stress and the coordination of DNA repair.
Introduction
Renal cell carcinoma (RCC) is associated with a 5-year survival of <10% for patients with metastatic disease at diagnosis.1 Clear cell RCC (ccRCC) is the most common histological subtype, accounting for 80–90% of RCCs. Exome sequencing studies of ccRCC have defined the mutational landscapes of these tumours, revealing that histone-modifying enzymes and chromatin-remodelling complexes are subject to recurrent mutations.2, 3 The most frequent genomic aberrations are the inactivation of the tumour-suppressor von Hippel-Lindau (VHL, >50% of cases)2, 3, 4, 5 and loss of heterozygosity (LOH) of chromosome 3p (>80% of ccRCCs),6 which encodes VHL as well as the candidate ccRCC tumour-suppressor genes PBRM1, BAP1 and SETD2.3, 7 Polybromo 1 (PBRM1 or BAF8), a SWI/SNF chromatin-remodelling complex component, and the deubiquitylase BAP1 are mutated in 40% and 10% of ccRCCs respectively.2, 3
SETD2 is estimated to be mutated in 4–8% of ccRCCs.2, 3, 5 We have previously observed multiple distinct SETD2 mutations in spatially separated regions of the same tumour occurring in the proximal branches of the tumour phylogenetic tree, indicating a strong selection pressure for SETD2 inactivation in a subset of ccRCCs around the time of branched evolution.4 SETD2 is the main methyltransferase responsible for trimethylation of histone-3 at lysine-36 (H3K36me3),8 and is recruited to sites of active transcription by interaction with the C-terminal domain of RNA polymerase II (RNA Pol II).9, 10
In this study, we used an integrated bioinformatics and functional approach to investigate the consequences of SETD2 loss-of-function in ccRCC.
Results
SETD2 mutations and LOH in ccRCC
Using data from The Cancer Genome Atlas (TCGA) Research Network, we assembled a ccRCC data set of 450 tumours with copy number data and 293 tumours with somatic mutation data (whole-exome sequencing data from the Broad institute consisting of 262 overlapping samples, Supplementary Table S1). MutSig analysis,11 which identifies significantly recurrent mutations based on variable mutation rates across the genome, gene size and replication timing, identified 14 genes that were significantly mutated in the data set suggestive of driver status, including SETD2 (Q-value=0, Supplementary Table S2).
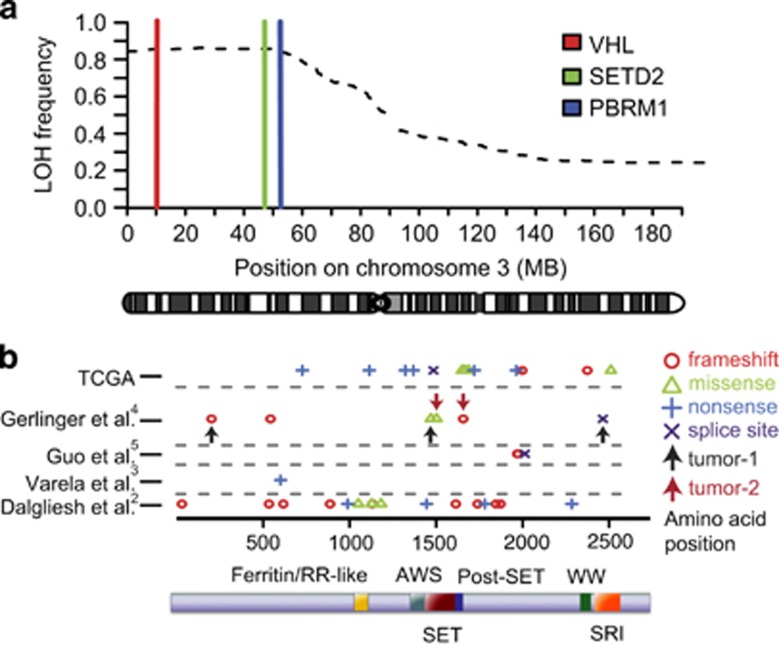
SETD2 is encoded on chromosome 3, and 3p LOH has previously been reported to be an early event in RCC.12, 13 In all, 98.8% (391/396) of TCGA samples with LOH at the VHL locus (3p25.3) also displayed LOH at the SETD2 locus (3p21.31) (Figure 1a and Supplementary Table S1). Non-synonymous SETD2 mutations or homozygous SETD2 deletions were found in 34 samples (Figure 1b), with paired DNA copy number data available for 30 samples. In total, 93.3% of these cases (28/30) showed LOH at the SETD2 locus. Ten of the 28 mutations were missense mutations (five were in the SET domain, one in the SRI domain and four in uncharacterized regions of the protein). One sample harboured homozygous deletion of SETD2. Therefore in total, both SETD2 alleles were mutated or deleted in 29 samples, collectively referred to as SETD2mut. The 210 samples with LOH at the SETD2 locus but no mutation of the gene are referred to as SETD2LOH.
Figure 1.
(a) Proportion of samples showing LOH along chromosome 3 using SNP 6.0 array data in 450 RCCs from TCGA. The genomic loci of VHL, SETD2 and PBRM1 are indicated. (b) Schematic of the locations of mutations reported in the SETD2 gene from published studies (2–5). Arrows indicate distinct mutations found within spatially separated regions of the same tumour.4
Chromosomal instability at H3K36me3 sites in SETD2 mutant ccRCC
Increasing evidence implicates transcriptional aberrations as one route to influence genome stability,14 we hence hypothesized that reduced H3K36me3 in the context of SETD2 bi-allelic aberrations may affect genomic stability in a site-specific manner. Genome-wide distributions of histone marks were obtained from publicly available ChIP-Seq data derived from two normal adult kidney samples (H3K36me3) and one fetal kidney (H3K27me3) from the ENCODE Consortium.15, 16, 17 We then mapped genomic H3K36me3 distributions to chromosomal breakpoint regions in the 29 SETD2mut and 210 SETD2LOH TCGA samples.
Chromosomal breakpoints were identified using single-nucleotide polymorphism (SNP) array data, the resolution of which only permits identification of breakpoint regions (between two genomic segments present at different copy numbers), rather than specific sites of breakage. Breakpoint regions were then defined using 16 different threshold combinations for minimum flanking segment length and maximum breakpoint region length, to take into account the occurrence of false positives in the predicted chromosomal aberrations (Supplementary Table S3 and Supplementary Methods). Using this cohort, we assessed the location of breakpoint regions, with respect to H3K36me3 sites in normal adult kidney, in the 210 SETD2LOH tumours and the 29 SETD2mut tumours (Figure 2a and Supplementary Figure S1a). We generated 5000 random breakpoint region profiles to estimate the expected background frequency distributions of H3K36me3 sites within breakpoint regions for SETD2mut versus SETD2LOH TCGA tumours, for each threshold combination.
Figure 2.
SETD2 mutation exposes H3K36me3 sites to chromosome breakage. (a) Schematic illustrating the mapping of chromosomal breakpoint regions to sites of H3K36me3/H3K27me3 (data from the ENCODE consortium) in both SETD2LOH and SETD2mut. Breakpoint regions, identified from SNP 6.0 array data, are the regions between two segments of a chromosome present at different allele-specific copy numbers. (b) Representative plots of observed H3K36me3 frequencies against expected frequencies in SETD2LOH and SETD2mut tumours, using a minimum segment length of 10 Mb and a maximum breakpoint region length of 20 kb.
SETD2mut tumours did not harbour more breakpoints on average (Supplementary Table S3) and did not show higher chromosomal complexity as measured by the weighted genome instability index18 (Supplementary Figure S1b). However, breakpoint regions from SETD2LOH samples were significantly depleted of H3K36me3 marks (12/16 threshold combinations with P<0.05; Supplementary Table S3 and Supplementary Figure S2). In contrast, such depletion was not observed when mapping the locations of breakpoint regions and H3K36me3 sites in SETD2mut tumours. Breakpoint regions in the SETD2mut samples were marginally enriched for H3K36me3 marks (3/16 threshold combinations with P<0.05; Supplementary Table S3 and Supplementary Figure S2). Representative plots of observed H3K36me3 frequencies against expected frequencies in SETD2LOH and SETD2mut samples are shown in Figure 2b. No depletion of H3K27me3, a histone mark found across the genome at similar frequencies to H3K36me3 (12.6% and 12.1%, respectively),15, 16, 17 could be observed in SETD2LOH breakpoint regions, suggesting that the depletion of H3K36me3 from breakpoint regions in these tumours is specific. In addition to the apparent association between chromosomal breakage and H3K36me3 sites, there was a moderately higher co-occurrence of point mutations with H3K36me3 marks in the SETD2mut samples compared with the SETD2LOH samples (Supplementary Table S4, P=0.04, one-tailed Fisher's test). Taken together, these data suggest that normally H3K36me3 sites are less prone to breakage than the rest of the genome in SETD2 wild-type cells, whereas following SETD2 bi-allelic mutation or deletion, this apparent site-specific protection is lost, increasing the probability of chromosome breaks at these sites to an equivalent or possibly higher level than the rest of the genome in SETD2 wild-type cells. The fact that H3K27me3 is not depleted in breakpoint regions from the SETD2LOH samples further suggests that this putative protection is specific to H3K36me3. These data suggested a potential role for SETD2 in the maintenance of genomic integrity, mediated through H3K36me3.
Reduced H3K36me3 and SETD2 expression are associated with DNA damage in vivo
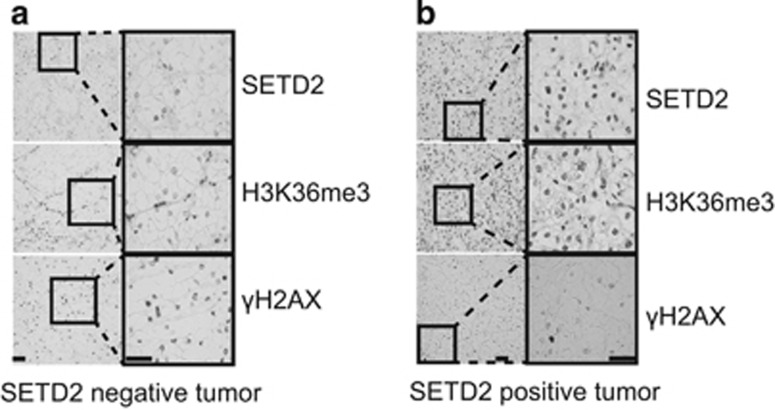
In light of the apparent protective role for SETD2 and the selection pressure for SETD2 inactivation in ccRCC as an intermediate event that occurs around the time of detectable branched ccRCC evolution, we explored the relationship between SETD2 mutation and genomic instability in vivo. We assessed SETD2 expression, H3K36me3 levels and DNA damage in a cohort of 202 samples from 100 RCC tumours, assayed by immunohistochemistry on tissue microarrays (Figures 3a and b and Supplementary Table S5). SETD2 mutation status was not available for this cohort. However, as SETD2 is the main H3K36 trimethylase8 and the majority of mutations identified to date result in frameshifts or premature stop codons in the N-terminal half of the gene, we used a C-terminal SETD2 antibody that would not detect SETD2 affected by protein truncations distal to the reported mutations, the majority of which occur before the C-terminal SRI domain (Figure 1b). Therefore, SETD2 and H3K36me3 immunohistochemical staining were used as surrogate markers for SETD2 loss-of-function. A highly significant correlation was observed between SETD2 and H3K36me3 immunohistochemical staining (P=3.8e-14, Cochran-Armitage test, Supplementary Figure S3a), suggesting that absence of H3K36me3 staining is a good surrogate for SETD2 loss-of-function.8 DNA damage was assessed by staining for phosphorylated histone 2 A.X (γH2AX), which marks ongoing DNA damage signalling and DNA double-strand breaks.19 Both SETD2 and H3K36me3 levels were significantly anti-correlated with γH2AX staining (P<0.05, Supplementary Figures S3b and c), suggesting that SETD2 loss-of-function may contribute to DNA damage and double-strand breaks. These data further suggest a role for SETD2 and H3K36me3 in the maintenance of genome stability.
Figure 3.
Reduced H3K36me3 is associated with elevated DNA damage in primary ccRCC tumours. (a) Images of tumour cells from a ccRCC case stained negatively for SETD2 and H3K36me3 but strongly positive for γH2AX (scale bar: 50 μm). (b) Images of tumour cells from a ccRCC case stained positive for SETD2 and H3K36me3, but weakly stained for γH2AX (scale bar: 50 μm).
SETD2 is required for efficient DNA repair
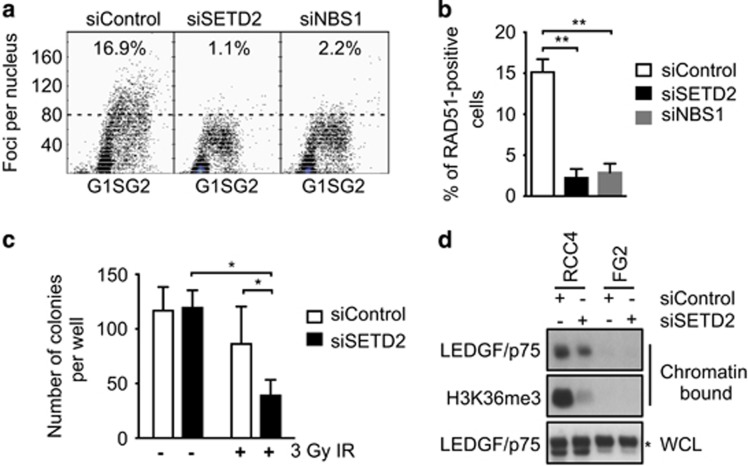
The transcriptional co-activator lens epithelium-derived growth factor (LEDGF/p75) contains a proline-tryptophan-tryptophan-proline domain, which has been shown to interact with H3K36me3.20 LEDGF is required for CtIP recruitment leading to RAD51 loading and DNA repair.21 We first investigated the proficiency of homologous recombination (HR) following SETD2 depletion in RCC cell lines by small-interfering RNA (siRNA). We used two alternative siRNA pools targeting SETD2, having excluded some sequences because of off-target effects (Supplementary Figures S4a and b). After 48 h both the pools and individual siRNA sequences efficiently silenced SETD2 and depleted H3K36me3 levels (Supplementary Figures sS4c and e). Consistent with a defect in HR, SETD2-depleted cells displayed reduced RAD51 foci formation, used as surrogate for detecting active HR, following induction of DNA damage with ionizing irradiation (IR) (Figure 4a and Supplementary Figure S5a). Using our newly developed quantitative method to assess RAD51 foci formation in large numbers of cells, we found that the proportion of RAD51-positive cells was reduced from 15.1% in control to 2.2% in SETD2-depleted cells and 2.9% in cells depleted of Nijmegen breakage syndrome 1, which was used as a positive control (both P=0.003, average from n= 3 independent experiments, Figure 4b). Similar results were obtained in U2OS cells (SETD2 wild-type) (Supplementary Figures S5b and c), and RCC-JW cells (SETD2 missense mutation, H3K36me3 competent) (Supplementary Figure S5d). In order to support the specificity of the RNA interference phenotype, we repeated these experiments in the RCC-FG2 line, which harbours bi-allelic aberrations in SETD2 (no detectable SETD2 protein and barely detectable H3K36me3, Supplementary Figures S4c and S5e). No decrease in RAD51 foci formation after IR was observed following SETD2 silencing in RCC-FG2. Consistent with a defect in HR, SETD2-depleted cells were also more sensitive to IR (P=0.035, two-tailed t-test, Figure 4c). As Rad51 foci formation and HR repair require CtIP-mediated double-strand break end resection, and CtIP recruitment to active-chromatin-associated double-strand breaks relies on the proline-tryptophan-tryptophan-proline domain-containing LEDGF,21, 22 we speculated that chromatin loading of LEDGF may depend on SETD2 function. Indeed, we observed reduced chromatin-bound LEDGF/p75 following SETD2 depletion in RCC cell lines, while there was no decrease in total cellular LEDGF levels (Figure 4d). Our data therefore support the hypothesis that the defective DNA repair in the absence of SETD2 could exacerbate the DNA damage observed in vivo. This conclusion is further supported by two recent studies, submitted while the manuscript was under preparation, reporting the link between SETD2, H3K36me3 and HR.20, 22
Figure 4.
Loss of SETD2 leads to impaired RAD51 foci formation and LEDGF association to chromatin. (a) A representative experiment of RAD51 foci formation following IR (8 Gy) in siRNA-transfected RCC4 cells. siRNA for Nijmegen breakage syndrome 1 was used as a positive control for impaired RAD51 foci formation. 45 An arbitrary threshold of 80 RAD51 foci was used. (b) Proportion of cells with >80 RAD51 foci (mean±s.e.m of three independent experiments Statistical test: two-tailed t-test). **P⩽0.01. (c) RCC4 cells were exposed to 0 (non-irradiated) or 3 Gy IR, allowed to form colonies over 16 days, before quantification of colony numbers (mean±s.e.m. of three independent experiments. Statistical test: two-tailed t-test). (d) Cellular fractionation was performed to examine chromatin-bound LEDGF/p75 in RCC4 and FG2 cells. Total levels of LEDGF/p75 in whole-cell lysates (WCL) are shown as control. * denotes a non-specific band.
SETD2 mutations are not associated with mutational signatures of microsatellite instability in ccRCC tumours or cell lines
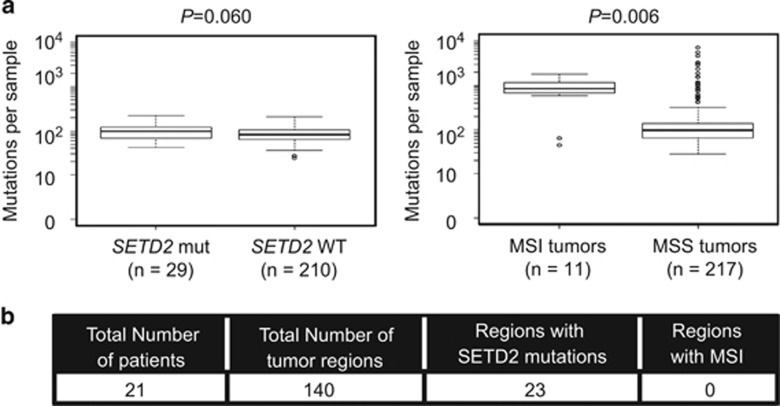
A recent study has described a novel role for SETD2 in mismatch repair and microsatellite instability (MSI) in a renal cancer cell line harbouring a SETD2 mutation.23 We decided to investigate the contribution of SETD2 mutation-induced MSI on genomic instability in vivo. As MSI is associated with a high frequency of point mutations, we first assessed mutational load in a cohort of SETD2mut tumours from the TCGA,24 in comparison with colon tumours with MSI. The mutational load in the SETD2mut tumours was substantially lower than that observed in MSI colon tumours (Figure 5a and Supplementary Figure S6). We next assessed MSI in an extended cohort of SETD2mut tumours, in comparison with colon tumours with MSI. We took advantage of a recently defined mutational signature associated with mismatch repair deficiency.25 Using the same data set as Alexandrov et al. (which included 281 of the 293 TCGA samples described above, as well as additional samples from other sources25), we investigated whether there was a relationship between SETD2 mutation status and the mismatch repair deficiency signature. Thirty-five out of 324 samples (10.8%) bore a SETD2 mutation. Only 2 of the 324 samples presented a clear mismatch repair deficiency signature, with one of them also bearing a SETD2 mutation. However, the sample with the SETD2 mutation presented LOH on the whole 3p arm, which also encodes the mismatch repair gene MLH1. This tumour also had a MLH1 frameshift insertion on the other allele, leading to the loss of both copies of MLH1. Therefore, it is likely that the MSI signature in this tumour is explained by loss of MLH1 function. In total, 30 samples harboured a SETD2 mutation in addition to SETD2LOH, of which 29 showed no evidence of mismatch repair deficiency according to mutational signatures. To extend our findings and to investigate the relationship between SETD2 mutational status and MSI in tumours, we assessed MSI in 21 tumours for which we had obtained multiple biopsies from each tumour and for which SETD2 mutation and chromosome 3p status were known for each biopsy (n=140; 23 regions with SETD2 mutations).4, 13 We examined five markers (D2S123, D5S346, D17S250, BAT25 and D11S904), including four established markers from the Bethesda panel for identification of MSI in colorectal cancer.26 As a positive control, we included five MSI positive colorectal cell lines. We observed no evidence of MSI across the five different markers (Figure 5b) including BAT25 (Supplementary Figures S7i and iv). Finally, we did not observe MSI over five markers in six ccRCC cell lines, SETD2-wild-type (RCC4, RCC-JW and UOK121) and SETD2 mutant (A498, RCC-FG2 and UOK143 cells; Supplementary Figure S7v). Taken together, our data suggest that SETD2 mutations are not associated with MSI in ccRCC.
Figure 5.
Absence of microsatellite instability in absence of SETD2. (a) Number of mutations in SETD2mut and SETD2LOH ccRCC samples (left) and MSI and MSS colorectal tumours (right). (b) Samples from ccRCC tumours were assessed by PCR for microsatellite instability over five microsatellite markers, and compared with five MSI colon cancer cell lines.
SETD2 depletion does not result in cryptic transcriptional initiation or altered splicing
SETD2 has been implicated in regulating the splicing machinery,27 and also in the suppression of cryptic transcription initiation within coding exons by coordinating nucleosome assembly following RNA Pol II elongation.28 We hypothesized that global transcriptional defects because of loss of SETD2-dependent trimethylation may contribute to the elevated DNA damage and genomic instability observed in the tumour samples. To assess this in ccRCC models, we first performed ChIP-Seq for H3K36me3 following SETD2 depletion to identify genes that were significantly dependent on SETD2 for trimethylation (Figure 6a). Having identified 2513 genes (false discovery rate (FDR)⩽0.05, logFC⩽−1.5 and logCPM≥1) as being dependent on SETD2 for H3K36me3, we then performed ChIP-Seq for RNA Pol II and gene body profiling for the distribution of RNA Pol II across these genes. We observed no global changes in RNA Pol II distribution following SETD2 depletion in RCC4 cells (Figure 6b).
Figure 6.
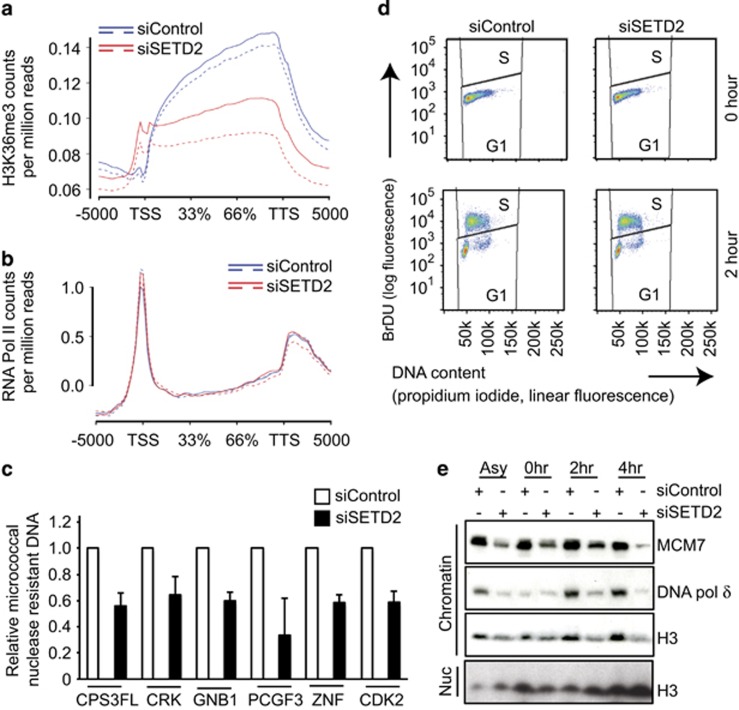
Altered nucleosome assembly induced by SETD2 silencing. (a) Gene body profile for H3K36me3 in RCC4 across all genes identifies genes most dependent on SETD2 for trimethylation (n=2). The graph plots the count per million mapped reads for H3K36me3 binding in siControl (blue) and siSETD2 (red) for all the genes in the Ensembl annotation compiled using ngs.plot (version 2.41.3).58 (b) The graph plots the per-sample log2 median-centred count per million mapped reads for RNA Pol II binding in siControl (blue) and siSETD2 (red) for significant 2513 genes with reduced H3K36me3 binding, compiled using ngs.plot (version 2.41.3)58 (n=2). (c) Micrococcal nuclease assays were performed after siRNA treatment. Quantitative PCR was carried out over a selection of genes most dependent on SETD2 for trimethylation. The graph shows the relative proportion of micrococcal nuclease-resistant DNA normalized against a control non trimethylated region. (d) RCC4 cells were treated with control or SETD2 siRNAs for 48 h then treated with a double thymidine block before being released into complete media containing 10 μM BrdU and analysed for cell cycle progression. (e) Cells were treated as in d and were harvested at the indicated time points and lysates were probed for the indicated proteins.
In the absence of global changes in RNA Pol II occupancy, we directly assessed the effect of SETD2 depletion on gene expression, by performing RNA-Seq in RCC4 cells. Surprisingly, only 326 genes (FDR⩽0.05) showed a significant differential expression between SETD2 siRNA-depleted versus non-depleted samples across the three experiments. The most significant upregulated and downregulated genes were validated by quantitative PCR (Supplementary Figure S8a). A significant overlap was found (Fisher's exact test, P-value=0.001211) between these genes and those determined to exhibit differential expression in the TCGA ccRCC24 data set (n=392) for SETD2mut (n=49) and SETD2wt (n=343) cohorts (Supplementary Figure S8b).
In order to examine whether RNA processing was defective following SETD2 depletion in RCC4 cells, we performed differential exon usage analysis using DEXSeq.29 and also calculated intron retention scores14 using our RNA-Seq data (Supplementary Figure 8c). We observed no evidence of differential exon usage or intron retention, arguing against a role for SETD2-dependent trimethylation as a global transcriptional regulator in this ccRCC model system.
SETD2 depletion results in altered nucleosome dynamics and DNA replication stress
Previous studies have suggested impaired loading of the FACT (Facilitates Chromatin Transcription) complex as well as changes in chromatin compaction following SETD2 depletion.14, 28 In order to study if chromatin architecture was changed following SETD2 depletion in RCC cells, we next investigated chromatin organization by micrococcal nuclease sensitivity assays. Quantitative PCR was performed over regions identified by ChIP-Seq as the most dependent on SETD2 for H3K36me3 trimethylation (Figure 6c). The relative amount of micrococcal nuclease-resistant DNA was calculated relative to a region that was not trimethylated in control cells. These data revealed that in the absence of SETD2, normally trimethylated regions had reduced nucleosome compaction.
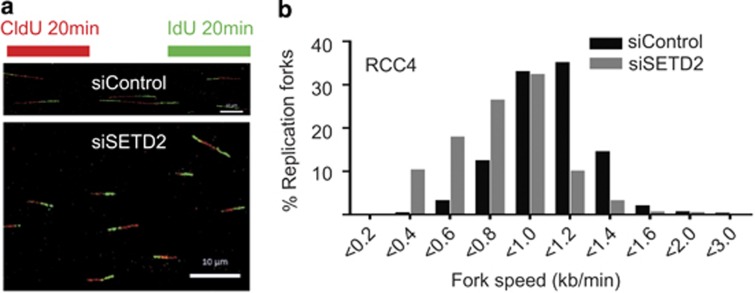
Histone dynamics have an important role in DNA replication, where the stability of the replication complex is dependent on the reassembly of nucleosomes behind the advancing fork.30, 31 In line with this, the FACT complex has been demonstrated to recruit the MCM complex to chromatin during replication.32, 33 In order to specifically study the role of SETD2 during S phase, SETD2- and control siRNA-treated cells were studied after release from a double thymidine block. SETD2-depleted cells exited the block and entered into S phase with similar dynamics as control-treated cells (Figure 6d). We observed reduced chromatin association of MCM7, DNA polymerase δ and histone H3, whereas overall protein levels of the replisome components were unaffected in RCC4, RCC-FG2, UOK121 and U2OS cell lines by SETD2 depletion, indicating that impaired nucleosome assembly may affect replication dynamics (Figure 6e and Supplementary Figure 8d). In support of this, RCC-FG2 cells, possessing SETD2 loss-of-function and absent H3K36me3, also exhibited reduced loading of replisome components onto chromatin (Supplementary Figure 8e). Nucleosome integrity has been demonstrated to have a key role in replisome stability during replication fork progression.30 Given the aberrant nucleosome occupancy and reduced chromatin association of several DNA replication factors (Figures 6c and e and Supplementary Figures 8d and e), we wished to directly assess any impact of SETD2 status on replication fork progression using DNA fibre assays. Replication fork progression was significantly slower following SETD2 depletion (Figures 7a and b) decreasing from 1.0 kb/min in control to 0.75 kb/min in SETD2-depleted cells, P=8.3e–104, two-tailed unpaired t-test with Welch's correction (Supplementary Figure S9a). In agreement with this finding, we observed significantly slower replication fork progression in A498 cells (0.73 kb/min), which have a homozygous truncating SETD2 mutation2 and reduced H3K36me3 levels (Supplementary Figure S4f), compared with RCC4 cells that have one SETD2 allele intact (1.1 kb/min, P=2.35e–75, two-tailed unpaired t-test with Welch's correction, Supplementary Figure S9b). RCC-FG2 cells, which are SETD2 deficient, also had a slower replication fork speed than RCC4 cells (0.85 kb/min), and no further reduction in replication speed was observed following depletion of SETD2 (Supplementary Figure S9c). Consistent with slower replication rates, we noted an accumulation of cells in S phase after SETD2 depletion (50% cells versus 36% in control cells, P=0.048, two-tailed t-test,) and an extended S phase duration (Supplementary Figures S10a and c). These results suggest that in the absence of SETD2, defective nucleosome assembly during S phase contributes to DNA replication fork instability leading to genomic damage.
Figure 7.
Replication fork progression is impaired following SETD2 depletion. RCC4 cells were treated with control or SETD2 siRNAs for 48 h, sequentially incubated for 20 min with CldU and IdU, followed by DNA fibre preparation. Representative DNA fibres are shown in: (a) (scale bar: 10 μm). (b) Distribution of replication fork progression rates in control versus SETD2-depleted cells.
Discussion
In this study, we provide several novel insights into the impact of SETD2 loss-of-function on nucleosome structure, replisome occupancy, replication fork progression and DNA repair by homologous recombination. These are all important cellular functions whose deregulation collectively could contribute to enhanced genomic instability, heterogeneity and adaptability of ccRCCs, thereby conceivably potentiating branched evolution in this disease.
First, in terms of DNA damage and repair, we have shown that tumours harbouring SETD2 bi-allelic aberrations display an altered distribution of both chromosomal breakpoints and mutations around H3K36me3 sites compared with tumours with at least one wild-type SETD2 allele (Figure 1). RNA interference experiments revealed a role for SETD2-dependent H3K36me3 in DNA repair by homologous recombination, mechanistically reflecting the requirement for H3K36me3-mediated recruitment of the LEDGF/CtIP complex20 critical for double-strand break end resection and thereby eventually for Rad51 loading and HR. These results complement other recent studies,22, 34, 35 collectively providing evidence for a function of SETD2-mediated H3K36 trimethylation in coordinating homologous recombination repair (Figures 4, 6 and 7).
Furthermore, in contrast to a recent in vitro study,23 we do not find an association between SETD2 loss-of-function and MSI in ccRCC in either the publicly available TCGA data set, in patient-derived tumour material or in SETD2mut ccRCC cell lines (Figure 5b and Supplementary Figure S7). The mutational load in SETD2mut tumours was substantially lower than that observed in MSI colon cancers (Figure 5a and Supplementary Figure S6) and we therefore conclude that there is no tangible evidence for an association between SETD2 mutations and MSI in ccRCC. It is possible that in vivo, in contrast to the reported in vitro findings, there is rapid adaptation to prevent MSI following SETD2 mutation. Another possibility is that MSI following SETD2 inactivation is confined to genomic loci normally marked by H3K36me3.23 Although no association was found with MSI, levels of H3K36me3 were inversely correlated with DNA damage in vivo (Figure 3), and we observed a shift in the distribution of chromosome breakpoints in SETD2mut tumours (Figure 2). Although in wild-type tumours, H3K36me3 sites were significantly depleted of chromosome breakpoints, this depletion was not observed in SETD2mut tumours, suggesting that loss-of-function of SETD2 may unmask sites that are normally protected by H3K36me3. However, we did not observe an overall increase in the number of breakpoints in SETD2mut tumours, as determined by SNP array analysis. This may be explained by relative confinement of the instability to H3K36me3 sites, and the fact that copy number changes identified by SNP array represent only those aberrations that are compatible with clonal expansion. Furthermore, along with others, we have previously found evidence for an optimal degree of genomic instability in tumours,36, 37, 38 suggesting that excessive genomic instability can be deleterious and result in autonomous cell lethality and that tumours may align to a ‘just-right threshold' of genomic aberrations sufficient to adapt to environmental pressures.12
Whereas SETD2 has been implicated in RNA processing defects,14, 28 we were unable to find any global transcriptional defects in the absence of SETD2 (Figure 6b and Supplementary Figure S8c). A possible explanation for the difference could be the systems used; a cell line derived from cervical cancer (HeLa)28 versus RCC4 cells derived from renal clear cell carcinoma, which harbours 3pLOH. Interestingly, no significant overlap was observed between genes that had lost H3K36 trimethylation and those exhibiting altered expression levels following SETD2 depletion, suggesting that SETD2-dependent trimethylation does not have a significant role in permitting transcription of genes neighbouring H3K36me3 sites in ccRCC cells. In contrast, there was a significant overlap between the genes deregulated following SETD2 siRNA in RCC4 cells and genes with differential expression in SETD2 mutant tumours versus non-mutant tumours in the TCGA ccRCC set, supporting the use of this approach to investigate the consequences of exclusive SETD2 loss (Supplementary Figure S8b). In conclusion, our data does not support a global transcriptional role for SETD2 in the system studied.
Interestingly, we find that in the absence of SETD2, chromatin compaction is impaired, nucleosome occupancy is aberrantly low and replication fork progression is slowed the latter phenomenon being shared by various conditions that evoke replication stress.18 The increased sensitivity to micrococcal nuclease in SETD2-depleted cells over normally trimethylated regions, and reduced chromatin association of histone H3 indicates that replication fork stability may be reduced by altered nucleosome reassembly (Figures 6c, e and 7).39 SPT16, a component of the FACT complex, previously demonstrated to interact directly with H3K36me3,28 has also been implicated in the recruitment of the MCM complex to chromatin during replication.32 DT40 cells with a conditional deletion of the FACT component SSRP1 exhibited S phase defects and impeded replication fork progression.33 In line with this, we observed that SETD2 depletion resulted in reduced chromatin levels of MCM7 and DNA polymerase δ as well as reduced replication fork progression and a prolonged S phase (Figures 4, 6 and 7), providing a potential explanation for the increased replication stress and DNA damage observed in tumours with reduced H3K36me3 levels.
Oncogene-induced replication stress is thought to occur widely across human malignancies,40 and replication stress has also been observed with the loss of tumour suppressors18, 41 As there are few described oncogenic mutations in renal cancer, loss of tumour suppressors may be particularly important in this tumour type. These studies suggest there may be a broader role for replication stress in the initiation of intratumour genetic heterogeneity, mediated through somatic events in distinct genes across different tumour types.
Our findings may have broader relevance to genetic heterogeneity across different malignancies, as SETD2 is recurrently mutated in 9% of non-small cell lung carcinomas,42 1 paediatric high-grade gliomas and 8% of adult high-grade gliomas,43 albeit usually in the absence of 3p LOH. In addition, the distribution of H3K36me3 is affected by the mutation of glycine-34 (G34V) on the histone variant H3.3, occurring in 60% of paediatric high-grade gliomas.39
Previously, we described that all recurrent somatic copy number aberrations in ccRCC other than 3pLOH are subclonal, present in some cells but not others in the branches of the tumour phylogenetic tree. We also found that SETD2 mutations often occur following the 3pLOH truncal event in the early branches of the tumour phylogenetic trees, coincident with the onset of tumour diversification.13
Consistent with the hypothesis that SETD2 loss-of-function contributes to branched evolution, in this study we find that H3K36me3 sites are enriched around breakpoint regions in SETD2 mutant tumours and SETD2 inactivation results in dysfunctional DNA replication and repair, fuelling subclonal diversification.
In summary, our study suggests that SETD2 mutations are not associated with MSI in renal cancer, and has revealed a relationship between nucleosome reassembly, replication stress, DNA repair by HR, chromosomal breakpoints and H3K36 trimethylation in ccRCC. This integrative approach may provide useful insights into the contribution of chromatin regulation to genomic instability more generally, by revealing how an altered histone mark unmasks fragile regions within the genome and, as a consequence, find order within chaotic cancer genomes.
Materials and methods
Cell lines
RCC4, A498 and U2OS cell lines used were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). RCC-FG2 and RCC-JW were purchased from Cell Lines Service (Eppelheim, Germany). UOK121 and UOK143 were a kind gift from Dr Linehan, Center for Cancer Research, NCI (Bethesda, MD, USA). All cell lines used in this study were maintained at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium with L-glutamine or RPMI 1640 media (Gibco/Life Technologies Ltd, Paisley, UK), supplemented with 10% fetal bovine serum and 1/10 000 units of penicillin–streptomycin (Sigma-Aldrich, Dorset, UK).
RNA interference
All siRNA (Dharmacon/GE Healthcare, Buckinghamshire, UK) transfections were performed at 40 nM final concentrations by reverse transfection with Lipofectamine RNAiMax (Invitrogen/Life Technologies Ltd, Paisley, UK). SETD2 (MU-012448) siRNA pool was made up of equal concentrations of siRNA 2 (5'-GCUCAGAGUUAACGUUUGA-3'), siRNA 3 (5'-GAAACCGUCUCCAGUCUGU-3') and siRNA 4 (5'-GGAGACAUUUGUAUGAGGA-3'). siRNA #1 was excluded in all experiments because of its nonspecific effects on other mitotic proteins (Supplementary Figure S4a). Additional siRNA oligos #7 (5'-GCUCAGAGUUAACGUUUGA-3') and #8 (5'-CCAAAGAUUCAGACAUAUA-3') were also used to deplete SETD2 levels. Non-targeting control siRNA was used as control in all experiments.
Protein extraction and western blotting
Total cell lysates were generated as described previously.18 Following sodium dodecyl sulphate–polyacrylamide gel electrophoresis, blots were probed with indicated antibodies diluted in 5% milk or bovine serum albumin in Tris-buffered saline. Antibodies used are as follows: SETD2 #ab31358, H3K36me3 #ab9050, total H3 #ab10799, RAD51 #ab213 (Abcam, Cambridge, UK). MAD2 #610678, LEDGF #611714 (BD Biosciences, San Diego, CA, USA), H2B #sc-8650, DNA polymerase δ, #sc-10784, MCM7, #sc-9966 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), horseradish peroxidase-conjugated anti-β-actin antibody #A3854 (Sigma-Aldrich) and horseradish peroxidase conjugated secondary antibodies (Dako, Cambridgeshire, UK). Immobilon Westerns (Millipore, Hertfordshire, UK) was used for detection.
MSI analyses
DNA was extracted according to manufacturer's protocols (DNeasy, Qiagen, Manchester, UK). The occurrence of microsatellite instability was investigated using seven markers (D2S123, D5S346, D5S652, D17S250, BAT25, BAT26 or D11S904).26, 44 Samples were defined as having MSI if ⩾2 microsatellite markers tested positive.24 PCR reactions were performed using two different DNA polymerase enzymes to exclude any enzyme-specific effects, and the products were subsequently subjected to fragment analysis using the ABI3130XL Genetic Analyzer (Life Technologies Ltd, Paisley, UK)
Thymidine block
Cells were treated with 2 mM thymidine for 18 h, washed with phosphate-buffered saline and released into complete media for 9 h. Cells were subsequently treated with a second thymidine block for a further 18 h. Cells were washed, released into complete media and harvested at the indicated time points.
DNA fibre assays
At 48-h posttransfection, cells were pulse labelled with CldU and IdU (Sigma-Aldrich) sequentially (20 min each). Assays were performed as described previously.18 Three independent experiments were performed and >200 double-labelled replication forks were counted per experiment.
Immunohistochemistry staining
For immunohistochemical analysis of archival formalin-fixed, paraffin-embedded human kidney tumour TMA, the tissue sections were de-paraffinized and processed for sensitive immunoperoxidase staining with the primary antibodies to SETD2 (Sigma-Aldrich HPA042451), H3K36me3 (Abcam ab9050) and phospho-histone H2AX Ser 139 (Millipore 05-636). The staining procedure was using the Vectastain Elite kit (Vector Laboratories, Peterborough, UK) and nickel sulphate enhancement without nuclear counterstaining, as described previously,45 followed by evaluation of the staining patterns by an experienced oncopathologist.
RAD51 foci formation assay
At 72-h post-siRNA transfection, cells were irradiated with 8 Gy IR (160 kV, 6 mA, 3 mm Alu filter), stained and scanned. For a detailed protocol, see Supplementary Information.
Colony formation assay
Cells were exposed to 0 (non-irradiated) or 3 Gy IR, then seeded at 500 cells per well and allowed to form colonies over 16 days, before quantification of colony numbers.
Patient tumour samples and tissue microarrays
For microsatellite analysis, we used DNA from ccRCC primary tumours (and corresponding normal tissue), as well as five MSI+ colorectal cell lines) available in the lab. All patients gave consent for use of their samples. Affymetrix SNP 6 array data were obtained for ccRCC (n=450) from TCGA Kidney ccRCC data set. Mutation data were obtained for ccRCC (n=403, resulting in 366 overlapping samples with both mutation and copy number data available) using the Broad GDAC firehose tool. ChIP-Seq data were retrieved from the Human Reference Epigenome Mapping Project (GEO accession: GSE19465), as part of the ENCODE Consortium.15, 16, 17 Tissue microarrays were provided by the Royal Marsden Hospital, London, UK (JL).46 The mutational loads and characterization of MSI (n=11) and MSS (n=217) colon tumours were retrieved using the Supplementary Information provided by a comprehensive analysis by TCGA.47 The relationship between SETD2 mutations and the mismatch repair deficiency mutational signature was assessed in the same cohort as Alexandrov et al.48
Micrococcal nuclease assays
At 72-h posttransfection, cells were harvested and processed for micrococcal nuclease sensitivity as described previously49 with minor modifications. Briefly, 100 μl of nuclei were incubated with 10 units of Micrococcal nuclease for 20 min at room temperature. An aliquot was analysed by real-time quantitative PCR. Primer sequences are available on request. The relative amount of Micrococcal nuclease-resistant DNA was calculated using the ▵▵Ct method. Values were further normalized to the relative amount of Micrococcal nuclease-resistant DNA in a non-trimethylated region.
Chromatin fractionation
Following trypsinization, cell pellets were washed in phosphate-buffered saline processed as detailed by others into nucleosolic and chromatin fractions.50
Chromatin immunoprecipitation
Chromatin immunoprecipitation was carried out on RCC4 cells as described previously5 using antibodies against H3K36me3 (ab9050, Abcam), RNA Pol II (sc-899, Santa Cruz) or rabbit immunoglobulin G at 4 °C overnight.
SNP array data normalization and copy number data analysis
The aroma R package (TumourBoost and CalMaTe)52, 53 was used to obtain logR and bi-allelic frequency values for all TCGA samples. All samples were analysed using hg19/Genome 37 coordinates. Sex chromosomes were excluded from the analysis. ASCAT (allele-specific copy number analysis of tumours) was run on all samples to obtain segmented allele-specific copy number profiles and ploidy.54 Segments consisting of <10 probes were discarded.
H3K36me3 ChIP-seq data for breakpoint analysis
ChIP-seq data were retrieved from the Human Reference Epigenome Mapping Project (GEO accession GSE19465). Samples GSM621634 and GSM773000 (adult kidney) were analysed using sample GSM621638 (adult kidney) as input control. Peaks were called using ChromaBlocks from the Repitools package,55 using ‘small' as preset and a FDR cut-off of 0.1. H3K36me3 sites were defined as the overlap between peaks from samples GSM621634 and GSM7733000. H3K27me3 sites were similarly derived from fetal kidney sample GSM621424 using sample GSM772733 as input control.
Differential binding analysis
ChIP-sequencing was carried out on the Illumina HiSeq 2500 platform and typically generated ~45 million 101- bp single-end reads per IP sample. Alignments were performed to the human genome (UCSC version hg19, University of California Santa Cruz Genomics Institute, Santa Cruz, CA, USA) using bwa (version 0.5.9-r16)56 permitting a maximum of three mismatches per read. Duplicate alignments were removed using the Picard MarkDuplicates program (picard-tools package version 1.81; http://picard.sourceforge.net) with default parameters. Differential binding analysis was performed against the Ensembl gene annotation (downloaded from UCSC on 15 April 2014) using edgeR (version 3.4.2)57 to assess the effect of SETD2 knockdown on H3K36me3 binding.
Statistics
For all experiments, the means of three experiments±s.e.m. were shown unless stated otherwise. Statistical significance of differences between means in immunofluorescence experiments was determined using Student's t-test (two-tailed). In immunohistochemistry analyses, statistical significance of association between different protein markers was computed using Cochran–Armitage test. DNA fibre assays were analysed using two-tailed unpaired t-test with Welch's correction.
Acknowledgments
We thank Dr Simon Boulton and our colleagues for critical discussions of this work. The results published here are in part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/ (dbGaP accession number: phs000178.v5.p5). We acknowledge the Encyclopedia of DNA Elements (ENCODE) Consortium, funded by the NHGRI, from which we obtained data for H3K36me3 and H3K27me3 analyses. We thank all funding boards for their continued support. The research leading to these results is supported by Cancer Research UK (XYG, RAB, EG, PM, PE, SG, C Santos, AJR, NM, PAB, AS and C Swanton), Breast Cancer Research Foundation (C Swanton and NK), Medical Research Council (ID: G0902275 to MG and C Santos; ID: G0701935/2 to AJR and C Swanton), the Danish Cancer Society (AMM, J Bartkova and J Bartek), the Lundbeck Foundation (R93-A8990 to J Bartek), the Ministry of the interior of the Czech Republic (grant VG20102014001 to MM and J Bartek), the National Program of Sustainability (grant LO1304 to MM and J Bartek), the Danish Council for Independent Research (grant DFF-1331-00262 to J Bartek), NIHR RMH/ICR Biomedical Research Centre for Cancer (JL), the EC Framework 7 (PREDICT 259303 to XYG, EG, PM, MG, TJ and C Swanton; DDResponse 259892 to J Bartek and J Bartkova and RESPONSIFY ID:259303 to C Swanton), UCL Overseas Research Scholarship (SG). C Swanton is also supported by the European Research Council, Rosetrees Trust and The Prostate Cancer Foundation. This research is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med 1996; 335: 865–875. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010; 463: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011; 469: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumour heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet 2012; 44: 17–19. [DOI] [PubMed] [Google Scholar]
- Larkin J, Goh XY, Vetter M, Pickering L, Swanton C. Epigenetic regulation in RCC: opportunities for therapeutic intervention? Nat Rev Urol 2012; 9: 147–155. [DOI] [PubMed] [Google Scholar]
- Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM et al. Histone methyltransferase gene SETD2 is a novel tumour suppressor gene in clear cell renal cell carcinoma. Cancer Res 2010; 70: 4287–4291. [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 2008; 27: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 2002; 277: 49383–49388. [DOI] [PubMed] [Google Scholar]
- Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA 2005; 102: 17636–17641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Birkbak NJ, Gerlinger M, McGranahan N, Burrell RA, Rowan AJ et al. Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol 2013; 230: 356–364. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014; 46: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 2014; 24: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004; 306: 636–640. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010; 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011; 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013; 494: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 2011; 13: 1161–1169. [DOI] [PubMed] [Google Scholar]
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet 2012; 8: e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sorensen CS et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol 2012; 19: 803–810. [DOI] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 2014; 21: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013; 153: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013; 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004; 96: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 2011; 18: 977–983. [DOI] [PubMed] [Google Scholar]
- Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J et al. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res 2013; 41: 2881–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 2012; 22: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J et al. Regulation of replication fork progression through histone supply and demand. Science 2007; 318: 1928–1931. [DOI] [PubMed] [Google Scholar]
- Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol 2014; 204: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J 2006; 25: 3975–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem 2011; 286: 30504–30512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 2014; 3: e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014; 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol 1999; 9: M57–M60. [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumourigenesis. Cancer Cell 2008; 14: 431–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prevent 2011; 20: 2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S et al. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 2013; 3: 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008; 319: 1352–1355. [DOI] [PubMed] [Google Scholar]
- Tort F, Bartkova J, Sehested M, Orntoft T, Lukas J, Bartek J. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumourigenesis. Cancer Res 2006; 66: 10258–10263. [DOI] [PubMed] [Google Scholar]
- Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012; 150: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 2013; 125: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altavilla G, Fassan M, Busatto G, Orsolan M, Giacomelli L. Microsatellite instability and hMLH1 and hMSH2 expression in renal tumours. Oncol Rep 2010; 24: 927–932. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K et al. DNA damage response as a candidate anti-cancer barrier in early human tumourigenesis. Nature 2005; 434: 864–870. [DOI] [PubMed] [Google Scholar]
- Suwaki N, Vanhecke E, Atkins KM, Graf M, Swabey K, Huang P et al. A HIF-regulated VHL-PTP1B-Src signaling axis identifies a therapeutic target in renal cell carcinoma. Sci Transl Med 2011; 3: 85ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. Micrococcal nuclease analysis of chromatin structure. Curr Prot Mol Biol 2005, Edited by Frederick M Ausubel [et al.]. Chapter 21: Unit 21.1. [DOI] [PubMed]
- Young DB, Jonnalagadda J, Gatei M, Jans DA, Meyn S, Khanna KK. Identification of domains of ataxia-telangiectasia mutated required for nuclear localization and chromatin association. J Biol Chem 2005; 280: 27587–27594. [DOI] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 2014; 28: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H, Neuvial P, Speed TP. TumourBoost: normalization of allele-specific tumour copy numbers from a single pair of tumour-normal genotyping microarrays. BMC Bioinform 2010; 11: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Estevez M, Aramburu A, Bengtsson H, Neuvial P, Rubio A. CalMaTe: a method and software to improve allele-specific copy number of SNP arrays for downstream segmentation. Bioinformatics 2012; 28: 1793–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W et al. Allele-specific copy number analysis of tumours. Proc Natl Acad Sci USA 2010; 107: 16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statham AL, Strbenac D, Coolen MW, Stirzaker C, Clark SJ, Robinson MD. Repitools: an R package for the analysis of enrichment-based epigenomic data. Bioinformatics 2010; 26: 1662–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 2014; 15: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.