Abstract
Investigating the susceptibility of oestrogen receptor-positive (ERpos) normal human breast epithelial cells (HBECs) for clinical purposes or basic research awaits a proficient cell-based assay. Here we set out to identify markers for isolating ERpos cells and to expand what appear to be post-mitotic primary cells into exponentially growing cultures. We report a robust technique for isolating ERpos HBECs from reduction mammoplasties by FACS using two cell surface markers, CD166 and CD117, and an intracellular cytokeratin marker, Ks20.8, for further tracking single cells in culture. We show that ERpos HBECs are released from growth restraint by small molecule inhibitors of TGFβ signalling, and that growth is augmented further in response to oestrogen. Importantly, ER signalling is functionally active in ERpos cells in extended culture. These findings open a new avenue of experimentation with normal ERpos HBECs and provide a basis for understanding the evolution of human breast cancer.
 Culturing normal primary breast cells that express the oestrogen receptor is difficult. Here, the authors isolate oestrogen receptor positive normal breast cells using the cell surface markers CD166 and CD117, and show that the cultures can be repeatedly passaged and retain oestrogen receptor protein expression.
Culturing normal primary breast cells that express the oestrogen receptor is difficult. Here, the authors isolate oestrogen receptor positive normal breast cells using the cell surface markers CD166 and CD117, and show that the cultures can be repeatedly passaged and retain oestrogen receptor protein expression.
Understanding the taxonomy and evolution of breast cancer has always relied heavily on the use of normal cell types as reference (for review see ref. 1). Nevertheless, ever since the first protocol for cultivation of normal human breast epithelial cells appeared three decades ago2, it has become increasingly clear that there are no protocols that support propagation of oestrogen receptor-positive (ERpos) cells. Thus, along with the appreciation of epithelial cell lineages in the human breast, primarily the luminal lineage and the basal/myoepithelial lineage, it became evident that the fastest growing cells in culture are of basal origin3,4. Moreover, when it was revealed that ERpos cells in situ accounted for an average of about 7% (mean 6.6%, ranging from 1.2 to 19.1% in a series of 15 normal breast samples) of the cells within the luminal epithelial lineage5, the chances of recovering these cells in culture without prospective isolation would in many cases be elusive. Thus, in culture medium that allowed luminal cells to be maintained after passaging, endogenous ER expression disappeared6,7. Likewise, even when employing freshly isolated small pieces of breast tissue, including the surrounding stroma thus preserving tissue architecture, steroid receptor expression is eventually lost8,9. As a consequence of this, the comparison of cancer with ‘normal', for example, the HMT-3522, MCF10A and 184B5 cell lines1,6,10, in cell-based assays has relied on normal cells lacking ER expression.
In an attempt to overcome the loss of receptor expression, ER has been ectopically introduced into such cell lines. This approach, however, has had a number of shortcomings, for example, instead of responding to oestrogen by increased proliferation as expected, the ER-transfected cells under standard culture conditions show growth inhibition11,12. Accordingly, most of our current knowledge of ER expression, regulation and action comes from breast carcinoma cell lines, whose relation to ERpos normal breast cells at best remains speculative.
Here we first identify the ERpos cells in situ and search for markers that allow their subsequent tracking in culture. We then describe culture conditions for primary ERpos cells in the presence of small-molecule inhibitors of transforming growth factor-beta (TGFβ) signalling. Importantly, these conditions also yield ERpos cells from luminal ERneg progenitors but not from basal cells. We envision that the present protocol will serve to bridge the existing gap of knowledge between normal human breast, which contain a small pool of ERpos cells and the overwhelming ER expression found in the majority of breast cancers.
Results
Identification and isolation of normal ERpos HBECs
To answer the long-standing question of whether loss of hormone receptors in culture is due to the loss of cells or the loss of receptor protein expression, it was necessary first to provide tools for cell tracking and sorting of the relevant cells. To unequivocally track ERpos HBECs at the single-cell level we screened our antibody library for surrogate markers with a long half-life, for example, cytokeratins13, in culture. In situ staining of more than 30 reduction mammoplasties revealed a surprising pattern with a monoclonal antibody (clone Ks.20.8) originally raised against cytokeratin 20, a simple epithelial cytokeratin with a very restricted expression pattern and not expressed in normal human breast14. The lack of true cytokeratin 20 expression in normal breast was here confirmed with two specific antibodies (listed in Table 1). Instead, Ks20.8 stained a subpopulation of luminal cells in a unique scattered pattern (Fig. 1a). Ks20.8 antibodies from four different suppliers (Table 1) revealed similar staining patterns (Supplementary Fig. 1a). The characteristic staining pattern led us to speculate that it indeed represented ERpos cells. While ER and Ks20.8 apparently co-localized in acini as well as in ducts (Supplementary Fig. 1b), the immunofluorescence staining was not sufficiently strong to allow quantification. For this purpose, it was necessary to apply dual antibody immunofluorescence. Since ER and progesterone receptor (PR) are expressed coordinately in essentially the same cells in the normal human breast15, we decided to enhance immunofluorescence staining of uncultured cells by mixing antibodies for ER and PR. This approach revealed that the scattered staining with Ks20.8 encompassed almost the entire population of hormone receptor-positive HBECs in acini as well as in ducts (Fig. 1b) with the most evident co-localization in foci with the strongest receptor expression (in four out of eight biopsies tested). The antibody screen unravelled a number of additional markers of Ks20.8pos HBECs some of which are well-known markers of ERpos cells, including activating enhancer-binding protein 2 beta (AP2β), a marker of luminal differentiation, GATA3 (ref. 16), a marker of cell survival/longevity, Bcl2, two TGFβ-mediated, epithelial–mesenchymal transition-related markers, N-glycan/CDw75 antigen17 and N-cadherin (reviewed in ref. 18), as well as the stem cell markers, ALCAM (CD166) (ref. 19) and the laminin receptor, 67LR (ref. 20; Supplementary Fig. 2).
Table 1. List of antibodies used for immunostaining and/or FACS analysis.
| Antibody | Clone | Company/Catalogue No. | Peroxidase | Fluorescence | FACS |
|---|---|---|---|---|---|
| AP2β | — | Santa Cruz, sc-8976 | — | 1:50 | — |
| BCL2 | 124 | Dako, M0887 | — | 1:25 | — |
| CDw75 | LN1 | NeoMarkers, MS-130-P | — | 1:10 | — |
| CD117 | 104D2 | Dako, M7140 | — | 1:50 | — |
| CD117 | K44.2 | Sigma, K0131 | — | 1:50 | — |
| CD117, PE | 104D2 | BD Biosciences, 332785 | — | — | 1:20 |
| CD166 | 3A6 | BioLegend, 343902 | — | 1:50 | — |
| CD166, Alexa Fluor 488 | 3A6 | AbD Serotec, MCA1926A488 | — | — | 1:20 |
| CD271, APC | ME20.4 | Cedarlane, CL10013APC | — | — | 1:50 |
| CD326, PerCP/Cy5.5 | 9C4 | BioLegend, 324214 | — | — | 1:10–1:20 |
| TGFβRII | MM0056–4F14 | Abcam, ab78419 | — | — | 1:20 |
| ERα | SP1 | Labvision, RM-9101-S | 1:25 | 1:10 | — |
| ERα | SP1 | Labvision, RM-9101-R7 | Prediluted, ready to use | — | — |
| ERα | 1D5 | Dako, M7047 | 1:100 | 1:25 | — |
| GATA3 | HG3–31 | Santa Cruz, sc-268 | — | 1:25 | — |
| Keratin 8 | M20 | Abcam, ab9023 | 1:50 | 1:50 | — |
| Keratin 8 | TS1 | Novocastra, NCL-CK8-TS1 | 1:50 | — | — |
| Keratin 20* | Ks20.8 | Dako, M7019 | 1:25 | 1:10 | — |
| Keratin 20* | Ks20.8 | Dako, IR777 | Prediluted, ready to use | — | — |
| Keratin 20* | Ks20.8 | Abcam, ab854 | 1:10 | — | — |
| Keratin 20* | Ks20.8 | Genemed, 61–0018–2 | 1:25 | — | — |
| Keratin 20* | Ks20.8 | Santa Cruz, SC-52320 | 1:25 | — | — |
| Keratin 20 | IT-Ks20.10 | Progen Biotechnik, 61054 | 1:25 | — | — |
| Keratin 20 | CK205 | Novocastra, NCL-CK20–543 | 1:50 | — | — |
| Keratin 8/18 | NCL-5D3 | Abcam, ab90102 | — | 1:50 | — |
| Keratin 14 | LL002 | NeoMarkers, MS-115-P | 1:300 | 1:25 | — |
| Keratin 15 | LHK15 | NeoMarkers, MS-1068-P | — | 1:25 | — |
| Keratin 18 | M9 | Monosan, MON3006 | 1:100 | — | — |
| N-cadherin | 32 | BD Transduction Laboratories, 610920 | — | 1:25 | — |
| PR | SAN27 | Vector Laboratories, VP-P987 | 1:100 | 1:25 | — |
| PR | SP2 | Labvision, RM-9102-S | — | 1:10 | — |
| PR | PgR 636 | Dako, M356901–2 | 1:50 | 1:10 | |
| 67-kDa laminin receptor | MLuC5 | Abcam, ab3099 | — | 1:50 | 1:50 |
*Originally described to recognize cytokeratin 20, but keratin 20 is not expressed in normal human breast14.
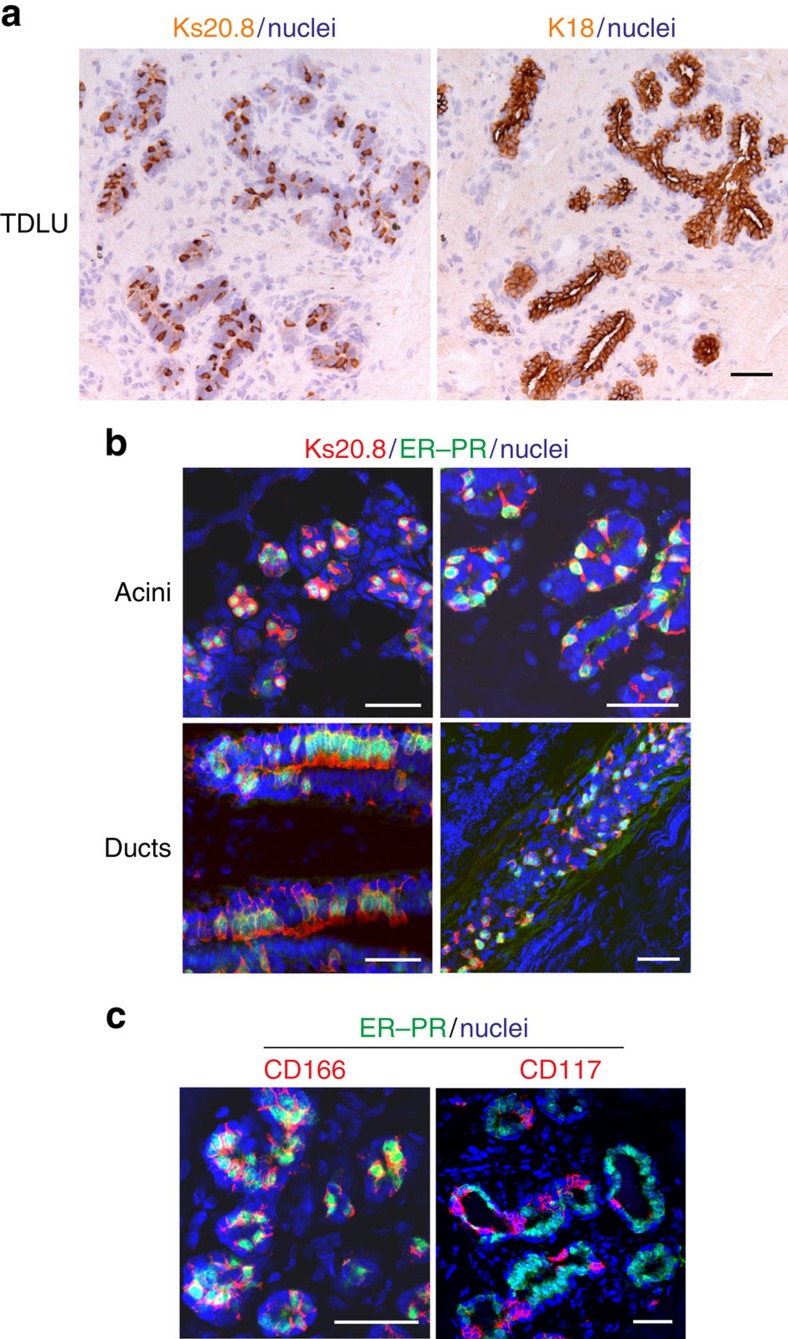
Figure 1. A unique staining signature Ks20.8pos/CD166high/CD117low is eligible for ERpos cell sorting and tracking.
(a) Serial cryostat sections of a normal human breast terminal duct lobular unit (TDLU) stained with immunoperoxidase against Ks20.8 (left) and K18 (right), and counterstained with haematoxylin. Note the characteristic scattered staining pattern with Ks20.8 against the uniform lineage-related staining of luminal cells with K18. Scale bar, 50 μm. (b) Multicolour imaging of normal human breast cryostat sections including acini and ducts from four different biopsies stained for Ks20.8 (red), hormone receptors ER–PR (SP1–SP2, green) and DAPI nuclear stain (blue). The Ks20.8pos compartment encompasses the ER–PR-expressing cells. Scale bar, 50 μm. (c) Multicolour imaging of normal human breast cryostat sections stained for CD166 (3A6, red), CD117 (K44.2, red), hormone receptors ER–PR (green) and DAPI stain (blue). While CD166 is highly expressed in hormone receptor-positive cells, CD117 shows the complementary pattern. Scale bar, 50 μm.
For cell sorting purposes we found that CD166 and CD117 made good candidate surface markers of potential ERpos and ERneg cells, respectively—again as revealed by enhanced multicolour immunofluorescence (Fig. 1c). The mutual exclusivity of CD117 and ER (Fig. 1c) confirms what has been reported by others21. Accordingly, we designed a fluorescence-activated cell sorting (FACS) protocol to first separate the basal cell population from the luminal cell population based on EpCAM (CD326) and NGFR (CD271) followed by sorting with CD166 and CD117 to further dissect the luminal compartment (Fig. 2a and Supplementary Fig. 3). This protocol yields three populations, the purity of which was assessed by staining smears with lineage and progenitor markers K14, K18 and K15 (ref. 22), ER–PR as well as the novel ERpos cell surrogate marker Ks20.8 (Fig. 2a). As expected, we found that ERpos/PRpos HBECs were highly enriched in the CD166high/CD117low gate (Fig. 2b). While immunofluorescence staining for ER alone evoked a cytoplasmic background staining, which prevented reliable quantification, the smears turned out to be ideal for accurate assessment of the level of co-localization of ER–PR and Ks20.8. Indeed, up to 90% of Ks20.8pos cells were also ER–PRpos (Supplementary Fig. 4) and up to 87% of ER–PRpos cells were Ks20.8pos. The separation of the three subpopulations was further validated by quantitative reverse transcription–PCR (qRT–PCR), which confirmed a high ER expression (ESR1) in the CD166high cells compared with the other subpopulations (Fig. 2c). An additional panel of markers further distinguished the two luminal subpopulations from the basal cell population (Supplementary Fig. 5). Importantly, we found that known ER signalling-related genes such as trefoil factor family-1 (TFF1; ref. 23) and growth regulation by oestrogen in breast cancer 1 (GREB1; ref. 24) were highly expressed in CD166high cells as compared with other HBECs (Supplementary Fig. 5). The degree of separation in the CD166/CD117 FACS analysis was, however, somewhat biopsy dependent. In a series of six biopsies originating from women between 19 and 44 years old, five exhibited a similar separation with 11–49% of the cells being CD166high/CD117low, while one biopsy apparently did not contain a CD117high population (Supplementary Fig. 6). As an alternative, CD117 could be replaced with the laminin receptor 67LR in the CD166 FACS to obtain enriched ERpos HBECs (80% increase in Ks20.8-positive cells in the 67LRhigh gate versus the 67LRlow gate; Supplementary Fig. 7). In conclusion, the combination of Ks20.8, CD166 and CD117 is a promising marker for ERpos HBEC tracking and sorting.
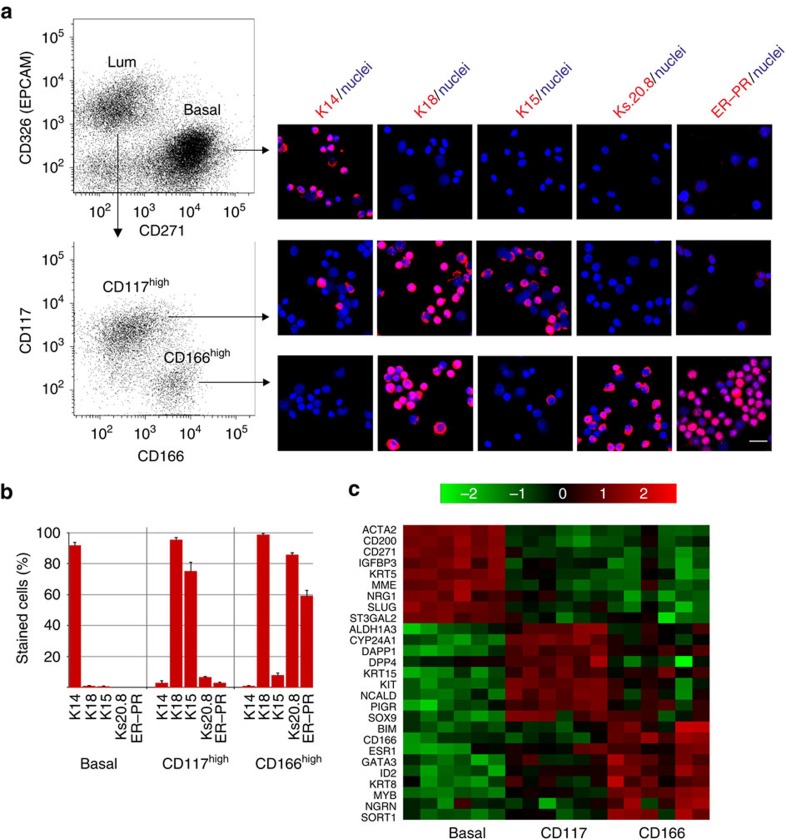
Figure 2. ERpos cells are purified and tracked by sequential CD326/CD271–CD166/CD117 FACS followed by multicolour staining and qRT–PCR.
(a) Multicolour flow cytometry of uncultured HBECs incubated with CD326/CD271/CD166/CD117 and visualized pairwise (left diagrams) to recover luminal cells (CD326high) and basal cells (CD271high) and from the luminal gate CD166high and CD117high cells. Smears of sorted cells were stained (right panel) with either of the markers against basal cells, cytokeratin K14; luminal cells, cytokeratin K18; luminal progenitors, cytokeratin K15; Ks20.8 or ER–PR and counterstained with DAPI nuclear stain. Hormone receptor-positive cells are observed primarily among CD166high cells. Scale bar, 50 μm. (b) Purity of sorted cells as determined by staining of smears followed by quantification of the percentage of cells stained with either of the markers cytokeratin K14, K18, K15, Ks20.8 or hormone receptors (ER–PR; 3 × 100 cells per slide, error bars indicate s.d.'s). (c) Heatmap representing qRT–PCR analysis of the relative gene expression of lineage markers in sorted basal cells (basal), CD117high luminal cells (CD117) and CD166high luminal cells (CD166) from six different biopsies. Data confirm lineage-specific transcriptional profiles of the three cell populations and restricts ER expression (ESR1) primarily to CD166high luminal cells. Colour bar indicates the fold difference of the relative gene expression in log2 scale.
ER protein is lost in culture
With the surrogate markers in hand we were now able to examine the behaviour of sorted ERpos HBECs in culture, potentially irrespective of ER expression. From the point of view that favourable conditions for luminal epithelial cells also apply to ERpos HBECs, we refined a protocol to permit ample colony formation of luminal cells at clonal density. As cell culture plastic we used Primaria, a substrate with a high content of nitrogen previously shown to promote adhesion of breast cells25. The growth medium ‘FAD2' was modified from previously described media for culturing keratinocytes or basal breast epithelium on mouse fibroblast feeders26,27. The basal medium is Ham's F12:DMEM (1:3) similar to that of Tan et al.27, but with less serum, that is, 5%, as in Liu et al.26 Under these conditions, we gauged for colony formation among the three FACS gated populations described above. When plated at a clonal density of 400 cells per cm2, indeed colony forming luminal cells from the CD117high gate were highly favoured over basal cells (Fig. 3a). However, it was also clear that the ERpos HBECs from the CD166high gate entirely failed to form colonies under similar conditions. However, a scrutiny of cultures at a higher magnification revealed that CD166high cells in fact did not disappear from the culture. Rather, they plated, survived and also stained with Ks20.8 as a testimony of their original identity, but they entirely refrained from growth and rapidly lost the ER protein (Fig. 3b). By comparison, other HBEC culture media, that is, M87A (ref. 28), MEGM or WIT-P-NC did not support plating of CD166high cells. Therefore, we conclude that failure of culturing ERpos HBECs is caused by both lack of growth and loss of ER expression under conditions otherwise favouring propagation of luminal epithelial cells.
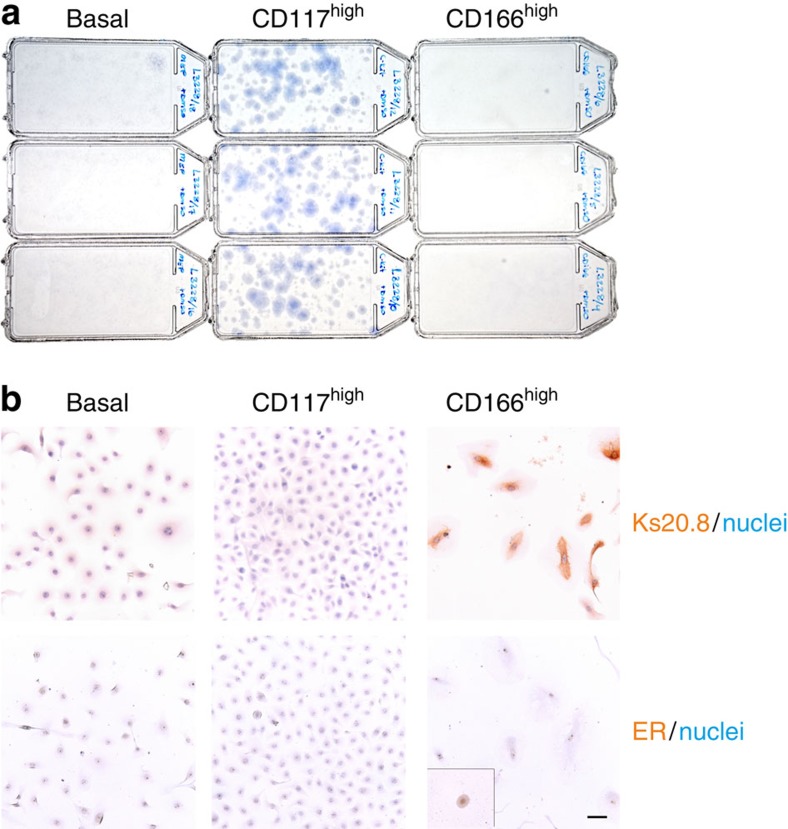
Figure 3. Loss of ERpos cells in culture is due to both lack of growth and down modulation of ER expression.
(a) T25 flasks in triplicate of basal cells (basal), CD117 high and CD166high ERpos luminal cells plated at a clonal density of 104 cells per flask (400 cells per cm2) and stained with haematoxylin after 14 days in culture. Note that only CD117high cells are colony forming at clonal density. (b) A higher magnification of cultures started at a higher cell density (3,000 cells per cm2) and immunoperoxidase stained at day 9 for Ks20.8 (upper row) or ER (SP1 prediluted, lower row; inset shows positive staining for ER at day 1 after plating) and counterstained with haematoxylin. Whereas ER expression is rapidly lost, the non-colony-forming cells of the CD166high cultures remain Ks20.8 positive and traceable in culture. Scale bar, 50 μm.
TGFβ inhibitors induce ER expression and release ERpos HBECs
We noted that earlier in vivo studies had implicated TGFβ1 signalling in the restraint of ERpos mammary epithelial cells29,30. Here we therefore examined three small inhibitor molecules of TGFβ signalling, RepSox, SB431542 and SD208, and combinations hereof for their ability to relieve a potentially negative regulation of ERpos HBEC growth in culture. We found that dual inhibition with SB431542 and RepSox, hereafter collectively termed TGFβR2i, recapitulated ER expression and stimulated ERpos HBEC colony formation in four out of four biopsies. Moreover, the cells maintained Ks20.8 reactivity, albeit more widespread than ER expression compared with what is seen in situ (Fig. 4a). ERpos cells from three different biopsies in low-density primary cultures in TGFβR2i (4,000 cells per cm2) exhibited a clonal capacity of 0.54%, 0.33% and 0.43%, respectively. By comparison, control conditions resulted in small, mostly abortive ERneg clones (Fig. 4b). In general, ER expression was particularly evident in the dense centre of proliferating colonies or at confluency. Two different sources of SP1 antibody gave similar results (Table 1). Moreover, staining was confirmed with the less-sensitive ER antibody 1D5 (ref. 31). The response to TGFβR inhibitors was reproducibly observed in 10 out of 10 biopsies.
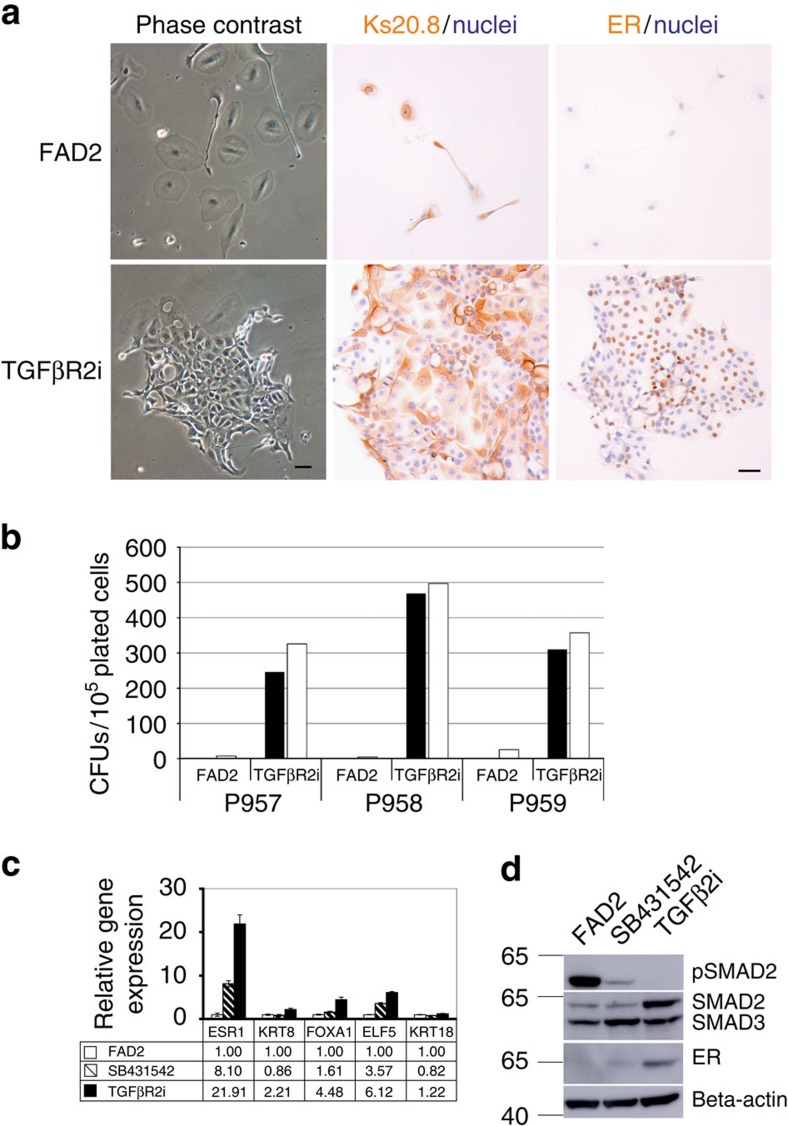
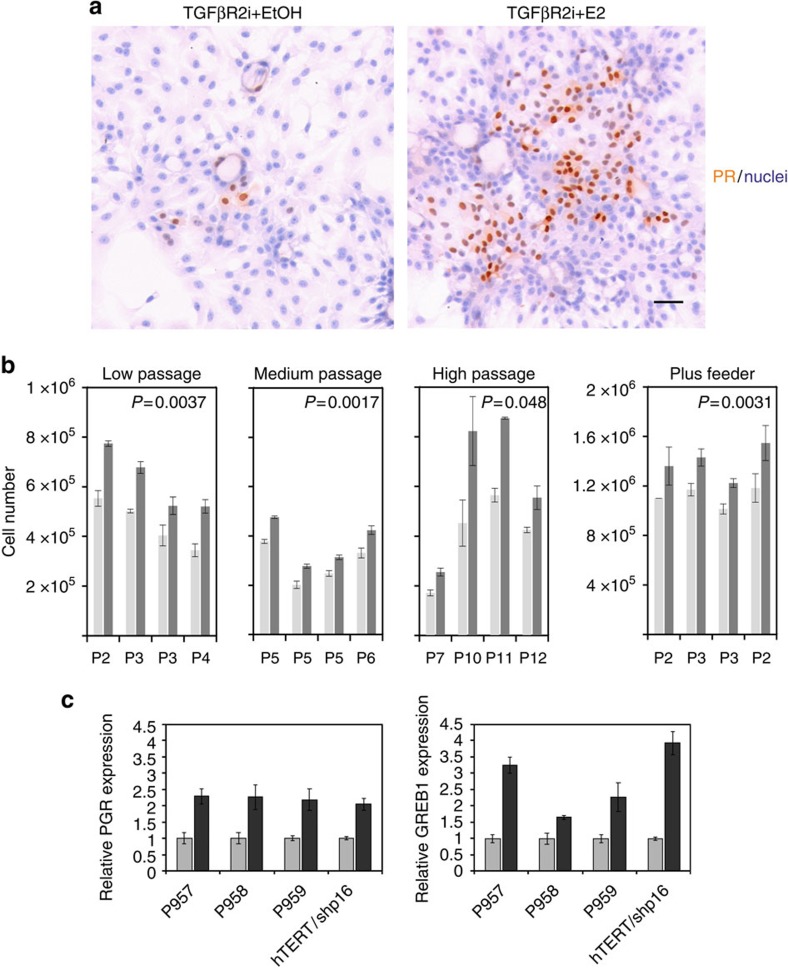
Figure 4. Relief of TGFβ-mediated negative regulation of growth releases ERpos cells from quiescence.
(a) Phase contrast micrographs (left column) after 8 days in culture and immunoperoxidase staining with Ks20.8 (middle column) and ER (SP1 prediluted, right column) of primary cultures of CD166high-derived cells in FAD2 (upper panel) or TGFβR2i (lower panel). Nuclei are counterstained with haematoxylin. Whereas Ks20.8pos cells remain quiescent and ER negative on FAD2, they are colony forming and ER positive in TGFβR2i. Scale bar, 50 μm. (b) Quantification at day 13 of ERpos (closed bar) or Ks20.8pos (open bar) colony-forming units (CFUs) derived from CD166high/CD117low cells from three consecutive biopsies (p957, p958 and p959) in FAD2 or TGFβR2i. In all three cases TGFβR2i supplied throughout 9–11 days supported colony formation of Ks20.8pos/ERpos cells. (c) qRT-PCR of ER (ESR1), K8 (KRT8), FOXA1, ELF5 and K18 (KRT18) of RNA extracted from second-passage cells cultured for 5 days in FAD2 (open bars), in FAD2 with SB421543 (shaded bars) and in TGFβR2i (solid bars), respectively. Note the collective upregulation of ER and ER-associated gene expression in TGFβR2i. Error bars indicate s.d. of three technical triplicates. (d) Western blotting of proteins extracted at day 6 from second-passage CD166high/CD117lowcells seeded at 4,000 cells per cm2 all cultured without EGF in FAD2 (left lane), in FAD2 with SB421543 (middle lane) and in TGFβR2i (right lane), respectively, incubated with antibodies recognizing phosphorylated SMAD2 (upper panel), SMAD2/3 (second panel), ER (third panel) and β-actin (lower panel). TGFβR2i inhibits pSMAD2 and upregulates ER protein expression.
In addition to ER, TGFβR2i induced the expression of keratin K8 and the luminal cell transcription factors forkhead box protein A1, FOXA1 and E74-like factor 5, ELF5 (for reviews, see refs 32, 33) as revealed at the transcriptional level (Fig. 4c). This expression pattern was confirmed using another biopsy. TGFβR2i also upregulated the transcription of a number of genes known to be downstream targets of ER or modulators of ER activity, such as TFF1 (ref. 23) and insulin growth factor-binding protein 5 (IGFBP5; ref. 34; Supplementary Fig. 8a). Moreover, induction of ER protein expression to a significant degree was specific to RepSox as twice the concentration of SB431542 or replacement of RepSox with another ALK5 kinase inhibitor SD208 was insufficient to induce K8 and ER (data not shown). RepSox alone was not as effective in inducing ER or PR as in combination with SB431542 (Supplementary Fig. 8b). Likewise, while SB431542 alone was capable of inducing increased expression of ER at the mRNA level (Fig. 4c), this translated to the protein level to a significant degree only in the presence of RepSox (Fig. 4d and Supplementary Fig. 9). These findings suggest that ER expression is controlled through specific inhibition of TGFβ signalling. The presence of TGFβR in HBECs as well as the inhibition of phosphorylated SMAD2 concurrent with inhibitor-induced ER expression supported this (Supplementary Fig. 10 and Fig. 4d). Indeed, TGFβR2i culture was a key to sustained ER protein expression as removal of TGFβR2i led to complete loss of ER protein expression as observed in four independent experiments. Thus, in a representative experiment the frequency of ERpos cells was reduced from 33.7% (s.d.±5.0%; 3 × 100 cells) in continuous TGFβR2i culture to 10.7% (s.d.±0.5%) after 3 days and to 0% after 5 days without TGFβR2i.
We next tested whether TGFβR2i would induce de novo ER expression in other subpopulations of luminal cells, for example, in the much more frequent CD117high-derived HBECs. Consistent with the presumed progenitor status of CD117high cells in the human breast35, clones emerged from CD117high cells that gained ER and stained positively for Ks20.8. This was, however, the only additional source we found of ERpos HBECs, since TGFβR2i failed to induce ER in basal cells, in breast fibroblasts or in the established normal cell breast lines MCF10A or HMT-3522. Thus, both from preexisting ERpos HBECs and from ERneg progenitors, TGFβR2i readily provides growing colonies of ERpos HBECs.
TGFβR2i allows growth of ERpos HBECs in serial subculture
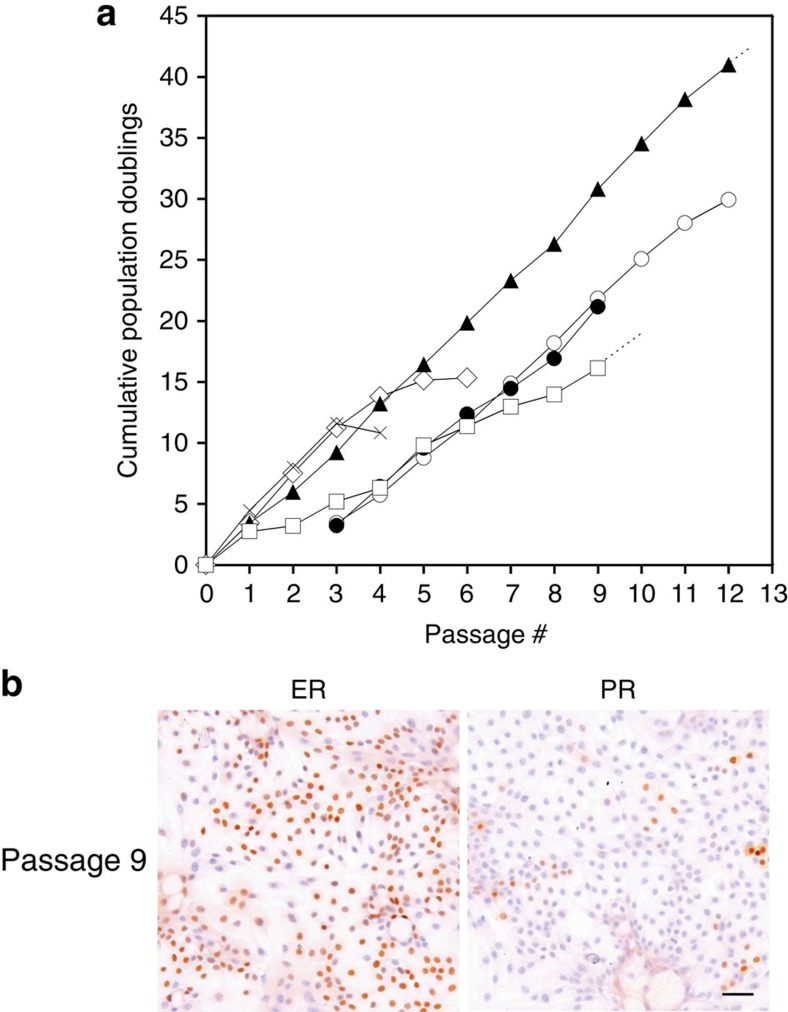
TGFβR2i appeared to support growth of ERpos HBECs also after passaging. To test this systematically, we plated CD166high-derived cells at a density of 6,400 cells per cm2 in primary culture, and the cultures were subsequently passaged at a density of 4,000 cells per cm2. On passaging, ER expression was particularly evident in the dense centre of proliferating colonies. Continuous proliferation under these conditions was maintained for up to six passages, corresponding to 15 population doublings (exemplified in Fig. 5a). Importantly, however, in the absence of RepSox, the cells could not be expanded beyond fourth passage (Fig. 5a). The lifespan and ER expression (ranging from 21 to 77% in second- to fourth-passage cultures, Table 2) were somewhat biopsy dependent, and in general proliferation slowed between fourth and sixth passages, and at the same time downregulation of ER expression was observed. While this narrowed the window of experimentation to up to fourth passage, the cells could easily and reproducibly be replaced with new cultures with similar luminal characteristics (Table 2). However, passage number could be increased by increasing seeding density. Thus, ERpos cells derived from three biopsies could be cultured for another two to three passages, that is, up to passage six to nine when passaged at a high density of 8,000–20,000 cells per cm2. Moreover, the lifespan of ERpos HBECs passaged at low seeding density could be further extended by initial plating on 3T3 feeders, which lead to proliferation for more than 10 passages, corresponding to more than 25 population doublings (Fig. 5a). Cells from a different sorting exhibited similar characteristics (Fig. 5a). We subsequently addressed whether TGFβR2i culture would allow for alternative approaches to extend the lifespan of normal breast-derived ERpos cells. ERpos HBECs were successfully transduced with pBABE-neo-hTERT and pLenti X2 hygro/shp16 constructs and have now been cultured for 4 months with weekly passages at 6,000 cells per cm2, exceeding 12 passages (Fig. 5a). Transduction of ERpos cells from a different biopsy and subsequent splitting at up to 1:4 supported the robustness of this approach. Of note, these long-term cultured cells exhibited a phenotype essentially similar to cultures with definitive lifespan, including a relatively high level of ER expression (Supplementary Fig. 11 and Table 2). Thus, while the proliferation of ERpos cells with definite lifespan is somewhat slow as it took more than 100 days to generate more than 25 population doublings, the present protocol nevertheless allows a considerable expansion of the ERpos cell population extending from low-passage cultures with low seeding density through medium-passage cultures with high seeding density to high-passage cultures of hTERT/shp16-tranduced cells, which provides a relatively wide window of experimentation. Of note, until senescence the cells maintain their expression of ER, Ks20.8 reactivity as well as—mainly in densely packed colonies—expression of PR (Fig. 5b). In conclusion, TGFβR2i readily supports serial subculture of a population of ERpos HBECs, that is, ERpos progenitors.
Figure 5. TGFβR2i allows efficient expansion of ERpos cells.
(a) Population doublings as a function of passage number calculated by continuous cell number recordings in triplicate cultures before confluency and plating at a fixed number of 4,000 cells per cm2 per flask at each split. TGFβR2i allows proliferation for up to six passages, corresponding to 15 population doublings (open diamond). If RepSox is omitted, the cells cannot be expanded beyond fourth passage (cross). Initial plating on 3T3 feeders with quantification starting in passage three extends proliferation to more than 10 passages, corresponding to more than 25 population doublings (open circle). Cells from a different sorting (albeit followed for a shorter period) exhibit similar extended proliferative capacity (closed circle). hTERT/shp16-transduced CD166high/CD117low cells subsequently passaged at a fixed number of 6,000 cells per cm2 at each split extended the proliferative capacity even further (closed triangle), and the cells have now been growing for more than 12 passages. hTERT/shp16-transduced CD166high/CD117low cells derived from a different biopsy, split at a ratio of up to 1:4, has so far been growing up to passage 9 (open square). (b) Even beyond 20 population doublings (passage 9), ERpos cells with definitive lifespan maintain ER and PR expression as shown by immunoperoxidase and haematoxylin staining (cells in ninth passage seeded at 4,000 cells per cm2 and stained at day 5 with SP1 prediluted and SAN27, respectively). Scale bar, 50 μm.
Table 2. Frequency of ERpos and lineage marker-positive cells as a function of passage number in culture.
| Passage # | Per cent ER+ cells | Per cent K8+ cells | Per cent K19+ cells | Per cent P63+ cells |
|---|---|---|---|---|
| 2 | 41/21/24/38/42 | 100/100/100/ND/100 | 44/47/100/ND/60 | 0*/5/0/ND/0* |
| 3 | 76†/47/22/50‡/26 | 100†/100/100/100‡/100 | 100†/41/62/100‡/76 | 0†/2/0/0‡/0 |
| 4 | 77†/26†/28/51†/23 | 100†/100†/100/100†/100 | 100†/79†/34/100†/76 | 0†/0†/0/0†/0 |
| 5 | 29†/72†/39/38/70/37 | 100†/100†/100/ND/ND/100 | 79†/100†/48/ND/29/99 | 0†/0*,†/0*/ND/0*/6 |
| 6 | 66†/27/42/22 | 100†/100/ND/97 | 100†/60/ND/69 | 0†/0*/ND/8 |
| 7 | 54†/28/30/25/24/45† | 100†/100/ND/100/96/ND† | 100†/69/ND/63/94/ND† | 0†/0*/ND/3/6/10† |
| 8 | 57†/22/34/18/45† | 100†/100/100/100/ND† | 100†/78/100/55/85† | 0†/0/0/0/12† |
| 9 | 52†/19/28/87 | 100†/100/100/100§ | 100†/100/100/14 | 0†/0/0/0 |
| 10 | 19 | ND | ND | ND |
ND, not done.
The numbers in black (roman font; not bold, italic or underlined entries) indicate low-density split, ∼4,000 cells per cm2.
Bold entries indicate high-density split, ∼8,000–20,000 cells per cm2.
Italic entries denote cells with extended lifespan on initial cultivation in BBMYAB and mouse feeders.
The entries that are underlined denote hTERT/shp16-transduced cells.
The percentage of ERpos and lineage marker-positive cells evaluated after immunoperoxidase staining of cultures and counterstained with haematoxylin. Aside from staining for ER (SP1, prediluted), the lineage markers included luminal keratin K8 (stained by M20 or TS1) and keratin K19, and myoepithelial P63. Each number refers to the frequency of stained cells among a total of 1,000 cells in randomly selected fields in a culture representing one biopsy. Cells were counted by use of a × 25 objective and a × 10 ocular equipped with a grid. Note that cultures remain essentially ERpos and luminal like. At the onset of senescence (starting at passage 4 in low-density split cultures and up to passage 6–9 in high-density split cultures) there is a tendency for the ER staining to fade out. High-density cultures from two biopsies maintained a high ER expression on omission of EGF and addition of E2.
*Very few, small foci or individual cells.
†EGF omitted, E2 added.
‡+E2 in passage 1.
§Stained in passage 11.
ERpos progenitors respond to oestrogen
As an ultimate test for a physiologically relevant ER expression we decided to assess the effects of added cognate ligand, that is, oestrogen (β-oestradiol). Accordingly, ERpos cells were exposed to 10−8 M of β-oestradiol or vehicle. The effect of oestrogen was tested in the absence of epidermal growth factor (EGF) since the transcriptional activation of ER can be induced by EGF-induced MAP kinase activity36. As a first indication of a functional ER, a higher focal expression of PR protein, a downstream target of ER signalling, was seen in the presence of oestrogen (Fig. 6a). Low-passage cultures (passage 2–4) were exposed to oestrogen or vehicle, and the cell number was quantified. As seen in Fig. 6b, significantly higher cell numbers were recorded in cultures with oestrogen (Fig. 6b). Medium-passage cultures (passage 5–6) reached by high seeding density exhibited a similar response, albeit in general exhibiting a slower growth (Fig. 6b). Likewise, a proliferative response was observed in high-passage culture of hTERT/shp16-transduced ERpos cells (passage 7–12; Fig. 6b). That the observed increase in cell number was indeed due to the specific effect of signalling through ER was verified on addition of the ER antagonist ICI182,780, which could abolish the effect completely (Supplementary Fig. 12). Long-term exposure to oestrogen further augmented the oestrogen response at the transcriptional level, as indicated by increased expression of PGR and GREB1 (Fig. 6c), as well as at the translational level as observed in 10 preparations from three biopsies tested in passage 2–6 (Supplementary Fig. 13a) with a significant overlap in ER and PR protein expression (Supplementary Fig. 13b). Thus, the cultured ERpos cells retain their ability to respond to oestrogen in a physiologically relevant manner. As an alternative approach we plated ERpos cells on fibroblast feeders, which have been used previously to reveal an oestrogenic response in mouse mammary cells37. ERpos cells were plated on confluent fibroblast feeders in TGFβR2i with or without oestrogen. At day 8, there was a significant increase in cell number with oestrogen as compared with the control (Fig. 6b). In the presence of fibroblasts, ERpos cells responded to oestrogen irrespective of the presence of EGF (Fig. 6b). These data imply that the stromal microenvironment beyond the influence of TGFβ is important in the regulation of the growth of ERpos cells.
Figure 6. ERpos cells in culture respond to oestrogen.
(a) Staining of PR (SAN27) in second-passage CD166high cells deprived of EGF and exposed to vehicle (EtOH) or oestrogen (E2) for 10 days. ERpos cells respond to oestrogen by increased expression of PR. (b) Quantification of cell number in four sets of pairwise triplicate cultures of EGF-deprived, oestrogen-stimulated CD166high low-passage cells grown for 4 days in second (P959) or third passage (P958), or 13 days in fourth passage (P959); medium-passage cells grown at high density before growth for 6 days (P958), 15 days (P957) and 14 days (P957 cultured without EGF since second passage) in fifth passage or cells with extended lifespan grown for 14 days in sixth passage; or high-passage cells (hTERT/shp16) in seventh passage grown for 13 days or in tenth to twelfth passage grown for 11, 15 and 11 days, respectively, in the presence of oestrogen (dark grey) or vehicle (light grey); or pairwise triplicate cultures of CD166high cells plus feeder grown for 8 days in the presence of oestrogen (dark grey) or vehicle (light grey). Second-passage cultures derived from two different biopsies (first and fourth set of bars) do not respond differently from third-passage cultures (second set of bars), irrespective of omission of EGF from TGFβR2i before co-culture (second set of bars)). Omission of EGF throughout the entire experimental period reduced the total cell number, but did not augment the response to oestrogen (third set of bars). Error bars indicate s.d.'s. In all cases, oestrogen treatment significantly increases the growth of ERpos cells, independent of passage group (by analysis of variance (ANOVA), P<0.05). The difference between experimental and control in each passage group is statistically significant by paired t-test (P values in each diagram). (c) Relative expression in triplicate of PGR and GREB1 from second (P957 and P959) and third passages (P958), or fourteenth passage long-term (hTERT/shp16) cultured ERpos cells exposed to vehicle (EtOH, grey bars) or oestrogen (E2, black bars) for 4 (P958 and P959), 22 (P957) or 13 (hTERT/shp16) days, respectively. Transcription of downstream target genes of ER signalling, PGR and GREB1 is in all cases significantly upregulated by oestrogen as assessed by two-tailed t-test, P<0.05. Error bars indicate s.d.'s.
In conclusion, we have developed a method to isolate, track and subculture ERpos and oestrogen-responsive cells from normal human breast.
Discussion
In spite of the fact that there are multiple protocols for enrichment, long-term cultivation and clonal growth of HBECs, none of them are able to isolate, track or support the growth of ERpos HBEC. This led us to screen our antibody repository for antibodies against human breast antigens eligible for flow cytometry and cell tracking, and to test whether the reluctant growth of ERpos HBEC in culture is due to inhibitory TGFβ signalling. Our present findings demonstrate that ERpos HBECs identified in situ by a panel of markers, including PR, AP2β, GATA3, Bcl2, CDw75, N-cadherin and 67LR, are enriched for in a CD326high/CD271low/CD166high/CD117low gate, and these cells express an antigen, unexpectedly decorated by an antibody, clone Ks20.8, originally raised against cytokeratin 20 (ref. 14). The distinction between ERpos cells, other luminal cells and basal cells is further supported by their relative expression of a panel of transcripts.
The prospective isolation and tracking of ERpos single cells from normal breast tissue hold promises for the future comparisons between normal, benign and malignant ERpos cells, which will hopefully shed some light on the evolution and pathogenesis of the most frequent form of human breast cancer. Being able to isolate and track the cells, however, would be of limited value if the ER expression was lost on culture. It has been anticipated that ERpos normal cells cannot divide and that this is why HBECs rapidly lose ER expression in culture38. We show here that ERpos cells can be released from growth restraint and sustained by TGFβR2i and that ERpos cells under these conditions can be expanded considerably. These findings may represent a paradigm shift in studying ER expression and function in the breast, in the future no longer relying exclusively on in vivo rodent models and human breast cancer cell lines. In addition, we believe the results answer the long-standing question of whether ERpos cells can self-renew, and further establishes that ERpos cells can be generated from ERneg progenitors39,40,41,42, here represented by CD117high luminal cells. Importantly, the response to TGFβR2i is specific to luminal cells only, as neither basal cells, fibroblasts nor ERneg normal breast cell lines are able to switch on ER in response to TGFβR2i. The fact that ERpos HBECs are here shown to be proliferating is in favour of the existence of ERpos progenitors. In general, however, most ERpos HBECs are considered to be close to the base in the hierarchy35. Thus, as has been hypothesized for breast cancer hierarchies43, our data could be interpreted in favour of the existence of bidirectionality also in the normal breast hierarchy, that is, on appropriate stimuli a subpopulation of apparently differentiated cells turn out in reality to be facultative progenitors.
The release of ERpos HBECs in culture by TGFβR2i is clinically relevant because it may help explain an enigmatic difference between the normal human breast and breast cancers. Thus, in the normal breast in vivo there is a strict dissociation between steroid receptor expression and proliferation15. In breast cancer and to a varying degree in precancerous breast lesions this negative association is lost44. It has been speculated that this may represent an important early change in the genesis of breast cancer either reflecting a failure to downregulate ER as cells enter the cell cycle or a failure to suppress division of ERpos cancer cells44. Our data indeed are in favour of the latter possibility because ERpos HBECs do divide if TGFβ signalling is perturbed, a very likely scenario in cancer (for review see ref. 45). While in primary breast cancer a normal-like TGFβ signalling is still in operation to restrain growth, in metastatic breast cancer TGFβ signalling has shifted to that of an epithelial–mesenchymal transition response45.
Another implication of the TGFβR2i culture protocol is that it represents a much-in-demand cell-based assay for oestrogen action on normal cells. It is already well established that oestrogen is a mitogen for ERpos breast cancer. However, its role in relation to ERpos HBECs has remained a mystery due to lack of ERpos cell culture models, and because ectopic expression of ER in basal cell lines under standard culture conditions has provided the paradoxical result of growth inhibition11,12. As a proof of principle, we here show that the TGFβR2i protocol serves as a physiologically relevant cell-based assay for oestrogen action, and moreover, in ERpos cells, several oestrogen-responsive genes downstream of ER are upregulated. Thus, the TGFβR2i protocol represents a cell-based assay for gauging oestrogen action reminiscent of its action in vivo. Intriguingly, in the presence of fibroblasts, growth of ERpos HBECs is stimulated by oestrogen irrespective of the presence of EGF. This implies that stromal factors modulate ER activity. With the relevant representatives of ERpos cells in hand, the complexity of this interaction can now be elucidated.
Our data further suggest that TGFβ signalling is key to the CD166high phenotype, and the exact mechanisms by which TGFβR2i generate ERpos cells clearly await further scrutiny. It cannot be excluded, however, that what we observe is part of a more general association between CD166 expression and TGFβ signalling. Thus, very recently others have found that TGFβ signalling is a main driver of the behaviour of CD166-expressing prostate cells46.
The present protocol is the result of several years of experimentation including many reduction mammoplasties from different donors until the present conditions were established. We cannot exclude that further improvements are possible. The data illustrate, however, that apparent post-mitotic cells in vivo, given the right conditions may multiply considerably, yet still exhibiting a definitive lifespan, and the approach moreover serves as a platform for extending the lifespan of ERpos cells into serial subculture.
While we have reproducibly recovered and propagated ERpos cells from all biopsies tested, we nevertheless wish to emphasize on a number of technicalities, which should be carefully observed when the protocol is applied. Biopsies inherently vary with respect to the frequency of ERpos cells5, which may eventually influence the definite number of ERpos cells that can be derived from a given biopsy. Biopsies also vary with respect to growth potential, but in our hands they all perform well up to at least passage 4 (Table 2). It should be kept in mind, however, that the protocol releases only a minority of the total number of initially quiescent ERpos cells, that is, up to 0.54% when plated at 4,000 cells per cm2 on sorting. Also, we strongly recommend that any attempts to validate Ks20.8 as well as co-expression of ER and PR by multicolour immunofluorescence are accompanied by immunoperoxidase staining to unravel the true overlap in staining. This is particularly important because we found that tracking of ERpos HBECs in smears must rely on either the ER/PR antibody mixture or the Ks20.8 surrogate marker specifically combined with the ER fixation protocol. Last, we would like to draw the attention to the fact that while the cultured CD166high/CD117low cells for the major part retain an elaborate profile of ERpos cells in situ, they may—again somewhat dependent of the biopsy of origin—display focal traits of basal or myoepithelial cells, such as p63, concurrently with luminal traits (Table 2).
That said, there are multiple implications of the present protocol. First, knowledge about how to turn on and off the ER expression in non-malignant breast epithelial cells may offer an alternative to selective oestrogen receptor modulators in prevention of breast cancer in women with elevated risk of disease47. Second, a reproducible source of normal human ERpos HBECs will represent a first step towards a physiological, cell-based screen for environmental oestrogenic activity and susceptibility of normal cells to breast cancer48. And third, the protocol may serve to test the functional role of recently identified single-nucleotide polymorphisms associated with increased risk of breast cancer49. In conclusion, we have provided a cell-based assay that allows normal breast ERpos cells to present themselves for investigation. These findings may fuel future advances in breast cancer prevention.
Methods
Tissue
Normal breast biopsies were collected with consent from women undergoing reduction mammoplasty for cosmetic reasons. The use of human material has been reviewed by the Regional Scientific Ethical Committees (Region Hovedstaden) and approved with reference to H-2–2011–052. Normal breast tissue was prepared as described25. In brief, normal breast specimens were minced with opposing scalpels and treated overnight at 37 °C with rotation in collagenase (Type 3, Worthington Biochemical Corporation, 900 U ml−1 in DMEM/F-12 (Life Technologies, catalogue no. 21041) with 2 mM glutamine (Sigma, G7513) and 50 μg ml−1 gentamycin (Biological Industries)) before differential centrifugation to aspirate dissolved fat and to isolate epithelial organoids and fibroblasts. These were then either used directly or frozen in liquid nitrogen for later use.
Fluorescence-activated cell sorting
To reveal epithelial cell composition and to isolate single cells, organoids from 13 biopsies were trypsinized, filtered through a 100-μm filter and resuspended in HEPES buffer supplemented with 0.5% BSA (bovine fraction V; Sigma-Aldrich) and 2 mM EDTA (Merck), pH 7.5. The suspended cells were incubated for 45 min at 4 °C in the presence of conjugated monoclonal antibodies EpCAM/CD326-PerCP cy5.5 (9C4, 1:20, BioLegend), NGFR (neurotrophin receptor, p75)/CD271-APC (ME20.4, 1:50, Cedarlane Laboratories) to separate basal and luminal cells, and activated leukocyte cell adhesion molecule CD antigen, ALCAM/CD166-AF488 (3A6, 1:20, AbD Serotec) and C-Kit/CD117-PE (104D2,1:20, BD) to separate luminal cells into luminal ERneg (CD166low/CD117high) cells and luminal ERpos (CD166high/CD117low) cells. On incubation, the cells were washed twice in HEPES/BSA/EDTA buffer and filtered through a 20-μm filter cup (Filcons) to prevent clogging of the FACS apparatus. Propidium iodide (Invitrogen) was added at a concentration of 1 μg ml−1, and the cells were analysed and sorted using a flow cytometer (FACSAria; BD Biosciences). An alternative way to isolate the two luminal subpopulations is to incubate for 30 min with EpCAM/CD326-PerCP cy5.5 and ALCAM/CD166-AF488 along with 67 kDa Laminin Receptor (MLuC5 1:50, Abcam) followed by 20 min incubation with secondary antibody Alexa Fluor 647 Goat Anti-Mouse IgM (1:500, Life Technologies).
To detect TGFβ receptors on the cell surface, third-passage CD166high/CD117low cells were incubated with monoclonal antibody TGFβRII (MM0056–4F14, 1:20, Abcam) followed by secondary antibody AF488 (IgG1, 1:500) and analysed by FACS. Overlay histograms were produced using Flowing Software 2.5.1 (University of Turku, Finland).
To establish fibroblast feeders, fourth-passage fibroblasts were incubated as described above with monoclonal antibodies CD26 (202–36, 1:200, Abcam) and conjugated CD105-AF488 (1:25, AbD Serotec) followed by secondary antibody AF647 (IgG2b, 1:500) to isolate CD105high/CD26low cells.
Cell culture
On sorting, the primary cell populations were plated in Primaria T25 flasks (#3813, Becton Dickenson) in the presence of ‘FAD2' ((DMEM, high glucose, no calcium, Life Technologies):Ham's F12 Nutrient Mixture (F12, Life Technologies) 3:1 v/v) with 2 mM glutamine, 0.5 μg ml−1 hydrocortisone, 5 μg ml−1 insulin, 10 ng ml−1 cholera toxin (Sigma-Aldrich), 10 ng ml−1 epidermal growth factor (Peprotech), 1.8 × 10–4 M adenine, 10 μM Y-27632 (Y0503, Sigma-Aldrich or 1683, Axon Medchem) and 5% fetal bovine serum (Sigma-Aldrich), modified from refs 26, 27). On plating, which could take up to 2 days for the luminal ERneg (CD166low/CD117high) cells and thus determined the time point for addition of TGFβR2i, a combination of the selective inhibitor of TGFβ type I receptor activin receptor-like kinase ALK5, ALK4 and ALK7, SB431542 (ref. 50) (10 μM, S4317, Sigma-Aldrich or 1661, Axon Medchem) and an inhibitor of autophosphorylation of ALK5, RepSox51,52 (25 or 50 μM, R0158, Sigma-Aldrich) was added. To test the specificity of the effect of TGFβR2i, in some experiments SB431542 alone or the double concentration of SB431542 was used instead of TGFβR2i, or RepSox was replaced by another TGFβ type I receptor activin receptor-like kinase ALK5 inhibitor, SD208 (1 μM, Tocris Bioscience). The vehicle, dimethyl sulfoxide (Sigma-Aldrich), was included in all experiments in appropriate concentrations for control cultures. Gentamycin (50 μg ml−1, Biological Industries) was added to the cultures throughout the first week after sorting, otherwise antibiotics were not included. To address whether propagation of ERpos cells was restricted to the FAD2 medium, CD166high/CD117low cells sorted from one of the biopsies were plated at a density of 4,000 cells per cm2 on Primaria six-well in either FAD2, M87A28, MEGM (LONZA) or WIT-P-NC (STEMGENT) medium including cholera toxin (100 ng ml−1) (ref. 53) and the cultures were observed for plating for 3 days. The possible induction of ER by TGFβR2i in other breast cells was further tested in MCF-10A54, HMT-3522 (ref. 55), fibroblasts purified from normal breast tissue25, CD117high cells purified from three different biopsies, as well as in basal cells isolated by FACS as described above and cultured on irradiated NIH-3T3 feeders26 before exposure to TGFβR2i for up to 7 days followed by staining for ER and Ks.20.8. Fibroblasts were routinely grown to confluency on collagen-coated T25 flasks (Nunc, 8 μg collagen per cm2, PureColl, CellSystems) in DMEM/F12, with 2 mM glutamine and 5% FBS before co-culture with CD166high/CD117low luminal cells. For phase contrast microscopy a Nikon Diaphot 300 microscope was used.
Cloning efficiency at low density in FAD2 was observed by plating the sorted basal cells, and the luminal populations CD166low/CD117high and CD166high/CD117low, at 400 cells per cm2 and culturing for 14 days, followed by fixation in methanol for 5 min at −20 °C and counterstaining of nuclei with haematoxylin. To assess the Ks20.8 and ER expression of the sorted populations and to further assess whether starting the culture at a higher density would change the expression, cells were seeded at 3,000 cells per cm2 and cultured for 9 days before immunocytochemical staining.
To quantify the frequency of Ks20.8high/ERpos colony-forming units CD166high/CD117low luminal cells from three different biopsies were plated at a density of 4,000 cells per cm2 with or without TGFβR2i and grown for 13 days before staining for ER and Ks20.8 followed by quantification using an ocular grid. The number of stained colonies defined as presence of either marker in three areas of each culture relative to the initial number of seeded cells was calculated.
Low-density split cultures were initially seeded at 6,400 cells per cm2 and grown for up to 15 days in primary culture in TGFβR2i or with SB431542 alone. Next, the cultures were trypsinized and seeded at 4,000 cells per cm2 in triplicate cultures, and subsequently passaged at the same density before the cultures reached confluency. Parallel cultures were stained to assess Ks20.8, ER and PR expression status. The number of cells was quantified manually using a counting chamber. Medium-passage cultures were passaged at high seeding density, that is, 8,000–20,000 cells per cm2. For extended cultivation of low-density cultures sorted luminal Ks20.8pos/ERpos (CD166high/CD117low) cells were first allowed to form colonies on irradiated NIH-3T3 feeders in modified breastoid base medium (BBM) without HEPES56 (DMEM/F12 supplemented with glutamine, 100 μM ethanolamine (Sigma , E0135), 1 μg ml−1 hydrocortisone (Sigma, H0888), 9 μg ml−1 insulin (Sigma, I6634), 5 μg ml−1 transferrin (Sigma, T1147), 5.2 ng ml−1 selenous acid (sodium salt, BD Biosciences, 354201), 20 ng ml−1 basic fibroblast growth factor (Peprotech, 100-18B) and 5nM amphiregulin (R&D Systems, 262-AR-100/CF)) with the addition of Y-27632, adenine and the serum replacement B27 (20 μl ml−1, Life Technologies), and subsequently sorted by FACS on incubation with CD326 and CD271 as described above to isolate CD326high/CD271high cells before plating and passaging in TGFβR2i culture. Population doublings were calculated as n=3.32(log UCY−log I)+X, where n=population doubling, UCY=cell yield, I=inoculum number and X= population doubling rate of inoculum.
To determine whether continuous TGFβR2i culture was needed to sustain ER and Ks20.8 expression, cells in the sixth passage were switched to FAD2 and the number of ERpos cells (n=3 × 100) was compared with parallel TGFβR2i cultures at day 3 and 5. Three similar experiments were performed with cloned hTERT/shp16-transduced cells (please see below).
As an alternative approach to extend the lifespan of Ks20.8pos/ERpos cells, the method by Kiyono et al.57 was adapted by introducing human telomerase (hTERT) and shRNA p16 (shp16) to second-passage CD166high/CD117lowcells. The viral constructs, pLENTi X2 Hygro/shp16 (w192–1, AddGene #22264, a gift from Eric Campeau) and pBABE-neo-hTERT58 (AddGene #1774, a gift from Robert Weinberg) were prepared as follows: lentiviral particles containing the shp16 construct were generated by transient co-transfection into HEK-293T cells using the calcium phosphate method with the vesicular stomatitis virus glycoprotein expressing construct pCMV–vesicular stomatitis virus glycoprotein as well as pol–gag construct pLP1 and rev construct pLP2. Retroviral hTERT particles were generated by transient co-transfection of the vector construct into a pantropic retroviral packaging cell line GP2–293 (Clonetech), which stably expresses the retroviral gag–pol genes. Medium was changed ∼16 h after transfection and viral media was collected ∼48 h later and stored at 4 °C for further purification. High-titre stocks of the virus were purified using a 20% sucrose gradient during ultracentrifugation with a Beckman SW32Ti rotor at 25,000 r.p.m. for 1.5 h. Purified viral solution was stored at −80 °C.
Before transduction, the cells were first treated with 200 mU ml−1 neuraminidase (N7885, Sigma) for 2 h at 4 °C, and then transduced using a high-viral titre59 containing the pBABE-neo-hTERT construct in TGFβR2i without FBS and incubated at 37 °C in 5% CO2 overnight. The culture medium was changed and the transduced cells underwent antibiotic selection for 10 days with 500 μg ml−1 G418 (Life Technologies). On confluency, the cells were passaged and underwent an identical transduction procedure with viral particles containing the pLenti X2 Hygro/shp16 construct and subsequent antibiotic selection with 100 μg ml−1 hygromycin (Sigma) for more than 2 weeks. The efficiency of the protocol was confirmed by hTERT/shp16 transducing second-passage CD166high/CD117low/Ks20.8pos/ERpos cells derived from a different biopsy.
To test the proliferative response to oestrogen in low-passage (p2–4) cultures, ERpos cells in TGFβR2i culture were seeded with 4,000 cells per cm2 in TGFβR2i with 25 μM RepSox and without EGF, supplemented with oestrogen (10–8 M, β-oestradiol, E2758, Sigma-Aldrich) and cultured for up to 13 days with medium change every other day. A primary culture from another biopsy was cultured 27 days in the presence of oestrogen and without EGF before splitting. The cells were passaged at 4,000 cells per cm2 in triplicate cultures with oestrogen or vehicle (96% ethanol). At day 4, the cultures were trypsinized and quantified using a counting chamber. Medium-passage (p5–6) cultures were passaged at 10,000–20,000 cells per cm2 before seeding at 4,000 cells per cm2, and the high-passage (p7–12) cultures including two hTERT/shp16-transduced populations derived from separate biopsies were passaged at 6,000 cells per cm2 or split at up to 1:4 before the growth experiment with a seeding density of 6,000 cells per cm2. The effect of an oestrogen antagonist (ICI182,780, Sigma-Aldrich), was tested in sixteenth passage, single-cell-cloned hTERT/shp16-transduced cells seeded at 4,000 cells per cm2 in TGFβR2i exposed to vehicle or oestrogen (10–10 M) and increasing concentrations of ICI182,780 (10–10, 10–9 and 10–8 M). At day 6, cultures were trypsinized and quantified in a cell counter (Roche Innovatis).
To assess whether the response of ERpos cells to oestrogen was modulated by the presence of fibroblasts, second- or third-passage ERpos cells, with or without the omission of EGF in the previous passage, were plated at a density of 5,600 cells per cm2 on confluent fibroblast feeders in triplicate culture. The following day, the culture medium was switched to TGFβR2i with oestrogen or vehicle. Under these conditions, fibroblasts did not grow. At day 8, the cultures were trypsinized and the total cell number was quantified. Whether a continuous lack of EGF influenced the response to oestrogen was tested in a similar set-up, where EGF was omitted from the medium during the entire experimental period.
The long-term effect of oestrogen on ER and PR expression in cultures without EGF was tested in 10 preparations of pairwise cultures in passage 2–6 derived from three different biopsies. The cultures were grown up to 26 days with oestrogen and vehicle, respectively, and subsequently stained by immunoperoxidase or immunofluorescence (see below).
RNA extraction and qRT–PCR
Total RNAs from sorted normal primary cells from eight biopsies were extracted using Trizol (Invitrogen) and were reverse transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). For general gene expression profiling, 2 ng of total cDNA was used to perform quantitative real-time PCR using the TaqMan Gene Expression Assays (Applied Biosystems) on CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The primers are listed in Supplementary Table 1, and real-time PCR conditions were the following: 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 30 s. To quantify cDNA concentration represented by cycle threshold (Ct) values, Cts were determined at the initial period of exponential amplification in triplicates and the different PCR runs were adjusted by inter-run calibrators (Bio-Rad CFX manager 3.0). Each gene expression level was to the mean of four reference gene expressions (GAPDH, HPRT1, TBP, and TFRC). The qRT–PCR data were then visualized as a heatmap using the statistical programme R (ref. 60) or as a bar graph using the values calculated by the 2−ΔΔCt method61. For qRT-PCR of ER, K8, FOXA1, ELF5 and K18, second-passage cultures of CD166high cells at a density of 8,000 cells per cm2 in FAD2, SB431542 or SB431542 switched to TGFβR2i culture conditions for 5 days before RNA extraction were used.
The expression of ER signalling and its downstream target genes were analysed in CD166high cells with or without TGFβR2i by using a RT2 Profiler PCR array (human oestrogen signalling, Qiagen) according to the manufacturer's instructions. Total 4 μg of RNA was used per array, which was performed in duplicates from two different biopsies. Three sets of E2- or ethanol (vehicle)-treated second- or third-passage ERpos cells or hTERT/shp16-transduced cells were further analysed in triplicate for PGR and GREB1 expression with the TaqMan Gene Expression Assays in the same condition mentioned above, using 20 ng of cDNA in each PCR reaction.
Western blotting
Whole-cell lysates were prepared for extraction by incubation in RIPA lysis buffer for 30 min at 4 °C with protease inhibitor cocktail (P8340, Sigma) and phosphatase inhibitor cocktail (Sigma, P5726). Proteins were separated by a 4–12% Novex Bis-Tris pre-cast polyacrylamide gradient gel (Life Technologies) and transferred to a polyvinylidene difluoride membrane using iBlot dry blotting system (Life Technologies). Molecular weight was indicated by using a pre-stained protein ladder (SM0671, Fermentas). After blocking 1 h in room temperature, the membrane was incubated with primary antibodies recognizing Smad2/3 (1:1,000, #3102, Cell Signaling), pSmad 2 (1:1,000, #3101, Cell Signaling), β-actin (1:5,000, A-5441, Sigma) or ER (1:500, NCL-ER-6F11/2, Novocastra) in blocking solution with gentle rocking overnight at 4 °C. Secondary antibodies conjugated with horseradish peroxidase (Dako) were incubated for 1 h at room temperature. Western blots were visualized using enhanced chemiluminescence solution (PerceECL 32106, Thermo Scientific) and a chemiluminescence imager (Amersham Image 600, GE Healthcare life sciences).
Immunohistochemistry and cytochemistry
Cryostat sections, smears of sorted cells and monolayer cultures were prepared and stained by immunoperoxidase or immunofluorescence, including negative controls without primary antibodies3,22,62. Specifically, cellular smears for ER–PR staining were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) followed by fixation in ice-cold methanol:acetone (1:1) for 10 min at −20 °C. Smears were then washed two times for 3 min and blocked for 10 min in 0.5% saponin (Sigma)/10% goat serum in PBS. All subsequent washing and incubation steps were performed in 0.5% saponin/10% goat serum. To visualize hormone receptor expression smears were incubated with anti-ER (SP1, 1:10) and anti-PR (SP2, 1:10) for 2 h at room temperature followed by 30 min with Alexa Fluor 568 Goat anti-Rabbit IgG (1:500, Invitrogen). For quantification of ER–PR and Ks20.8 expression, smears were fixed as described above and subsequently incubated with anti-ER (SP1, 1:10), anti-PR (SP2, 1:10) and Ks20.8 (1:10) for 2 h at room temperature followed by incubation for 30 min with Alexa Fluor 568 Goat anti-Rabbit IgG (1:500, Invitrogen) and Alexa Fluor 488 Goat anti-Mouse IgG2a (1:500, Invitrogen). Slides were mounted with ProLong Gold antifade reagent with 4,6-diamidino-2-phenylindole (DAPI; Life Technologies), and quantification of stained smears was routinely based on 3 × 100 cells, directly observed or evaluated in random micrographs obtained with a × 20 objective in the confocal microscope.
Antibodies are listed in Table 1. Many antibodies stain independently of fixation procedure, but of note, to stain for ER and PR, cultures were rinsed in PBS, pH 7.4, or sections were air dried before fixation, for 5 min at room temperature, in 3.7% formaldehyde, two rinses in PBS, fixation in methanol:acetone (1:1) for 5 min at 20 °C, two rinses in PBS, permeabilization in 0.1% Triton X-100 in PBS, twice for 7 min, rinse in PBS and kept wet before application of UltraV Block (Thermo Scientific). ER and PR expression in cultures with long-term oestrogen exposure was assessed by immunoperoxidase for ER (SP1, prediluted) and PR (SAN27 or PgR636) or by double-labelling immunofluorescence for ER (SP1, 1:10) and PR (PgR636, 1:10) with Alexa Fluor 488 Goat anti-Rabbit IgG (1:500) and Alexa Fluor 568 Goat anti-Mouse IgG1 (1:500) as secondary antibodies.
To stain for K15, UltraV Block was substituted for 10% normal goat serum in PBS. Immunofluorescence and peroxidase stainings were evaluated, quantified and photographed using a laser-scanning microscope (LSM 510 or LSM700; Carl Zeiss MicroImaging, Inc.) and bright-field microscopes (Laborlux S or DM5500B, Leica), respectively. For quantification of immunoperoxidase staining of ER, K8, K19 and p63, nuclei were counterstained with haematoxylin and counted in randomly selected fields using an ocular grid and given as the percentage of stained cells of a total of 1,000 cells evaluated with a × 25 objective.
Additional information
How to cite this article: Fridriksdottir, A. J. et al. Propagation of oestrogen receptor-positive and oestrogen-responsive normal human breast cells in culture. Nat. Commun. 6:8786 doi: 10.1038/ncomms9786 (2015).
Supplementary Material
Supplementary Figures 1-13 and Supplementary Table 1
Acknowledgments
We gratefully acknowledge the expert technical assistance from Tove Marianne Lund, Lena Kristensen and Kristina Mærsk, and Keld Ottosen for photographic assistance. We also thank Benedikte Thuesen, Københavns Privathospital and the donors for providing the normal breast biopsy material, and Vera Timmermans Wielenga, Pathology Department, Rigshospitalet for confirming the normalcy of the tissue. This work was supported by Novo Nordisk Foundation (to DANSTEM), Danish Cancer Society R2-A356–09-S2, Danish Research Council 08-045450 (to L.R.-J.) and 10-092798 (to DANSTEM), Toyota-Fonden Denmark, and Kirsten and Freddy Johansens Fond.
Footnotes
Author contributions A.J.F., R.V., J.K., O.W.P. and L.R.-J. conceived and designed the experiments and wrote the paper; A.F.J., R.V., M.C.K., J.K., B.M.H. and L.R.-J. carried out the laboratory experiments; stainings were evaluated and photographed by R.V., M.C.K., O.W.P. and L.R.-J., and J.K. performed statistical analysis for heatmap presentation.
References
- Petersen O. W. & Polyak K. Stem cells in the human breast. Cold Spring Harb. Perspect. Biol. 2, 1–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S. L., Ham R. G. & Stampfer M. R. Serum-free growth of human mammary epithelial cells: rapid clonal growth in defined medium and extended serial passage with pituitary extract. Proc. Natl Acad. Sci. USA 81, 5435–5439 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen O. W. & van Deurs B. Growth factor control of myoepithelial-cell differentiation in cultures of human mammary gland. Differentiation 39, 197–215 (1988). [DOI] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J. et al. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. J. Cell. Sci. 94, 403–413 (1989). [DOI] [PubMed] [Google Scholar]
- Petersen O. W., Høyer P. E. & van Deurs B. Frequency and distribution of estrogen receptor-positive cells in normal, nonlactating human breast tissue. Cancer Res. 47, 5748–5751 (1987). [PubMed] [Google Scholar]
- Rønnov-Jessen L., Petersen O. W. & Bissell M. J. Cellular changes involved in conversion of normal to malignant breast: the importance of the stromal reaction. Physiol. Rev. 76, 69–125 (1996). [DOI] [PubMed] [Google Scholar]
- Kang K.-S. et al. Expression of estrogen receptors in a normal human breast epithelial cell type with luminal and stem cell characteristics and its neoplastically transformed cell lines. Carcinogenesis 18, 251–257 (1997). [DOI] [PubMed] [Google Scholar]
- Graham J. D. et al. DNA replication licensing and progenitor numbers are increased by progesterone in normal human breast. Endocrinology 150, 3318–3326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos T. et al. Progesterone/RANKL is a major regulatory axis in the human breast. Sci. Transl. Med. 5, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- Stampfer M. R. & Bartley J. C. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo(a)pyrene. Proc. Natl Acad. Sci. USA 82, 2394–2398 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundholt B. K., Briand P. & Lykkesfeldt A. E. Growth inhibition and growth stimulation by estradiol of estrogen receptor transfected human breast epithelial cell lines involve different pathways. Breast Cancer Res. Treat. 67, 199–214 (2001). [DOI] [PubMed] [Google Scholar]
- Zajchowski D. A., Sager R. & Webster L. Estrogen inhibits the growth of estrogen receptor-negative, but not estrogen receptor-positive, human mammary epithelial cells expressing a recombinant estrogen receptor. Cancer Res. 53, 5004–5011 (1993). [PubMed] [Google Scholar]
- Denk H., Lackinger E., Zatloukal K. & Franke W. W. Turnover of cytokeratin polypeptides in mouse hepatocyes. Exp. Cell Res. 173, 137–143 (1987). [DOI] [PubMed] [Google Scholar]
- Moll R., Löwe A., Laufer J. & Franke W. W. Cytokeratin 20 in human carcinomas. A new histodiagnostic marker detected by monoclonal antibodies. Am. J. Pathol. 140, 427–447 (1992). [PMC free article] [PubMed] [Google Scholar]
- Clarke R. B., Howell A., Potten C. S. & Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 57, 4987–4991 (1997). [PubMed] [Google Scholar]
- Asselin-Labat M. L. et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 (2007). [DOI] [PubMed] [Google Scholar]
- Lu J. et al. β-Galactoside α2,6 sialyltransferase 1 promotes transforming growth factor-β-mediated epithelial-mesenchymal transition. J. Biol. Chem. 289, 34627–34641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock M. J., Shintani Y., Maeda M., Fukumoto Y. & Johnson K. R. Cadherin switching. J. Cell Sci. 121, 727–735 (2008). [DOI] [PubMed] [Google Scholar]
- Levin T. G. et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 139, 2072–2082 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri C. et al. The metastasis-associated 67-kDa laminin receptor is involved in G-CSF-induced hematopoietic stem cell mobilization. Blood 108, 2476–2484 (2006). [DOI] [PubMed] [Google Scholar]
- Westbury C. B. et al. Genome-wide transcriptomic profiling of microdissected human breast tissue reveals differential expression of KIT (c-Kit, CD117) and oestrogen receptor-alpha (ERalpha) in response to therapeutic radiation. J. Pathol. 219, 131–140 (2009). [DOI] [PubMed] [Google Scholar]
- Villadsen R. et al. Evidence of a stem cell hierarchy in the adult human breast. J. Cell Biol. 177, 87–101 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio M. C. & Chambon P. The pS2 gene, mRNA, and protein: a potential marker for human breast cancer. Cancer Cells 2, 269–274 (1990). [PubMed] [Google Scholar]
- Ghosh M. G., Thompson D. A. & Weigel R. J. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60, 6367–6375 (2000). [PubMed] [Google Scholar]
- Rønnov-Jessen L. & Petersen O. W. Induction of α-smooth muscle actin by transforming growth factor-β1 in quiescent human breast gland fibroblasts. Implications for myofibroblast generation in breast neoplasia. Lab. Invest. 68, 696–707 (1993). [PubMed] [Google Scholar]
- Liu X. et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180, 599–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D. W. M. et al. Single-cell gene expression profiling reveals funcional heterogeneity of undifferentiated human epidermal cells. Development 140, 1433–1444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J. C. et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 69, 7557–7568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewan K. B. R. et al. Proliferation of estrogen receptor-α-positive mammary epithelial cells is restrained by transforming growth factor-β1 in adult mice. Am. J. Pathol. 167, 409–417 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S. et al. Molecular profiling of human mammary gland links breast cancer risk to a p27(+) cell population with progenitor characteristics. Cell Stem Cell 13, 117–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogina G. et al. Comparison of anti-estrogen receptor antibodies SP1, 6F11, and 1D5 in breast cancer: lower 1D5 sensitivity but questionable clinical implications. Am. J. Clin. Pathol. 138, 697–702 (2012). [DOI] [PubMed] [Google Scholar]
- Augello M. A., Hickey T. E. & Knudsen K. E. FOXA1: master of steroid receptor function in cancer. EMBO J. 30, 3885–3894 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes S. R., Hilton H. N. & Ormandy C. J. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res. 8, 207–216 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire W. L. Jr et al. Regulation of insulin-like growth factor-binding protein (IGFBP) expression by breast cancer cells: use of IGFBP-1 as an inhibitor of insulin-like growth factor action. J. Natl Cancer Inst. 84, 1336–1341 (1992). [DOI] [PubMed] [Google Scholar]
- Lim E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–915 (2009). [DOI] [PubMed] [Google Scholar]
- Kato S. et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270, 1491–1494 (1995). [DOI] [PubMed] [Google Scholar]
- Haslam S. Z. Mammary fibroblast influence on normal mouse mammary epithelial cell responses to estrogen in vitro. Cancer Res. 46, 310–316 (1986). [PubMed] [Google Scholar]
- Duss S. et al. An ostrogen-dependent model of breast cancer created by transformation of normal mammary epithelial cells. Breast Cancer Res. 9, R38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M. et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14, R134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J. E. & Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspective. Genes Dev. 28, 1143–1158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeth G. et al. Aldehyde dehydrogenase and estrogen receptor define a hierarchy of cellular differentiation in the normal human mammary epithelium. Breast Cancer Res. 16, R52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt L. M. et al. Human breast progenitor cell numbers are regulated by WNT and TBX3. PLoS ONE 9, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P. B. et al. Stochastic state transitions give rise to phenotypic equlibrium in populations of cancer cells. Cell 146, 633–644 (2011). [DOI] [PubMed] [Google Scholar]
- Shoker B. S. et al. Estrogen receptor-positive proliferating cells in the normal and precancerous breast. Am. J. Pathol. 155, 1811–1815 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses H. & Barcellos-Hoff M. H. TGF-β biology in mammary development and breast cancer. Cold Spring Harb. Perspect. Biol. 3, a003277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. G. et al. ALCAM/CD166 is a TGF-β-responsive marker and functional regulator of prostate cancer metastasis to bone. Cancer Res. 74, 1404–1415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J. et al. Selective oestrogen receptor modulators in prevention of breast cancer: an updated meta-analysis of individual participant data. Lancet 381, 1827–1834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis S. et al. High-fat or ethinyl-oestradiol intake during pregnancy increases mammary cancer risk in several generations of offspring. Nat. Commun. 3, 1053–1061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sal-lari R. et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 44, 1191–1200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laping N. J. et al. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of TGF-β1 type I receptor kinase activity: SB-431542. Mol. Pharmacol. 62, 58–64 (2002). [DOI] [PubMed] [Google Scholar]
- Gellibert F. et al. Identification of 1,5-naphthyridine derivatives as a novel series of potent and selective TGF-β type I receptor inhibitors. J. Med. Chem. 47, 4494–4506 (2004). [DOI] [PubMed] [Google Scholar]
- Ichida J. K. et al. A small-molecule inhibitor of TGF-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell 5, 491–503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince T. A. et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell 12, 160–170 (2007). [DOI] [PubMed] [Google Scholar]
- Soule H. D. et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 (1990). [PubMed] [Google Scholar]
- Briand P., Petersen O. W. & van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell. Dev. Biol. 23, 181–188 (1987). [DOI] [PubMed] [Google Scholar]
- Pasic L. et al. Sustained activation of the HER1-ERK1/2-RSK signaling pathway controls myoepithelial cell fate in human mammary tissue. Genes Dev. 25, 1641–1653 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono T. et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396, 84–88 (1998). [DOI] [PubMed] [Google Scholar]
- Counter C. M. et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl Acad. Sci. USA 95, 14723–14728 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines W. C., Yaswen P. & Bissell M. J. Modelling breast cancer requires identification and correction of a critical cell lineage-dependent transduction bias. Nat. Commun. 6, 6927–6937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H. & Bertone P. HTqPCR: high-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics 25, 3325–3326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-2-ΔΔCt method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Rønnov-Jessen L., Celis J. E., van Deurs B. & Petersen O. W. A fibroblast-associated antigen: characterization in fibroblasts and immunoreactivity in smooth muscle differentiated stromal cells. J. Histochem. Cytochem. 40, 475–486 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-13 and Supplementary Table 1