Abstract
Due to the emergence and rapid spread of drug resistance in bacteria, there is an urgent need for the development of novel antimicrobials. SecA, a key component of the general bacterial secretion system required for viability and virulence, is an attractive antimicrobial target. Earlier we reported that systematical dissection of a SecA inhibitor, Rose Bengal (RB), led to the development of novel small molecule SecA inhibitors active against E. coli and B. subtilis. In this study, two potent RB analogs were further evaluated for activities against methicillin-resistant Staphylococcus aureus (MRSA) strains and for their mechanism of actions. These analogs showed inhibition on the ATPase activities of S. aureus SecA1 (SaSecA1) and SecA2 (SaSecA2), and inhibition of SaSecA1-dependent protein-conducting channel. Moreover, these inhibitors reduce the secretion of three toxins from S. aureus and exert potent bacteriostatic effects against three MRSA strains. Our best inhibitor SCA-50 showed potent concentration-dependent bactericidal activity against MRSA Mu50 strain and very importantly, 2–60 fold more potent inhibitory effect on MRSA Mu50 than all the commonly used antibiotics including vancomycin, which is considered the last resort option in treating MRSA-related infections. Protein pull down experiments further confirmed SaSecA1 as a target. Deletion or overexpression of NorA and MepA efflux pumps had minimal effect on the antimicrobial activities against S. aureus, indicating that the effects of SecA inhibitors were not affected by the presence of these efflux pumps. Our studies show that these small molecule analogs target SecA functions, have potent antimicrobial activities, reduce the secretion of toxins, and have the ability to overcome the effect efflux pumps, which are responsible for multi-drug resistance. Thus, targeting SecA is an attractive antimicrobial strategy against MRSA.
Keywords: SecA-dependent secretion, SecA inhibitors, SecA-liposomes, Rose Bengal analogs, MRSA, efflux
Graphical abstract
1. Introduction
Due to the widespread emergence of bacterial resistance to currently marketed antibiotics, the need for the development of new therapeutic agents is urgent. Methicillin-resistant Staphylococcus aureus (MRSA) is one of the major drug-resistant bacterial pathogens, causing serious hospital- and community-acquired infections [1–5]. As the prototype of clinical Gram-positive multidrug resistant (MDR) bacteria, MRSA is the focus of numerous mechanistic and therapeutic studies. In addressing infections by drug resistant bacteria, such as MRSA, it is important to consider issues beyond simple potency. Specifically, antimicrobials capable of inhibiting virulence factor production and overcoming the negative effect of efflux pumps on potency are important traits in addition to bacteriostatic and bactericidal effects. However, currently, there are no antimicrobials that have all three traits together, although the benefits of attacking all three mechanisms using one antimicrobial are obvious. We have previously published the design and synthesis of a novel class of thiouracil containing SecA [6] inhibitors and extensively reviewed all known SecA inhibitors so far [7, 8]. In this study, the ability for SecA inhibitors to take on this three-pronged approach has been explored.
SecA is an indispensable ATPase of the general protein translocation machinery present in bacteria. It is responsible for the secretion of many vital proteins and essential for bacterial growth [9–11]. SecA also plays important roles in bacterial virulence, being involved in the secretion of many toxins and other virulence factors [12, 13]. Moreover, in addition to interacting with membrane protein SecYEG in soluble form, SecA is also involved in forming a membrane protein-conducting channel [14, 15]. Therefore inhibitors might be able to directly act on SecA without having to enter into cells and thus may bypass the negative effect of efflux pumps. SecA is highly conserved in bacteria and has no counterpart in mammalian cells [11, 16], thus providing an ideal target for developing broad-spectrum antimicrobial agents. We have recently developed Rose Bengal (RB) [17] and its analogs [18] as SecA inhibitors using SecA from E. coli and B. subtilis as models. In this study, we focus on examining several key issues in evaluating the scope of applications of these inhibitors. First, we were interested in seeing whether these SecA inhibitors would be effective against the clinically important pathogenic MRSA. This is very important because of MRSA’s role in mortality in hospital- and community-acquired infections. Second, most antibiotics available do not have the intrinsic ability to attenuate virulence factor secretion. As a result, sometimes the control of infection does not always correlate with the control of bacterial pathogenicity. We hypothesize that SecA inhibitors can inhibit virulence factor secretion and plan to evaluate this point using our most potent inhibitors. Third, efflux pumps are well-known to attenuate the effectiveness of antibiotics by reducing their intracellular concentrations, and are responsible for multi-drug resistance. This is a widespread problem in drug-resistant bacteria such as MRSA. There has been a long-standing interest in the field to find approaches to nullify the effect of efflux pumps to no avail. We hypothesize that SecA inhibitors would have the intrinsic ability to overcome the effect of efflux pumps because SecA is mainly a membrane target and can be accessible by direct diffusion of the inhibitor into the membrane without the need of enhanced intracellular concentrations. We plan to probe this issue using our available inhibitors. If proven to be true, this would be the first case that one can use a single inhibitor to achieve the effect of (1) bacterial inhibition, (2) virulence factor secretion attenuation, and (3) overcoming the effect of efflux pumps.
2. Materials and Methods
2.1 Bacterial strains and culture condition
All bacteria used were S. aureus. ATCC 35556 and ATCC 6538 were from the American Type Culture collection. Mu50, Mu3, and N315 were kindly provided by C.-D. Lu, and Newman strain from Z. Eichenbaum of Georgia State University. Five efflux pump-related strains 8325-4 [19], K1758 (NorA−), K2361 (NorA++), K2908 (MepA−), K2068 (MepA++) [19, 20] were kindly provided by GW Kaatz at Wayne State University School of Medicine. All strains were grown on Luria-Bertani (LB) broth or agar plates at 37 °C.
2.2 Chemical compounds
Rose Bengal was from Sigma-Aldrich. The syntheses of SCA-41 and SCA-50 (corresponding to 22a and 22c respectively) have been described previously [18]. The synthesis of SCA-254 is described in the Supplemental Information section. DMSO was used to prepare stock solutions, which were further diluted for assays. All control samples without treatment contained DMSO at the same concentration.
2,3 Protein preparations
The SaSecA1 and SaSecA2 genes were amplified from S. aureus ATCC 35556. SaSecA1 gene was cloned into pET-21d and SaSecA2 gene was cloned into pET-29a, and both genes were over-expressed in BL21λDE3 at 20 °C with 0.5 mM IPTG. SaSecA1 and SaSecA2 were purified with HisTrap affinity column and Superdex-200 column (GE Healthcare).
2.4 Intrinsic SecA ATPase activity assay
The ATPase activity was determined by malachite green colorimetric method described previously [17, 18, 21]. In this assay, the reactions were carried out at 25 °C for 3 hr (SaSecA1) or 30 min (SaSecA2). IC50 is the concentration of the compound that inhibits 50% ATPase activities.
2.5 SecA-liposome ion-channel activity assays in the oocyte
Liposomes were prepared as described previously [14–18]. Oocytes were obtained from live frog Xenopus laevis (Xenopus Express, Inc) and injected with sample mixtures as described previously [14, 22]. Briefly, 50 nl sample mixtures containing 120 ng liposomes, 120 ng SecA, 14 ng proOmpA, 2 mM ATP, 1 mM Mg(OAC)2, and different concentrations of inhibitors were injected into dark animal pole side of oocytes (average volume: 500 nl) using Nanoject II injector (Drummond Scientific Co., Broomall, PA). After injection, the oocytes were incubated at 23 °C for 3 hr; then the ion current was recorded continuously for 1 min. IC50 is the concentration of the compound that inhibits 50% ion-channel activities of SaSecA1.
2.6 SaSecA1-liposome alone in vitro proOmpA translocation
Inhibition of in vitro translocation activity of SaSecA1 was determined by using the liposome alone-proOmpA translocation system [14]. Translocation mixtures per 0.1 ml contained 50 mM Tris-HCl (pH 7.6), 20 mM NH4Cl, 25 mM KCl, 2 mM Mg(OAc)2, an ATP regenerating system, 1.5 µg SaSecA1, 100 ng SecB, 120 ng of proOmpA, and 120 µg of liposomes. After incubation with different concentrations of inhibitors at 30 °C for 1.5 hr, the translocation of purified proOmpA was determined by its inaccessibility to proteinase K as described previously [14]. The inhibition effect was evaluated by quantitation of translocated proOmpA with Western blots. IC50 is the concentration of the compound that inhibits 50% in vitro translocation activit ies of SaSecA1.
2.7 Toxin secretion
S. aureus Mu50 was grown in LB broth at 37 °C. Inhibitors were added to the mid-log phase. Culture was collected after treating with inhibitor at various time points. The supernatant and cell pellet were separated by centrifugation and the supernatant was further filtered through 0.45-µm syringe disks (Fisher Scientific). Western blots with specific toxin antibodies were used to detect the amount of toxins in the supernatant. S. aureus antibodies for α-hemolysin were obtained from Sigma-Aldrich; for toxin shock syndrome toxin-1 (TSST-1) from Acris-Antibodies; and for enterotoxin B from Abcam.
2.8 Pull down assay
S. aureus ATCC 6538 was grown in LB medium at 37 °C. Overnight cells were harvested by centrifugation and washed with TBST buffer (0.1 M Tris-HCl pH 7.0, 0.15 M NaCl, 0.1% Tween-20). Cell pellets were washed three times with the same buffer and re-suspended with TBST buffer containing EDTA-free cocktail protease inhibitors. The cells were broken with French Press at 10,000 psi. Unbroken cells were removed by 5,000 rpm centrifugation. Whole cell lysates were treated with PureProteome Streptavidin Magnetic Beads (Purchased from Millipore) at 4 °C for 1 hr to remove non-specific binding. The supernatant was mixed with or without 400 µM SCA-254 with gentle rolling at 4 °C for 1 hr. The beads were washed with TBST buffer for 3 times. After washing, beads were mixed with SDS sample buffer, and then boiled for 15 min. Cross-reacting EcSecA antibody (lab stock) was used to detect SaSecA1 by Western blots.
2.9 Bacteriostatic effect
Bacteriostatic effects were tested according to the guidelines of the Clinical and Laboratory Standards Institute [23]. This assay was performed in a 96-well microtiter plate in triplicates in three separate experiments as described previously [18] at 37 °C with shaking at 250 rpm for 24 hrs. MIC is the lowest concentration of inhibitor, at which cells were not able to grow.
2.10 Bactericidal effect
Bactericidal effect was determined under normal room light condition as described previously [18]. Log-phase growing cells (OD600 ≈ 0.5) were mixed with different concentrations of inhibitors, and then incubated in an Eppendorf Thermonmixer R (Brinkmann instruments) at 37 °C with shaking (1,000 rpm) for 1 hr. Cultures were serially diluted with LB broth and spreaded on LB plates, and incubated at 37 °C overnight for colony forming units (CFU). Bactericidal effect was determined by the reduction of CFU as described previously [18].
2.11 Effects of light on bactericidal result
Compounds were added into early log phase culture (OD600 ≈ 0.8) of S. aureus ATCC 6538 grown in glass tubes, and then the mixture was incubated at 37 °C (shaking at 250 rpm) in dark or with 15W fluorescent light. After 2 hr, cultures were serially diluted with LB broth and CFU were similarly determined. Bactericidal effect was determined by the reduction of CFU as described previously [18].
2.12. Molecular docking
Docking was done using SYBYL-X 2.0 by following the standard procedures provided in the manual. Surflex-dock method was used to dock ligands into SecA crystal structure (PDB ID: 2FSG) using automatic docking. Standard parameters given in SYBYL X 2.0 were employed for energy minimization of SecA crystal structure. Protein preparation was done using AMBER7 FF99 force field and the default parameters in Surflex-Dock (SFXC). A maximum of 20 docking poses were generated for each molecule.
3. Results and Discussions
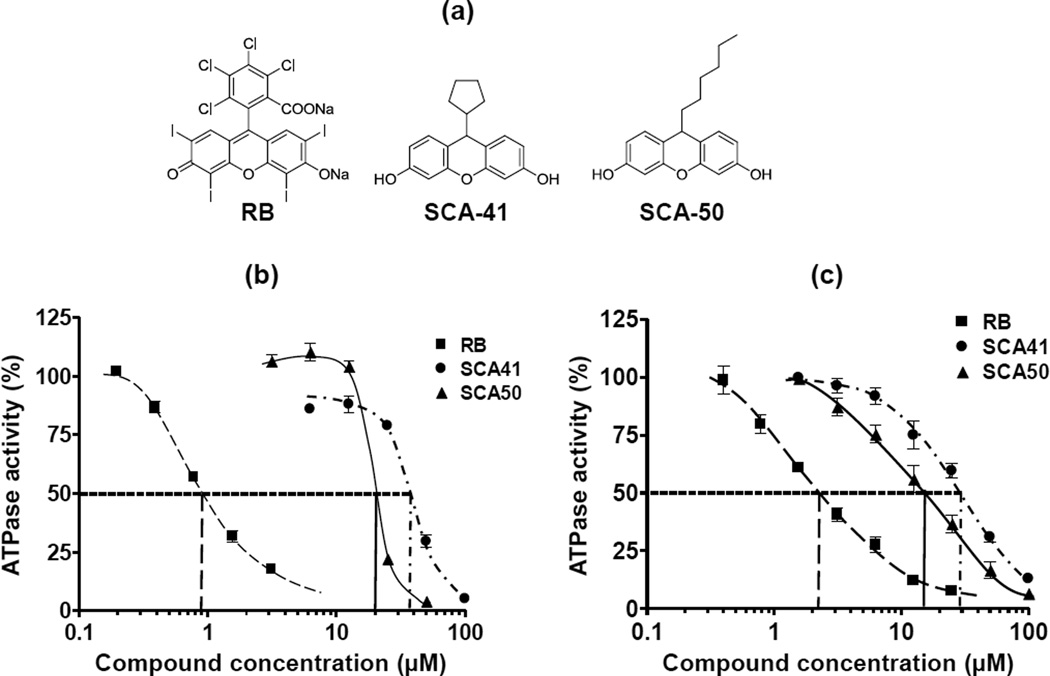
SecA is essential for cell growth in all bacteria, and is required for Sec-dependent secretion of proteins including extracellular toxins and other virulence factors. Therefore, targeting S. aureus SecA might achieve the effect of decreasing bacterial survivability and reducing virulence. In our previous studies, EcSecA and BsSecA were used as model systems to develop new SecA inhibitors. RB was found as a potent SecA inhibitor through random screening [17], and a series of novel SecA inhibitors were further developed from systematic dissections of RB [18]. Figure 1a shows the structures of RB and two RB analogs developed from our previous study. These two RB analogs have low molecular weight (RB MW, 1017 daltons, SCA-41, 282 daltons, and SCA-50, 298 daltons) and display potent antimicrobial activities against two model bacteria strains E. coli NR698 and B. subtilis 168 [18]. In the current study, we evaluate the inhibitory effects of these RB analogs against pathological MRSA strains and examined the functional effect on SecA inhibition in SaSecAs.
Figure 1. Inhibition on the ATPase activities of SaSecA1 and SaSecA2.
(a) Structures of RB (MW, 1017 daltons) and RB analogs, SCA-41 (MW, 282 daltons) and SCA-50 (MW 298 daltons). (b) Inhibition of the intrinsic ATPase activity of soluble SaSecA1 (n = 6). (c) Inhibition of the intrinsic ATPase activity of SaSecA2 (n = 6). The ATPase activity without inhibitor was defined as 100%.
3.1 Inhibition of the ATPase activities of SaSecA1 and SaSecA2
Two SecA homologues have been identified in S. aureus [13]: SaSecA1 is the essential housekeeping SecA, while SaSecA2 is only involved in the secretion of adhesin-SraP [13]. The RB analogs SCA-41 and SCA-50 were tested for their inhibitory effects on the intrinsic ATPase activities of SaSecA1 and SaSecA2. The intrinsic ATPase activity without inhibitor was defined as 100% and IC50 is the concentration of the inhibitor, at which 50% inhibition of ATPase activity was achieved. As shown in Figure 1b and 1c, the IC50 value of SCA-41 was 35 µM for SaSecA1 and 30 µM for SaSecA2; for SCA-50, the IC50 value was 20 µM for SaSecA1 and 15 µM for SaSecA2. Such results indicate that these two compounds might have dual targets in S. aureus, which could increase the chance of combating infection and reduce the occurrence of mutational resistance. Though their inhibitory effects for intrinsic ATPase were not as good as that of RB (IC50 ≈ 1 µM against SaSecA1, 2 µM against SaSecA2), RB is much bigger, and also has other targets and limitations (see below). Because SecA is a multi-functional protein, exists in multi-forms, and more importantly functions in the membrane, potency of SecA inhibitors needs to be evaluated by multiple assays, especially related to membrane activities, as discussed earlier [17, 18].
3.2 Inhibition on the activities of SecA-dependent protein-conducting channels
SecA exerts its essential functions in the membrane, not in its soluble form. However, currently there is no suitable in vitro system for studying protein translocation using membrane and a precursor protein from S. aureus. To assess the inhibitors’ effect, we have developed two very sensitive assays that mimic the in vivo protein translocation activity for testing the effectiveness of the SecA inhibitors. These two assays involve using SecA-liposomes interacting with protein precursor proOmpA. One is a very sensitive electrophysiological assay that opens the SecA-specific channels in oocytes; the other is an in vitro protein translocation SecA-liposome alone assay [14]. However, it needs to be noted that such assays are only applicable to SecA1, but not the soluble SecA2. Thus, there will be no side-by-side comparisons of inhibitory activities against SecA1 and SecA2 in such experiments.
3.2.1 Inhibition on ion-channel activity
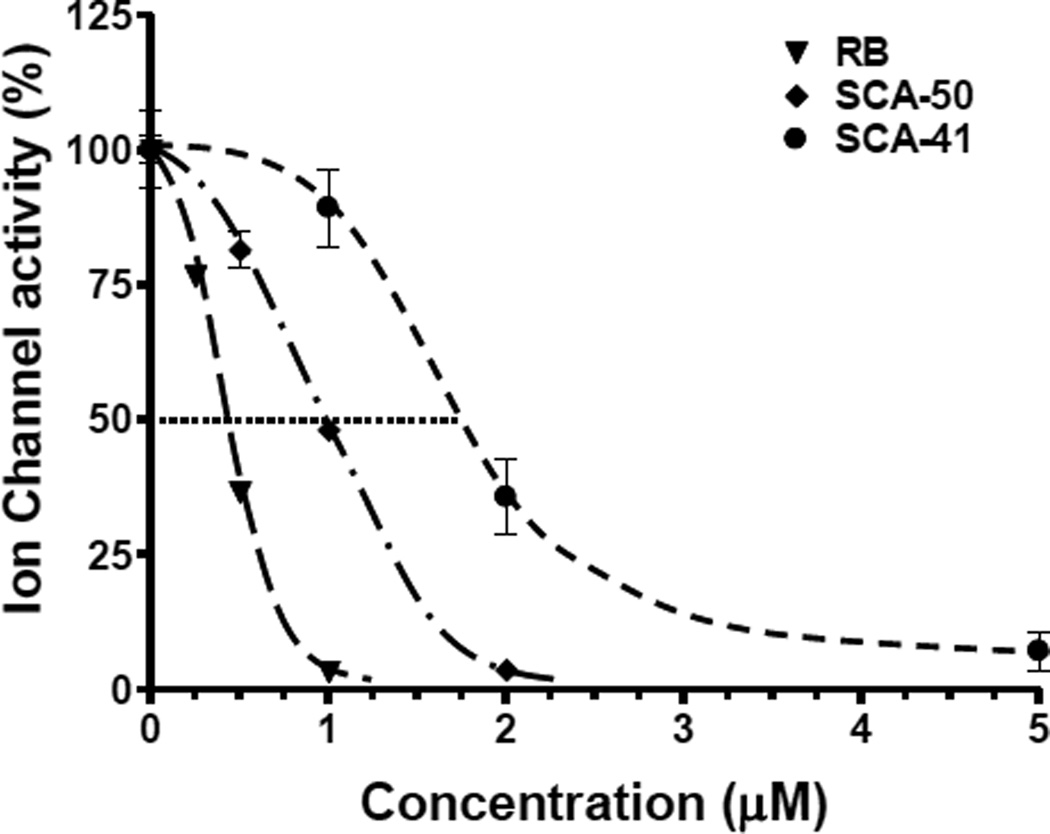
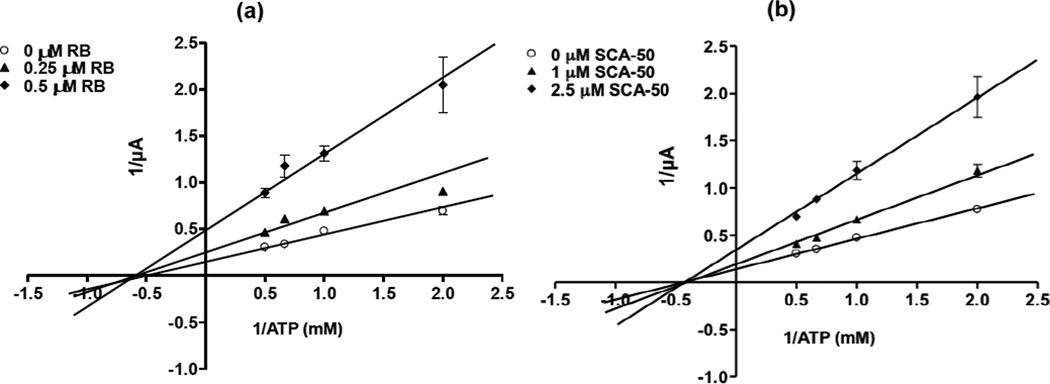
To assess the efficacy of the inhibitors against the functions of SaSecA1 in membrane environments, a sensitive semi-physiological assay in oocytes [22] was applied to evaluate SecA-liposomes ion-channel activities [14, 18, 22]. SaSecA1-liposome was injected simultaneously into oocytes in the presence or absence of an inhibitor. RB and two analogs displayed potent inhibition against the ion-channel activity of SaSecA1 (SaSecA2 has no channel activity, data not shown). The ion-channel activity of SaSecA1 was defined as 100%, and IC50 is the concentration of inhibitor, at which 50% inhibition of its ion-channel activity was achieved. The IC50 value of RB was 0.5 µM (Fig. 2), which is closed to its IC50 value in ATPase inhibition. The IC50 value of SCA-50 and SCA-41 were 1 µM and 2 µM (Fig. 2), respectively, which are much lower than the IC50 values against the intrinsic ATPase activity of SaSecA1 (Fig.1), indicating that these two compounds exert better inhibition in a membrane environment. Kinetic studies showed that both SCA-50 and RB inhibit translocation related ATPase activity of SaSecA1 in a non-competitive manner in the liposomes-oocytes (Fig. 3), suggesting that the inhibition is mainly a conformational effect with SecA ATP binding site in the membranes. These results also indicate intrinsic ATPase activity of soluble SecA may not be an appropriate single test for the purpose of drug discovery screening.
Figure 2. Inhibition of the ion-channel activity of SaSecA1.
SaSecA1-liposomes ion-channel activity was determined in oocytes with or without inhibitors (n = 20–30). The ion-channel activity without inhibitors was defined as 100%.
Figure 3. Non-competitive inhibition of the membrane ATPase activity of SaSecA1.
SaSecA1-liposomes ion-channel activity was determined in the oocytes with different concentration of ATP and SCA-50 (n = 9–10). (a) Inhibition of RB. (b) Inhibition of SCA-50.
3.2.2 Inhibition of in vitro protein translocation activity
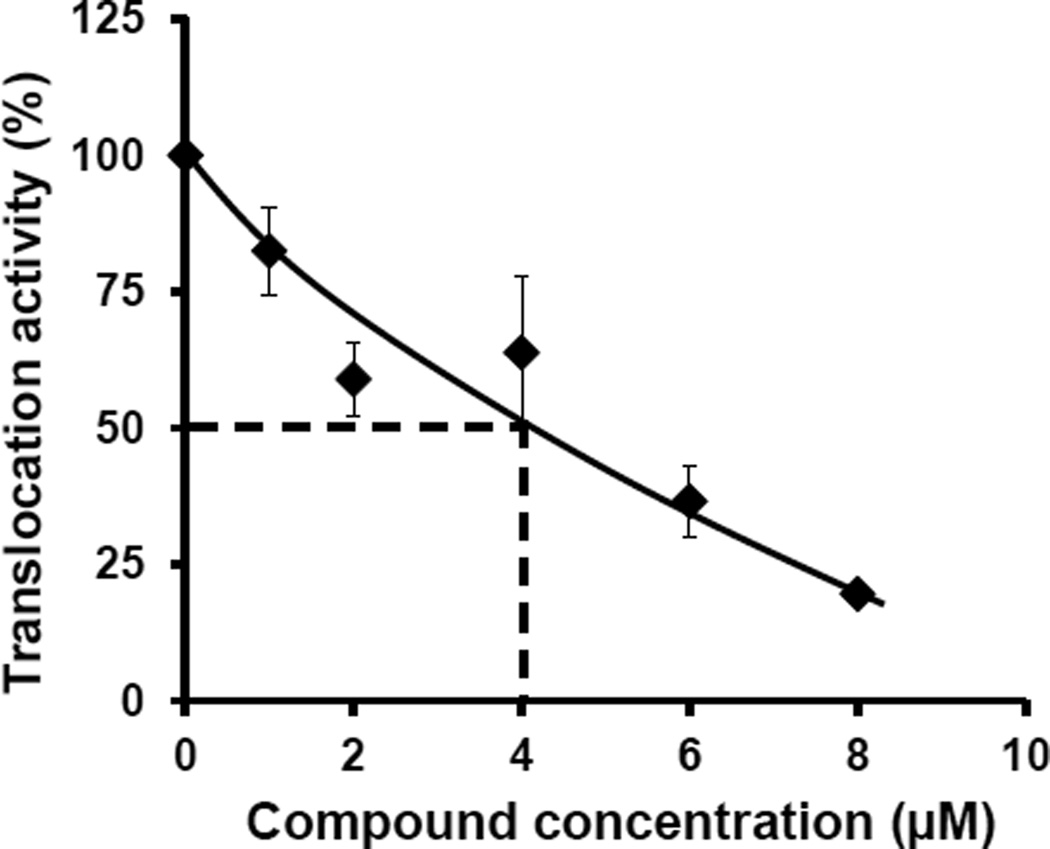
SecA is essential for translocation of proteins containing the Sec-dependent signal peptide to traverse membranes. Therefore, the most important in vitro functional assay for evaluation of SecA inhibitor is protein translocation across membranes. In this study, a SecA alone in vitro liposome translocation system without SecYEG [14] was applied to determine the inhibitory effect of SCA-50 against SaSecA1-dependent proOmpA translocation. The in vitro translocation activity of SaSecA1 without inhibitor was defined as 100%, and IC50 was the concentration of inhibitor, at which 50% inhibition of its in vitro translocation activity was achieved. Figure 4 shows that SCA-50 inhibited in vitro translocation activity of SaSecA1 with an IC50 value of about 4 µM, indicating potent inhibition of SaSecA1-mediated protein translocation.
Figure 4. Inhibition of in vitro translocation activity of SaSecA1.
Liposomes-SaSecA1-dependent proOmpA translocation activity was determined at 30 °C with or without SCA-50 (n = 3). The in vitro translocation activity of SaSecA1 without inhibitor was defined as 100%.
3.3 Validation of SaSecA1 as a target by protein pull-down assay
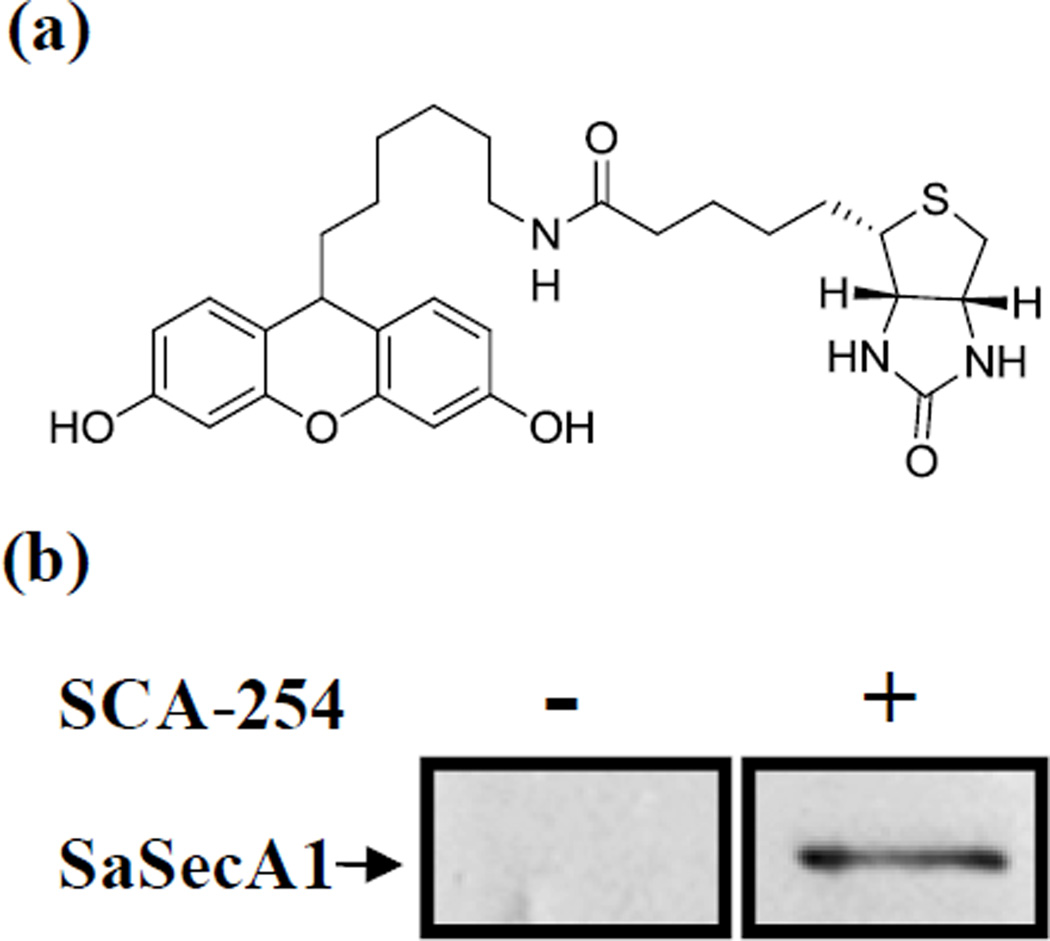
To further examine the interaction between RB analogs and SecA in biological matrices, a biotinylated SCA-50 analog (SCA-254) was synthesized and used to carry out the protein target pull-down assay (Fig. 5a). Enzymatic assays showed that SCA-254 retained reasonable inhibition effect on the ATPase activity of EcSecAN68 (IC50 ≈ 90 µM). We used EcSecAN68 to check its ATPase activity because this is the intrinsic and unregulated ATPase activity and SaSecAN68 is not available. All the other available SecA forms are regulated with an inhibitory regulatory domain. Thus we chose the EcSecAN68 assay, which is least likely to cause artifact. Of course, this is a judgment call, and there is never a “perfect model” system.
Figure 5. Validation SaSecA1 as a drug target with pull down assay.
(a) Structure of SCA-254. (b) Whole cell lysate of S. aureus ATCC 6538 was mixed with or without SCA-254 (biotinylated SCA-50 analog) for 1 hr, then Streptavidin magnetic beads were used to pull out proteins interact with SCA-254. The interaction between SaSecA1 and SCA-254 was examined by Western blot with SecA antibody.
Western blot results showed that SCA-254 could recognize and pull down SaSecA1 from whole cell lysates (Figure 5b), while control experiments did not yield the same results. Such results demonstrate the specific interactions between SecA and the inhibitor, further verifying that SaSecA1 is a target of this group of inhibitors. However, Coomassie Brilliant Blue staining showed SaSecA1 was not the only protein pulled out from whole cell lysates (data not shown). Therefore we cannot rule out the possibility that SCA-50 might have other targets besides SecA. Affinity modifications of small molecule typically result in many more non-specific binding proteins than actual targets; therefore the results observed are not uncommon in not being able to confirm the singularity of the inhibitor’s target. Furthermore, spiking SecA2 into the mixture did allow the pull down of SecA2, suggesting that the relative abundance might be the reason that SecA2 was not observed in the pull down assay using whole cell lysates.
3.4 Inhibition of the secretion of Sec-dependent secretory toxins
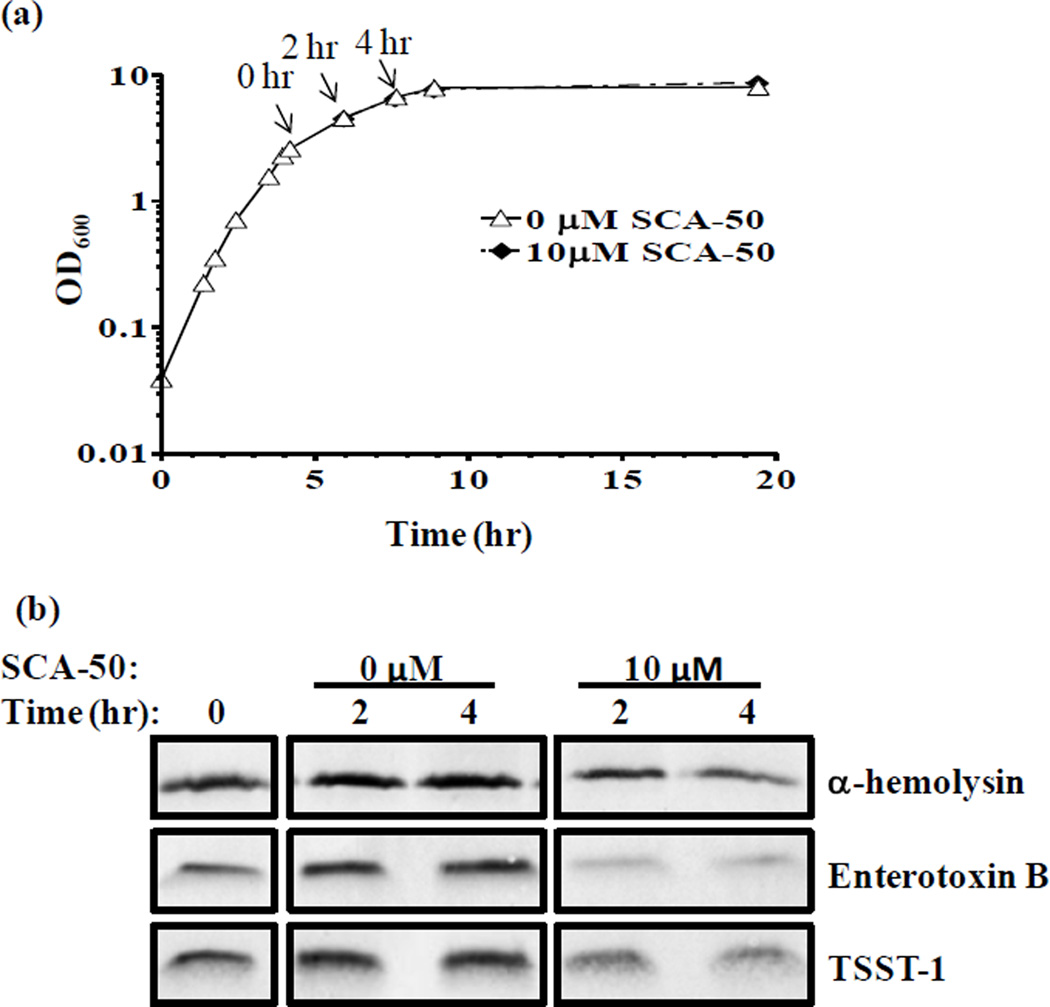
Sec-system is responsible for the secretion of more than 20 toxins or virulence factors in S. aureus [12]. These toxins play important roles in the pathogenesis of S. aureus infection. We used inhibitors at concentrations that do not affect the OD during growth to avoid complications. The MIC of SCA-50 is 12.5 µM (4 µg/ml) against S. aureus Mu50; however the compound was added to dilute early-log phase bacteria culture (OD600 ≈ 0.05) in the MIC assay. When adding into mid-log phase of S. aureus Mu50 (OD600 ≈ 2.6), SCA-50 at 10 µM didn’t obviously change the OD600 reading when compared with control within the first 15 hr (Fig. 6a), indicating that general protein synthesis was not affected. However, 10 µM SCA-50 reduced the amount of many extracellular proteins in the supernatant (data not shown). To determine whether SCA-50 could inhibit the secretion of specific S. aureus toxins, secretions of α-hemolysin, enterotoxin B, and TSST-1 were examined with or without the RB analogs. Each of these three secreted toxins contains a Sec-dependent signal peptide; thus they are expected to secret through the Sec-system. The amounts of toxins in the supernatant at different time points were determined by using specific toxin antibodies. Western blot results showed that 10 µM SCA-50 significantly decreased the amount of α-hemolysin, enterotoxin B, and TSST-1 in the supernatant (Fig. 6b). Therefore, the reduction of these toxins in the supernatant is likely due to inhibition of secretion alone. These results indicate that SCA-50 can inhibit the in vivo function of SaSecA1 in protein secretion, further validating SaSecA1 as a target in vivo.
Figure 6. Inhibition of the secretion of S. aureus toxins.
SCA-50 or same volume of DMSO as control was added to of S. aureus Mu50 at time point 0 hr as indicated. (a) Growth curve. (b) Western blot analysis the amount of toxins in the supernatant.
Moreover, α-hemolysin, enterotoxin B, and TSST-1 play important roles in the pathogenesis of S. aureus infection. In addition to these toxins, there are other twenty toxins containing Sec-dependent signal peptide, which might be secreted through the Sec-dependent pathway. All these toxins play important roles in the pathogenesis of S. aureus infection by promoting adhesion, colonization, and spread in host tissue, protecting bacteria from environment toxic condition or from host immune defense system, and causing serious host cell damage or toxic syndrome [12]. Therefore, the SecA inhibitors are expected to reduce virulence of S. aureus by blocking secretion of toxins and other virulence factors.
3.5 Bacteriostatic effects of RB analogs against MRSA strains
SecA is essential for the growth and viability of bacteria. Thus, antimicrobial activities are the most important evaluation for SecA inhibitors. RB has been reported to possess antimicrobial activities against S. aureus [24, 25]. We determined the antimicrobial effect of the RB analogs against S. aureus, including three MRSA strains (N315, Mu3, and Mu50). These inhibitors showed potent bacteriostatic effects against all S. aureus strains tested with MIC values of 3.7 to 25.6 µg/ml (Table 1). Most interestingly, the bacteriostatic effects of the tested RB analogs were more effective than that of RB in all strains tested, presumably reflecting the improved permeability of the smaller molecule of the RB analogs. MIC values of the RB analogs against MRSA had no obvious difference in MIC value against a non-MRSA strain (ATCC 6538). Three MRSA strains used in this study are agr group II, which were defective in agr function, resulting in less toxin secretion [26–28]. To further examine whether antibacterial activity of our inhibitors could be affected by the function of agr, S. aureus Newman strain with functional agr gene was used for comparison. The MIC value of this agr functional strain is similar to those three agr inactivated strains; therefore the agr gene did not affect the bacteriostatic effect of these RB analogs.
Table 1.
Bacteriostatic effect against various S. aureus strains
| Comp. | Mu50 | N315 | Mu3 | ATCC6538 | Newman |
|---|---|---|---|---|---|
| RB | 42 ± 8 | 21 ± 4 | 59 ± 22 | 42 ± 8 | 51 |
| SCA-41 | 14 | 14 | 14 | 11 | 7 |
| SCA-50 | 4 | 4 | 4 | 4 | 4 |
For MIC values (µg/ml), n = 6–9; no variation in other test.
S. aureus Mu50 is a MRSA strain with multi-drug resistance (MDR) and intermediate level resistance to vancomycin [29]. As shown in Table 2, our best SecA inhibitors were far more potent in their antimicrobial activity against Mu50 than the majority of commonly used antibiotics examined. MIC of SCA-50 is 4 µg/ml, which is lower than that of RB and is not only 250 fold lower than that of ampicillin as expected, but also that of kanamycin, erythromycin, and rifampicin against a MRSA strain. The MIC values of polymyxin B, norfloxacin, and tetracycline, are 7 to 60 fold higher than that of SCA-50. More importantly, for vancomycin, which is considered the last resort antibiotic against MRSA, its MIC is two-fold higher than that of SCA-50 (Table 2). Thus, the RB analogs described in this study are very potent inhibitors against MRSA strains.
Table 2.
Comparison of the bacteriostatic effect of SecA inhibitors with other antibiotics against S. aureus Mu50
| Antibiotics | Bacteriostatic effect MIC (µg/ml) |
|---|---|
| RB | 42 ± 8 |
| SCA-50 | 4 |
| Vancomycin | 8 |
| Ampicillin | 1000 |
| Kanamycin | 1000 |
| Polymxin B | 31 |
| Tetracycline | 63 |
| Erythromycin | >1250 |
| Norfloxacin | 250 |
| Rifampicin | >1000 |
For MIC values, n = 6; no variation in other tests.
3.6 The bactericidal effect and photooxidation
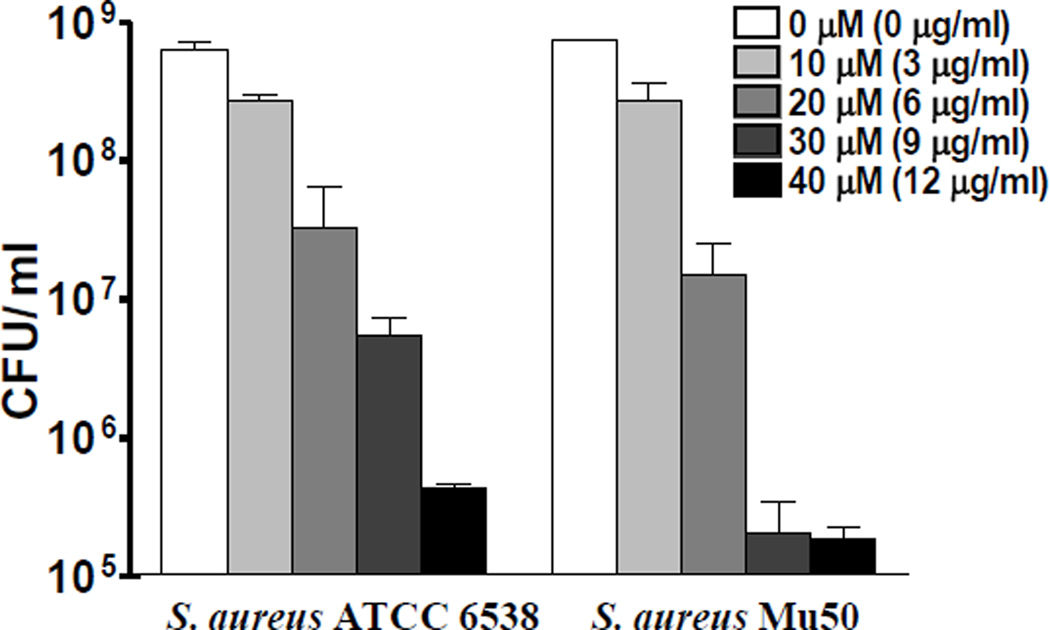
SCA-50 is our most potent compound in this class of inhibitors; thus it was further evaluated for bactericidal activity against MRSA strain Mu50 and MSSA 6538. SCA-50 showed bactericidal activity in a concentration-dependent manner against both strains: 30 µM in 1 hr was enough to achieve 2-log units of CFU reduction of MSSA 6538 and more than 3-log units of CFU reduction of MRSA Mu50 (Fig. 7).
Figure 7. Bactericidal effects of SCA-50.
Bactericidal effects were determined by counting CFU after 1 hr treatment with different concentration of SCA-50.
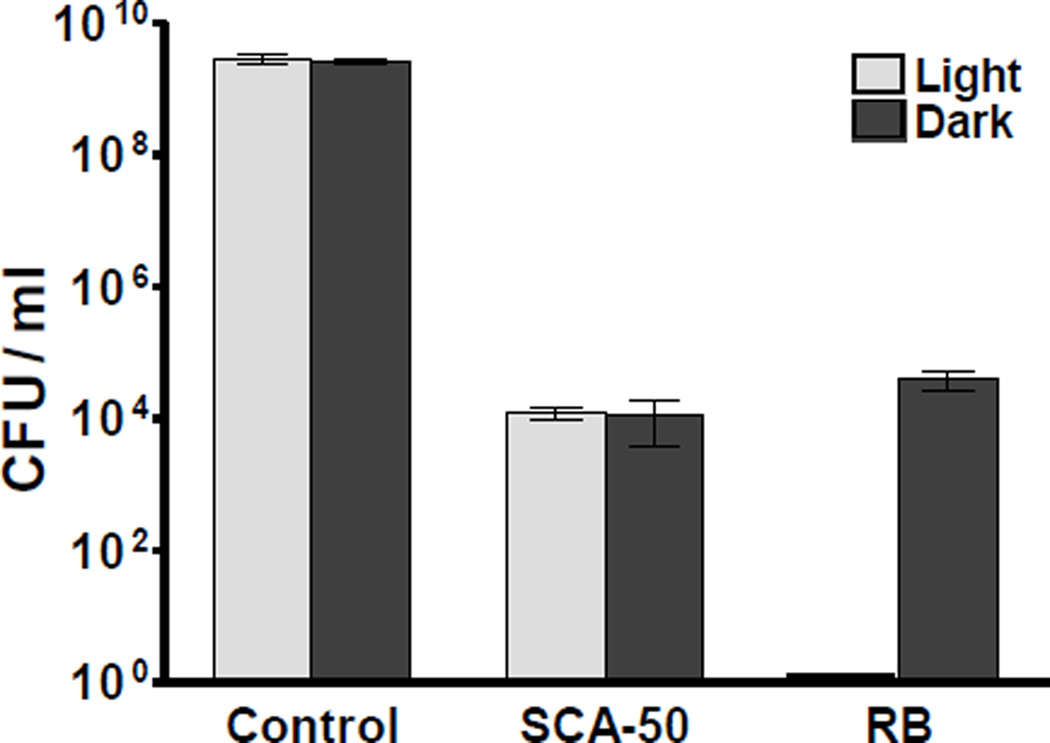
Previous studies showed that at least part of RB’s antimicrobial activities is due to photooxidation [30, 31]. Therefore, we further determined whether the activities of our smaller RB analogs are related to photooxidation. Bactericidal effects of SCA-41 and SCA-50 were tested with ATCC 6538 in the dark and under room light in comparison with RB. Under light, RB at 40 µM eliminated all 9 log of CFU, while in the dark, its bactericidal effect was reduced by 5 log units of CFU (Fig. 8), confirming that photooxidation contributes to RB’s antimicrobial activity. However the bactericidal effects of SCA-50 were not obviously affected by light (Fig. 8). Similar results were obtained with SCA-41 (data not shown). These results indicate that the antimicrobial activities of SCA-50 and SCA-41, unlike that of their parent compound RB, are not due to photooxidation.
Figure 8. Photooxidation does not contribute to the bactericidal effects of SCA-50.
Early log phase cells of S. aureus ATCC 6538 were treated with 40 µM of inhibitor for 2 hr with or without exposure to light. Control was absence of inhibitor, and bactericidal effects were determined by the reduction of CFU comparing inhibitor treatment with control.
3.7 Bypassing the negative effects of efflux pumps
Previous studies showed that SecA functions as a membrane protein and is involved in forming a protein-conducting channel that spans the entire membrane [15, 32–36]. It was reported that the majority of SecA in Streptoccous pyogenes [36, 37] and Streptococcus agalactiae [38] is present in the membranes as an ‘Exporter’. We also found that the majority of SecA1 in S. aureus is in the membrane fraction (data not shown). Thus, in Gram-positive bacteria, inhibitors may be able to directly access SecA from the extracellular matrix and exert their effect without having to enter the cells. If so, targeting SecA may allow an intrinsic way of bypassing the negative effect of efflux pumps in bacteria, which is a major mechanism for the development of multi-drug resistance [39–44]. Strains Mu50 and N315 are resistant to QacA efflux-mediated antibiotics [43, 45]. Our SecA inhibitors showed similar efficacy against strains Mu50 and N315 as against those without QacA efflux pump, suggesting that QacA-mediated efflux has no effect on the inhibitory effect of these SecA inhibitors. NorA and MepA are two efflux pumps of S. aureus with 23% or 4% frequencies of overexpression [20, 46]. We further determined whether overexpression of NorA or MepA may affect the antimicrobial activities of SCA-41 and SCA-50. Thus the antimicrobial effects of NorA or MepA deletion or overexpression mutants were compared with their parental strain 8325-4. Overexpression or deletion NorA or MepA had no significant effect on the MIC values of SCA-50 and SCA-41 (Table 3). Previous studies showed that RB is a substrate of NorA [47], which we confirmed (Table 3). Therefore RB analogs decrease the level of drug resistance in S. aureus. Such results indicate that the effects of SecA inhibitors were not affected by the presence of efflux pumps, suggesting that these two compounds either are not substrates of the tested efflux pumps or can bypass the effects of efflux pumps. In any case, these SecA analogs are effective inhibitors regardless of the levels of efflux pumps tested.
Table 3.
Bacteriostatic effects against S. aureus efflux strains.
| Compounds | WT 8325-4 |
NorA− K1758 |
NorA++ K2361 |
MepA− K2908 |
MepA++ K2068 |
|---|---|---|---|---|---|
| RB | 13 | 22 | 35 | 11 | 15 |
| SCA-50 | 4 | 4 | 4 | 4 | 4 |
For MIC (µg/ml) values, n = 6; no variation as tested.
SecA functions as a membrane protein forming a trans-membrane channel [14, 15, 33, 36, 37] and thus provides the possibility for inhibitors to reach this target without entering into the cell. In addition, SecA is involved in the secretion or assembly of many membrane proteins; therefore blocking SecA’s function probably inhibits directly or indirectly the assembly of efflux pumps. If the same concept is demonstrated in more bacterial strains with various efflux pumps, this would be the very first approach to the development of new broad-spectrum antimicrobials that have the intrinsic ability to overcome the negative effects of efflux. Given the wide-spread nature of efflux in bacteria and its importance in drug-resistance, antimicrobial agents with intrinsic abilities to overcome efflux will be extremely useful in combating bacterial drug resistance.
3.8 A putative binding site
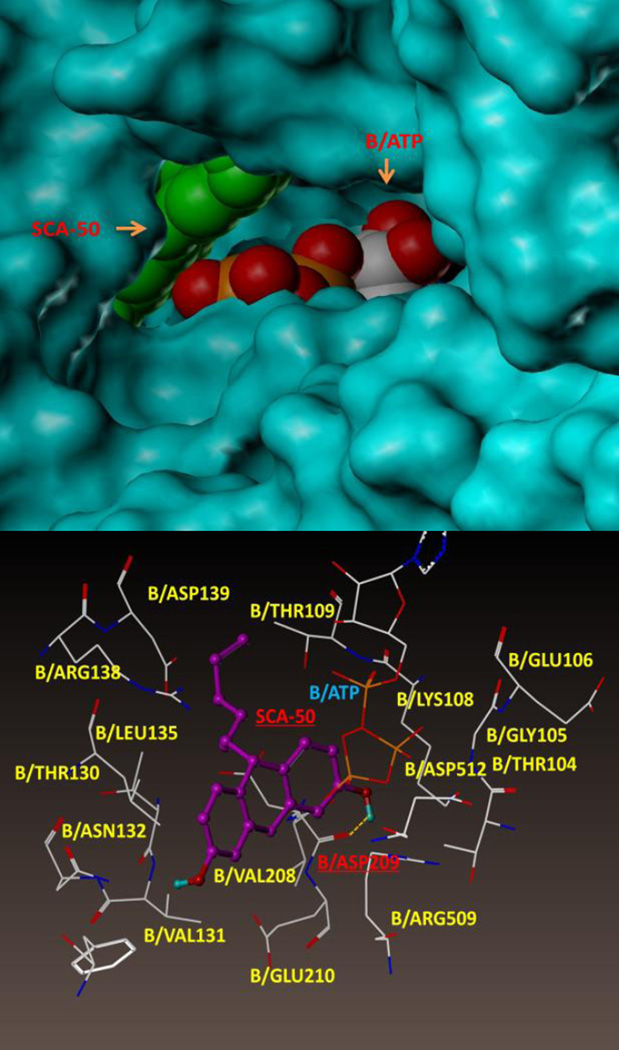
In order to achieve an initial understanding of the possible binding site, we conducted an extensive docking search using the SYBYL program. We propose that SCA-50 binds outside the ATP pocket of chain B, and is stacked against the entrance as shown in Figure 9. The binding pocket is defined by amino acid residues B/Arg 138, B/Leu 135, B/Thr 130, B/Asn 132, B/Val 208, B/Asp 209, B/ASP 512, and B/Lys 108. This site has the best docking score, and is consistent with the experimental data showing non-competitive inhibition. However, this is a hypothetical binding site. More experimental evidence is needed to validate this.
Figure 9.
Molecular modeling of SCA-50’s binding site in E. Coli SecA. Top panel shows SCA-50 partially blocks the ATP entrance in chain B. Bottom panel shows the amino acid residues in the binding pocket.
4. Conclusions
Our studies show that the small molecule RB analogs target SecA functions and have antimicrobial effects against MRSA with higher potency than most available antimicrobial agents including vancomycin. They also reduce the secretion of toxins and their potency is not affected by the presence of efflux pumps. The results obtained demonstrated an important proof of concept, i.e., targeting SecA could achieve antimicrobial effect through three mechanisms, which are not seen with any single class of antimicrobial agents.
Supplementary Material
Acknowledgments
We thank Dr. GW Kaatz from Wayne State University School of Medicine and Drs. CD Lu and Z. Eichenbaum of Georgia State University for kindly providing S. aureus strains. This work was supported in part by Molecular Basis of Diseases fellowships to J.J., and Y.H.H., the Center of Biotechnology and Drug Design at Georgia State University, and the National Institutes of Health (AI104168).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jinshan Jin, Email: jshjin@gmail.com.
Jianmei Cui, Email: jcui@gsu.edu.
Arpana Sagwal Chaudhary, Email: Sagwal.arpana@gmail.com.
Ying-Hsin Hsieh, Email: yhsieh3@gsu.edu.
Krishna Damera, Email: hzhang11@student.gsu.edu.
Hao Zhang, Email: kdamera@gsu.edu.
Hsiuchin Yang, Email: hyang@gsu.edu.
Binghe Wang, Email: wang@gsu.edu.
Phang C. Tai, Email: biopct@gsu.edu.
References
- 1.Calfee DP. Curr Opin Infect Dis. 2012;25:385. doi: 10.1097/QCO.0b013e3283553441. [DOI] [PubMed] [Google Scholar]
- 2.Kallen AJ, Mu Y, Bulens S, Reingold A, Petit S, Gershman K, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Patel PR, Fridkin SK MRSA Investigators of the Emerging Infections Program Active Bacterial Core surveillance. JAMA. 2010;304:641. [Google Scholar]
- 3.Klein E, Smith DL, Laxminarayan R. Emerg Infect Dis. 2007;13:1840. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. JAMA. 2007;298:1763. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 5.Moellering RC., Jr J Antimicrob Chemother. 2012;67:4. doi: 10.1093/jac/dkr437. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary AS, Jin J, Chen W, Tai PC, Wang B. Bioorg Med. Chem. 2015;23:105. doi: 10.1016/j.bmc.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhary A, Chen W, Jin J, Tai PC, Wang B. Future Med. Chem. 2015;7:989. doi: 10.4155/fmc.15.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Chaudhary A, Cui J, Jin J, Hsieh YH, Yang H, Huang YJ, Tai PC, Wang B. Chin. J. Pharm. Sci. 2012;21:526. [Google Scholar]
- 9.Driessen AJ, Nouwen N. Annual review of biochemistry. 2008;77:643. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 10.Natale P, Bruser T, Driessen AJ. Biochimica et biophysica acta. 2008;1778:1735. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt MG, Rollo EE, Grodberg J, Oliver DB. J. Bacteriol. 1988;170:3404. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sibbald MJ, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJ, Raangs GC, Stokroos I, Arends JP, Dubois JY, van Dijl JM. Microbiol Mol Biol Rev. 2006;70:755. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siboo IR, Chaffin DO, Rubens CE, Sullam PM. J Bacteriol. 2008;190:6188. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh YH, Zhang H, Lin BR, Cui N, Na B, Yang H, Jiang C, Sui SF, Tai PC. J Biol Chem. 2011;286:44702. doi: 10.1074/jbc.M111.300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HW, Chen Y, Yang H, Chen X, Duan MX, Tai PC, Sui SF. Proc Natl Acad Sci U S A. 2003;100:4221. doi: 10.1073/pnas.0737415100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapoport TA, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 17.Huang YJ, Wang H, Gao FB, Li M, Yang H, Wang B, Tai PC. ChemMedChem. 2012;7:571. doi: 10.1002/cmdc.201100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J, Jin J, Hsieh Y-hH, Yang H, Ke B, Tai PC, Wang B. ChemMedChem. 2013 doi: 10.1002/cmdc.201300216. [DOI] [PubMed] [Google Scholar]
- 19.Kaatz GW, Moudgal VV, Seo SM. J Antimicrob Chemother. 2002;50:833. doi: 10.1093/jac/dkf224. [DOI] [PubMed] [Google Scholar]
- 20.Kaatz GW, McAleese F, Seo SM. Antimicrob Agents Chemother. 2005;49:1857. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, Huang YJ, Gundala SR, Yang H, Li M, Tai PC, Wang B. Bioorg Med Chem. 2010;18:1617. doi: 10.1016/j.bmc.2009.12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin BR, Gierasch LM, Jiang C, Tai PC. J. Membr. Biol. 2006;214:103. doi: 10.1007/s00232-006-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CLSI. 2011 [Google Scholar]
- 24.Decraene V, Pratten J, Wilson M. Curr. Microbiol. 2008;57:269. doi: 10.1007/s00284-008-9188-7. [DOI] [PubMed] [Google Scholar]
- 25.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Lancet. 1997;350:1670. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 26.Saravia-Otten P, Muller HP, Arvidson S. J Bacteriol. 1997;179:5259. doi: 10.1128/jb.179.17.5259-5263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. Mol Microbiol. 2001;40:1439. doi: 10.1046/j.1365-2958.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 28.Sakoulas G, Eliopoulos GM, Moellering RC, Jr, Wennersten C, Venkataraman L, Novick RP, Gold HS. Antimicrob Agents Chemother. 2002;46:1492. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inbaraj JJ, Kukielczak BM, Chignell CF. Photochem Photobiol. 2005;81:81. doi: 10.1562/2003-11-04-ra-002.1. [DOI] [PubMed] [Google Scholar]
- 30.Demidova TN, Hamblin MR. Antimicrob Agents Chemother. 2005;49:2329. doi: 10.1128/AAC.49.6.2329-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Lu L, Zhu S, Li Y, Cai W. Curr. Microbiol. 2006;52:1. doi: 10.1007/s00284-005-0040-z. [DOI] [PubMed] [Google Scholar]
- 32.Carlsson F, Stalhammar-Carlemalm M, Flardh K, Sandin C, Carlemalm E, Lindahl G. Nature. 2006;442:943. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Brown T, Tai PC. J Bacteriol. 1998;180:527. doi: 10.1128/jb.180.3.527-537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Xu H, Tai PC. J Biol Chem. 1996;271:29698. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 35.Kanei M. Nihon Saikingaku Zasshi. 1983;38:645. [PubMed] [Google Scholar]
- 36.Rosch JW, Caparon MG. Mol Microbiol. 2005;58:959. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- 37.Brega S, Caliot E, Trieu-Cuot P, Dramsi S. PLoS One. 2013;8:e65832. doi: 10.1371/journal.pone.0065832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikaido H, Zgurskaya HI. Curr Opin Infect Dis. 1999;12:529. doi: 10.1097/00001432-199912000-00001. [DOI] [PubMed] [Google Scholar]
- 39.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TA, Kaatz GW. Antimicrob Agents Chemother. 2007;51:3235. doi: 10.1128/AAC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Lancet. 2001;357:1225. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 41.Levy SB. Symp Ser Soc Appl Microbiol. 2002;65S [PubMed] [Google Scholar]
- 42.Markham PN, Neyfakh AA. Curr Opin Microbiol. 2001;4:509. doi: 10.1016/s1369-5274(00)00243-5. [DOI] [PubMed] [Google Scholar]
- 43.Van Bambeke F, Balzi E, Tulkens PM. Biochem Pharmacol. 2000;60:457. doi: 10.1016/s0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Eric Ballard C, Zheng SL, Gao X, Ko KC, Yang H, Brandt G, Lou X, Tai PC, Lu CD, Wang B. Bioorg Med Chem Lett. 2007;17:707. doi: 10.1016/j.bmcl.2006.10.094. [DOI] [PubMed] [Google Scholar]
- 45.Li XZ, Nikaido H. Drugs. 2009;69:1555. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tegos GP, Masago K, Aziz F, Higginbotham A, Stermitz FR, Hamblin MR. Antimicrob Agents Chemother. 2008;52:3202. doi: 10.1128/AAC.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price CT, Kaatz GW, Gustafson JE. Int J Antimicrob Agents. 2002;20:206. doi: 10.1016/s0924-8579(02)00162-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.