Abstract
Congenital hearing loss is an important clinical problem because, without early intervention, affected children do not properly acquire language and consequently have difficulties developing social skills. Although most newborns in the US are screened for hearing deficits, even earlier diagnosis can be made with prenatal genetic screening. Genetic screening that identifies the relevant mutated gene can also warn about potential congenital defects in organs not related to hearing. We will discuss efforts to identify new candidate genes that underlie the Branchiootorenal spectrum disorders in which affected children have hearing deficits and are also at risk for kidney defects. Mutations in two genes, SIX1 and EYA1, have been identified in about half of the patients tested. To uncover new candidate genes, we have used the aquatic animal model, Xenopus laevis, to identify genes that are part of the developmental genetic pathway of Six1 during otic and kidney development. We have already identified a large number of potential Six1 transcriptional targets and candidate co-factor proteins that are expressed at the right time and in the correct tissues to interact with Six1 during development. We discuss the advantages of using this system for gene discovery in a human congenital hearing loss syndrome.
Keywords: Branchiootorenal, Eya1, Mcsr1, Six1, Sobp, 2G4/Eb1, Xenopus, Zmym2
Introduction
A very important use of animal models in biomedical research is to discover new genes involved in human diseases and characterize the functions of the proteins that they encode in normal and disease states. Xenopus laevis, the South African clawed frog, has been a leading model for uncovering the molecules that control normal embryonic development in vertebrates because the embryos are large, produced in abundance, and highly accessible for experimental manipulations, which facilitates their use in biochemical and molecular analyses (Gilchrist, 2012; Harland and Grainger, 2011; Khokha, 2012). Now that the Xenopus genome has been sequenced, shown to have high synteny with human, and can be genetically modified (Blitz et al., 2013; Hellsten et al., 2010; Nakayama et al., 2013; Nakayama et al., 2014), they also are ideal for modeling human developmental defects and congenital syndromes. In this review, we will discuss a hearing loss syndrome in which we have exploited the experimental resources of Xenopus and of Drosophila to discover new genes that may underlie human congenital disorders that affect ear development.
Branchiootorenal Spectrum Disorders
Hearing loss is the most prevalent birth defect in developed countries (Hilgert et al., 2009); it occurs in 1/500 newborns and increases to 3.5/100 by adolescence (Morton and Nance, 2006; Smith et al., 2014). Half of the cases are due to genetic causes, and over 400 genetic syndromes that include hearing loss have been described (Toriello et al., 2004). It is very important to diagnose congenital hearing loss as early as possible because without intervention these children have difficulty acquiring language skills. As a consequence of their impaired communication, they also often suffer from social isolation. According to the National Institutes of Health, about 98% of infants born in hospitals in the United States are now functionally screened for hearing loss before being discharged because early interventions, including cochlear implants, otologic surgery and intensive speech therapy, have proven successful in ameliorating the language and social problems that these children face.
Branchiootorenal spectrum disorders are the second most common type of autosomal dominant syndromic hearing loss (Smith, 2014). Affected individuals have a 50% chance of transmitting the disorder to each of their children, penetrance is estimated to be 100%, and prevalence is estimated to be about 1:40,000 (Chang et al., 2004; Fraser et al., 1980). Two syndromes, Branchiootic syndrome (BOS) and Branchiootorenal (BOR) syndrome, comprise this spectrum. Both are characterized by: second branchial arch malformations including fistulas and cysts; external ear malformations including preauricular pits, lop-ear deformities, cup-ear deformities, preauricular tags, and atresia or stenosis of the external auditory canal; middle ear malformations including ossicle deformities, ossicle fixation and malformed middle ear space; and inner ear malformations including cochlear hypoplasia, enlarged cochlear and vestibular ducts, and semicircular canal hypoplasia (Fig. 1). BOR is diagnosed when kidney malformations are additionally detected. Although this is a fully penetrant, autosomal dominant syndrome, patients present with highly variable degrees of malformations, even when comparing right and left sides of the same individual (Fig. 1). Hearing loss is a hallmark, but its extent is highly variable and can be conductive, sensori-neural or mixed in type. Many of the craniofacial abnormalities can be surgically corrected, but patients need interventions for their hearing impairments as early as possible to promote language acquisition. BOR patients additionally require close monitoring for renal function because they may eventually require kidney transplantation or dialysis.
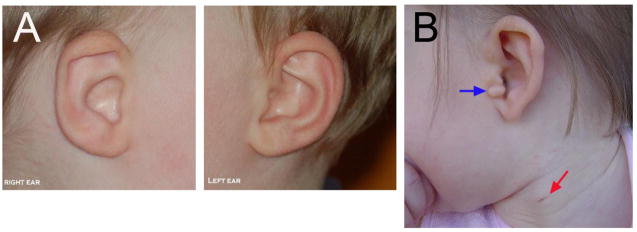
Figure 1.
External phenotypes of BOS/BOR patients. (A) The shapes of the two external ears of a patient are asymmetric. The right ear shows an affected phenotype whereas the left ear appears normal. (B) A patient showing a preauricular tag (blue arrow) and a second branchial arch fistula (red arrow). (Images downloaded from: http://pl.wikipedia.org/wiki/Zespół_skrzelowo-uszno-nerkowy and used under the terms of the Creative Commons Attribution-Share Alike 3.0 Unported license).
Mutations in two genes have been identified in about half of patients with BOS/BOR (reviewed in Moody and Saint-Jeannet, 2014; Smith, 2014). The gene encoding SIX1, a homeodomain transcription factor, is mutated in about 4% of patients diagnosed as BOS3 (Online Mendelian Inheritance in Man [OMIM] #608389) or BOR3 (Smith, 2014). The gene encoding EYA1, a co-factor protein that binds to SIX1 and modifies its transcriptional activity, is mutated in about 40% of patients diagnosed as BOS1 (OMIM #602588) or BOR1 (OMIM #113650). There is a report that the related SIX5 protein is causative for BOR2 (Hoskins et al., 2007), but Six5 is not normally expressed in otic precursors (reviewed in Brugmann and Moody, 2005) and a recent study disputes the reported mutation (Krug et al., 2011). Identifying gene mutations in BOS/BOR patients is of diagnostic importance because their variable hearing losses can be difficult to detect by the standard methods used in newborns and potential kidney defects can be life threatening. For these reasons, screening for mutations in SIX1 and EYA1 are often included in genetic screens (e.g., the OtoSCOPE panel, University of Iowa; the Branchiootorenal Spectrum panel at Cincinnati Children’s Hospital Medical Center). Yet current prenatal/perinatal testing cannot detect the mutated gene in over half of BOS/BOR cases. Since genetic screening is considered a best practice to identify at-risk BOS/BOR children and begin early treatment (Smith, 2014), we initiated screens in Xenopus to identify potential candidate genes that are genetically and functionally related to Six1 during ear (otic) development. One approach has been to identify genes that are regulated by Six1, and therefore are part of a gene regulatory network that controls ear and kidney development. A second approach has been to identify novel cofactor proteins that modify Six1 activity. The rationale is that new genes discovered in the Six1-regulated pathway controlling ear and kidney development are high priority candidates for functional characterization and ultimate inclusion in BOS/BOR genetic screening.
Why use Xenopus?
Xenopus has been a preferred vertebrate model for discovering new genes that control developmental processes and for elucidating their cellular and protein functions (Gilchrist, 2012; Harland and Grainger, 2011; Khokha, 2012; Wheeler and Brändli, 2009; see also http://www.xenbase.org/anatomy/intro.do). A female can lay up to a few thousand eggs at one time, and they develop externally and are very large (~1400μm in diameter compared to ~700μm for zebrafish and ~100μm for mouse). These attributes mean that it is possible to easily collect enough material at specific developmental times or from specific regions of the embryo to perform high-throughput, in vivo biochemical analyses of gene regulation and protein function. In addition, embryonic manipulations, such as single cell or tissue isolation, culture, and transplantation, are quite feasible. The individual cells of the early embryo (up 2- through 32-cells) can be identified and their fates have been mapped by lineage tracing techniques. These maps are used to target manipulations to the progenitors of specific tissues, such as the neural crest that becomes the middle ear, the placodes that become the inner ear and the mesoderm that becomes the kidney. These identified progenitors can be microinjected with a variety of molecules, including proteins and nucleic acids. Targeted microinjection of mRNAs can be used for gain-of-function of the encoded protein or expression of the protein in an ectopic location. Microinjection of dominant-negative constructs or antisense morpholino oligonucleotides (MOs) that block RNA splicing or protein translation allows one to perform loss-of-function experiments in specific lineages. In addition, transgenesis techniques for testing the regulations of gene expression are well worked out. A drawback of Xenopus laevis, the most commonly used species, has been its lack of easy genomic mutation due to the genome being allotetraploid. However, the introduction of Xenopus tropicalis into the laboratory has overcome this problem, as this species is diploid and has a generation time similar to mouse. For both species, genome editing techniques to make customized gene mutations is being very successfully applied. Thus, Xenopus is a versatile and powerful animal model in which one can rapidly test the function of genes and proteins (wild-type or mutant from frog or any other species) in an in vivo system.
Furthermore, because Xenopus is a tetrapod, it shares a close evolutionary history with mammals. The Xenopus and human genomes are highly syntenic (Hellsten et al., 2010; http://www.xenbase.org/entry/doNewsRead.do?id=136) and many proteins, including Six1 and Eya1, are highly conserved at the amino acid level. Transcriptome comparisons show that the mature inner ear of human and Xenopus express many of the same genes (Powers et al., 2012). In addition, as a tetrapod the otic anatomy of Xenopus evolved for land-based hearing. While frogs do not have external ears, the mechanics of sound transmission is very similar to that in humans (Mason et al., 2009; Van Dijk et al., 2011). They detect sound by deflections of a tympanic membrane, and these are transmitted through an air-filled middle ear by two ossicles (derived from the cranial neural crest; Sandell, 2014) to the inner ear sensory epithelium (derived from the otic placode/otocyst). The amphibian inner ear is comprised of 5 vestibular end-organs, two auditory end organs and one acoustico-vestibular sacculus that function similarly to the mammalian inner ear. In fact, the auditory organs detect frequencies in the same range as the mammalian cochlea (Elepfandt et al., 2000; Schoffelen et al., 2008; Van Dijk et al., 2011). Thus, novel genes in the Xenopus Six1 pathway are likely to be highly relevant to human ear development and related congenital disorders.
Six family genes
SIX1 is one of 6 vertebrate transcription factors highly related to Drosophila Sine oculis (SO). SO has been studied in great detail because it is a major player in the formation of the Drosophila visual system. In fact, SO contributes to the development of every visual component, including the photosensitive organs of the larva (Bolwig’s organ), the fly (compound eyes and ocelli) and the processing centers in the brain (optic lobes) (Cheyette et al., 1994; Blanco et al., 2010a; Piñeiro et al., 2014; Serikaku and O’Tousa, 1994). In the developing compound eye, where its function is best understood, SO controls the expression of critical factors at every step of organ formation, thereby contributing from the specification of retinal progenitors to the differentiation of the photoreceptor neurons and their accessory cells (Hayashi et al., 2008; Pauli et al., 2005; Pignoni et al., 1997; Yan et al., 2003; Zhang et al., 2006; Zhou et al., 2014).
All SO/Six proteins contain a highly conserved Six-type homeodomain, which binds DNA, and an N-terminal domain, called the Six domain (SD), that interacts with co-factor proteins (Kawakami et al., 2000; Kobayashi et al., 2001; Pignoni et al., 1997). The Six genes from fly and vertebrates are highly conserved and have been grouped into three subfamilies (Six1/Six2; Six4/Six5; Six3/Six6) on the basis of sequence variation in both homeodomain and SD (Kawakami et al., 2000; Seo et al., 1999). The presence of Drosophila representatives in each class (SO=Six1/2; Six4; Optix=Six3/6) shows that their origin predates the last common ancestor of bilaterians. Interestingly, all three fly genes are located on the same chromosome arm, and in humans, mouse, Xenopus, chicken and zebrafish, one member of each class (Six1, Six4 and Six6) are also clustered on the same chromosome. Other vertebrate family members map to different sites. In humans, mouse and Xenopus, Six2 and Six3 are clustered on another chromosome, and Six5, in contrast, is located on a separate chromosome. It has been proposed that the differences in their arrangements on the chromosomes may account for minor expression and functional differences during development (Moody and Saint-Jeannet, 2014).
Three of the Six genes (Six1, Six2, Six4) have overlapping expression patterns in the cranial placodes that give rise to the sensory organs of the vertebrate head, including the otic placode that gives rise to the inner ear and to the auditory-vestibular sensory ganglion; they also are expressed in the developing kidney (reviewed by Brugmann and Moody, 2005; Saint-Jeannet and Moody, 2014). There is very little information on the function of Six2 or Six4 in otic development (Ozaki et al., 2001; Self et al., 2006), although a role for Six4 is suggested because Six1/Six4-double null mutant mice have more severe defects than either single mutant (Chen et al., 2009; Grifone et al., 2005; Zou et al., 2006a). To our knowledge, no human syndromes have been assigned to mutations in SIX2 or SIX4. In contrast, several studies in frog, fish, chicken and mouse demonstrate that Six1 has a central role in cranial placode development. Six1 loss-of-function in Xenopus and chick results in reduced expression of several placode genes and defects in otic development (Brugmann et al., 2004; Christophorou et al., 2009; Schlosser et al., 2008). In zebrafish, Six1 knock-down results in loss of inner ear hair cells (Bricaud and Collazo, 2006; Bricaud and Collazo, 2011). Six1-null mice show defects in the olfactory placode, inner ear and cranial sensory ganglia (Chen et al., 2009; Ikeda et al., 2007; Ikeda et al., 2010; Konishi et al., 2006; Laclef et al., 2003; Ozaki et al., 2004; Zheng et al., 2003; Zou et al., 2004); whereas Six1-heterozygous mice have hearing loss due to cochlear defects (Zheng et al., 2003). Six1 gain-of-function studies in Xenopus and chick further show that this factor expands placode domains at the expense of the adjacent epidermal, neural crest and neural plate regions (Brugmann et al., 2004; Christophorou et al., 2009).
Mutations in SIX1 have been identified in about 4% of BOS/BOR patients. Nine mutations in BOS3 patients from 16 unrelated families have been reported to date. Seven are missense mutations in the SD and two are missense or deletion mutations in the homeodomain (Fig. 2) (Ito et al., 2006; Kochhar et al., 2008; Noguchi et al., 2011; Ruf et al., 2004; Sanggaard et al., 2007). The mutations either disrupt the interaction with Eya1 or the ability to bind to DNA (Patrick et al., 2009; Ruf et al., 2004). In zebrafish, expression of mutant Six1 mRNA that carries a BOS patient mutation (R110W) interferes with the Six1-Eya1 interaction that promotes cell proliferation and hair cell formation (Bricaud and Collazo, 2011). The Catweasel (Cwe) mouse mutant harbors a missense mutation in the SD (Fig. 2; Bosman et al., 2009) that is similar to a mutation in a BOR family whose only reported defects are auditory (Mosrati et al., 2011). Heterozygous-Cwe mice have an ectopic row of hair cells in the cochlea and homozygous-Cwe mice have fewer hair cells in the cochlea, semicircular canals and utricle.
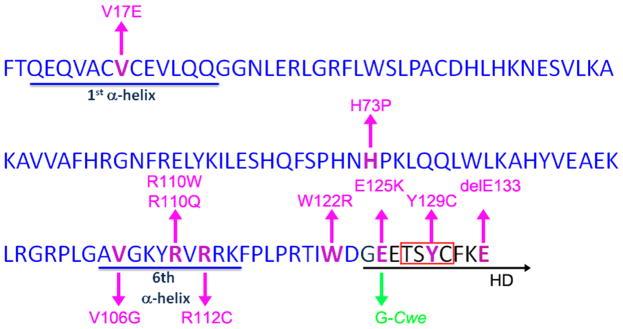
Figure 2.
The amino acid sequence of the Six domain (SD) and the homeodomain (HD) in human and Xenopus laevis Six1 are identical. The amino acids comprising the SD are blue and those comprising the N-terminus of the HD are black and are underlined in black. The 1st and 6th of the six α-helices in the SD are underlined in blue. The known human mutations are noted in magenta using the human Six1 amino acid numbers; seven are in the SD and three are in the HD. The Cwe mutation is marked in green. Most of the known human mutations occur in the 1st α-helix, 6th α-helix or the N-terminal region of the HD. The red box in the HD denotes the tetrapeptide used to discriminate the three Six subfamilies (Seo et al., 1999).
Although Six1 is a key regulator of placode, and therefore ear, development, mutations in Six1 only account for a small number of BOS/BOR patients. Therefore, it is important to identify additional components of the Six1 genetic network linked to ear formation. Interestingly, mutations in the cofactor Eya account for a much larger fraction of cases than Six1 (Smith, 2014). It is possible that the lower representation of Six1 mutations in BOS/BOR patients reflects the pleiotropic role of the DNA-binding factor, whose loss would present with more complex phenotypes or lead to catastrophic outcomes. In this context, loss-of-function of single targets or partners of Six1 would impair some but not all Six1 functions and result in viable but affected individuals. For this reason, we decided to focus on identifying the factors that either carry out the Six1 program (i.e., transcriptional targets) or modify its activity (i.e., co-factors). Eya1 is an excellent example of both: it is up-regulated by Six1 (Brugmann et al., 2004) and it modifies the transcriptional activity of Six1 (Ikeda et al., 2002; Li et al., 2003; Silver et al., 2003). We review here current knowledge on both Six and Eya genes, and describe how we are using Xenopus and Drosophila to identify new candidates for BOS/BOR causative genes.
Six1 transcriptional targets
Six1 and Eya1 are expressed in the developing placodes, including the otic placode, and the developing kidney (Neilson et al., 2010; Ohto et al., 1998; Pandur and Moody, 2000; Xu et al., 1999; Xu et al., 2003). Genes that act downstream of Six1 in the ear and kidney precursors are high priority candidates for novel BOS/BOR-causing mutations because they carry out the Six1 developmental program. Originally linked in the regulation of eye development in Drosophila, Pax, Six and Eya genes, as well as Fox genes, are all involved in kidney and placode development in vertebrates (Fig. 3; reviewed in Bhattacharyya and Bronner-Fraser, 2004; Brodbeck and Englert, 2004; Grocott et al., 2012; Moody and LaMantia, 2015). Experimental studies indeed show that Pax and Fox genes play several roles in placode development. For example, Pax2 and Pax8 are expressed in the otocyst and kidney as well as other organs, and Pax2 can interact with Eya in ear development (Zou et al., 2006b). Knock down of Pax2 in several animal models have ear defects, and while patients with PAX2 mutations present primarily with eye and renal defects (OMIM 167409), some also have sensorineural deafness (reviewed in Grocott et al., 2012). Patients with PAX8 mutations and Pax8-null mice present primarily with hypothyroidism and kidney defects (OMIM 167415), whereas evidence from mouse, chick and zebrafish suggest that Pax2 and Pax8 cooperate in ear development (reviewed in Grocott et al., 2012). Although all placodes are initially specified to express Pax6 (Bailey et al., 2006), patients with PAX6 mutations present primarily with ocular and brain defects. While Foxi1 and Foxi3 are important for otic placode development in animal models (Khatri and Groves, 2013; Nissen et al., 2003; Ohyama and Groves, 2004; Solomon et al., 2003), only patients with FOXI1 mutations present with deafness and vestibular defects (OMIM 601093). In mouse, Foxg1 is expressed in all placodes and plays a role in auditory and cerebral cortex development (Duggan et al., 2008; Hatini et al., 1999; Hwang et al., 2009; Kawauchi et al., 2009; Pauley et al., 2006). However, patients with FOXG1 mutations present with Rett Syndrome and severe intellectual disability (OMIM 164874). Thus, while mutations in some of these genes are associated with syndromes affecting the ear or the kidney, none of them affect both or phenocopy BOS/BOR. Similarly, other studies have experimentally placed SoxB1 members (e.g., Sox2), Irx1, Tbx1, and Tbx5 downstream of Six1 in placode development (Brugmann et al., 2004; Schlosser and Ahrens, 2004; Schlosser et al., 2008; reviewed in Grocott et al., 2012; Streit, 2004). However, the transcriptional relationships of these genes to Six1 have not yet been clearly delineated, and human mutations in these genes are associated with tissues other than ear and kidney. Therefore, a search for additional transcriptional targets is warranted.
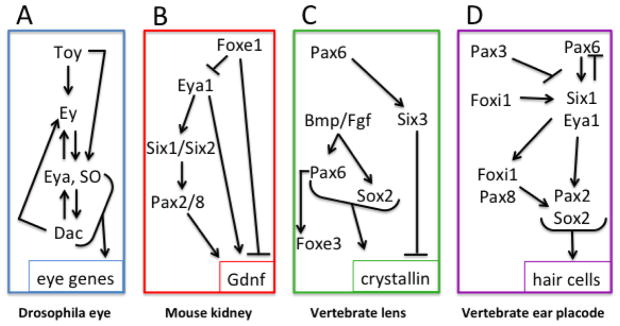
Figure 3.
Four different tissues use a combination of Pax, Six, Eya and Fox transcription factors during development. Note that the epistatic relationships between these genes are specific to the tissue. (A) In Drosophila eye, a combination of SO, Eya and Dac is required to initiate expression of eye-specific genes. Toy and Ey, which are homologues of vertebrate Pax6, act upstream. (Based on Brodbeck and Englert, 2004) (B) In developing mouse kidney, a combination of Six, Eya and Pax up-regulate an important kidney signaling factor, Gdnf. Foxe1 inhibits kidney differentiation. (Based on Brodbeck and Englert, 2004) (C) In developing vertebrate lens, Pax6 up-regulates Six3, which is required for lens formation but inhibits terminal differentiation. Bmp and Fgf signaling are required to maintain Pax6 expression and initiate Sox2 expression, which in combination up-regulate crystalline genes. Pax6 acts upstream of Foxe3, which maintains lens cells in an undifferentiated state. (Based on Kenyon et al., 1999; Bhattacharyya and Bronner-Fraser, 2004) (D) In vertebrate placode development, Pax6 is expressed in pre-placodal ectoderm upstream of Six1 and Eya1, but must be down-regulated in the ear placode. Pax3 expression in the neural border zone promotes neural crest formation and represses placode formation. Early Foxi1 expression promotes placodes, and later Foxi1 expression, along with Pax2 and Pax8 promotes otic placode formation. A combination of Pax2 and Sox2 promotes hair cell genes. (Based on Grocott et al., 2012; Moody and LaMantia, 2015).
The extensive studies of eye development in Drosophila have been a rich source of factors associated with SO/Six function; however, very little is known about any potential role in the auditory or renal systems of the fly. Nonetheless, SO directly regulates several genes (reviewed in Jusiak et al., 2014a, b) whose vertebrate homologues are involved in placode development: eyeless (vertebrate Pax6, mentioned above), dachshund (vertebrate Dach1/2, which binds Eya), atonal (vertebrate Atoh family, members of which are involved in placode neurogenesis), prospero (vertebrate Prox1/2 are involved in lens placode development), and hedgehog (vertebrate Shh is involved in adenohypophyseal placode development) (reviewed in Saint-Jeannet and Moody, 2014). Ear defects have not been identified in mouse null mutants of Dach, Prox or Shh genes. Although Atoh1 is required for ear development in animal models, no human mutations have yet been described (OMIM 601461). A ChIP-Seq analysis of SO binding to DNA isolated from developing fly eye-antennal imaginal discs identified nearly 6,000 putative SO target genes, over half of which do not have a described function in eye development (Jusiak et al., 2014b). Perhaps some of these genes may be relevant to the transcriptional regulation of vertebrate placode development, based on the sometimes surprisingly robust conservation of regulatory networks between flies and vertebrates. With the similar goal of screening for novel Six1 targets, we performed a microarray expression assay on Xenopus Six1-expressing ectodermal explants, and identified 72 genes that were significantly up-regulated and 58 genes that were significantly down-regulated (Yan et al., 2015). As in the fly ChIP-Seq study, most candidates are of unknown function, but over 30 of these genes are expressed in placodes (including otic) and in kidney precursors. We are currently performing loss-of-function and gain-of-function assays to investigate their potential involvement in BOS/BOR pathologies.
Six1 co-factors
An important aspect of regulation of gene expression by SO/Six proteins involves their association with co-factors that do not bind DNA themselves but influence the transcriptional activity of the complex (reviewed in Saint-Jeannet and Moody, 2014). Experiments with fly and vertebrate SO/Six proteins show that an association with Eya co-factors promotes transcriptional activation, whereas recruitment of Groucho (Gro)/Groucho-related (Grg) co-factors results in transcriptional repression (Ikeda et al., 2002; Li et al., 2003; Silver et al., 2003; Zhu et al., 2002). In the frog embryo, expressing an activating Six1 protein (SD and homeodomain fused to the viral VP16 activation domain) induced the same phenotypes as co-expression of wild-type Six1 with Eya1. In contrast, expressing a repressive Six1 protein (SD and homeodomain fused to the fly Engrailed repressive domain) induced the same phenotypes as co-expression of wild-type Six1 with Gro (Brugmann et al., 2004). Similarly, co-expression of Six1 and Eya2 in chick embryos up-regulates placode genes (Christophorou et al., 2009). Interestingly, in the zebrafish otocyst, combined Six1/Eya1 activation of target genes is required for hair cell formation, whereas combined Six1/Gro repression of target genes is required for auditory-vestibular ganglion neuron formation (Bricaud and Collazo, 2011).
Eya and other co-factors bind to the N-terminal Six domain (SD) of SO/Six proteins (Kawakami et al., 2000; Kobayashi, et al., 2001; Pignoni et al., 1997) (Fig. 2). The crystal structure of the human SIX1 SD bound to the EYA2-Eya domain (ED) shows that the SD is folded into six α-helices and it predominantly interacts with the EYA2-ED via the first α-helix (Patrick et al., 2013). Strikingly, the amino acid sequence of the first α-helix (Fig. 2) is 100% identical in fly, frog, mouse and human. One of the known BOR mutations occurs in the first α-helix (V17E) and this single amino acid substitution inhibits Eya binding (Patrick et al., 2009; Ruf et al., 2004). The majority of the other known BOS/BOR mutations occur in the sixth α-helix of the SD or just C-terminal to it in the homeodomain (HD) (Patrick et al., 2009; Ruf et al., 2004; Sanggaard et al., 2007). The SIX1/EYA2-ED crystal structure predicts that the sixth α-helix could contact the major groove of DNAand suggests that mutations in the sixth α-helix might interfere with DNA binding (Patrick et al., 2013). However, conflicting biochemical data have been reported, showing sixth α-helix mutations to result in impaired (Ruf et al., 2004) or normal (Patrick et al., 2009) DNA binding. Since mutations in the sixth α-helix also alter the specificity of co-factor binding in fly (Kenyon et al., 2005b) and disrupt Eya binding in yeast (Ruf et al., 2004), this α-helix also is likely to have a role in SO/Six association with co-factors. Another mutation, W122R, lies between the sixth α-helix and the homeodomain (Fig. 2). The functional consequence of this mutant has yet to be determined, however since tryptophan (W) is found at this position in all species and all Six proteins (Six1-Six6) studied to date it is likely to have an important functional role. We predict that since W122 is in the unstructured flexible linker region between the SD and HD, it is likely to be an exposed residue. If this is the case, it may bind to other hydrophobic residues mediating protein-protein interactions. The nearby Y129C mutation has been predicted to interfere with EYA1 binding and with binding to target genes (Patrick et al., 2009 Ruf et al., 2004), but it had no discernable effect when expressed in wild-type zebrafish (Bricaud and Collazo, 2011). Clearly, our understanding of the consequences of the BOS/BOR mutations in SIX1 is rudimentary. It will be important to carry out a more nuanced analysis of the interactions between cofactor proteins and SIX1 disease variants, in order to understand the contributions of SIX1-based complexes to ear development and pathology. For example, mutations that cause disruption of fewer interactions or more subtle changes in binding affinities may be associated with the milder congenital defects of the ear that present in some BOR/BOS patients.
The focus of SO/Six co-factor research has been primarily on Eya and Gro/Grg family members because they are known to bind to Six proteins, affect their activity and are expressed in the same tissues, the ear and kidney being of particular relevance to BOS/BOR (Bajoghli et al., 2005; Bane et al., 2005; Brugmann and Moody, 2005; Brugmann et al., 2004; Kobayashi et al., 2001; Li et al., 2010; Neilson et al., 2010; Ohto et al., 1999; Ozaki et al., 2002; Zhu et al., 2002). But, SO/Six1 proteins also can interact with other proteins. Extensive yeast two-hybrid assays in fly identified more than 25 proteins that interact with SO (Anderson et al., 2014; Giot et al., 2003; Kenyon et al., 2005a; Pignoni et al., 1997). As expected, Eya and Gro have the highest interaction scores, but several other proteins are likely to also act as co-factors. We took advantage of this data set to screen for potential vertebrate Six1 co-factors using Xenopus. Because the amino acid sequence of the SD of fly SO is highly conserved in vertebrate Six1 and the frog SD amino acid sequence is identical to human (Fig. 2), we predicted that proteins that bind to fly SO are likely to bind to vertebrate Six1, and thereby be relevant to BOS/BOR. Our approach was to first perform BLAST analyses to identify genes in the known Xenopus transcriptome (this was done before the Xenopus genome sequence was available) that are likely homologues of the fly co-factors. We identified 33 genes with high sequence similarity to 20/25 fly SO-interactors. We next performed in situ hybridization (ISH) analyses for 20 of those genes (representing 11 of the fly factors), and found that 16 are expressed in the otic placode/otocyst and 11 in the nephric mesoderm (Neilson et al., 2010). We then considered several criteria to nominate a gene as a potential candidate for BOS/BOR: 1) high similarity in protein structure to the human proteins; 2) expression in the developmental rudiment of the inner ear (otic placode/otocyst) and/or middle ear (neural crest-derived branchial arches); 3) requirement for normal ear development; 4) binding to the Six1 protein; and 5) ability to affect Six1 transcriptional activity. We are using these criteria to begin a molecular and functional characterization of five novel candidate co-factors: Sobp (Sine oculis binding protein); Zmym2/Zfp198; Zmym4/MGC132098; 2G4/Ebp1; and Mcrs1/LOC100049093 (Microspherule-1 protein).
Sobp is a FCS-type zinc finger protein that contains two proline-rich domains (including Box 3) and two nuclear localization signals (NLS) that are highly conserved from fly to human (Fig. 4). Sobp also contains a cluster of SUMO-interacting motifs of unknown function (Sun and Hunter, 2012). Fly Sobp is comprised of 813 amino acids, Xenopus of 871 and human of 873; human and Xenopus Sobp are 82.5% identical at the amino acid level. In the fly eye field, Sobp expression is restricted to the developing neuronal region where cells acquire their specific fate and differentiate accordingly (Kenyon et al., 2005a). In frog, Sobp is expressed in the neural tube, and several placodes including the otic placode. It is not expressed in the neural crest derived branchial arch mesoderm that gives rise to the middle ear, and thus cannot contribute to BOS/BOR middle ear defects. It also is not expressed in the kidneys, and therefore cannot contribute to BOR renal dysfunction (Neilson et al., 2010). It has a similar expression pattern in mouse (Chen et al., 2008). Ectopic expression of Sobp in fly retina progenitors maintains them in an undifferentiated, proliferative state (Kenyon et al., 2005b), but it is unknown how this gain-of-function phenotype relates to Sobp’s physiological role since the effect of loss-of-function is still unknown. Important insights, instead, have come from two naturally occurring mutations in mouse Sobp that result in inner ear deformities affecting auditory and vestibular functions (Chen et al., 2008). In humans, a mutation in SOBP caused craniofacial abnormalities in a family with hearing loss in one member (Birk et al., 2010). Both the mouse and human mutations result in truncated proteins that are missing the C-terminal NLS; in addition, part of the proline-rich domain and Box 3 are deleted by the mouse mutation that causes the more severe phenotype (Fig. 4). The function of the proline-rich domain is not yet known. However, proline-rich regions mediate protein/protein interactions and are often found in zinc finger proteins (Gerber et al., 1994; Kay et al., 2000; Morgan and Rubenstein, 2013; Williamson, 1994). Some studies show that these domains participate in transcriptional regulation (Han and Maney, 1993). We have knocked-down Sobp translation in wild-type Xenopus embryos using targeted microinjections of MOs, and find that gene expression in the otic placode is severely disrupted and the otocyst is smaller or malformed, indicating an early role in ear development. Ectopic expression of Sobp by targeted mRNA microinjection down-regulates neural crest genes, suggesting that gain-of-function may antagonize middle ear formation. In fly, Sobp has been shown to directly bind to SO (Giot et al., 2003; Kenyon et al., 2005b) and alter its activity (Kenyon et al., 2005a, and unpublished). However, similar data for vertebrate Sobp have yet to be published.
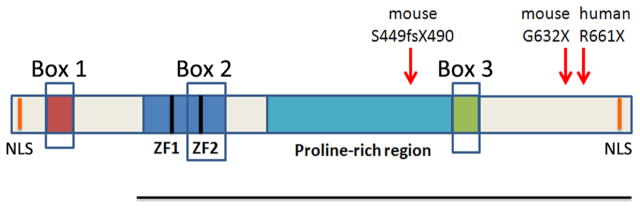
Figure 4.
Schematic of the domains present in Sobp. Sobp has N-terminal and C-terminal nuclear localization signals (NLS) indicated by orange lines. Box 1 includes 20 amino acids that are identical in Xenopus tropicalis and human SOBP proteins. Sobp has 2 FCS-type Zinc finger domains (ZF1 and ZF2), a proline-rich region of 245 amino acids and Box 3 (a 25 amino acid domain identical in Xenopus tropicalis and human Sobp that contains multiple prolines). The amino acid location of the known mouse and human Sobp mutations are indicated by red arrows. Each mutation introduces a stop codon that truncates the protein (Chen et al., 2008; Birk et al., 2010). The black line represents the portion of the Drosophila Sobp protein used in Y2H assays to demonstrate SO/Sobp binding (Kenyon et al., 2005a). Box 2 includes one of the FCS-type ZF domains in fly and sequence similarities to Box 2 and Box 3 were used to identify Zmym2 and Zmym4 (Neilson et al., 2010).
Zmym2 and Zmym4 were identified in our screen by their sequence similarity in the Box 2 and Box 3 regions present in the yeast 2-hybrid fly Sobp construct used to demonstrate SO/Sobp binding (Kenyon et al., 2005a). Zmym2 and Zmym4 have several FCS-type zinc fingers, a proline-rich Box 3-like domain, and a C-terminal NLS. They are vertebrate-specific proteins ranging from ~1200 amino acids to ~1500 amino acids. Xenopus Zymy2 is 71.3% identical and Zymy4 is 76.6% identical to their human homologues at the amino acid level. In both Xenopus (Neilson et al., 2010) and mouse (Gray et al., 2004), Zmym2 and Zmym4 are expressed in the otic placode, otocyst and branchial arches, and thus could contribute to both inner and middle ear development. Zmym4 is also expressed in the kidneys. Little is known about the function of these proteins during development. However, Zmym2 is part of a transcriptional co-repressor complex that down-regulates E-cadherin (Gocke and Yu, 2008). Our preliminary data show that both Zmym2 and Zmym4 are required for proper formation of the otocyst; their phenotypes after MO-mediated knock-down are similar to those of Xenopus Sobp. Increased expression of Zmym2 in the otic precursor region by targeted mRNA injection expands neural crest and neural plate genes and reduces placode genes. Whether Zmym2 or Zmym4 directly bind to Six1 or alter its activity has yet to be determined.
The proliferation-associated 2G4/Ebp1 protein emerged as a potential BOS/BOR candidate based on its high similarity to Drosophila CG10576, an SO-binding partner in flies (Giot et al., 2003). Structural studies indicate that 2G4/Ebp1 proteins are homologous to type II methionine aminopeptidases based on their “pita bread” fold structure, yet lack the associated enzymatic activity (Kowalinski et al., 2007; Monie et al., 2007). Instead, its fold is thought to interact with other proteins enabling its ability to regulate cell proliferation and differentiation (Figeac et al., 2014). In addition to the fold, 2G4/Ebp1 proteins possess an extended C-terminus containing distinct motifs involved in RNA and protein binding (Kowalinski et al., 2007, Monie et al., 2007; Squatrito et al., 2004). In mammals, 2G4/Ebp1 can interact with a diverse range of proteins including the epidermal grown factor receptor Erb3, the androgen receptor (AR), Sin3A (involved with histone deacetylation), protein kinase R, the serine/threonine kinase AKT and the cell cycle regulator Rb (Xia et al., 2001 Yoo et al., 2000; Zhang et al., 2008). 2G4/Ebp1 can also interact with rRNA, influencing ribosome assembly in the nucleolus (Squatrito et al., 2004), as well as mRNAs, including those encoding AR (Zhou et al., 2011). In mammalian cell culture, it associates with the inactive form of ErbB-3 and translocates to the nucleus following receptor activation (Yoo et al., 2000). The multiplicity of 2G4/Ebp1 functions in controlling cell growth depends upon isoform expression. Mammalian 2G4/Ebp1 genes code for two major isoforms: p42 and p48 (Liu et al., 2006). The longer p48 is more abundantly expressed and can localize either to the nucleus or the cytoplasm; the shorter isoform remains mostly in the cytoplasm (Liu et al., 2006). The two isoforms can be functionally distinct; p48 has been shown to inhibit apoptosis and promote cell survival whereas the shorter isoform inhibits cell proliferation (Liu et al., 2006). Several types of cancers demonstrate atypical expression and/or activity of 2GF/Ebp1 (Kim et al., 2011; Lu et al., 2011; Zhou et al., 2010). In frog embryos, 2G4/Ebp1 is expressed in the otic placode/otocyst, neural crest/branchial arches and kidney (Neilson et al., 2010). In mouse it is expressed in the branchial arches and the otocyst (Gray et al., 2004). Most importantly, loss of 2G4/Ebp1 in frog (by targeted MO microinjection) interferes with otocyst formation, and increased 2G4/Ebp1 (by targeted mRNA microinjection) has effects similar to those described above for the Zmym proteins. Ebp1-deficient mice are 30% smaller compared to wild type littermates and show cellular hallmarks associated with growth retardation, but specific effects on the ear were not reported (Zhang et al., 2008). In fly, CG10576 was identified in a few genome-wide studies (Bidet et al., 2003; Blanco et al., 2010b), and when over-expressed in embryos it causes neurogenic patches to form within developing muscle (Bidet et al., 2003). We are currently determining whether 2G4 binds directly to Six1 and/or affects Six1 transcription.
Mcrs1 contains a forkhead-associated domain, which is a phosphopeptide binding domain found in some forkhead transcription factors (Li et al., 2000). This domain forms a sandwich of two anti-parallel β-sheets that binds phosphorylated serine, threonine and tyrosine residues to mediate protein-protein interactions. In frog, Mcrs1 is expressed in the otic placode, otocyst, neural crest/branchial arches and kidney. In mouse, Mcrs1 also is expressed in the branchial arches and the otocyst (Gray et al., 2004). In mammalian cancer cells Mcrs1 has been implicated in proliferation (Shi et al., 2009; Wu et al., 2012; Zhong et al., 2013), and shown to function as a transcriptional repressor (Hsu et al, 2014). Mcrs1 also associates with the Fragile X mental retardation protein (FMRP) and may be involved in escorting silent ribonucleoparticles from the cell body of neurons to their distant synapses for translation (Davidovic et al., 2006). The fly homologue (dMcrs2/Rcd5/CG1135) codes for a multifunctional protein implicated in centrosome function, transcriptional regulation and cell cycle control. It is part of a transcriptional complex that recruits RNA polymerase II to gene promoters (Andersen et al., 2010). Loss-of-function phenotypes generated by P-element insertion demonstrate early larval lethality (http://flybase.org/reports/FBgn0263832.html). Raja and colleagues (2010) determined that dMCRS2 is a direct binding partner of several other proteins comprising the non-specific lethal (NSL) complex that positively influences transcription. Mutations in mammalian Mcrs1 have yet to be reported, but MO-mediated knock-down in frog results in significant otocyst defects. We are currently determining whether Mcrs1 binds directly to Six1 and/or affects Six1 transcription.
These data encourage us to continue the functional characterization of these five proteins because they are normally expressed in ear and kidney progenitor tissues, their loss affects otic development, and they are highly similar to the human proteins. However, importantly, their interactions with Six1 still need to be elucidated. Further, the effects of both loss- and gain-of-function on the neural crest progenitors of the middle ear and on the developing kidney need to be detailed to determine if they phenocopy any of BOS/BOR malformations.
Summary and future directions
BOS/BOR patients present with variable defects in inner and middle ear, which are derived from the otic placode and neural crest, respectively; a subset also have kidney defects. While genetic screening is considered the best approach to identify newborns at risk for hearing loss and kidney dysfunction, only two causative genes (SIX1, EYA1) have been identified and they only account for about half of the patients. Therefore, identifying additional genes that interact with Six1/Eya1 during normal development may uncover new causative genes that can be added to genetic screenings for at-risk newborns. We used the powerful Drosophila model to identify putative BOS/BOR candidate genes, and are now harnessing the biochemical and embryological advantages of Xenopus to determine whether these candidates are functionally required for Six1 activity during otic development. Functional testing of these candidates in an aquatic animal model with high genetic, protein and functional similarity to human is likely to rapidly uncover high priority candidates for patient genome sequencing. With this approach we hope to uncover additional genes that are diagnostic for Branchiootorenal spectrum disorders.
Acknowledgments
Grant support:
NIH R01 DE022065 (SAM)
NSF IOS-0817902 (SAM)
NIH R03 HD055321 (KMN)
NIH R01 DE016289 (DA)
NIH R01 EY013167 (FP)
In no case was the funding source involved in this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen DS, Raja SJ, Colombani J, Shaw RL, Langton PF, Akhtar A, Tapon N. Drosophila MCRS2 associates with RNA polymerase II complexes to regulate transcription. Mol Cell Biol. 2010;47:4744–4755. doi: 10.1128/MCB.01586-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, Weasner BP, Weasner BM, Kumar JP. The Drosophila Wilm’s tumor1-associating protein (WTAP) homolog is required for eye development. Dev Biol. 2014;390:170–180. doi: 10.1016/j.ydbio.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AP, Bhattacharyya S, Bronner-Fraser M, Streit A. Lens specification is the ground state of all sensory placodes, from which FGF promotes olfactory identity. Dev Cell. 2006;11:505–517. doi: 10.1016/j.devcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Bajoghli B, Aghaallaei N, Czerby T. Groucho corepressor proteins regulate otic vesicle outgrowth. Dev Dyn. 2005;233:760–771. doi: 10.1002/dvdy.20398. [DOI] [PubMed] [Google Scholar]
- Bane BC, Van Rybroek JM, Kolker SJ, Weeks DL, Manaligod JM. EYA1 expression in the developing inner ear. Ann Otol Rhinol Laryngol. 2005;114:853–858. doi: 10.1177/000348940511401108. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Bronner-Fraser M. Hierarchy of regulatory events in sensory placode development. Curr Opin Genet Dev. 2004;14:520–526. doi: 10.1016/j.gde.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Bidet Y, Jagla T, Da Ponte JP, Dastugue B, Jagla K. Modifiers of muscle and heart cell fate specification identified by gain-of-function screen in Drosophila. Mech Dev. 2003;120:991–1007. doi: 10.1016/s0925-4773(03)00182-5. [DOI] [PubMed] [Google Scholar]
- Birk E, Har-Zahav A, Manzini CM, Pasmanik-Chor M, Kornreich L, Walsh CA, Noben-Trauth K, Albin A, Simon AJ, Colleaux L, Morad Y, Rainshtein L, Tischfield DJ, Wang P, Magai N, Shoshani N, Rechavi G, Gothelf D, Maydan G, Shohat M, Basel-Vanagaite L. SOBP is mutated in syndromic and non-syndromic intellectual disability and is highly expressed in the brain limbic system. Am J Hum Genet. 2010;87:694–700. doi: 10.1016/j.ajhg.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Pauli T, Seimiya M, Udolph G, Gehring WJ. Genetic interactions of eyes absent, twin of eyeless and orthodenticle regulate sine oculis expression during ocellar development in Drosophila. Dev Biol. 2010a;344:1088–1099. doi: 10.1016/j.ydbio.2010.05.494. [DOI] [PubMed] [Google Scholar]
- Blanco E, Ruiz-Romero M, Beltran S, Bosch M, Punset A, Serras F, Corominas M. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev Biol. 2010b;10:94. doi: 10.1186/1471-213X-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, Cho KWY. Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system. genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman EA, Quint E, Fuchs E, Hrabe de Angelis M, Steel KP. Catweasel mice: a novel role for Six1 in sensory patch development and a model for branchio-otic-renal syndrome. Dev Biol. 2009;328:285–296. doi: 10.1016/j.ydbio.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricaud O, Collazo A. The transcription factor six1 inhibits neuronal and promotes hair cell fates in the developing zebrafish (Danio rerio) inner ear. J Neurosci. 2006;26:10438–10451. doi: 10.1523/JNEUROSCI.1025-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricaud O, Collazo A. Balancing cell numbers during organogenesis: Six1a differentially affects neurons and sensory hair cells in the inner ear. Dev Biol. 2011;357:191–201. doi: 10.1016/j.ydbio.2011.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck S, Englert C. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–255. doi: 10.1007/s00467-003-1374-z. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Moody SA. Induction and specification of the vertebrate ectodermal placodes: precursors of the cranial sensory organs. Biol Cell. 2005;97:303–319. doi: 10.1042/BC20040515. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- Chang EH, Menezes M, Meyer NC, Cucci RA, Vervoort VS, Schwartz CE, Smith RJ. Branchio-oto-renal syndrome: the mutation spectrum in EYA1 and its phenotypic consequences. Hum Mutat. 2004;23:582–589. doi: 10.1002/humu.20048. [DOI] [PubMed] [Google Scholar]
- Chen B, Kim EH, Xu PX. Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev Biol. 2009;326:75–85. doi: 10.1016/j.ydbio.2008.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Montcouquioi M, Calderon R, Jenkins NA, Copeland NG, Kelley MW, Noben-Trauth K. Jac1/Sobp, encoding a nuclear zinc finger protein, is critical for cochlear growth, cell fate and patterning of the organ of Corti. J Neurosci. 2008;28:6633–6641. doi: 10.1523/JNEUROSCI.1280-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Christophorou NA, Bailey AP, Hanson S, Streit A. Activation of Six1 target genes is required for sensory placode formation. Dev Biol. 2009;336:327–336. doi: 10.1016/j.ydbio.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Davidovic L, Bechara E, Gravel M, Jaglin XH, Tremblay S, Sik A, Bardoni B, Khandjian RW. The nuclear Microspherule protein 58 is a novel RNA-binding protein that interacts with fragile X mental retardation protein in polyribosomal mRNPs from neurons. Hum Mol Genet. 2006;15:1525–1538. doi: 10.1093/hmg/ddl074. [DOI] [PubMed] [Google Scholar]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 2008;28:5229–5239. doi: 10.1523/JNEUROSCI.1134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elepfandt A, Eistetter I, Flelig A, Gunther E, Hainich M, Hepperle S, Traub B. Hearing threshold and frequency discrimination in the purely aquatic frog Xenopus laevis (Pipidae): measurement by means of conditioning. J Exp Biol. 2000;203:3621–3629. doi: 10.1242/jeb.203.23.3621. [DOI] [PubMed] [Google Scholar]
- Figeac N, Serralbo O, Marcelle C, Zammit PS. ErbB3 binding protein-1 (ebp-1) controls proliferation and myogenic differentiation of muscle stem cells. Dev Biol. 2014;386:135–151. doi: 10.1016/j.ydbio.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Fraser FC, Sproule JR, Halal F. Frequency of the branchio-oto-renal (BOR) syndrome in children with profound hearing loss. Am J Med Genet. 1980;7:341–349. doi: 10.1002/ajmg.1320070316. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Shcaffner W. Transcriptional activation modulated by homopolymorphic proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Gilchrist MJ. From expression cloning to gene modeling: the development of Xenopus gene sequence resources. Genesis. 2012;50:143–154. doi: 10.1002/dvg.22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RS, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gocke CB, Yu H. Znf198 stabilizes the lsd1-corest-hdac1 complex on chromatin through its mym-type zinc fingers. PLoS One. 2008;3:1–12. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;293:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: definition of minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V, Xin Y, Balas G, Lai E. Dynamics of placodal lineage development revealed by targeted transgene expression. Dev Dyn. 1999;215:332–343. doi: 10.1002/(SICI)1097-0177(199908)215:4<332::AID-AJA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Xu C, Carthew RW. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development. 2008;135:2787–2796. doi: 10.1242/dev.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichman DS, Dubchak I, Amaya E, Detter DC, Fletcher R, Gerhard DS, Goodstein D, Grave T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Willis A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Smith RJH, Van Camp G. Function and expression pattern of nonsyndromic deafness genes. Curr Mol Med. 2009;9:546–564. doi: 10.2174/156652409788488775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins BE, Cramer CH, Silvius D, Zou D, Raymond RM, Orten DJ, Kimberling WJ, Smith RJ, Weil D, Petit C, Otto EA, Xu PX, Hildebrandt F. Transcription factor Six5 is mutated in patients with Branchio-Oto-Renal Syndrome. Am J Hum Genet. 2007;80:800–804. doi: 10.1086/513322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Che CH, Hsu TI, Hung JJ, Ko JL, Zhang B, Lee YC, Chen HK, Chang WC, Lin DY. The 58 Kda Microspherule Protein (Msp58) represses human telomerase reverse transcriptase (Htert) gene expression and cell proliferation by interacting with telomerase transcriptional element-interacting factor (Teif) Biochim Biophys Acta. 2014;1843:565–579. doi: 10.1016/j.bbamcr.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Hwang CH, Simeone A, Lai E, Wu DK. Foxg1 is required for proper separation and formation of sensory cristae during inner ear development. Dev Dyn. 2009;238:2725–2734. doi: 10.1002/dvdy.22111. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ookawara S, Sato S, Ando Z, Kageyama R, Kawakami K. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev Biol. 2007;311:53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Kageyama R, Suzuki Y, Kawakami K. Six1 is indispensable for production of functional progenitor cells during olfactory epithelial development. Int J Dev Biol. 2010;54:1453–1464. doi: 10.1387/ijdb.093041ki. [DOI] [PubMed] [Google Scholar]
- Ito T, Noguchi Y, Yashima T, Kitamura K. SIX1 mutation associated with enlargement of the vestibular aqueduct in a patient with branchio-oto syndrome. Laryngoscope. 2006;116:797–799. doi: 10.1097/01.mlg.0000209096.40400.96. [DOI] [PubMed] [Google Scholar]
- Jusiak B, Wang F, Karandikar UC, Kwak SJ, Wang H, Chen R, Mardon G. Genome-wide DNA binding pattern of the homeodomain transcriptional factor Sine oculis (So) in the developing eye of Drosophila melanogaster. Genom. Data. 2014a;2:153–155. doi: 10.1016/j.gdata.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusiak B, Karandikar UC, Kwak SJ, Wang F, Wang H, Chen R, Mardon G. Regulation of Drosophila eye development by the transcription factor Sine oculis. PLoS One. 2014b;9:e89695. doi: 10.1371/journal.pone.0089695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes-structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Kim J, Santos R, Wu HH, Lander AD, Calof AL. Foxg1 promotes olfactory neurogenesis by antagonizing Gdf11. Development. 2009;136:1453–1464. doi: 10.1242/dev.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Kenyon KL, Li DJ, Clouser C, Tran S, Pignoni F. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn. 2005a;234:497–504. doi: 10.1002/dvdy.20442. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Yang-Zhou D, Cai CQ, Tran S, Clouser C, Decene G, Ranade S, Pignoni F. Partner specificity is essential for proper function of the SIX-type homeodomain proteins Sine oculis and Optix during fly eye development. Dev Biol. 2005b;286:158–168. doi: 10.1016/j.ydbio.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Moody SA, Jamrich M. A novel forkhead gene mediates early steps during Xenopus lens formation. Development. 1999;126:5107–5116. doi: 10.1242/dev.126.22.5107. [DOI] [PubMed] [Google Scholar]
- Khatri SB, Groves AK. Expression of the Foxi2 and Foxi3 transcription factors during development of chicken sensory placodes and pharyngeal arches. Gene Expr Patterns. 2013;13:38–42. doi: 10.1016/j.gep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK. Xenopus white papers and resources: folding functional genomics and genetics into the frog. Genesis. 2012;50:133–142. doi: 10.1002/dvg.22015. [DOI] [PubMed] [Google Scholar]
- Kim CK, Nguyen TL, Joo KM, Nam DH, Park J, Lee KH, Cho SW, Ahn JY. Negative regulation of p53 by the long isoform of ErbB3 binding protein Ebp1 in brain tumors. Cancer Res. 2010;70:9730–9741. doi: 10.1158/0008-5472.CAN-10-1882. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, Yamamoto M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev Biol. 2001;232:315–326. doi: 10.1006/dbio.2001.0185. [DOI] [PubMed] [Google Scholar]
- Kochhar A, Orten DJ, Sorensen JL, Fischer SM, Cremers CW, Kimberling WJ, Smith RJ. SIX1 mutation screening in 247 branchio-otic-renal syndrome families: a recurrent missense mutation associated with BOR. Hum Mutat. 2008;29:565. doi: 10.1002/humu.20714. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Ikeda K, Iwakura Y, Kawakami K. Six1 and Six4 promote survival of sensory neurons during early trigeminal gangliogenesis. Brain Res. 2006;1116:93–102. doi: 10.1016/j.brainres.2006.07.103. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Bange G, Bradatsch B, Hurt E, Wild K, Sinning I. The crystal structure of EBP1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 2007;581:4450–4454. doi: 10.1016/j.febslet.2007.08.024. [DOI] [PubMed] [Google Scholar]
- Krug P, Moriniere V, Marlin S, Koubi V, Gabriel HD, Colin E, Bonneau D, Salomon R, Antignac C, Heidet L. Mutation screening of the EYA1, SIX1, and SIX5 genes in a large cohort of patients harboring branchio-oto-renal syndrome calls into question the pathogenic role of SIX5 mutations. Hum Mutat. 2011;32:183–190. doi: 10.1002/humu.21402. [DOI] [PubMed] [Google Scholar]
- Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Li J, Lee GI, Van Doern SR, Walker JC. The FHA domain mediates phosphoprotein interactions. J Cell Sci. 2000;23:4143–4149. doi: 10.1242/jcs.113.23.4143. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Li Y, Manaligod JM, Weeks DL. EYA1 mutations associated with the branchio-otic renal syndrome result in defective otic development in Xenopus laevis. Biol. Cell. 2010;102:277–292. doi: 10.1042/BC20090098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ahn JY, Liu X, Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. PNAS USA. 2006;103:10917–10922. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhou H, Chen W, Zhang Y, Hamburger AW. The ErbB3 binding protein EBP1 regulates ErbB2 protein levels and tamoxifen sensitivity in breast cancer cells. Breast Cancer Res Treat. 2011;126:27–36. doi: 10.1007/s10549-010-0873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M, Wang M, Narins P. Structure and function of the middle ear apparatus of the aquatic frog, Xenopus laevis. Proc Inst Acoust. 2009;31:13–21. [PMC free article] [PubMed] [Google Scholar]
- Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SA, LaMantia AS. Transcriptional regulation of cranial sensory placode development. Curr Top Dev Biol. 2015;111:301–350. doi: 10.1016/bs.ctdb.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SA, Saint-Jeannet JP. Development of the Pre-placodal ectoderm and cranial sensory placodes. In: Moody SA, editor. Principles of Developmental Genetics. 2. Elsevier Inc; 2014. pp. 331–356. [Google Scholar]
- Morgan AM, Rubenstein E. Proline: The distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS ONE. 2013;8:e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Nance WE. Newborn hearing screening - a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- Mosrati MA, Hammami B, Rebeh IB, Ayadi L, Dhouib L, Ben Mahfoudh K, Hakim B, Charfeddine I, Mnif J, Ghorbel A, Masmoudi S. A novel dominant mutation in SIX1, affecting a highly conserved residue, results in only auditory defects in humans. Eur J Med Genet. 2011;54:e484–e488. doi: 10.1016/j.ejmg.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KW, Grainger RM. Cas9-based genome editing in Xenopus tropicalis. Methods Enzymology. 2014;546:355–375. doi: 10.1016/B978-0-12-801185-0.00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson KM, Pignoni F, Yan B, Moody SA. Developmental expression patterns of candidate co-factors for vertebrate Six family transcription factors. Dev Dyn. 2010;239:3446–3466. doi: 10.1002/dvdy.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess SM. Zebrafish foxi modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–2554. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Noguchi Y, Ito T, Nishio A, Honda K, Kitamura K. Audiovestibular findings in a branchio-oto syndrome patient with a SIX1 mutation. Acta Otolaryngol. 2011;131:413–418. doi: 10.3109/00016489.2010.543146. [DOI] [PubMed] [Google Scholar]
- Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K. Tissue and developmental distribution of Six family gene products. Int J Dev Biol. 1998;42:141–148. [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga S, Ozaki H, Sato S, Kawakami K. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–646. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Watanabe Y, Takahashi K, Kitamura K, Tanaka A, Urase K, Momoi T, Sudo K, Sakagami J, Asano M, Iwakura Y, Kawakami K. Six4, a putative myogenin gene regulator, is not essential for mouse embryonic development. Mol Cell Biol. 2001;21:3343–3350. doi: 10.1128/MCB.21.10.3343-3350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki H, Watanabe Y, Ikeda K, Kawakami K. Impaired interactions between mouse Eya1 harboring mutations found in patients with branchio-oto-renal syndrome and Six, Dach, and G proteins. J Hum Genet. 2002;47:107–116. doi: 10.1007/s100380200011. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, Okamura HO, Kitamur K, Muto S, Kotaki H, Sudo K, Horai R, Iwakura Y, Kawakami K. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. 2000;96:253–257. doi: 10.1016/s0925-4773(00)00396-8. [DOI] [PubMed] [Google Scholar]
- Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization for six SIX1 Branchio-otic-renal syndrome mutations. J Biol Chem. 2009;284:20781–20790. doi: 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR Syndrome. Nature Struct Mol Biol. 2013;20:447–453. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development. 2005;132:2771–2782. doi: 10.1242/dev.01841. [DOI] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2480. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Piñeiro C, Lopes CS, Casares F. A conserved transcriptional network regulates lamina development in the Drosophila visual system. Development. 2014;141:2838–2847. doi: 10.1242/dev.108670. [DOI] [PubMed] [Google Scholar]
- Powers TR, Virk SM, Trujillo-Provencio C, Serrano EE. Probing the Xenopus laevis inner ear transcriptome for biological function. BMC Genomics. 2012;13:225. doi: 10.1186/1471-2164-13-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, Akhtar A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol. Cell. 2010;38:827–841. doi: 10.1016/j.molcel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RMJ, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–27. doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L. Neural crest cells in ear development. In: Trainor PA, editor. Neural Crest Cells. Evolution, Development and Disease. Elsevier; 2014. pp. 167–188. [Google Scholar]
- Sanggaard K, Rendtorff ND, Kjaer KW, Eiberg H, Johnsen T, Gimsing S, Dyrmose J, Nielsen KO, Lage K, Tranebjaerg L. Branchio-otic-renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur J Hum Genet. 2007;15:1121–1131. doi: 10.1038/sj.ejhg.5201900. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffelen RL, Segenhout JM, Van Dijk P. Mechanics of the exceptional anuran ear. J Comp Physiol A. 2008;194:417–428. doi: 10.1007/s00359-008-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self M, Lsagutin OV, Bowling B, Hendrix J, Cal Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, Curtiss J, Mlodzik M, Fjose A. Six class homeobox genes in drosophila belong to three distinct families and are involved in head development. Mech Dev. 1999;83:127–123. doi: 10.1016/s0925-4773(99)00045-3. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Chen S, Jin H, Xu C, Dong G, Wang W, Zhang H, Lin W, Zhang J, Davidovic L, Yao L, Fan D. Downregulation of MSP58 inhibits growth of human colorectal cancer cells via regulation of the cyclin D1-cyclin-dependent kinase 4-p21 pathway. Cancer Sci. 2009;100:1585–1590. doi: 10.1111/j.1349-7006.2009.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJH. Branchiootorenal Spectrum Disorders. Gene Reviews. 2014 ( http://www.ncbi.nlm.nih.gov/books/NBK1380/)
- Smith RJH, Shearer AE, Hildebrand MS, Van Camp G. Deafness and Hereditary Hearing Loss Overview. Gene Reviews. 2014 ( http://www.ncbi.nlm.nih.gov/books/NBK1434/)
- Solomon KS, Logsdo JM, Fritz A. Expression and phylogenetic analyses of three zebrafish Foxi class genes. Dev Dyn. 2003;228:301–307. doi: 10.1002/dvdy.10373. [DOI] [PubMed] [Google Scholar]
- Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- Streit A. Early development of the cranial sensory nervous system: from a common field to individual placodes. Dev Biol. 2004;276:1–15. doi: 10.1016/j.ydbio.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Sun H, Hunter T. Poly-small ubiquitin-like modifier (poly-SUMO)-binding proteins identified through a string search. J Bol Chem. 2012;287:42071–42083. doi: 10.1074/jbc.M112.410985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriello HV, Reardon W, Gorlin RJ. Hereditary Hearing Loss and its Syndromes. Oxford University Press; 2004. [Google Scholar]
- Van Dijk P, Mason MJ, Schoffelen RL, Narins PM, Meenderink SW. Mechanics of the frog ear. Hear Res. 2011;273:46–58. doi: 10.1016/j.heares.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GN, Brändli AW. Simple vertebrate models for chemical genetics and drug discovery screens: Lessons from zebrafish and Xenopus. Dev Dyn. 2009;238:1287–1308. doi: 10.1002/dvdy.21967. [DOI] [PubMed] [Google Scholar]
- Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang ZG, Qin HZ, Zhang J, Gao GD, Lin W, Wang J, Zhang J. Downregulation of MSP58 suppresses cell proliferation in neuroblastoma cell lines. NeuroReport. 2012;23:932–936. doi: 10.1097/WNR.0b013e328359566e. [DOI] [PubMed] [Google Scholar]
- Xia X, Cheng A, Lessor T, Zhang Y, Hamburger AW. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J Cell Physiol. 2001;187:209–217. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Ranganathan R, Maynard T, Streit A, Moody S. Microarray identification of novel genes downstream of Six1, a critical factor in cranial placode, somite and kidney development. Dev Dyn. 2015;244:181–210. doi: 10.1002/dvdy.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yoo JY, Wang XW, Rishi AK, Lessor T, Xia XM, Gusstafson TA, Hamburger AW. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br J Cancer. 2000;82:683–690. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu Y, Zhou H, Lee M, Liu Z, Hassel BA, Hamburger AW. Alterations in cell growth and signaling in ErbB3 binding protein-1 (Ebp1) deficient mice. BMC Cell Biol. 2008;9:69. doi: 10.1186/1471-2121-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Zhang X, Li B, Chen CS, Ji GL, Li SX, Bi DQ, Zhao QC, Shi H. Expression of MSP58 in hepatocellular carcinoma. Med Oncol. 2013;30:539–549. doi: 10.1007/s12032-013-0539-2. [DOI] [PubMed] [Google Scholar]
- Zhou H, Mazan-Mamczarz K, Martindale JL, Barker A, Liu Z, Gorospe M, Leedman PJ, Gartenhaus RB, Hamburger AW, Zhang Y. Post-transcriptional regulation of androgen receptor mRNA by an ErbB3 binding protein 1 in prostate cancer. Nucleic Acids Res. 2010;38:3619–3631. doi: 10.1093/nar/gkq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhang Y, Hamburger AW. EBP1 inhibits translation of androgen receptor mRNA in castration resistant prostate cancer cells. Anticancer Res. 2011;31:3129–3135. [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang T, Jemc JC, Chen Y, Chen R, Rebay I, Pignoni F. Onset of atonal expression in Drosophila retinal progenitors involves redundant and synergistic contribution of Ey/Pax6 and So binding sites within two distant enhancers. Dev Biol. 2014;386:152–164. doi: 10.1016/j.ydbio.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Davenport J, Grifone R, Maire P, Xu PX. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev Biol. 2006a;293:499–512. doi: 10.1016/j.ydbio.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas of the inner ear. Dev Biol. 2006b;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]