Abstract
Amphibian vertebrates are important models in regenerative biology because they present exceptional regenerative capabilities throughout life. However, it takes considerable effort to rear amphibians to juvenile and adult stages for regeneration studies and the relatively large sizes that frogs and salamanders achieve during development make them difficult to use in chemical screens. Here we introduce a new tail regeneration model using late stage Mexican axolotl embryos. We show that axolotl embryos completely regenerate amputated tails in 7 days before they exhaust their yolk supply and begin to feed. Further, we show that axolotl embryos can be efficiently reared in microtiter plates to achieve moderate throughput screening of soluble chemicals to investigate toxicity and identify molecules that alter regenerative outcome. As proof of principle, we identified integration 1 / wingless (Wnt), transforming growth factor beta (Tgf-β), and fibroblast growth factor (Fgf) pathway antagonists that completely block tail regeneration and additional chemicals that significantly affected tail outgrowth. Furthermore, we used microarray analysis to show that inhibition of Wnt signaling broadly affects transcription of genes associated with Wnt, Fgf, Tgf-β, epidermal growth factor (Egf), Notch, nerve growth factor (Ngf), homeotic gene (Hox), rat sarcoma/mitogen-activated protein kinase (Ras/Mapk), myelocytomatosis viral oncogene (Myc), tumor protein 53 (p53), and retinoic acid (RA) pathways. Punctuated changes in the expression of genes known to regulate vertebrate development were observed; this suggests the tail regeneration transcriptional program is hierarchically structured and temporally ordered. Our study establishes the axolotl as a chemical screening model to investigate signaling pathways associated with tissue regeneration.
Keywords: Axolotl, regeneration, chemical screening, Wnt, C59
1. Introduction
Fish and amphibians provide efficient models to identify chemicals that alter biological processes (Tomlinson et al., 2012; Tal et al., 2010). Their embryos and larvae can be reared in microtiter plates to rapidly screen well-characterized or novel compounds for effects at molecular, cell, developmental, or whole-organism levels. An area of particular interest is tissue regeneration, the breadth and capacity of which is highest in non-mammalian vertebrates. While regeneration assays have been established for early stages of Xenopus and zebrafish (reviewed by Slack et al., 2008; Gemberling et al., 2013), comparable assays have not been established for the more highly regenerative Mexican axolotl. The development of an early stage assay for the axolotl will reveal the extent to which regeneration programs are conserved among these disparate, highly regenerative models. Also, an early stage assay is needed because it takes considerable time and effort to rear axolotls to body sizes and ages that are typically used in experiments. Axolotls are generally reared for several months or even for more than a year before they are used to study tissue regeneration (Voss et al 2012). If an early stage regeneration assay were established for the axolotl, it would increase the pace of experimentation, increase efficiency of animal use, and allow more robust experimental designs.
Here we introduce an early stage tail regeneration assay using axolotl embryos. There are several advantages of this model. First, the assay uses ~20 day old (i.e. post fertilization) embryos that have sufficient yolk reserves to fully regenerate an amputated, distal tail tip in 7 days. The approximately 1 cm embryos are efficiently reared in microtiter plates with out need for feeding. Below, we detail the normal regeneration process at a gross morphological level and then describe regenerative outcomes after screening 33 chemicals, the majority of which are known inhibitors of major signaling pathways. Finally, we used microarray analysis to explore one of the positive hits, C59 a chemical inhibitor of Wnt ligand secretion (Proffitt et al., 2013). These results show that Wnt signaling affects multiple signaling pathways associated with tail regeneration.
2.1 Materials and methods
2.1 Ethical procedures
According to Public Health Service policy, use of pre-feeding stage axolotls does not require a protocol approved by an Institutional Animal Care and Use Committee. The embryos that were used in this study were treated according to the same ethical standards that apply to feeding axolotls.
2.2 Regeneration assay
Mexican axolotl embryos were obtained from the Ambystoma Genetic Stock Center at University of Kentucky. Stage 42 (Bordzilovskaya et al 1989) embryos were manually hatched, administered benzocaine anesthesia (0.2 g in 10 ml ethanol / liter water) and photographed. For each chemical that was tested, four embryos were administered tail amputations. Additionally, control groups of embryos with tail amputations were reared in artificial pond water (43.25 g NaCl, 0.625 g KCl, 1.25 g MgSO4, 2.5 g NaHCO3, and 1.25 g CaCl per 50 liters charcoal filtered municipal water). In administering amputations, the distal most 2 mm of tail tissue was removed with a sterile razor blade and photographs were taken to document the regeneration process. Images were captured using an Olympus microscope with 0.5x objective lens and DP400 camera. Embryos were reared at 18–19 C in 12-well microtiter plates, one embryo per well. Each well contained 2.0 ml of artificial pond water and 10 μM of chemical, although different concentrations were tried for some chemicals (Table 1). All chemicals were purchased from Selleckchem.com (TX, USA) and diluted in 0.1% DMSO. The solutions were changed on days 3 and 5 post-amputation (DPA), and experiments were terminated on 7 DPA. At this time embryos were euthanized by prolonged exposure to 10x benzocaine (0.4 g in 10 mls ethanol / liter water).
Table 1.
Targeted pathway, chemical, and experimental result. All chemicals were tested at 10 uM unless otherwise noted:
| Pathway/Process | Chemical | Result Prob |
|---|---|---|
| Iron chelation | ciclopirox | Toxic |
| Myosin II ATPase | (−)-blebbistatin | Toxic |
| PI3K | wortmannin | Toxic |
| Protein secretion / trafficking | brefeldin A | Toxic |
| Ros Inhibitor | apocynin | Toxic |
| Wnt | tideglusib | Toxic |
| Wnt | TWS119 | Toxic |
| WNT/GSK3 | BIO3 | Toxic |
| EGFR | gefitinib | NS |
| FGFR | danusertib | NS |
| FGFR | AZD4547 | NS |
| MMP | doxycycline | NS |
| Multiple biological effects | dexamethasone2 | NS |
| Notch | semagacestat | NS |
| Notch | MG-132 | NS |
| Protein translation | azithromycin | NS |
| Stat1 | fludarabine | NS |
| TNFa | lenalidomide | NS |
| V-ATPase H+ Inhibitor | concanamycin A6 | NS |
| Wnt | ICG-001 | NS |
| Wnt | KY02111 | NS |
| Wnt | agonist I | NS |
| Wnt | nicotine | NS |
| Apoptosis | NS3694 | < 0.05 |
| Hedgehog | vismodegib | < 0.05 |
| Multiple biological effects | luteolin | < 0.05 |
| Wnt | agonist II SKL | < 0.05 |
| Wnt | XAV-9391 | < 0.05 |
| Wnt | C59 | < 0.01 |
| Wnt | IWR-1-endo | < 0.01 |
| Fgfr | BGJ398 | < 0.002 |
| Multiple biological effects | dexamethasone Na phosphate | < 0.002 |
| Tgfβ/activin | SB-505124 | < 0.002 |
| Wnt | C594,5 | < 0.002 |
40.0 uM;
25.0 uM;
15.0 uM;
5.0 uM;
2.5 uM;
1.0 uM.
Prob is the probability that the proportional change in post-amputation body length is significantly different between control and chemically treated embryos. NS = not significant. The probability value 0.002 is the Bonferroni correction for multiple testing.
2.3 Microarray Experiment
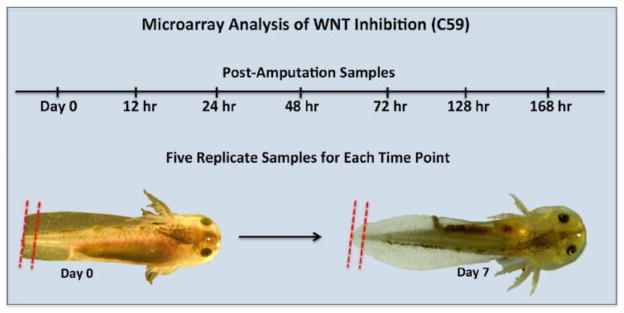
Two hundred and sixty embryos were administered tail amputations and reared as described above. Exactly 1 mm of the distal tail tip was removed from 20 embryos immediately after tail amputation to obtain Day 0 samples. Tissues from four embryos were pooled into a 1.5 ml tube with 0.5 ml of RNA-later (Qiagen). Thus, this procedure yielded 5 replicate pools of tissue. The remaining 240 embryos were equally split between two treatments – one group was reared in artificial pond water and the other was reared in artificial pond water with 5 uM of C59. The tissue sampling and pooling procedure described above was used to create 5 replicates for each of 6 samples −12, 24, 48, 72, 120, and 168 hours post amputation (HPA). The tissue samples were maintained at 4 C in RNA later prior to RNA isolation using first the Trizol method, and then a Qiagen minikit with on-the-column DNAse treatment of contaminating DNA. Microarray hybridization using an Ambystoma Affymetrix array (Huggins et al., 2010) was performed by the University of Kentucky Microarray Core Facility. The raw microarray data (.CEL files) and the microarray annotations are available at Sal-Site (Smith et al., 2005).
2.4 Statistical Analyses
The proportional increase in body length (Day 7 body length – post-amputation body length / post-amputation body length) was compared between chemically treated and control embryos using Student’s t-test. For the microarray experiment, several quality control methods were used to examine expression values across the 65 GeneChips in the experiment. Box plots were generated in Expression Console (Affymetrix, Santa Clara, CA) to examine the consistency of expression across arrays, and principal components analysis and Mahalanobis distances were calculated in JMP to examine array clustering in multivariate space. One of the GeneChips (12 hr non-treated) was identified as an outlier and removed from the experiment. All of the retained GeneChips were normalized using Affymetrix Expression Console software to accomplish robust multichip averaging (RMA) (Irizarry et al., 2003). Student’s t-test was performed separately for each time point to identify probe sets that yielded significantly different average expression values as a function of treatment. These lists were further filtered using a false discovery rate of α = 0.05 and by requiring a 1.5 fold difference between treatment means.
3. Results
3.1 Tail regeneration assay
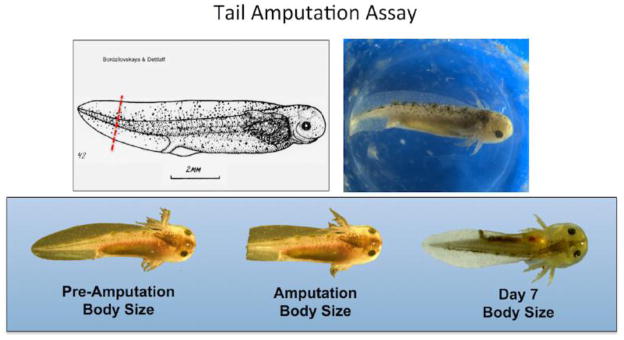
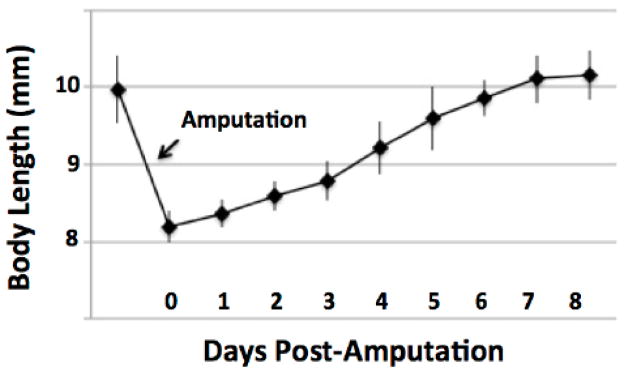
Late stage embryos are larval in appearance; they have external gills, upper and lower tail fins, and are capable of swimming (Fig. 1). They have a large store of yolk in their gut that is sufficient to sustain growth and complete tail regeneration prior to the onset of feeding behavior. Benzocaine anesthesia rendered embryos motionless in 1–2 minutes. Under anesthesia, embryos were photographed in a thin film of water or on top of wet paper towels, and 2 mm of the distal tail tip was removed using a sterile razor blade. Every 24 hrs through 7 DPA, embryos were anesthetized and photographed to characterize rate and pattern of growth (Fig. 2). Our observations of embryo tail regeneration suggest it is morphologically similar to larval tail regeneration; however, studies will be needed to determine if the embryo model is histologically equivalent (Monaghan et al 2007). We noticed that a wound epithelium formed over the cut surface by 12 hours post amputation (HPA). At this time, soft tissues in the tail fins had retracted proximally by approximately 0.5 mm relative to the end of the notochord. At 1 DPA, tail tissue extended beyond the cut end of the notochord and after this time total body length increased linearly, although the slope of the growth profile was a bit steeper between 3 and 5 DPA. By 5 DPA the tail had a rounded appearance and by 7 DPA the tail appeared as it was before amputation. The growth profile plateaued between 7 and 8 DPA, and at these times the average body length was greater than the pre-amputation average body length. These results show that axolotl embryos are capable of regenerating their tails to pre-amputation tail length in approximately 7 days.
Figure 1. The axolotl tail amputation assay uses developmental stage 42 (Bordzilovskaya et al. 1989) embryos that are approximately 1 cM in total body length.
The proportional increase in body size is determined 7 days post-amputation from images that document total body length.
Figure 2. Plot showing the average increase in body length after amputating approximately 2 mm from the distal tail tip of an axolotl embryo.
The error bars are standard deviations of the mean.
3.2 Chemical screen
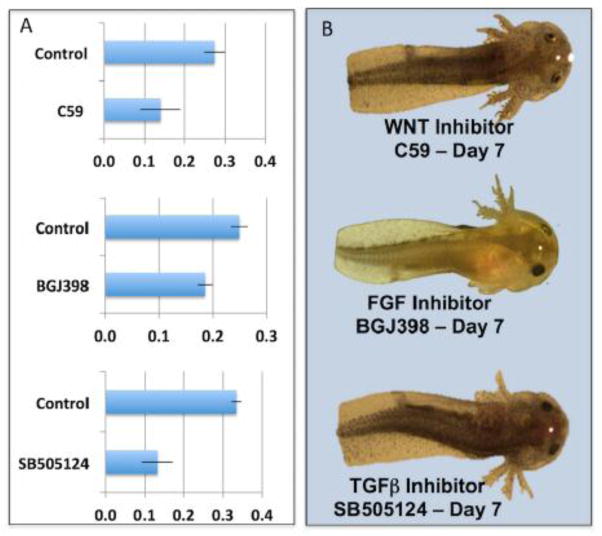
After establishing the time to complete regeneration, a screen was performed to identify chemicals that alter tail outgrowth after amputation. Amputated and non-amputated axolotl embryos were photographed and arrayed among wells of microtiter plates that contained either artificial pond pond water or artificial pond water and a commercially available chemical (Table 1). We note that the mortality and regeneration outcomes reported below are specific to the concentration of chemical that was used, which was a single dose for almost all chemicals. In other words, had we tested a range of concentrations for each chemical we may have obtained different outcomes. Overall, 33 chemicals were tested for an effect on tail outgrowth. Eight of the chemicals were toxic and caused the death of all amputated and non-amputated embryos – tideglusib (Domínguez et al., 2012), TWS119 (Ding et al., 2003), wortmannin (Wymann et al., 1996), ciclopirox ethanolamine (Niewerth et al., 2003), apocynin (Heumüller et al., 2008), brefeldin A (Fugiwara et al., 1988), blebbistatin (Kovács et al., 2004), and BIO (Meijer et al., 2003). After 7 days of development, embryos were photographed and the proportional increase in post-amputation body length was calculated and compared to control embryos. We note that the proportional increase in post-amputation body length is highly correlated with tail lengthening during regeneration. The advantage of using the former measurement in a chemical genetic screen is that there is less ambiguity in defining the two ends of an embryo than there is in defining the proximal end of the tail. Once significant growth differences are identified statistically in a screen, treatment effects can be evaluated further by referencing photographic records. No significant differences in growth were observed between treated and control embryos for 15 chemicals. Significant variation in post-amputation body size was observed among individuals for the remainder of the chemicals; given that only 4 individuals were assayed per chemical treatment, we report the results for different probability thresholds. Notably, four chemicals were statistically significant at a highly conservative Bonferroni probability correction for multiple testing (p < 0.002) and photographs showed that these chemicals completely inhibited tail regeneration - C59 (Proffitt et al., 2013), BGJ398 (Guagnano et al., 2011), SB505124 (DaCosta et al., 2004)(Fig. 3), and an aqueous-soluble prodrug form of dexamethasone, dexamethasone sodium phosphate (Samtani et al., 2005). While dexamethasone may affect multiple signaling pathways, C59, BGJ398, and SB505124 are known to specifically inhibit Wnt, Fgf, and Tgf signaling pathways, respectively.
Figure 3. Panel A shows the average proportional increase in body size at day 7 for embryos that were administered tail amputations and reared with or without (controls) chemical inhibitors of Wnt (C59), Fgf (BGJ398), and Tgf (SB505124) signaling pathways.
The error bars are standard deviations of the mean. The difference between control and treatment mean is significant for each chemical treatment (Students 2-tailed T test, p < 0.001). Panel B shows representative pictures of the effects of the three chemicals on embryos at day 7.
3.3 Identification of transcripts that changed as a result of inhibiting Wnt signaling
Previous studies have established that inhibition of Wnt signaling blocks regeneration of amputated anuran tails and fish fins (Lin and Slack, 2008; Stoick-Cooper et al., 2007; Kawakami et al., 2006). However, very little is known about the downstream gene targets of Wnt signaling. To identify transcriptional targets of Wnt signaling, we used custom Affymetrix GeneChips and performed a microarray analysis. Axolotl embryo tails were amputated and individuals were reared in the presence or absence of 5 uM C59, a chemical that inhibits PORCN and thus the secretion of Wnt ligand from cells (Proffitt et al., 2013; Chen et al., 2009). Exactly 1 mm of tissue was collected from the distal tip of amputated tails throughout the first week of regeneration − 12, 24, 48, 72, 120, and 168 HPA. We note that the 1 mm of tissue collected from control animals after 72 HPA arises wholly from the regenerating portion of the tail (Fig. 4). In contrast, tissue collected from regeneration-inhibited C59 embryos was sampled from the tail stump at all time points. Thus, the tissue collected between control and C59 treated embryos differed quantitatively and qualitatively with respect to the amount of stump verses regenerating tissue collected over time.
Figure 4. Design of the microarray experiment and tissue sampling strategy.
The red dotted lines show the amount of tissue collected at Day 0 and Day 7. During the time course, samples from control embryos are progressively enriched for regenerating tissue relative to embryos reared in C59
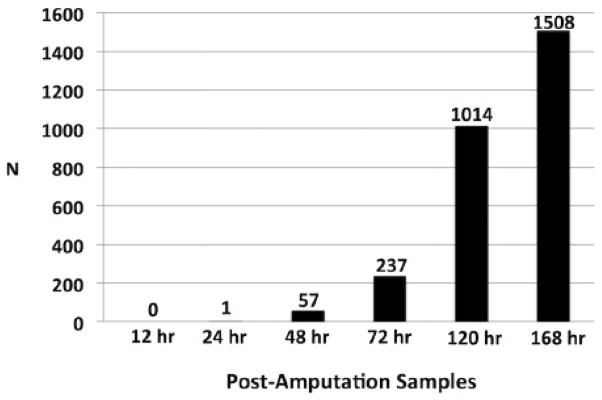
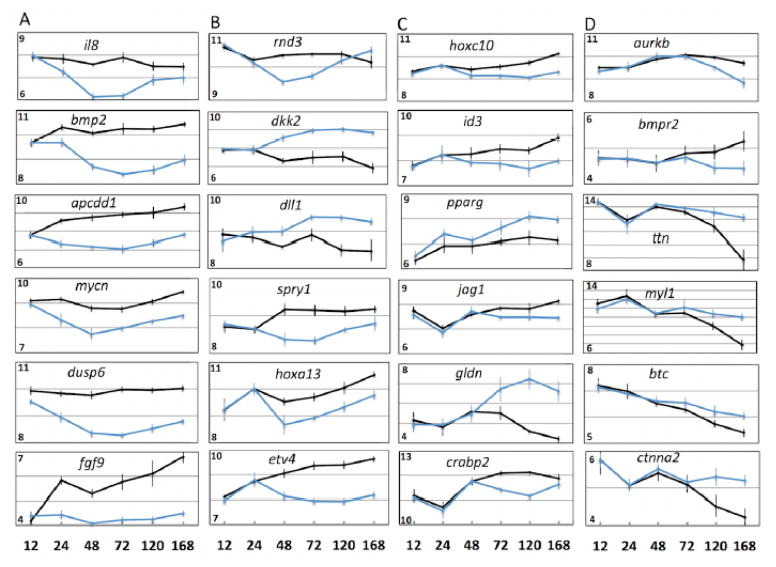
T-tests were performed for each post-amputation time point that total RNA was sampled to identify probe sets that were expressed differently between control and C59-treated embryos. Probe sets were retained for enrichment analysis if they met statistical (FDR corrected p-value < 0.05) and fold change (> 1.5 fold difference between treatment and control mean estimates) criteria. None of the probe sets met both criteria for the first post-amputation time point (12 HPA) and only one probe set (fgf9) met both criteria at 24 HPA; however, we note significant differences in expression for 24 genes at 24 HPA. After this time, the number of genes identified as differently expressed, meeting both statistical and fold change criteria, increased for each subsequent sample. A total of 57, 237, 1014, and 1508 probe sets were identified for the 48, 72, 120, and 168 HPA samples (Fig. 5). Twenty-three of the probe sets identified at 48 hr, including fgf9 and genes associated with Wnt, Tgf-β, Ffg, Egf, Ngf, and Hox signaling, were also identified as differentially expressed for all subsequent sample times (Table 2). In several cases, gene expression profiles diverged between control and C59-treated axolotls at specific sample times (Fig. 6a–d). For example, genes that encode Wnt (dkk2), Fgf (spry1), and Notch (dll1, rnd3) pathway proteins were expressed differently at 48 HPA and remained different throughout the remainder of the time course. At 72 HPA, genes associated with Notch (jag) and retinoic acid (RA) signaling (crabp2) diverged in expression, along with genes that associate with cell proliferation (pparg) and patterning (hoxc10) (Fig. 6c). Genes associated with Egf signaling (btc), Bmp signaling (bmpr2), cell proliferation (aurkb, ctnna2), and skeletal muscle (ttn, myl1) diverged at 120 and 168 HPA (Fig. 6d). These results show that inhibition of WNT signaling altered transcription during tail regeneration, with relatively few but important signaling factors affected as early as 24–48 HPA post amputation, and considerably more genes affected at latter time points. The observation of punctuated patterns of gene expression, where genes were expressed differently between control and C59-treated embryos at specific post-amputation times, suggests the regeneration transcriptional program is hierarchically structured and temporally ordered.
Figure 5.
The number of genes that were identified as statistically significant and > 1.5 fold differently expressed between control and C59-treated embryos at each time tissues were collected for RNA isolation.
Table 2.
Probe sets that were differently expressed at 48, 72, and 120 hrs post-amputation. The bolded genes were also significant at 168 hrs.
| Probeset | Gene ID |
|---|---|
| axo01743-f_at | APCDD1 |
| axo02472-f_at | HAPLN3 |
| axo04593-f_at | ETV4 |
| axo07001-f_at | BMP2 |
| axo07256-f_at | EPAS1 |
| axo07306-r_at | IL8 |
| axo07473-f_at | AREG |
| axo07816-f_at | FGF9 |
| axo08059-r_at | LAMB1 |
| axo08282-f_at | NGFR |
| axo09045-f_at | PHLDA2 |
| axo10046-f_at | DYNC1I1 |
| axo11014-r_at | MYCN |
| axo12159-f_at | SLC2A1 |
| axo13739-f_at | ANKRD1 |
| axo13755-f_at | DKK2 |
| axo18467-f_at | DUSP6 |
| axo18474-f_at | HOXC8 |
| axo23290-r_at | unknown |
| axo29791-f_at | FGFR3 |
| axo29801-f_at | AXIN1 |
| axo11014-r_at | MYCN |
| axo27228-f_at | INHBB |
| axo29536-f_at | SP7 |
| axo29905-f_at | SP7 |
| axo07974-f_at | INHBB |
| axo27128-f_at | WNT5A |
| axo28029-f_at | CYP26B1 |
| axo01779-r_at | TMEM92 |
| axo03170-f_at | PRICKLE2 |
| axo07700-f_at | CTGF |
| axo07929-r_at | HMOX1 |
| axo08144-f_at | MAS1 |
| axo08178-f_at | MMP3 |
| axo10448-r_at | LHX2 |
Figure 6. Expression profiles for genes that diverged significantly in expression between control and C59-treated embryos at 24 HPA (A), 48 HPA (B), 72 HPA (C), and 120 and168 HPA (D).
The numbers on the y-axes of each plot show the range of log (2) expression values. The error bars are standard deviations of the mean.
3.4 Enrichment analysis
The probe sets identified at 48 HPA significantly enriched the developmental process Gene Ontology (GO) category (N = 19; p = 4.62−5) (Table 3). These probe sets correspond to growth factors (areg, fgf9, bmp2, inhbb, ctgf), transcription factors (hoxc8, mycn, mas1, lhx2, etv4), signaling molecules (prickle2, dusp6, spry2, dkk2, ankrd1, il8, bmp2, appcd1), and proteins that function in the extracellular matrix (mmp1, mmp3, hapln3, lamb1) to regulate ectodermal (N = 10; p = 7.86−4), mesodermal (N = 10, p = 9.90−4), and nervous system (N = 9; p = 1.16−2) development. These results show that inhibition of Wnt pathway signaling affected the transcription of proteins that regulate fundamental developmental processes.
Table 3.
List of significantly enriched biological processes using genes that were differently expressed between control and C59-treated embryos. Asterisks show when biological processes were identified as significant.
| Biological Process | 48 hrs | 72 hrs | 120 hrs | 168 hrs |
|---|---|---|---|---|
| system development | * | * | * | * |
| developmental process | * | * | * | * |
| ectoderm development | * | * | * | * |
| mesoderm development | * | * | * | * |
| cellular process | * | * | * | |
| cell communication | * | * | * | |
| nervous system development | * | * | * | |
| angiogenesis | * | * | ||
| embryo development | * | |||
| B cell mediated immunity | * | |||
| digestive tract mesoderm development | * | |||
| female gamete generation | * | |||
| pattern specification process | * | |||
| segment specification | * | |||
| multicellular organismal process | * | * | * | |
| muscle organ development | * | * | * | |
| single-multicellular organism process | * | * | * | |
| skeletal system development | * | * | * | |
| anatomical structure morphogenesis | * | * | ||
| cell cycle | * | * | ||
| cellular component morphogenesis | * | * | ||
| cellular component organization | * | * | ||
| cellular component organization or biogenesis | * | * | ||
| chromosome segregation | * | * | ||
| cytokinesis | * | * | ||
| mitosis | * | * | ||
| muscle contraction | * | * | ||
| biological adhesion | * | |||
| cell adhesion | * | |||
| cell differentiation | * | |||
| cell-cell adhesion | * | |||
| cell-matrix adhesion | * | |||
| DNA replication | * | |||
| system process | * |
A total of 193 of the 237 probe sets identified at 72 HPA were not expressed differently between the control and treatment groups at 48 HPA. These probe sets further enriched the GO categories identified at 48 HPA, including all of the developmental process categories listed above (Table 3). For example, while 19 genes were identified for the developmental process GO at 48 HPA, 48 were identified at 72 HPA. These additional genes provided greater resolution of enriched processes, including pattern (N = 11; p = 4.40−4) and segment specification (N = 7; p = 3.08−3), and muscle organ development (N = 12; p = 1.93−3). Notably, the 72 hr group included tgfb1, nov, ntf3, ereg, hoxb7, hoxb9, meis2, msx2, emx2, fyn, sp7, lef1, gli3, smad7, crabp2, wif1, mmp2, and dlx6. The expression profiles for these genes suggest the vast majority, excepting genes associated with muscle tissue (see below), were lowly expressed in axolotls that were treated with C59. Thus, transcript levels were observed to be higher in control embryos for an increasing number of developmental regulatory proteins as the tail regeneration program progressed. Because later tissue samples in control animals were biased for regenerating tissue, it is likely that these genes are intimately associated with a normal tail regeneration transcriptional response.
More probes sets having significantly different levels of expression were identified at 120 (N = 1014) and 168 HPA (N = 1508). In addition to general developmental process ontologies, cell cycle (168 HPA; N = 125; p = 1.14−4) associated ontologies were identified as significantly enriched (Table 3). Of 103 non-redundant genes annotated to the cell cycle ontology, 83 were expressed more highly in control axolotls. These genes encode proteins for DNA replication (pola2, pole2, prc1, top2a, rpa1, rpa2, pcna), chromosome condensation, assembly and segregation (kif11, kif20a, kif22, kif23, kif26a, kif26b, kif2c, kif4a, kifc1, mcm3, mcm4, mcm6, mcm7, smc2, smc4, aurka, aurkb, mad2l1), and cell cycle regulation (cdk7, ccna2, ccnb1, ccnb3, ccne2, ccni, gtse1, plk1). Cell cycle annotated genes that were expressed more highly in axolotls treated with C59 were associated with negative regulation of growth (gas2, gas6, sesn1, relb) and mitosis (cdkn1b). Also genes that function in muscle contraction (myh1, myh2, myh3, myh4, myh6, acta2, acta2) and that annotated to the muscle contraction ontology were more highly expressed in C59-treated embryos. Again, these expression patterns and annotations likely reflect differences in tissue sampling; for later post amputation samples, newly, regenerated tail tissue was collected from controls, while stump tissue was sampled from C59-treated individuals. The relative abundance of transcripts associated with proliferating cells was higher in control, regenerating tissue samples. Conversely, the relative abundance of transcripts associated with differentiated cell types, or negative regulation of cell proliferation, was higher in the C59-treated, non-regenerating tissue. These results show that quantitative differences in transcript abundances, that were associated with regenerative and non-regenerative tissues, can be reliably diagnosed with the axolotl tail regeneration model.
3.5 Identification of Wnt pathway targets between axolotl and zebrafish
A transgenic approach was used recently to inhibit Wnt pathway signaling and identify downstream transcriptional targets during zebrafish fin regeneration (Wehner et al., 2014). Specifically, heat shock was applied to induce the expression of axin1 and dkk2, two antagonists of Wnt-pathway signaling. Then, RNA was isolated from tissues collect at 6, 48, and 96 HPA and microarray analysis was performed to identify genes that differed significantly from control fish. Of the 1274 non-redundant genes identified as differently expressed between C59-treated and control axolotl embryos, 387 were identified in the zebrafish fin regeneration experiment (Supplemental File 1). This includes 14 of the genes from Table 1 that were differently expressed at all post-amputation time points in this study (axin1, bmp2, ctgf, cyp26b1, dusp6, etv4, hapln3, inhbb, lamb1, mycn, prickle2, and wnt5a). These results suggest that many of the same genes are downstream transcriptional targets of Wnt pathway signaling in zebrafish and axolotl regeneration models.
4. Discussion
Our study establishes a new amphibian tail regeneration model using Mexican axolotl embryos. We show that this model provides an efficient means to identify water-soluble chemicals that affect tail outgrowth during regeneration. Almost certainly, this model could be adapted to identify chemically induced changes in tissue-specific transgene reporter lines, or to directly test genes for required functions using morpholinos or transgenics. Further, we showed that microarray analysis of C59-treated and control embryos efficiently identified downstream effects of Wnt signaling on genes and biological processes that are associated with regenerative and non-regenerative tissues. We observed precise estimates of transcript abundance and dynamic expression profiles for developmental regulatory genes that are generally lowly expressed and difficult to estimate when using heterogeneous tissue. Below we discuss the significance of the new axolotl model, the chemicals that were identified, and insights gained about the transcriptional program underlying tail regeneration.
4.1 Conservation of signaling mechanisms among amphibians and fish
Non-mammalian vertebrates provide important models for the study of regeneration. The use of chemical inhibitors and transgenic approaches is well established in the zebrafish fin and Xenopus tail regeneration models (Tal et al., 2010; Taylor et al., 2010; Slack et al., 2008). In several cases, the same tools and experimental approaches have been used to investigate signaling pathways and identify conserved mechanisms of regeneration. In zebrafish and Xenopus, Wnt signaling precedes Fgf signaling and disruption of either pathway blocks blastema formation [Lin and Slack, 2008; Chen et al., 2009; Wehner et al., 2014). In addition to identifying Wnt and Fgf inhibitors, our screen also identified Tgf-β signaling (SB-505124) as important in axolotl tail regeneration. Ho and Whitman (2008) used SB-505124 and SB-431542 to show that TGF signaling is required at different times during Xenopus tail regeneration, first during wound closure and later during tail outgrowth. Our results support the idea that several of the major signaling pathways required for regeneration are shared among amphibian and fish models. It therefore seems likely that insights about signaling from the highly regenerative axolotl will help bridge animal models and advance understanding of regeneration mechanisms.
4.2 Wnt pathway and gene expression
While considerable progress has been made in regenerative biology by targeting well-characterized genes and signaling pathways, there is need to explore more fully the complexity of regeneration using global, genomic methods. Our study shows that global gene expression analysis can be used as an efficient follow-up approach to chemical genetic screening of axolotl embryos. By comparing 1 mm slices of heterogeneous tail tissue between regenerating and chemically-inhibited, non-regenerating axolotl embryos, we identified precise temporal changes in gene expression that allowed us to detail underlying biological processes and signaling pathways. To our knowledge, this is the first study to use microarray analysis to investigate tail regeneration after applying a chemical inhibitor of the Wnt signaling pathway. We expected to observe differences in gene expression because C59, a potent inhibitor of Wnt ligand secretion (Chen et al., 2009), completely inhibited axolotl tail regeneration. Additionally, we only collected tissue from the distal tail tip, and thus later post-amputation samples contained a greater number of regenerating cells in comparison to non-regenerating samples. As a result of this sampling design, cell cycle transcripts and transcripts that are expressed highly in developing and regenerating appendages (e.g. dact2, msx2) were quantitatively higher in the 120 and 168 HPA samples collected from control embryos, and conversely, transcripts associated with differentiated muscle cell phenotypes were lower. These patterns were also observed in a microarray study of anuran tail regeneration that associated transcriptional changes with histological and immunological characterizations of proliferating and differentiated cell types (Love et al., 2011). Quantitative differences in the abundance of transcripts between regenerative and non-regenerative tissue samples likely correlate with a higher relative abundance of proliferative cell types in the former, and differentiated cell types in the latter.
Discrete changes in gene expression were also observed in our study. After tail or limb amputation in juvenile axolotls, hundreds of genes typically show transcript abundance changes in just a few hours (Monaghan et al., 2007, 2009, 2013). These early gene expression changes have been attributed to processes that occur during wound healing, including innate immune responses and re-epithelialization. We observed (but did not report) gene expression changes between the Day 0 and 12 hour samples, including a > 4 fold up regulation of leptin, which is commonly observed as highly up regulated in regeneration studies. However, none of these genes were differently expressed between C59-treated and control axolotl embryos at 12 hr. This suggests that Wnt signaling is relatively delayed after limb amputation and not required for the early, wound healing response. This is consistent with findings from Xenopus that show Tgf-β signaling functions upstream of Fgf signaling (and thus presumably Wnt-signaling) to regulate formation of the wound epidermis (Ho and Whitman, 2008); also in zebrafish there is normal wound closure after Wnt-inhibition, although the basal layer of the wound epidermis does not pattern correctly for proper signaling interactions to be established with underlying progenitor cells (Stoick-Cooper et al., 2007). At 24 HPA, transcript abundances increased significantly for several genes, with fgf9 showing the largest fold change. The expression profile for fgf9 was unique because it increased throughout regeneration in control axolotls but remained low and flat in C59-treated axolotls. In fact, the Day 0 transcript abundance estimate was consistent with background. In other words, fgf9 appeared to be transcriptionally inactive in controls until some time between 12 and 24 HPA. In Xenopus, fgf9 and other Fgf genes show increasing transcriptional abundances in the distal tail tip during regeneration (Stoick-Cooper et al., 2007) and Wnt signaling in the zebrafish and Xenopus is known to activate fgf20, which is required for blastema formation (Whitehead et al., 2005; Love et al., 2013). We speculate that fgf9 is an important, early target of Wnt signaling in the axolotl and it’s function maybe analogous to fgf20 in other vertebrates. In zebrafish spinal cord, Fgf signaling is associated with proliferation and differentiation of radial glia, which form a bridge to facilitate axon extension across a spinal cord transection injury (Goldshmit et al., 2012). It will be important to determine the source and spatial organization of cells that secrete Wnts and express fgf9 in the axolotl model.
In addition to fgf9, 24 other probe sets met the statistical threshold at 24 hr but not the 1.5 fold change threshold; these included 11 probe sets (apcdd1, tmem92, hapln3, bmp2, il8, areg, fgf9, has2, mycn, dusp6 inhbb, and axin) that met both threshold criteria at all subsequent time points. We speculate that these genes were sampled at a time when regenerative and non-regenerative tissues were in the early process of transcriptional divergence. The fact that these genes were regulated differently between regenerative and non-regenerative tissues at all subsequent time points suggest they are required for tail regeneration, a hypothesis that can now be tested directly using axolotl transgenics. Additionally, cyp26a, egr1, chsy1, rhou, and nr2f were identified as differently regulated at 24 HPA. Overall, this indicates that multiple signaling pathways are activated between 12 – 24 HPA, including Wnt, Ffg, Bmp, Tgf-β, Myc, Ras/Mapk, Egf, Notch, and RA.
4.3 Relationship of the Axolotl Tail Regeneration Model to Xenopus and Zebrafish
It is important to have multiple animal models because each provides unique opportunities to investigate different facets of tissue regeneration and collectively they allow conserved mechanisms to be identified. With respect to this later point, our results support several findings from studies of Xenopus tail and zebrafish fin regeneration, the most important of which is the requirement of Wnt signaling for axolotl tail regeneration. We found that inhibition of Wnt signaling affected the expression of genes from multiple signaling pathways, and several hundred of these genes were also identified in a recent study that characterized Wnt transcriptional targets from the zebrafish fin model (Wehner et al., 2014). These results suggest that Wnt signaling plays a primary role in orchestrating a complex and evolutionarily conserved transcriptional response that is shared among different examples of tissue regeneration. We also note that many of the same genes were identified as differently expressed among zebrafish larval, juvenile, and adult fin regeneration models (Yoshinari et al., 2009; Mathew et al., 2009).
This suggests that the axolotl embryo model will likely provide mechanistic insights that translate to latter life stages and possibly even other regeneration paradigms, such as the limb. The axolotl embryo tail regeneration model is unique from other models in at least two important ways. First, this model provides a context for investigating the role of the immune response, cellular metabolism, and aging on tissue regeneration. Loss of regenerative ability is associated with aging and maturation of physiological systems, including the immune system (Seifert and Voss, 2013). It was recently shown that 7 day post-hatchling axolotl larvae, approximately 10–14 days older than the embryos used in this study, are capable of lens regeneration but lose this potential as they age (Suetsugu-Maki et al., 2012). It was subsequently shown that young and older axolotl larvae show quantitatively different gene expression patterns, with the former showing higher expression of transcripts associated with cell proliferation and the latter showing higher expression of transcripts associated with cellular differentiation, negative regulation of cell proliferation, and immune system function (Sousounis et al., 2014). The immune system is very rudimentary in axolotl embryos. The spleen, which is the site where erythrocytes, lymphocytes, and thrombocytes are produced (Charlemagne, 1972), only starts to differentiate at the stage that tail amputations were performed in this study. We observed that at the time of amputation the spleen was colorless, however at 168 HPA the spleen was red in color. The spleen does not fully mature until several weeks after hatching and axolotls are not capable of synthesizing and secreting immunoglobulins until 56–70 days post-hatching (Fellah et al., 1989). We did not observe gene expression changes that were consistent with a robust immune response, which is typically identified as an enriched gene ontology in studies of tissue regeneration. Similarly, we did not identify transcriptional changes that were indicative of a change in metabolism within regenerating tissues, excepting the expression of leptin and a glucose transporter (slca1) that are similarly up-regulated in the Xenopus tail regeneration model (Love et al., 2011). It is possible that the availability of yolk as an energy supply in the axolotl embryo model negates the need to shift metabolic strategy to recruit and sustain proliferation in progenitor cells. Regardless, these results suggest that a mature immune response and overt changes in cellular metabolic strategy are not required for tail regeneration in axolotl embryos. These and other insights that are likely to arise from the axolotl model highlight the value of having multiple animal models to resolve mechanisms of regeneration.
Supplementary Material
Genes identified as significantly differently expressed between C59 treated and control axolotl embryos and between Wnt-inhibited and control zebrafish. The number 1 references the experimental treatments in the zebrafish study (Wehner et al. 2014) where genes were identified as significant.
Acknowledgments
This research was funded by the Army Research Office (56157-LSMUR), the National Institutes of Health (R24OD010435), the National Center for Advancing Translational Sciences (UL1TR000117) and the National Science Foundation (DBI-0951484) through its support of the Ambystoma Genetic Stock Center at the University of Kentucky. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the ARO, NIH, or NSF.
Footnotes
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the ARO, NIH, or NSF.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bordzilovskaya NP, Dettlaff TA, Duhon ST, Malacinski GM. Developmental-stage series of axolotl embryos. In: Armstrong JB, Malacinski GM, editors. Developmental Biology of the Axolotl. Oxford University Press; New York: 1989. pp. 201–219. [Google Scholar]
- Charlemagne J. Morphological studies of blood cell differentiation in the axolotl, Ambystoma mexicanum Shaw. Z Zellforsch Mikrosk Anat. 1972;123:224–239. [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, Roth MG, Amatruda JF, Chen C, Lum L. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Love NR, Amaya E. Tadpole tail regeneration in Xenopus. Biochem Soc Trans. 2014;42:617–623. doi: 10.1042/BST20140061. [DOI] [PubMed] [Google Scholar]
- Fellah JS, Vaulot D, Tournefier A, Charlemagne J. Ontogeny of immunoglobulin expression in the Mexican axolotl. Development. 1989;107:253–263. doi: 10.1242/dev.107.2.253. [DOI] [PubMed] [Google Scholar]
- Gebruers E, Cordero-Maldonado ML, Gray AI, Clements C, Harvey AL, Edrada-Ebel R, de Witte PA, Crawford AD, Esguerra CV. A phenotypic screen in zebrafish identifies a novel small-molecule inducer of ectopic tail formation suggestive of alterations in non-canonical Wnt/PCP signaling. PLoS One. 2013;8:e83293. doi: 10.1371/journal.pone.0083293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611–620. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmit Y, Sztal TE, Jusuf PR, Hall TE, Nguyen-Chi M, Currie PD. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J Neurosci. 2012;32:7477–7492. doi: 10.1523/JNEUROSCI.0758-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DM, Whitman M. TGF-β signaling is required for multiple processes during Xenopus tail regeneration. Dev Biol. 2008;315:203–216. doi: 10.1016/j.ydbio.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins P, Johnson CK, Schoergendorfer A, Putta S, Stromberg AJ, Voss SR. Identification of thyroid hormone responsive genes from the brain of the Mexican axolotl (Ambystoma mexicanum) Comp Biochem Phys Part C: Pharm Tox. 2012;155:128–135. doi: 10.1016/j.cbpc.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Martí M, Dubova I, Izpisúa Belmonte JC. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Slack JM. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol. 2008;316:323–335. doi: 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Love NR, Chen Y, Bonev B, Gilchrist MJ, Fairclough L, Lea R, Mohun TJ, Paredes R, Zeef LA, Amaya E. Genome-wide analysis of gene expression during Xenopus tropicalis tadpole tail regeneration. BMC Dev Biol. 2011;11:70. doi: 10.1186/1471-213X-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew LK, Sengupta S, Franzosa JA, Perry J, La Du J, Andreasen EA, Tanguay RL. Comparative expression profiling reveals an essential role for raldh2 in epimorphic regeneration. J Biol Chem. 2009;284:33642–33653. doi: 10.1074/jbc.M109.011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Athippozhy A, Seifert AW, Putta S, Stromberg AJ, Maden M, Gardiner DM, Voss SR. Gene expression patterns specific to the regenerating limb of the Mexican axolotl. Biology Open. 2012;1:937–948. doi: 10.1242/bio.20121594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Epp L, Putta S, Page RB, Walker J, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. Microarray and DNA Sequence Analysis of Transcription During Nerve-Dependent Limb Regeneration. BMC Biology. 2009;7:1–12. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Walker JA, Beachy CK, Voss SR. Microarray analysis of early gene expression during natural spinal cord regeneration in the salamander Ambystoma mexicanum. J Neurochem. 2007;101:27–40. doi: 10.1111/j.1471-4159.2006.04344.x. [DOI] [PubMed] [Google Scholar]
- Moriya S, Che XF, Komatsu S, Abe A, Kawaguchi T, Gotoh A, Inazu M, Tomoda A, Miyazawa K. Macrolide antibiotics block autophagy flux and sensitize to bortezomib via endoplasmic reticulum stress-mediated CHOP induction in myeloma cells. J Oncol. 2013;42:1541–50. doi: 10.3892/ijo.2013.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Westerfield M. Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis Model Mech. 2014;7:739–743. doi: 10.1242/dmm.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA, Hannoush RN, Virshup DM. Pharmacological inhibition of the wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73:502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- Seifert AW, Voss SR. Revisiting the relationship between regenerative ability and aging. BMC Biology. 2013;11:2. doi: 10.1186/1741-7007-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JMW, Lin G, Chen Y. The Xenopus tadpole: a new model for regeneration research. Cell Mol Life Sci. 2008;65:54–63. doi: 10.1007/s00018-007-7431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Walker JA, Kump DC, Samuels AK, Monaghan JR, Weisrock DW, Staben C, Voss SR. Sal-Site: Integrating new and existing ambystomatid research and information resources. BMC Genomics 2005. 2005;6:181. doi: 10.1186/1471-2164-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousounis K, Athipozzhy AT, Voss SR, Tsonis PA. Plasticity for axolotl lens regeneration is associated with age-related cellular differentiation of iris and ontogeny of the immune system. Regeneration. 2014;1:47–57. doi: 10.1002/reg2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu-Maki R, Maki N, Nakamura K, Sumanas S, Zhu J, Del Rio-Tsonis K, et al. Lens regeneration in axolotl: new evidence of developmental plasticity. BMC Biol. 2012;10:103. doi: 10.1186/1741-7007-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tanguay RL. Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish – a mini-review. Gerontology. 2010;56:231–240. doi: 10.1159/000259327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson ML, Hendry AE, Wheeler GN. Chemical genetics and drug discovery in Xenopus. Methods Mol Biol. 2012;917:155–66. doi: 10.1007/978-1-61779-992-1_9. [DOI] [PubMed] [Google Scholar]
- Wehner D, Cizelsky W, Vasudevaro MD, Ozhan G, Haase C, Kagermeier-Schenk B, Röder A, Dorsky RI, Moro E, Argenton F, Kühl M, Weidinger G. Wnt/b-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Reports. 2014;6:467–481. doi: 10.1016/j.celrep.2013.12.036. [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 310:1957–60. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Yoshinari N, Ishida T, Kudo A, Kawakami A. Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev Biol. 2009;325:71–81. doi: 10.1016/j.ydbio.2008.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes identified as significantly differently expressed between C59 treated and control axolotl embryos and between Wnt-inhibited and control zebrafish. The number 1 references the experimental treatments in the zebrafish study (Wehner et al. 2014) where genes were identified as significant.