Summary
Acute Myelogenous Leukemia (AML) is an aggressive cancer that strikes both adults and children, and is frequently resistant to therapy. Thus, identifying signals needed for AML propagation is a critical step toward developing new approaches for treating this disease. Here we show that Tetraspanin3 is a target of the RNA binding protein Musashi2, which plays a key role in AML. We generated Tspan3 knockout mice which were born without overt defects. However, Tspan3 deletion impaired leukemia stem cell self-renewal and disease propagation, and markedly improved survival in mouse models of AML. Additionally, Tspan3 inhibition blocked growth of AML patient samples suggesting that Tspan3 is also important in human disease. As part of the mechanism, we show that Tspan3-deficiency disabled responses to CXCL12/SDF-1, and led to defects in AML localization within the niche. These identify Tspan3 as an important regulator of aggressive leukemias and highlight a role for Tspan3 in oncogenesis.
Introduction
Acute Myelogenous Leukemia (AML) is a cancer marked by the rapid and uncontrolled growth of immature cells of the myeloid lineage (Shipley and Butera, 2009). Because it is a heterogeneous disease involving a wide array of chromosomal translocations and/or mutations, response to therapy differs widely between subclasses of AML. For example, while leukemias with Flt3 mutations or MLL-translocations are generally associated with poor prognosis in both adults and children, those driven by PML/RAR translocations respond well to therapy (Chen et al., 2011; Fernandez et al., 2009; Krivtsov and Armstrong, 2007; Roboz, 2012; Zeisig et al., 2012). However, despite improvements in therapy for some subtypes of AML, current treatments which include chemotherapy and bone marrow transplantation, remain ineffective for a vast majority of AML patients. Thus, identifying new approaches to more effectively target common regulators of therapy resistant AML remains critically important.
In an effort to identify pathways that mediate the aggressive growth of AML and other hematologic malignancies, we have focused on stem cell programs that are subverted to drive the oncogenic state. One important regulator of such programs is the RNA binding protein Musashi. Musashi 2 (Msi2) has been shown to predict poor prognosis in patients with Chronic Myelogenous Leukemia (CML), and is critical for progression to the blast crisis phase of the disease (Ito et al., 2010). Msi2 is also highly expressed in several AML lines and can serve as an indicator of poor outcome (Byers et al., 2011; Kharas et al., 2010). The fact that multiple hematologic malignancies require Msi2 suggested that identifying stem cell programs triggered by Msi2 could lead to the discovery of pathways important for establishing and sustaining disease. Genome wide expression analysis of Msi-deficient cancer stem cell from blast crisis CML and de novo AML identified genes commonly regulated in both leukemias. This strategy identified Tetraspanin 3 (Tspan3), a recently identified member of the tetraspanin family, as a key downstream target of Msi2 and a potential functional element in myeloid leukemia.
The tetraspanin (tetraspan or TM4SF) family forms a large group of integral membrane proteins possessing four membrane-spanning domains separated by short intracellular and extracellular domains, as well as one long extracellular domain (Hemler, 2005). Tetraspanins interact with each other and with a variety of different receptors and signaling molecules to organize supramolecular complexes in membranes. Although tetraspanins are expressed across a wide variety of cells and tissue types and are involved in diverse cellular processes such as cell adhesion, proliferation, and immune responses (Wright et al., 2004), many tetraspanins remain understudied and the roles they play in normal stem cell biology and in disease remain unknown. This is particularly true of Tspan3, which has been studied in context of oligodendrocyte migration (Tiwari-Woodruff et al., 2001), and about which little else is known. The regulation of Tspan3 by Msi2 in AML led us to test its role in leukemia development and propagation.
Expression analysis showed that Tspan3 is expressed in the hematopoietic stem/progenitors as well as in leukemia, and its pattern of expression closely mirrors that of Msi2. To test the requirement for Tspan3 in cancer, we generated Tspan3 knockout mice. These mice were born healthy and showed no overt defects in development or homeostasis. While the loss of Tspan3 did not affect normal hematopoiesis, it blocked AML self-renewal and propagation in vitro and in vivo. Further, the inhibition of Tspan3 in patient samples led to decreased colony formation in vitro, and leukemic growth in primary patient derived xenografts. Finally, Tspan3 deletion triggered defects in chemokine responsiveness and AML cell migration, which may contribute mechanistically to the defects observed. Collectively, our data identify a critical role for Tspan3 in de novo AML, and suggest that Tspan3 may be valuable as a therapeutic target.
Results
Identification of genomic programs important for myeloid leukemia
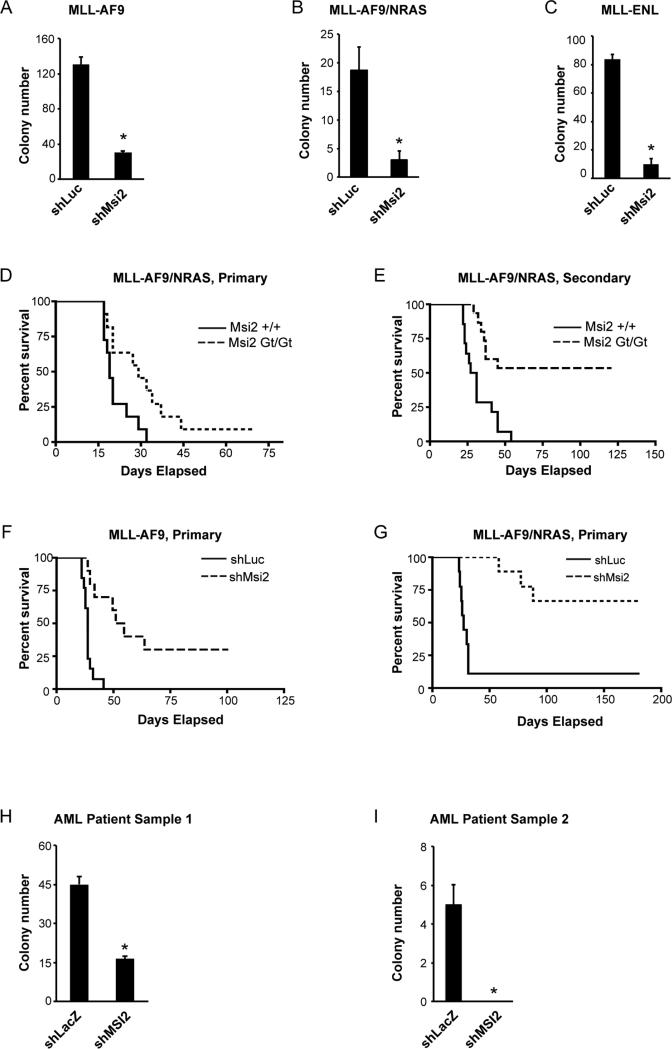
Our previous studies demonstrated that Msi2 is critical for the development of blast crisis CML (Ito et al., 2010). In addition, consistent with published work with AML cell lines (Kharas et al., 2010), we found that loss of Msi2 impaired primary AML growth as well. shRNA-mediated knockdown of Msi2 led to suppression of colony formation induced by MLL-AF9, MLL-AF9/NRAS, MLL-ENL, MLL-ENL/NRAS, and MLL-AF10 (Figures 1A-1C and Figures S1A-D). Loss of Msi2 also blocked the in vivo propagation of MLL-AF9and MLL-AF9/NRAS-driven AML (Figures 1D-1G and Figures S1E-H), as well as primary patient-derived AML cells (Figures 1H, I). Given this shared genetic dependence of two aggressive hematologic malignancies on Msi2, we postulated that analysis of the common molecular programs downstream of Msi2 could identify programs generally important for the propagation of myeloid leukemia.
Figure 1. MLL-driven primary Acute Myelogenous Leukemia is dependent on Msi2.
(A-C) Colony-forming ability of established MLL-AF9 (A), MLL-AF9/NRAS (B) and MLL-ENL (C) leukemia. cKit+ leukemic cells (A and B) or Mac1+ cells (C) were transduced with either firefly luciferase shRNA as a control (shLuc) or Msi2 shRNA (shMsi2), sorted and plated in methylcellulose media to assess primary colony formation. Representative data from two to four independent experiments are shown. Error bars represent s.e.m. *p<0.05. (D) Survival curve of mice receiving MLL-AF9/NRAS-infected control or Msi2 null KLS cells. Data shown is from three independent transplant experiments (n=11; p=0.016). (E) Survival curve of mice receiving established MLL-AF9/NRAS cKit+ leukemic cells derived from wild type or Msi2 null mice. Data shown is from three independent transplant experiments (control, n=14; Msi2 null, n=15; p<0.001). (F and G) Survival curve of mice receiving MLL-AF9 cells (F) or MLL-AF9/NRAS cells (G) infected with either control shLuc or shMsi2. (F, n=10, p<0.0001; G n=9, p=0.0011). (H and I) Patient-derived AML samples were infected with either a control virus (shLacZ) or MSI2 knockdown lentivirus (shMSI2), sorted and plated in methylcellulose media to assess primary colony formation. Error bars represent s.e.m. *p<0.05.
See also Figure S1.
Genome wide expression analysis was performed on wild type and Msi2 null leukemic stem cells from BCR-Abl/Nup98-HoxA9-driven blast crisis CML (bcCML), and from MLL-AF9 driven de novo AML (Figure 2A). Subsequently, genes that were significantly changed (FDR <0.05) following loss of Msi2 in bcCML and in de novo AML were compared and categorized into four groups: concordant (blue), bcCML specific (magenta), AML specific (red) and discordant (i.e. opposing directions, green) (Figure 2B). Of these, 319 probes formed a core set of concordantly-regulated genes in both malignancies (Figure 2C). A gene ontology (GO) analysis on gene sets concordantly regulated in both bcCML and MLL-AF9 leukemia revealed changes in many pathways, the most significant of which were regulation of cell differentiation, cell surface receptor-linked signaling pathways and immune system processes (Figures S2A-C). Within these groups, we found many signals such as Flt3, Sox4, and Pdgfrβ, as well as genes regulating apoptosis such as Bid and Bcl2a1, indicating that Msi2 controls, either directly or indirectly, expression of genes integrally linked to oncogenesis (Figures 2D, E).
Figure 2. Genomic scale analysis of shared programs in AML and bcCML stem cells.
(A) A schematic of the strategy used for genome wide gene expression analysis of Msi2-deficient AML and bcCML. (B) Representation of concordantly and discordantly regulated genes in wild type and Msi2 null bcCML and AML leukemic cells. (C) Venn diagrams displaying the intersection of probe sets that are differentially regulated in wild type and Msi2 null bcCML and AML. (D and E) Heat maps indicating commonly dysregulated genes in both bcCML and MLL-AF9 leukemia (D) and top-ranked dysregulated oncogenic signals (E); selected genes linked to oncogenesis are shown. (F) Expression level of Tspan3 in different cell populations from mouse bone marrow were analyzed by RT-PCR. Representative data from two independent experiments is shown. (G) Expression level of Tspan3 in control and Msi2 null MLL-AF9 leukemia cells determined from microarray analysis; n=3, *p<0.05. (H) cKit+ leukemia cells from MLL-AF9/NRAS-driven AML were infected with either a control or Msi2 shRNA virus and Tspan3 expression was analyzed by RT-PCR; n=2, *p<0.05. (I) Wild type KLS cells were infected with either a control virus or Msi2-expressing virus, and expression levels of Tspan3 were analyzed by real-time PCR. Representative data from two independent experiments is shown. (J) Graph shows relative enrichment in TSPAN3, ACTIN and IGF2 mRNA levels by real-time PCR following RNA-immunoprecipitation with anti-Flag antibody from MV411 AML cells expressing Flag-GFP or Flag-Msi2. (K) Colony-forming ability of cKit+ cells from MLL-AF9/NRAS leukemia infected with either shLuc and Vector, shMsi2 and Vector, or shMsi2 and Tspan3. Representative data from two independent experiments is shown. Error bars represent s.e.m. *p<0.05.
See also Figure S2.
Generation of Tspan3 knockout mice
The whole genome expression analysis not only identified Msi2 as a key upstream regulator of genes implicated in cancer but was of particular value as a database that could be mined to identify aditional regulatory signals in this disease. One differentially-expressed gene that had not previously been implicated in hematologic malignancies or in any other primary cancer was Tetraspanin 3 (Tspan3), a member of the tetraspanin family of four pass transmembrane proteins. We focused specifically on this because as a cell surface protein it would be more amenable to targeting via biologics in the long term.
In the normal hematopoietic system Tspan3 expression was most highly expressed in stem and progenitor cells (Figure 2F), and was critically dependent on the presence of Msi2. Inhibition of Msi2 led to a 5-6 fold reduction in Tspan3 levels (Figures 2G, H and Figure S2D), and ectopic overexpression of Msi2 in stem/progenitors triggered a rise in Tspan3 (Figure 2I). Importantly, TSPAN3 RNA could be specifically immunoprecipitated with Msi2 in a RIP-PCR analysis of human AML cells (Figure 2J) suggesting that Msi2 may be a direct regulator of Tspan3; genes including IGF2 and ACTIN were not immunoprecipitated by Msi2 and served as negative controls (Figure 2J). This together with an earlier report showing that the Msi1 can bind Tspan3 RNA in HEK-293T cells (de Sousa Abreu et al., 2009) suggests that Tspan3 is a direct target of both Msi1 and Msi2. Finally, ectopic overexpression of Tspan3 resulted in partial but significant rescue of the colony forming ability of Msi2 null cells (Figure 2K), indicating that Tspan3 can act functionally downstream of Msi2 in regulating AML growth. The partial rescue may be because the impact of Msi2 on leukemogenesis is likely to be mediated collectively by programs triggered by multiple genes (Figure 2C), with Tspan3 contributing to specific aspects of disease propagation.
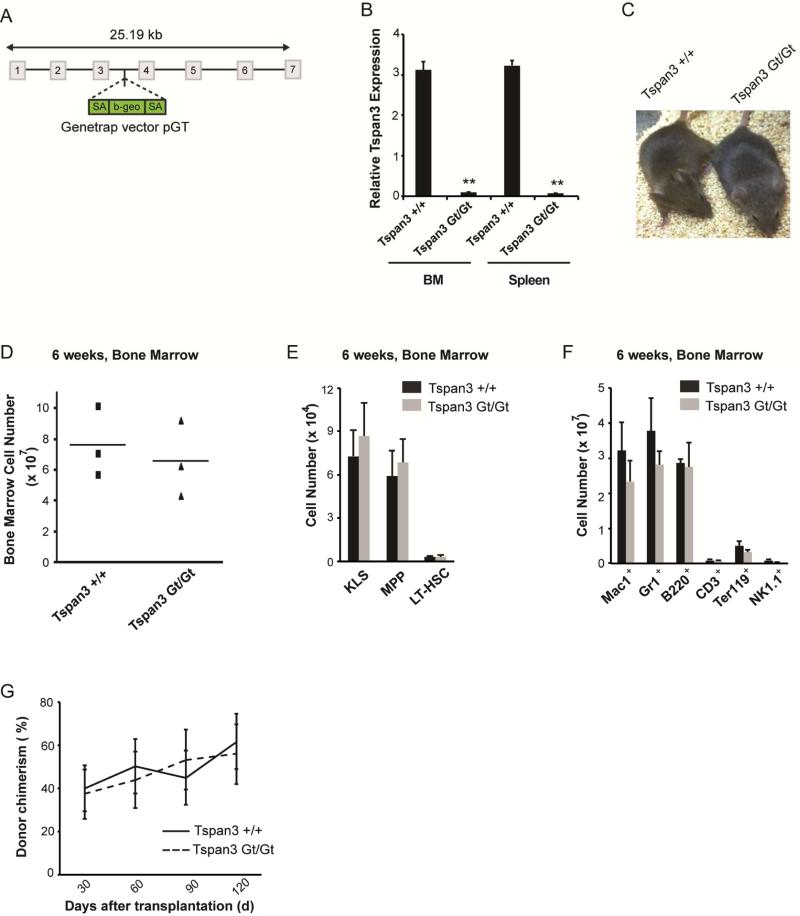
To functionally define the role of Tspan3 in AML, we generated knockout mice from Tspan3 genetrap embryonic stem cells (Figure 3A). Tspan3 mRNA expression in these mutants (denoted as Gt/Gt) was nearly undetectable (Figure 3B). Tspan3 null mice were viable and did not display any overt developmental defects; moreover, they had comparable numbers of total bone marrow cells in adult life (6 weeks, Figure 3C-3F). Importantly, the Tspan3 null HSCs showed no functional defects in long-term bone marrow reconstitution following transplantation or in homing (Figure 3G and Figure S3A). However, Tspan3 null mice displayed defects in hematopoiesis as they aged. Specifically, older mice exhibited a 40-60% reduction (except Ter119+ cells, which were reduced by 27%) in the numbers of progenitors and differentiated cells (Figures S3B, C). This suggests that Tspan3 plays a role in protecting hematopoietic cell function during aging.
Figure 3. Generation and analysis of Tspan3 knockout mice.
(A) Schematic diagram of integration site of gene trap vector used to generate Tspan3 null mice. (B) Spleen and whole bone marrow cells were isolated from wild type and Tspan3 null mice, and RT-PCR analysis was performed to determine expression of Tspan3. (C) Image of 2 month old wild type and Tspan3 null mice. (D) Total cellularity of bone marrow from 6 week old wild type and Tspan3 null mice. (E and F) Bone marrow cells from wild type or Tspan3 null mice were analyzed for hematopoietic lineage development; n=3. (G) Average donor chimerism in lethally irradiated recipients transplanted with wild type or Tspan3 null LT-HSCs (500 LT-HSCs/mouse); n=6 recipients per cohort. Data shown are from two independent experiments. Error bars represent s.e.m.
See also Figure S3.
Tspan3 is required for AML development and propagation in mouse models
To define the role of Tspan3 in leukemia, hematopoietic stem/progenitor cells (KLS) from wild type or Tspan3 null mice were infected with MLL-AF9/NRAS and transplanted (Figure 4A). Whereas 90% of recipients transplanted with wild type cells died of leukemia and only 10% survived, 53% of those transplanted with Tspan3 null cells survived, indicating a 5-fold increase in survival (Figure 4B). The cKit+ Gr1− leukemic stem cell population expressed Tspan3 at higher levels relative to bulk leukemia, suggesting Tspan3 may be specifically important for these cells (Figure 4C). To directly test whether loss of Tspan3 impaired self-renewal of leukemic stem cells, we serially transplanted cKit+ cells from wild type or Tspan3 null tumors. As shown in Figure 4D, 100% of the mice transplanted with wild type leukemia cells developed leukemia within 69 days, whereas only 31% of mice transplanted with Tspan3 null cKit+ cells developed leukemia. Tspan3 loss not only significantly reduced the incidence of leukemia but also markedly increased the latency of disease (Figure 4D). These data indicate that Tspan3 is required for maintaining the self-renewal of leukemic stem cells and tumor-propagating ability of de novo AML.
Figure 4. Loss of Tspan3 impairs the development and propagation of MLL-driven acute myelogenous leukemia.
(A) Experimental strategy to generate MLL-AF9/NRAS-driven leukemia from wild type or Tspan3 null mice. (B) Survival curve of mice receiving MLL-AF9/NRAS-infected wild type or Tspan3 null KLS cells. Data shown are from four independent experiments (wild type, n=19; Tspan3 null, n=15; p=0.0152). (C) Relative expression of Tspan3 mRNA in cKit+ Gr1− cells isolated from MLL-AF9/NRAS and in bulk leukemia (p<0.05) (D) Survival curve of mice receiving established MLL-AF9/NRAS cKit+ leukemic cells derived from wild type or Tspan3 null mice. Data shown are from three independent experiments (wild type, n=15; Tspan3 null, n=16; p<0.0001). (E and F) Survival curve of mice receiving established MLL-AF9 (E) or MLL-AF9/NRAS cells (F) infected with either control shLuc or shTspan3 virus; (E, n=19, p=0.02; F, n=7 for control, n=10 for shTspan3, p=0.0011). (G) MLL-AF9/NRAS-driven leukemic cKit+ cells were obtained from wild type or Tspan3 null leukemic mice, plated in methylcellulose in the presence of the indicated cytokines for 7 days and colony formation assessed. (H) Relative expression of indicated genes in Msi2 null cKit+Sca1−Lin− MLL-AF9 or Tspan3 null cKit+ MLL-AF9/NRAS leukemia as compared to their respective wild-type controls. Genes were selected from the genomic analysis of Msi2 null leukemic cells shown in Figure 2; n=3.
Error bars represent s.e.m., *p<0.05.
See also Figure S4.
To determine whether Tspan3 is also required in established AML, and to rule out the possibility that impaired AML propagation in Tspan3 knockouts could be due to developmental defects, we used shRNA to knock down Tspan3 in established leukemia (Figure S4A). AML cells were transduced with control or Tspan3 shRNA and transplanted (Figures 4E, F). Whereas almost all mice transplanted with control leukemia succumbed to disease, leukemia formation was significantly impaired with Tspan3 inhibition (Figure 4E), with only 50-68% of mice transplanted with Tspan3-knock down MLL-AF9 or MLL-AF9/NRAS AML succumbing to disease. Consistent with this, Tspan3 deficient leukemia cells were less able to form colonies in vitro (Figure 4G). These data indicate that Tspan3 is critical for MLL-driven leukemia, and that its inhibition can impair propagation of established AML in vitro and in vivo. Interestingly, not only was the impact of Tspan3 deletion on AML growth strikingly similar to that seen with Msi2 inhibition (Figure 1), but Tspan3 loss also phenocopied Msi2 loss at a molecular level. Thus, genes selected from the expression analysis of Msi2−/− AML cells were retested on Tspan3−/− AML cells. These genes, which included Flt3, Sox4 and Pecam1, reflected the same pattern of dysregulation as seen with Msi2 deletion (Figure 4I). Collectively these data, together with complementation analysis presented earlier (Figure 2K) support the possibility that Tspan3 is a functional downstream mediator of Msi2 in AML.
To define if Tspan3 may be a more general regulator of AML driven by distinct alleles, we tested the role of Tspan3 in a non-MLL driven model of AML. To this end, we infected control or Tspan3 null HSCs with a virally-delivered AML-ETO9a allele together with mutant NRas and tested leukemic growth in vitro and in vivo. Tspan3 deletion showed a strong trend towards improved survival (P=0.06) and led to an increase in median survival in vivo: specifically, while the median survival of control mice was 24 days, the deletion of Tspan3 in AML cells extended latency to 43 days, thus almost doubling the survival time (Figures S4B, C). Moreover, to determine the repopulating ability of leukemic cells, colony forming ability of AML-ETO9a/NRas-driven leukemic cells obtained from control or Tspan3 null leukemia was determined. Leukemic cells from Tspan3 null leukemia formed five-fold less colonies as compared to controls, suggesting that Tspan3 is needed for self-renewal in vitro (Figure S4D). These data collectively indicate that the influence of Tspan3 is not limited to MLL-AF9-driven de novo AML but may be broader across other classes of the disease.
Tspan3 is required for a normal response to the chemokine CXCL12/SDF-1
To gain insight into the mechanisms by which Tspan3 controls AML propagation, we first examined cell proliferation and apoptosis. In vivo BrdU delivery indicated that Tspan3-null leukemic cells proliferate less (Figures S4E, F). Further, no difference in apoptosis was seen using annexin V binding, although a mild increase in the frequency of cells undergoing necrosis was noted (Figures S4G, H).
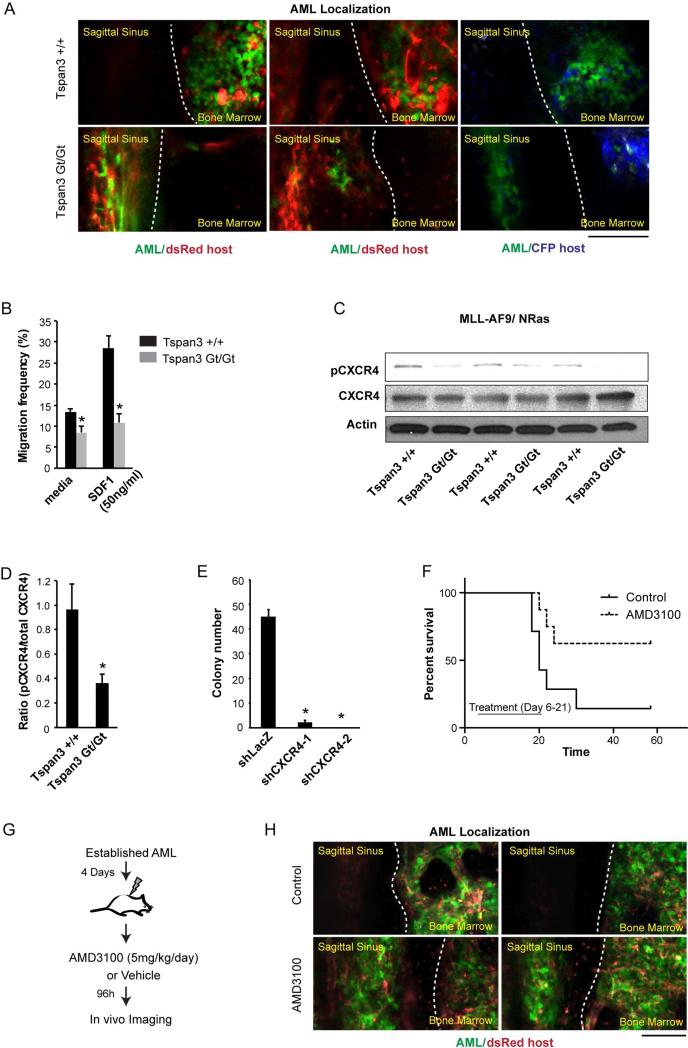
The fact that neither the defects in proliferation or cell survival in Tspan3 null cells were as severe as defects observed in AML propagation (Figures 4A-4G) suggested that Tspan3 may operate through additional mechanisms in context of the microenvironment in vivo. We thus tracked the spatial distribution of wild type and Tspan3 null leukemic cells within the bone marrow through in vivo imaging in real time. While control AML cells localized to an area with vascular beds known to be rich in SDF-1 on both sides of the central sinus within the calvarium (Colmone et al., 2008; Paxinos and Franklin, 2012; Sipkins et al., 2005), Tspan3 null AML cells were located predominantly in the central sinus distant from SDF-1 enriched areas (Figure 5A). Consistent with these defects in vivo, Tspan3 deletion impaired the capacity of leukemic cells to migrate toward SDF in vitro. While 28% of the wild type leukemia cells migrated towards SDF-1 within 4 hours in a Boyden chamber assay, 2.5 fold fewer Tspan3 null cells (11%) migrated within the same period of time (Figure 5B). Defects in chemokine responsiveness were couple with defects in receptor activation, as loss of Tspan3 led to significantly reduced levels of activated (Ser339 phosphorylated) CXCR4 (Figures 5C, D and Figure S5A). To test if CXCR4 acts functionally downstream of Tspan3, we inhibited CXCR4 using shRNAs and AMD3100, a small-molecule inhibitor of CXCR4-signaling. Inhibition of CXCR4 through either strategy resulted in a marked loss of colony forming ability of cKit+ AML cells (Figure 5E and Figure S5B). Further, in vivo delivery of AMD3100 showed significantly prolonged survival (Figure 5F), and profoundly altered the in vivo localization of AML cells (Figures 5G, H), phenocopying the effects of Tspan3 deletion.
Figure 5. Tspan3 is required for normal migration and SDF responsiveness of AML cells.
(A) In vivo image of bone marrow region in mouse calvarium. MLL-AF9/NRAS-driven leukemic cells obtained from wild type or Tspan3 null leukemia were injected retro-orbitally into sublethally irradiated dsRed or CFP mice and the calvarial bone marrow analyzed 8 to 15 days afterward to assess localization within the bone marrow niche. The images shown were acquired in a single plane. Tspan3Gt/Gt images focus on central sinusoid region to show aberrant enrichment of cells. (B) Migration of wild type and Tspan3 null MLL-AF9/NRAS leukemic cells to SDF1 was measured 4 hours after exposure. Error bars represent s.e.m., *p<0.05. (C and D) Protein lysates from wild type and Tspan3 null -driven leukemia were analyzed by western blot for phosphorylated CXCR4 (C) and band intensity was quantified using Image J (D). (E) Colony-forming ability of cKit+ MLL-AF9/NRAS leukemia transduced with either firefly luciferase shRNA as a control (shLuc) or two independent CXCR4 shRNAs (shCXCR4). (F) Survival curve of mice transplanted with established cKit+ MLL-AF9/NRAS leukemic wild type or Tspan3 null cells. Mice were treated with 5mg/kg/day AMD3100 or vehicle (water) for 15 days, starting 6 days post-transplant (n=8 for each cohort, data compiled from two independent experiments), p<0.05 (G) Experimental scheme for imaging leukemia localization in vivo following CXCR4 inhibition (E) In vivo image of bone marrow region (red) showing defects in localization of MLL-AF9/NRAS- leukemia cells (green) following AMD3100 treatment. Dotted white line demarcates the boundary between central sinusoid and bone marrow regions. Images are maximum intensity projections of on average 60μm z-stacks. Scale bar represents 100μm. Error bars represent s.e.m., *p<0.05.
See also Figure S5.
CXCR4 expression could also rescue the defects triggered by Tspan3 or Msi2 deletion. Thus over-expression of active CXCR4 partially rescued in vitro colony formation and in vivo chimerism of Tspan3−/− cells (Figures S5C, D), as well as the colony forming defects caused by the loss of Msi2 (Figure S5E). Since CXCR4 rescued Tspan3 null colony forming defects in the absence of stromal cells in vitro, we tested whether AML cells themselves were a source of SDF. Immunofluorescence analysis indicated that 14% of primary MLL-leukemia cells expressed SDF1 protein (Figure S5F), suggesting that CXCR4 mediated signaling may be sustained in these cells through an autocrine loop. Collectively, these data suggest that Tspan3 may influence AML growth, at least in part, by controlling CXCR4-mediated chemokine responsiveness.
Tspan3 is required for growth of human myeloid leukemia
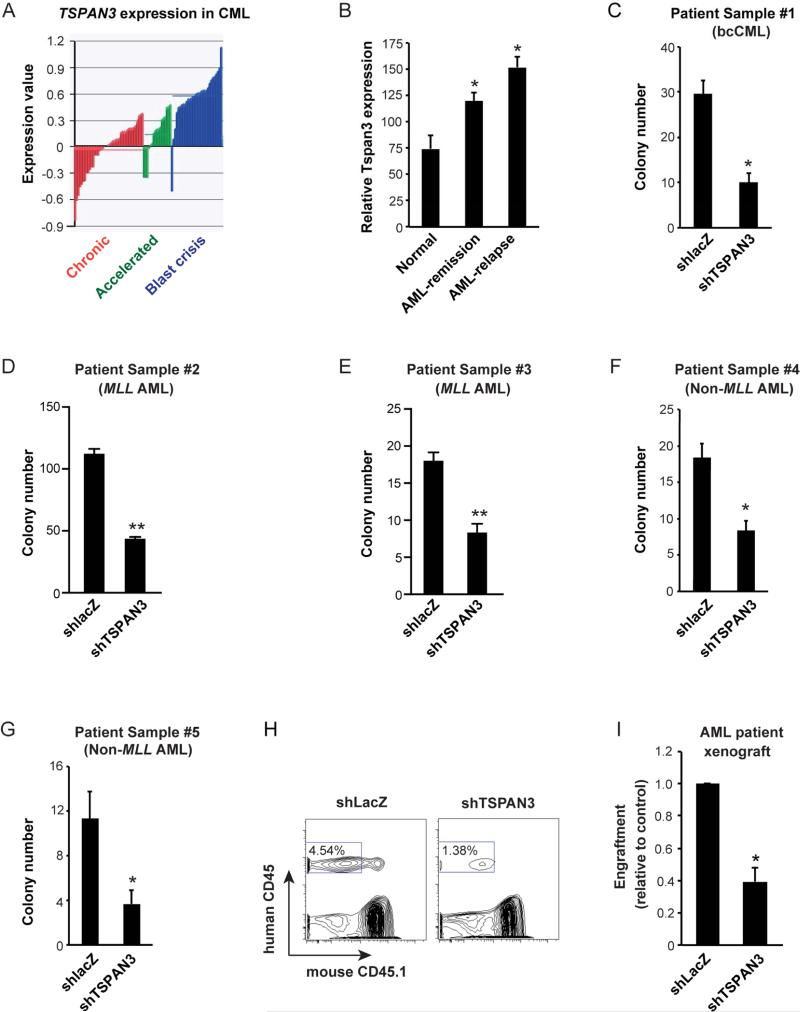
To define whether Tspan3 is relevant for human disease, we analyzed its expression in patient samples of both bcCML and de novo AML. Tspan3 expression increased with CML progression(Figure 6A), displaying a remarkable parallel with the rise in Msi2 expression (Ito et al., 2010). Further, Tspan3 expression was significantly higher in relapsed AML (Figure 6B) compared to AML in remission or healthy samples (Yagi et al., 2003). Blocking Tspan3 through shRNA delivery (Figure S6) significantly impaired colony-formation of both bcCML patient samples in vitro as well as de novo patient samples with MLL-rearrangements (Figures 6C-E). In addition, consistent with data from mouse models of non-MLL driven AML, Tspan3 inhibition blocked growth of patient samples with non-MLL rearrangements both in vitro (Figures 6F, G) as well as in xenografts (Figures 6H-I). These results strongly suggest that Tspan3 is important for the propagation of a broad range of human myeloid leukemias.
Figure 6. TSPAN3 is required for growth of human myeloid leukemia in vitro and in xenografts.
(A) Expression of TSPAN3 is shown in bone marrow and peripheral blood samples from 42 chronic phase (CP, red bars), 17 accelerated phase (AP, green bars), and 31 blast crisis (BC, dark blue bars) CML patient samples. Gene expression data were obtained using the Rosetta platform; data are expressed as the log10 ratio of the normalized expression of TSPAN3 in each patient sample compared to its expression in a pool of CP CML patients. (B) Analysis of gene expression data with increased Tspan3 in normal, AML-remission and AML-relapse samples. (C-G) Patient-derived bcCML (C), AML carrying MLL-translocations (D and E) and AML without MLL-translocations (F and G) were infected with either control (shlacZ) or TSPAN3 knockdown lentivirus (shTSPAN3), sorted and plated in methylcellulose media to assess colony formation. (H and I) Peripheral blood chimerism in NSG mice transplanted with primary patient AML samples following TSPAN3 knockdown. Patient-derived leukemic samples were infected with either control (shlacZ) or TSPAN3 knockdown lentivirus (shTSPAN3), transplanted into sublethally irradiated NSG recipients, and chimerism determined after 2 months. Data from two independent experiments are displayed relative to control set at 1. Error bars represent s.e.m. n=4, *p<0.05.
See also Figure S6.
Discussion
To identify the mechanisms that drive aggressive leukemias, we have focused on stem cell pathways that become reactivated in cancer. In particular, to define pathways important in this malignancy we have mapped signals activated by Msi2, a key regulator of leukemic growth. This led to identification of the tetraspanin superfamily member Tspan3 as a critical regulator of de novo AML. We found that loss of Tspan3 significantly impairs AML propagation in mouse models both in vitro and in vivo. Further, inhibition of Tspan3 in patient-derived AML samples inhibits their growth in vitro and in xenografts, indicating that Tspan3 is functionally important for human disease as well. As a possible mechanism, we found that Tspan3 null AML exhibit a marked inability to respond to the chemokine SDF-1 and subsequently mislocalize within the niche.
Msi2's role in primary AML, coupled with its role in bcCML, indicates that Msi2 is a broadly important regulator of aggressive hematologic malignancies. Genome wide analysis to compare gene expression patterns in leukemic stem cells between Msi2-deficient bcCML and AML revealed genes that were altered by loss of Msi2 specifically in either bcCML or AML, but also identified a cluster of genes regulated concordantly in both cancers. The concordantly regulated gene cluster includes key factors and signaling pathways through which Msi2 could impact cell fate and represents a valuable resource for continued mining of pathways and genes that are critical for leukemia stem cell maintenance and that could be targeted in the long term. Importantly, these data show that several genes critical in solid cancers, such as Flt3, Pdgfrb, and ErbB3, are extinguished upon Msi2 loss, raising the intriguing possibility that activation of these pathways may depend on Msi2 expression in solid cancers as well.
A key finding reported here is that Tspan3 is critically required for de novo AML propagation. Tspan3 belongs to the Tetraspanin superfamily, an understudied family of proteins that includes 34 members in the mouse and 33 members in humans (Garcia-Espana et al., 2008). These cell surface molecules play a wide variety of roles in distinct cells and tissues. Some Tspan family members have been associated with tumor development (Boucheix et al., 2001; Hemler et al., 1996); for example, CD9 and CD231 have been identified as markers for leukemias, and CO-029 and SAS have been implicated in carcinomas and sarcomas (Jankowski et al., 1994; Szala et al., 1990). Tspan3 itself may play a role in other cancers: for example, it is expressed in pancreatic cancer, and its inhibition blocks the growth of a colon cancer cell line in vitro (Moss et al., 2007). Our work showing that Tspan3 plays a critical role in leukemia suggests that it will be important to investigate whether Tspan3 is important for the growth of primary solid cancers in vivo as well.
Our work also makes a connection between Tspan3 and SDF-CXCR4 chemokine signaling. Consistent with this, Tspan3 loss led to remarkable defects in SDF-1 responsiveness and CXCR4 activation, and CXCR4 activation complemented defects triggered by loss of Msi2 or Tspan3. Our own data as well as significant prior work suggests that CXCR4 can have a powerful influence on AML. Our findings that CXCR4 inhibition profoundly impairs MLL driven AML, complements previous work showing that blocking CXCR4 reduces AML burden driven by other alleles (Uy et al., 2012; Zeng et al., 2009). Further, SDF-1/CXCR4 signaling has been shown to influence not only the proliferation, survival, and differentiation of AML (Chen et al., 2013; Nervi et al., 2009; Sison et al., 2013; Tavor et al., 2008; Tavor et al., 2004; Zeng et al., 2009) but also the interaction between AML and the microenvironment (Sugiyama et al., 2006; Tavor et al., 2004). Finally, CXCR4 has been previously shown to be associated with poor prognosis (Christiansen et al., 2005; Rombouts et al., 2004; Spoo et al., 2007). By collectively identifying CXCR4 as an important element in controlling leukemia propagation, these data underscore the importance of CXCR4's link with Tspan3 in multiple contexts. How Tspan3 influences SDF/CXCR4 signaling is not yet clear; however, the fact that tetraspanin-enriched microdomains (TEMs) facilitate membrane fusion and signal transduction suggests that Tspan3 could modulate the activity of CXCR4 in spatially-defined TEMs as well. Thus, enabling chemokine responsiveness may be a significant part of the mechanism by which Tspan3 controls myeloid leukemia, and elucidating these links will be an important area for further work.
Previously, several other signaling molecules have been shown to be important in de novo AML propagation. Loss of the FOXO and HOX transcription factors or the transcriptional co-activator beta-catenin impairs AML growth in vivo (Faber et al., 2009; Sykes et al., 2011; Wang et al., 2010), and deletion of PBX-3, which increases the DNA binding activity of HOX proteins, leads to similar results (Li et al., 2013). A role for the Polycomb group member Bmi-1 in the proliferation of AML stem/progenitor cells has also been demonstrated (Lessard and Sauvageau, 2003). How these and other pathways are integrated with each other and with Tspan3 and whether they act in a hierarchy will be important to define in order to fully map the molecular networks that regulate myeloid leukemia.
The development of the Tspan3 knockout mouse reported here has significance beyond the findings in this work. Because so little is known about Tspan3 function in vivo, these mice represent a valuable resource for defining the genetic role of Tspan3 in other systems. Although our data did not reveal any overt defects, detailed analyses of specific tissues are needed to assess any impact on normal function. For example, Tspan3 has been implicated in migration of oligodendrocytes, the myelin-producing cells of the central nervous system (Tiwari-Woodruff et al., 2001). Thus, analysis of brain and spinal cord function will be essential to determine if any defects in myelination or neurotransmission occur in Tspan3 null mice. In addition to its use in understanding the role of Tspan3 in normal cellular function, Tspan3 null mice has the potential to be invaluable in identifying the role of Tspan3 in other hematologic malignancies and solid cancers.
Our finding that Tspan3 is necessary for the growth and progression of AML in distinct mouse models as well as in diverse patient-derived AML samples suggests that inhibition of Tspan3 signaling may be an effective strategy for blocking leukemia growth. This is a particularly attractive possibility because the cell surface expression of Tspan3 makes it well suited for antibody-mediated therapy. Rituximab and TRU-016, which target the Tspan-like molecule CD20 and the Tspan family member CD37 respectively, represent precedents for Tspan targeting by monoclonal antibodies (Robak et al., 2010; Robak et al., 2009). Rituximab is in use clinically for the treatment of chronic lymphocytic leukemia, follicular lymphoma, and diffuse large B-cell lymphoma, and TRU-016 is being tested in trials against chronic lymphocytic leukemia. The fact that Tspan3 knockout mice are viable, and do not display overt developmental defects, provides a potential therapeutic window for anti-Tspan3 immunotherapy. Collectively, our findings raise the possibility that targeting Tspan3 could provide a new approach for therapy of aggressive leukemias.
Experimental Procedures
See Supplemental Information for more extensive methods.
Mice
C57BL6/J mice were used as transplant donors, B6-CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) and C57BL6/J were used as transplant recipients. Human samples were transplanted in NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ). All mice were 1-5 months of age unless otherwise specified. Msi2 mutant mice, B6;CB-Msi2Gt(pU-21T)2Imeg have been described previously (Ito et al., 2010). Mice were bred and maintained in the animal care facility at University of California San Diego. All animal experiments were performed according to protocols approved by Duke University and University of California San Diego Institutional Animal Care and Use Committees.
Generation of Tspan3 knockout mice
Tspan3 knockout mice were generated from mouse embryonic stem cell line NPX312 (strain 129/Ola; Baygenomics, http://www.mmrrc.org). The gene trap vector (pGT1 TMpfs) was inserted between exons 3 and 4. These cells were injected into C57BL/6 blastocysts, which were then implanted into pseudo pregnant females. High-contribution chimeras were obtained. Chimeric mice were backcrossed to C57BL/6 mice and knockout mice were generated by breeding offspring of heterozygous mice. Experiments were performed with mice of mixed genetic background.
Generation and analysis of leukemic mice
Bone marrow KLS cells were sorted and cultured overnight in RPMI media (Invitrogen) supplemented with 20% fetal bovine serum, stem cell factor (SCF, 100 ng/ml, R&D Systems), IL3 and IL6 (10ng/ml, R&D Systems). Subsequently, cells were retrovirally infected with MSCV-MLL-AF9-IRES-GFP (or tNFGR) to generate MLL-AF9 leukemia, MSCV-MLL-AF9-IRES-GFP (or tNFGR) and MSCV-NRAS-IRES-YFP to generate MLL-AF9/NRAS leukemia, or MSCV-AML-ETO9a-IRES-huCD2 and MSCV-NRAS-IRES-YFP to generate AML-ETO9a/NRAS leukemia. Doubly infected or unsorted cells collected 48 hours post-infectionb were transplanted retro-orbitally into B6 recipients. Recipients were sublethally irradiated (4-6 Gy) for MLL-driven leukemia, and lethally irradiated for AML-ETO9a-driven leukemia (9.5Gy). For secondary transplantation, GFP+/YFP+ and cKit+ cells were sorted from primary transplanted mice and 1,000-3,000 cells transplanted per mouse. Diseased mice were analyzed as previously described (Zimdahl et al., 2014).
In vivo imaging of leukemia cells
MLL-AF9/ NRas leukemic WT and Tspan null cells (50,000 to 200,000 cells/mouse) were injected into sublethally irradiated Actin-dsRed or Actin-CFP mice. The calvarial bone marrow was imaged 8 to 15 days after transplants. Images were acquired by the Leica LAS AF 2.7.3 software with TCS SP5 upright DM600 CFS Leica confocal system using the HCX APO L 20x/1.00 W Leica Plan Apochromat objective.
Genome wide expression analysis
Lin− bcCML cells and Lin−Sca1−cKit+ AML cells were FACS-sorted and total cellular RNA purified. RNAs were amplified, labeled, hybridized onto Affymetrix GeneChip Mouse Genome 430 2.0 Arrays and raw hybridization data collected (Asuragen Inc., Austin, TX). Expression level data were normalized using a multiple-loess algorithm as previously described (Sasik et al., 2004). Probes whose expression level exceeds a threshold value in at least one sample were considered detected. The threshold value is found by inspection from the distribution plots of log2 expression levels. Detected probes were sorted according to their q-value, which is the smallest false discovery rate (FDR) (Klipper-Aurbach et al., 1995) at which the gene is called significant. An FDR value of α is the expected fraction of false positives among all genes with q ≤α. FDR was evaluated using Significance Analysis of Microarrays and its implementation in the official statistical package samr (Tusher et al., 2001). To prevent unwarranted variances, the percentile of standard deviation values used for the exchangeability factor s0 in the regularized t-statistic was set to 50. The probe list, sorted by q-value in ascending order, was translated into Entrez gene ID's and parsed so that where several different probes represent the same gene, only the highest-ranking probe was kept for further analysis. The sorted list of genes was subjected to a non-parametric variant of the Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005), in which the p-value of a gene set was defined as the minimal rank-order p-value of a gene in the gene set rather than the Kolmogorov-Smirnov statistic as in GSEA. Briefly, let rk be the k-th highest rank among a gene set of size N. The rank-order p-value pk of this gene is the probability that among N randomly chosen ranks without replacement, the k-th highest rank will be at least rk. The p-value of a gene set was defined as the smallest of all pk. Finding the p-value of a gene set of size N requires calculation of N rank-order p-values; however, there is no need to adjust the p-values for the number of genes tested as the tests are highly statistically dependent. A Bonferroni adjustment of gene set p-values for the number of gene sets tested was performed. Gene sets with adjusted p-values ≤ 0.05 were reported. Heatmaps were created using in-house hierarchical clustering software and the colors qualitatively correspond to fold changes.
Statistical analysis
The statistical analysis was carried out using the T-test, Mann Whitney test and Logrank test.
Supplementary Material
Highlights.
Expression analysis implicates Tetraspanin 3 (Tspan3) in leukemia
Tspan3 loss impairs AML propagation and improves survival in mouse disease models
The chemokine response of AML cancer cells is impaired by Tspan3 deletion
Tspan3 knockdown impairs human AML growth in xenografts
Acknowledgements
We are grateful to Jonathan Yang for technical support, Marcie Kritzik for help with manuscript preparation. We would also like to thank Scott Armstrong for the MLL-AF9 construct; Scott Lowe for the AML-ETO9a construct and Dong-Er Zhang for experimental advice; Warren Pear and Ann Marie Pendergast for the BCR-ABL construct; Gary Gilliland for the NUP98-HOXA9 construct. J.B. is supported by a postdoctoral fellowship from the National Cancer Center, T.I. is a recipient of a California Institute for Regenerative Medicine interdisciplinary stem cell training program fellowship, T.K. is supported by a postdoctoral fellowship from the Japanese Society for the Promotion of Science, C.S.K. and N.L. received support from the NIH Pharmacological Sciences Training Program (T32 GM007752), and H.Y.K was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A1A2A16052133). This work was also supported by a Lymphoma and Leukemia Society Scholar Award and the University of California San Diego Moores Cancer Center NCI Core Grant, P30CA23100, as well as by NIH grants DK63031, HL097767 and DP1 CA174422 awarded to T.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.Y.K. and J.B. planned, designed and performed the majority of experiments and helped write the manuscript. T.I. carried out all genomic scale analysis of wild type and Msi2 null AML and bcCML. A.B., T.K., J.W., N.K.L. and C.S.K. provided experimental data and help. D.R., C.C. and V.G.O. provided primary leukemia patient samples and experimental advice. R.S. and G.H. carried out all bioinformatics analysis on microarray data. T.R. planned and guided the project and wrote the manuscript.
Accession Number
The GEO accession number for the data associated with this paper is GSE69512.
References
- Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert reviews in molecular medicine. 2001;2001:1–17. doi: 10.1017/S1462399401002381. [DOI] [PubMed] [Google Scholar]
- Byers RJ, Currie T, Tholouli E, Rodig SJ, Kutok JL. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118:2857–2867. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cortes J, Estrov Z, Faderl S, Qiao W, Abruzzo L, Garcia-Manero G, Pierce S, Huang X, Kebriaei P, et al. Persistence of cytogenetic abnormalities at complete remission after induction in patients with acute myeloid leukemia: prognostic significance and the potential role of allogeneic stem-cell transplantation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2507–2513. doi: 10.1200/JCO.2010.34.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. The Journal of clinical investigation. 2013;123:2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen DH, Andersen MK, Desta F, Pedersen-Bjergaard J. Mutations of genes in the receptor tyrosine kinase (RTK)/RAS-BRAF signal transduction pathway in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2005;19:2232–2240. doi: 10.1038/sj.leu.2404009. [DOI] [PubMed] [Google Scholar]
- Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le SY, Shapiro BA, Penalva LO. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. The Journal of biological chemistry. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, Racevskis J, Dewald GW, Ketterling RP, Bennett JM, et al. Anthracycline dose intensification in acute myeloid leukemia. The New England journal of medicine. 2009;361:1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Espana A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nature reviews Molecular cell biology. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Hemler ME, Mannion BA, Berditchevski F. Association of TM4SF proteins with integrins: relevance to cancer. Biochimica et biophysica acta. 1996;1287:67–71. doi: 10.1016/0304-419x(96)00007-8. [DOI] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski SA, Mitchell DS, Smith SH, Trent JM, Meltzer PS. SAS, a gene amplified in human sarcomas, encodes a new member of the transmembrane 4 superfamily of proteins. Oncogene. 1994;9:1205–1211. [PubMed] [Google Scholar]
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nature medicine. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Medical hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, Jiang X, Hong GM, Kunjamma RB, Ren H, et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121:1422–1431. doi: 10.1182/blood-2012-07-442004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AC, Doran PP, Macmathuna P. In Silico Promoter Analysis can Predict Genes of Functional Relevance in Cell Proliferation: Validation in a Colon Cancer Model. Translational oncogenomics. 2007;2:1–16. [PMC free article] [PubMed] [Google Scholar]
- Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, Prior JL, Piwnica-Worms D, Bridger G, Ley TJ, DiPersio JF. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–6214. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. Paxinos and Franklin's the Mouse Brain in Stereotaxic Coordinates) 2012 [Google Scholar]
- Roboz GJ. Current treatment of acute myeloid leukemia. Current opinion in oncology. 2012;24:711–719. doi: 10.1097/CCO.0b013e328358f62d. [DOI] [PubMed] [Google Scholar]
- Rombouts EJ, Pavic B, Lowenberg B, Ploemacher RE. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood. 2004;104:550–557. doi: 10.1182/blood-2004-02-0566. [DOI] [PubMed] [Google Scholar]
- Sasik R, Woelk CH, Corbeil J. Microarray truths and consequences. Journal of molecular endocrinology. 2004;33:1–9. doi: 10.1677/jme.0.0330001. [DOI] [PubMed] [Google Scholar]
- Sipkins DA, Wei X, Wu JW, Runnels JM, Cote D, Means TK, Luster AD, Scadden DT, Lin CP. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison EA, McIntyre E, Magoon D, Brown P. Dynamic Chemotherapy-Induced Upregulation of CXCR4 Expression: A Mechanism of Therapeutic Resistance in Pediatric AML. Molecular cancer research : MCR. 2013;11:1004–1016. doi: 10.1158/1541-7786.MCR-13-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoo AC, Lubbert M, Wierda WG, Burger JA. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood. 2007;109:786–791. doi: 10.1182/blood-2006-05-024844. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, Ferraro F, Mercier F, Singh H, Brumme KM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szala S, Kasai Y, Steplewski Z, Rodeck U, Koprowski H, Linnenbach AJ. Molecular cloning of cDNA for the human tumor-associated antigen CO-029 and identification of related transmembrane antigens. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6833–6837. doi: 10.1073/pnas.87.17.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavor S, Eisenbach M, Jacob-Hirsch J, Golan T, Petit I, Benzion K, Kay S, Baron S, Amariglio N, Deutsch V, et al. The CXCR4 antagonist AMD3100 impairs survival of human AML cells and induces their differentiation. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22:2151–5158. doi: 10.1038/leu.2008.238. [DOI] [PubMed] [Google Scholar]
- Tavor S, Petit I, Porozov S, Avigdor A, Dar A, Leider-Trejo L, Shemtov N, Deutsch V, Naparstek E, Nagler A, Lapidot T. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer research. 2004;64:2817–2824. doi: 10.1158/0008-5472.can-03-3693. [DOI] [PubMed] [Google Scholar]
- Tiwari-Woodruff SK, Buznikov AG, Vu TQ, Micevych PE, Chen K, Kornblum HI, Bronstein JM. OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes. The Journal of cell biology. 2001;153:295–305. doi: 10.1083/jcb.153.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM, Kulkarni S, Abboud CN, Cashen AF, Stockerl-Goldstein KE, et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012;119:3917–3924. doi: 10.1182/blood-2011-10-383406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MD, Moseley GW, van Spriel AB. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue antigens. 2004;64:533–542. doi: 10.1111/j.1399-0039.2004.00321.x. [DOI] [PubMed] [Google Scholar]
- Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, Mizutani S, Imashuku S, Ohki M, Ichikawa H. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- Zeisig BB, Kulasekararaj AG, Mufti GJ, So CW. SnapShot: Acute myeloid leukemia. Cancer cell. 2012;22:698–698. e691. doi: 10.1016/j.ccr.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O, Levis M, Rubin JB, Negrin RR, Estey EH, et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–6224. doi: 10.1182/blood-2008-05-158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimdahl B, Ito T, Blevins A, Bajaj J, Konuma T, Weeks J, Koechlein CS, Kwon HY, Arami O, Rizzieri D, et al. Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nature genetics. 2014;46:245–252. doi: 10.1038/ng.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.