Abstract
DNA methylation and histone modifications are epigenetic marks implicated in the complex regulation of vertebrate embryogenesis. The cross-talk between DNA methylation and Polycomb-dependent H3K27me3 histone mark has been reported in a number of organisms [1], [2], [3], [4], [5], [6], [7] and both marks are known to be required for proper developmental progression. Here we provide genome-wide DNA methylation (MethylCap-seq) and H3K27me3 (ChIP-seq) maps for three stages (dome, 24 hpf and 48 hpf) of zebrafish (Danio rerio) embryogenesis, as well as all analytical and methodological details associated with the generation of this dataset. We observe a strong antagonism between the two epigenetic marks present in CpG islands and their compatibility throughout the bulk of the genome, as previously reported in mammalian ESC lines (Brinkman et al., 2012). Next generation sequencing data linked to this project have been deposited in the Gene Expression Omnibus (GEO) database under accession numbers GSE35050 and GSE70847.
Keywords: DNA methylation, Polycomb, Embryogenesis, Zebrafish
| Specifications | |
|---|---|
| Organism/cell line/tissue | Zebrafish (Danio rerio) mixture of AB and Tübingen strains |
| Sex | N/A |
| Sequencer or array type | HiSeq 2000 Sequencing System — Illumina |
| Data format | Raw data: FASTQ format Analyzed data: BED format (MACS2 peaks) |
| Experimental factors | Dome, 24 hpf and 48 hpf embryos |
| Experimental features | DNA methylation (MethylCap-seq) and H3K27me3 (ChIP-seq) profiles of zebrafish embryogenesis |
| Consent | N/A |
| Sample source location | N/A |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. MethylCap-seq and ChIP-seq procedures
Zebrafish embryos were collected at the following stages: blastula (dome), pharyngula (24 hpf) and hatching (48 hpf). For Methylated DNA Affinity Capture (MethylCap-seq) [1], [8] we harvested N = 500 dome embryos and N = 100 24 hpf and 48 hpf embryos, whereas for ChIP-seq these numbers were incremented tenfold. Detailed explanations for both procedures were previously described [9], [10].
2.2. Genome alignment and data processing
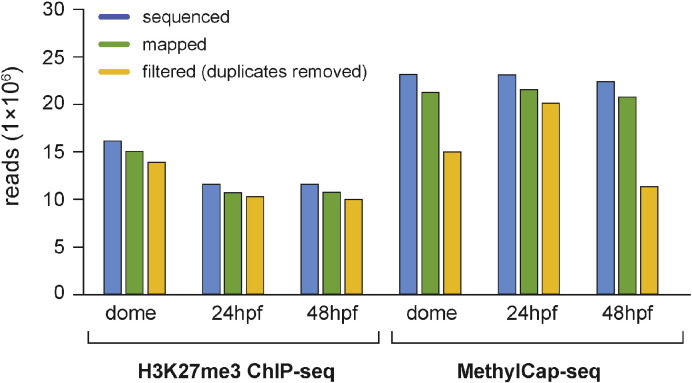
MethylCap-seq and ChIP-seq libraries were sequenced on a HiSeq 2000 Sequencing System (Illumina, CASAVA v1.8.2), generating an average of 18.6 million reads per sample (Fig. 1). The sequenced data were mapped to the zebrafish (danRer7/zv9) genome using Bowtie2 (v2.1.0) [11] with default settings (end-to-end alignment) resulting in an average mapping efficiency of 92% (Fig. 1). Mapped reads in SAM format were converted to BAM using Samtools (v0.1.19) < view >, < sort >, and < index > commands [12]. The aligned reads in BAM format were filtered for duplicates using Samtools (v0.1.19) < rmdup > (-r) command (Fig. 1). After duplicate removal, BAM files were converted to BED format using Bedtools (v2.18) < bamToBed > command [13]. The reads in BED format were summed in 10 bp intervals to create a WIG file using the sum_bed.pl script as previously described [14] and visualized in the UCSC genome browser [15].
Fig. 1.
Sequencing output and mapping efficiency for MethylCap-seq and H3K27me3 ChIP-seq data expressed as N reads (× 106).
2.3. Peak calling
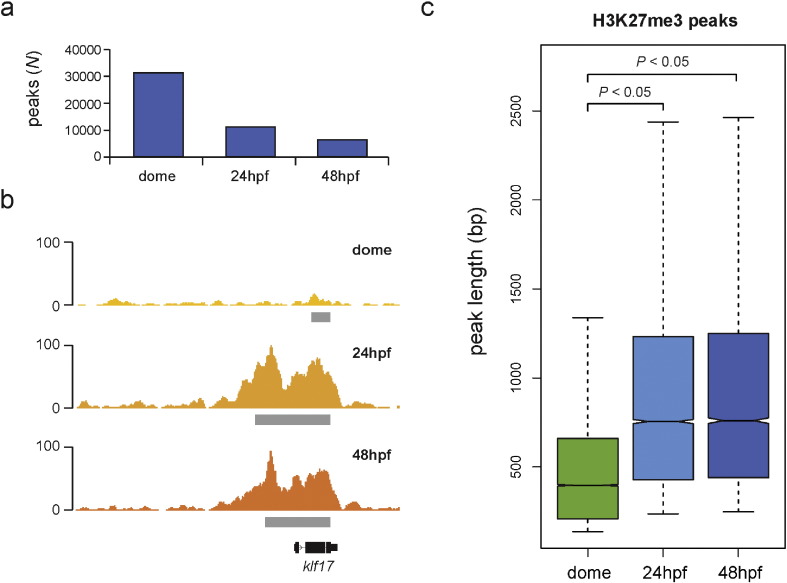
Sites of genomic enrichment (peaks) were called using the MACS algorithm (v2.1.0, https://github.com/taoliu/MACS/) [16] with default settings (callpeak) except for <-g 1.5e9 > and < –broad >. The <-g > option specifies the size of the zebrafish genome whereas the <–broad > option activates broad peak calling i.e. it clusters neighboring enriched regions into a broad region with a defined cut-off. This option is appropriate to use when expecting larger peak sizes which is often the case with H3K27me3 peaks [1], [2], [3], [4], [5], [6], [7]. The analysis resulted in the following number of peaks: N (dome) = 31,988, N (24 hpf) = 11,431, N (48 hpf) = 6436 (Fig. 2a). The peak files for H3K27me3 (dome) and H3K27me3 (48 hpf) are deposited in GEO (GSE70847) whereas the peak file for H3K27me3 (24 hpf) (GEO entry: GSE35050) is provided as Supplementary Table 1. We observe a significant (Kruskal–Wallis test, Dunn's multiple comparison test, P < 0.05) greater size distributions of H3K27me3 (24 hpf) and H3K27me3 (48 hpf) peaks when compared to H3K27me3 (dome) peaks (Fig. 2b, c) consistent with a previous report that demonstrated a developmental increase in H3K27me3 signal during Xenopus tropicalis embryogenesis [6].
Fig. 2.
Identification of H3K27me3 peaks. a) Number of identified H3K27me3 peaks. b) Example of developmental increase in H3K27me3 peak size. Gray boxes correspond to MACS2 peaks. c) Boxplots (outliers not shown) representing size distributions of H3K27me3 peaks at three developmental stages. The statistical significance of differences in size distributions was assessed by a Kruskal–Wallis test and Dunn's multiple comparison test, (P < 0.05).
2.4. Mean genomic profiles of DNA methylation enrichment over H3K27me3 peaks and CpG islands
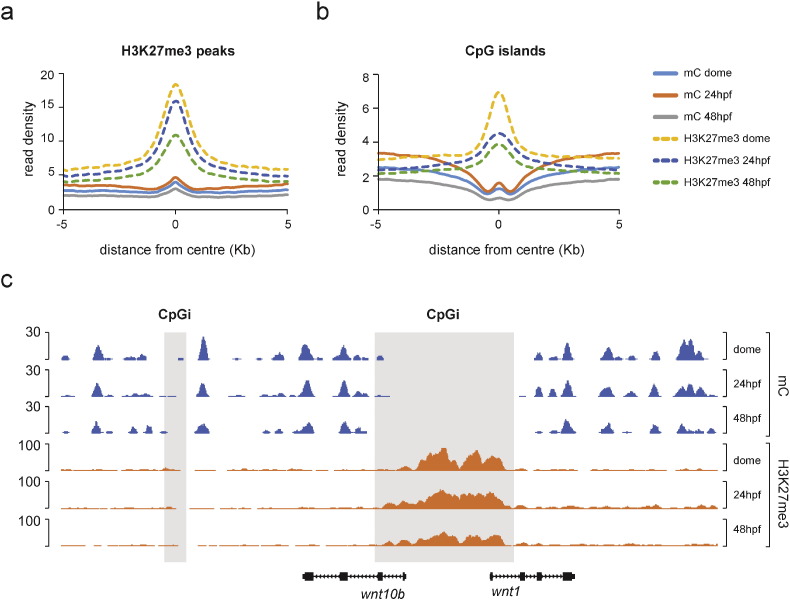
To explore the genomic relationships of DNA methylation and H3K27me3, we superimposed the DNA methylation signal (mapped reads in BED format) from dome, 24 hpf and 48 hpf embryos over H3K27me3 peaks using seqMINER (v1.3.3) [17] with default settings (5 kb upstream/downstream extension, 200 bp read extension, wiggle step = 50 bp, percentile threshold = 75%) (Fig. 3a). DNA methylation and H3K27me3 signals are generally compatible within H3K27me3 24 hpf peaks and similar genomic profiles were detected for H3K27me3 (dome) and H3K27me3 (48 hpf) peaks (Supplementary Fig. 1a-c). Next, we wanted to investigate the relationships between DNA methylation and H3K27me3 in CpG islands, major regulatory elements associated with vertebrate promoters [18]. To that end, we used a dataset that corresponds to CpG islands (also called non-methylated islands or NMIs) identified in 24 hpf zebrafish embryos through CxxC Bio-CAP profiling (GEO entry: GSE43512, sample: GSM1064697) [18]. Average profiles of DNA methylation and H3K27me3 over these regulatory elements identified a strong antagonism between DNA methylation and H3K27me3 at all the examined stages (Fig. 3b, c).
Fig. 3.
Genomic profiles of DNA methylation (mC) and H3K27me3 expressed as mean read density in a) H3K27me3 (24 hpf) peaks and b) CpG islands. c) An example of the DNA methylation/H3K27me3 antagonism in the wnt10b/wnt1 locus.
3. Conclusions
In the present study we describe genome-wide DNA methylation and Polycomb (H3K27me3) signatures of early zebrafish embryogenesis. We conclude that both marks can simultaneously exist within the sites of H3K27me3 genomic enrichment, except for CpG islands. CpG islands are enriched in H3K27me3 and strongly depleted of DNA methylation, as previously reported in X. tropicalis embryos and mouse embryonic stem cells [1], [2]. Our study extends these findings to the zebrafish (Danio rerio) model organism thereby suggesting the existence of an evolutionarily conserved, developmental chromatin state.
The following are the supplementary data related to this article.
Genomic profiles of DNA methylation (mC) and H3K27me3 expressed as mean read density in a) H3K27me3 (dome), b) H3K27me3 (24 hpf), and c) H3K27me3 (48 hpf) peaks.
BED file corresponding to MACS2 peaks for the H3K27me3 (24 hpf) dataset.
Animal procedures
All animal experiments were conducted following the guidelines established and approved by the local governments and the Institutional Animal Care and Use Committee, always in accordance with best practices outlined by the European Union.
Acknowledgments
Spanish and Andalusian government grants BFU2013-41322-P and BIO-396 to J.L.G.S. supported this work. O.B. is supported by an Australian Research Council Discovery Early Career Researcher Award —DECRA (DE140101962).
References
- 1.Bogdanovic O. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res. 2011;21(8):1313–1327. doi: 10.1101/gr.114843.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkman A.B. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22(6):1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irimia M. Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. 2012;22(12):2356–2367. doi: 10.1101/gr.139725.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landan G. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat. Genet. 2012;44(11):1207–1214. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- 5.Potok M.E. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153(4):759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heeringen S.J. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res. 2014;24(3):401–410. doi: 10.1101/gr.159608.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura R. Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates. Development. 2014;141(13):2568–2580. doi: 10.1242/dev.108548. [DOI] [PubMed] [Google Scholar]
- 8.Brinkman A.B. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52(3):232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanovic O. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22(10):2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogdanovic O. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods. 2013;62(3):207–215. doi: 10.1016/j.ymeth.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oti M., Kouwenhoven E.N., Zhou H. Genome-wide p63-regulated gene expression in differentiating epidermal keratinocytes. Genom Data. 2015;5:159–163. doi: 10.1016/j.gdata.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent W.J. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 2014;1150:81–95. doi: 10.1007/978-1-4939-0512-6_4. [DOI] [PubMed] [Google Scholar]
- 17.Ye T. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39(6):e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long H.K. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:e00348. doi: 10.7554/eLife.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic profiles of DNA methylation (mC) and H3K27me3 expressed as mean read density in a) H3K27me3 (dome), b) H3K27me3 (24 hpf), and c) H3K27me3 (48 hpf) peaks.
BED file corresponding to MACS2 peaks for the H3K27me3 (24 hpf) dataset.