Abstract
Sepsis is an especially common affliction in the elderly and despite its increased prevalence and mortality in older people, the immune response of the elderly during septic shock appears similar to that of younger patients. In the original study we conducted a global gene expression analysis of circulating neutrophils from elderly and young septic patients, as well as from age-matched healthy controls, to better understand how elder individuals respond to severe infectious insult (Pellegrina et al., 2015). Here we provide additional details pertaining processing and statistical analysis of the microarray data. Raw and normalized datasets linked to this project have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE67652.
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human neutrophils extracted from blood samples |
| Sex | 22 males and 2 females |
| Sequencer or array type | Agilent DNA SurePrint G3 Human Gene Expression 8x60k v2 Microarray Kit |
| Data format | normalized data: TSV table with an ID column and a log_ratio column (log2 Cy3/Cy5 ratio) |
| Experimental factors | Samples are from 4 groups: Adults and elderly with sepsis and adults and elderly healthy controls |
| Experimental features | Neutrophils were collected from blood samples using a Ficoll gradient. Total RNA was isolated using TRIzol and used to generate Cy3 labeled targets (Agilent Low Input Quick Amp Labeling Kit — cat # 5190-2306). A common reference (Universal Human Reference RNA — Agilent cat # 740000) was labeled with Cy5 dye. Equimolar amounts of Cy3/Cy5 labeled targets were combined and hybridized to microarrays. |
| Consent | Not applicable |
| Sample source location | Blood samples were collected from septic patients and healthy donors at the Hospital das Clínicas Intensive Care Unit, University of Sao Paulo, Brazil. |
1. Direct link to deposited data
All samples are separately available in: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67652.
2. Experimental design, materials and methods
2.1. Study design
The current study was a prospective cohort study. Neutrophils were isolated from blood samples obtained from 6 aged septic patients (age range 65 to 78 years old), 6 young septic patients (age range 22 to 35 years old), 6 healthy aged volunteers (age range 60 to 82 years old) and 6 healthy younger individuals (age range 20 to 35 years old). Total RNA was isolated and used in microarray hybridizations. More details can be found in the paper focused on the transcriptome analysis of this dataset and in a discussion of the functional implications of the results to the mechanisms of sepsis in the elderly [1].
A two color microarray experiment design was employed, in which Cy3-labeled targets from each sample were co-hybridized to individual arrays along with Cy-5 labeled common reference RNA targets to allow normalization and comparison across the different samples [2].
2.2. Data filtering and processing
After hybridization and washing steps according to the manufacturer's protocol, microarray slides were scanned using the SureScan Microarray Scanner (Agilent, USA) and images were processed using the Feature Extraction (FE) Software v.12 (Agilent, USA) [3]. Besides the signal intensity from each fluorophore detected in each spot, the FE Software provides additional information, such as each signal's standard deviation, the background signal intensity, Cy3/Cy5 log2 ratio values and also some booleans that result from tests to evaluate the quality of the signal measured in each array element. Of those booleans, we considered the ‘Well Above Background’ (WAB) test, a t-test that compares how different is the signal detected by each probe from the local background, considering the mean pixel intensities as well as their standard deviation, and a 99% confidence interval. The WAB test returns “0” if the signal is too weak and is indistinguishable from the background and “1” if it is significantly higher from the background. To be considered for further analysis, any given spot should be consistently detected in at least one experimental group in both Cy3 and Cy5 channels. If a spot was detected in at least five out of six samples of any given group with a valid WAB measurement (i.e. boolean “1”) it was kept, otherwise it was discarded. This arbitrary rule was created to not exclude spots that were consistently detected in one group but not in the others.
The next step was to address the fact that many spots are replicated in the array. Replicated spots contain probes that interrogate the same transcript. After the WAB intensity signal filtering, C3/Cy5 log2 ratios from the remaining replicated probes were averaged using a custom script that searched for probes with the same value in the “Probe name” column from the FE output files. After data filtering, 16,698 probes interrogating different gene transcripts remained out of the 58,717 represented in the array (28% of total).
2.3. Statistical analysis
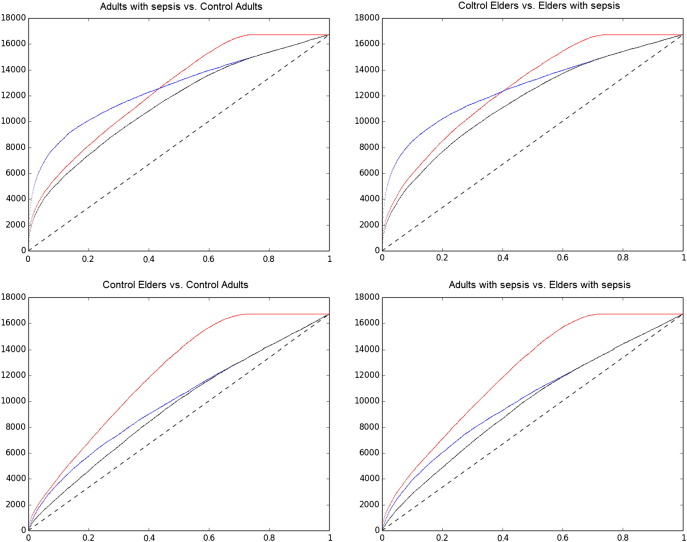
We used the normalized data to perform statistical analyses aimed to identify genes differentially expressed in septic patients and affected by aging. In our original study we performed two sets of differential gene expression analyses: one to identify genes deregulated in sepsis in young and elderly subjects compared to matched healthy controls, and a second that searched for genes deregulated in septic or healthy elderly subjects compared to matched young controls [1]. For each analysis, two different approaches were used to estimate the statistical significance of differential gene expression, namely Significance Analysis of Microarrays (SAM) [4] and RankProduct (RP) [5], both using publicly available R packages [6]. It is very important to note that while SAM uses means and standard deviations to compare gene expression between sample groups [4], RP sorts each sample's gene expression measurements and compares, for each gene, how differently they are ranked in each sample group [5]. Fig. 1 show the distributions of genes according to p-values calculated using SAM or RP in different sample group comparisons. Each graph shows, for a given group comparison, how many genes have a p-value smaller than a certain number (Fig. 1).
Fig. 1.
The number of genes identified as significantly differentially expressed (vertical axis) for a given p-value threshold (horizontal axis) according to SAM (blue line), RP (red line) or both approaches (black line) according to both. The dashed black line represents the random uniform distribution of p-values given the number of genes tested.
Note that the black lines only touch the colored lines (meaning that one of the algorithms is strictly more permissive than the other) at very high, nonsignificant p-values. Interestingly, at significant p-values the stricter algorithm varies according to the sample group comparison (Fig. 1, RP in upper panels, SAM in lower panels). Conceivably, each algorithm will produce a number of false positives, but since they are intrinsically different those will not be the same [7]. With that in mind we opted to consider to the functional analysis described in our original paper only genes identified as differentially expressed with a p ≤ 0.01 in both methods [1].
Acknowledgments
This work is supported by grants and fellowships from FAPESP, the Sao Paulo Research Foundation (grant 2012/03677-9 to MCM, fellowship 2014/03150-6 to DVSP) and CNPq, the National Council for Scientific and Technological Development (grant 470539/2013-15 to FPS, established investigator fellowship to EMR).
References
- 1.Pellegrina D.V.S., Severino P., Vieira Barbeiro H., Maziero Andreghetto F., Tadeu Velasco I., Possolo de Souza H. Septic shock in advanced age: transcriptome analysis reveals altered molecular signatures in neutrophil granulocytes. PLoS One. 2015;10(6):e0128341. doi: 10.1371/journal.pone.0128341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peixoto B.R., Vencio R.Z., Egidio C.M., Mota-Vieira L., Verjovski-Almeida S., Reis E.M. Evaluation of reference-based two-color methods for measurement of gene expression ratios using spotted cDNA microarrays. BMC Genomics. 2006;7:35. doi: 10.1186/1471-2164-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quackenbush J. Microarray data normalization and transformation. Nat. Genet. 2002;32:496–501. doi: 10.1038/ng1032. (Suppl.) [DOI] [PubMed] [Google Scholar]
- 4.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong F., Breitling R., McEntee C.W., Wittner B.S., Nemhauser J.L. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 6.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadota K., Nakai Y., Shimizu K. Ranking differentially expressed genes from Affymetrix gene expression data: methods with reproducibility, sensitivity, and specificity. Algorithms Mol. Biol. 2009;4:7. doi: 10.1186/1748-7188-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]