Abstract
Lung cancer is the leading cause of cancer death worldwide, and has a five-year survival rate of 18% [1]. MARK2 is a serine/threonine-protein kinase, and is a key component in the phosphorylation of microtubule-associated proteins [2], [3]. A recent study published by Hubaux et al. found that microtubule affinity-regulating kinase 2 (MARK2) showed highly frequent DNA and RNA level disruption in lung cancer cell lines and independent non-small cell lung cancer (NSCLC) cohorts [4]. These alterations result in the acquisition of oncogenic properties in cell lines, such as increased viability and anchorage-independent growth. Furthermore, a microarray-based transcriptome analysis of three short hairpin RNA (shRNA)-mediated MARK2 knockdown lung adenocarcinoma cell lines (GEO#: GSE57966) revealed an association between MARK2 gene expression and cell cycle activation and DNA damage response. Here, we present a detailed description of transcriptome analysis to support the described role of MARK2 in promoting a malignant phenotype.
Keywords: MARK2, Microarray, Affymetrix, Gene set enrichment analysis, Lung cancer
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Sex | Male, female |
| Sequencer or array type | Affymetrix GeneChip Human PrimeView Array (Affymetrix, Santa Clara, CA, USA) |
| Data format | Raw data: Affymetrix .CEL files |
| Experimental factors (i.e. tumor vs. normal, any pretreatment of samples) |
Lung adenocarcinoma cell lines: NCI-H1650, NCI-H1993, NCI-H1693 |
| Experimental features | Gene expression profiles of shRNA-mediated MARK2 knockdown and corresponding control lung cancer cell lines using Affymetrix Human PrimeView expression microarrays. |
| Consent | Cell Lines: Not applicable Patient samples: Written informed consent; approved by the University of British Columbia — BC Cancer Agency Research Ethics Board. |
| Sample source location (City, Country of model organism and/or Latitude & Longitude (& GPS coordinates) for collected samples if applicable) | Cell Lines: ATCC®, Manassas, Virginia USA; Patient samples: BC Cancer Agency Research Centre, Vancouver, British Columbia, Canada |
Direct link to deposited data
1. Experimental design, materials and methods
1.1. Cell lines and study design
In the investigation of the potential oncogenic role of MARK2 in lung cancer, three lung adenocarcinoma cell lines (NCI-H1650, NCI-H1993, NCI-H1693) with high endogenous expression of MARK2 were selected for further gene expression manipulation studies. Cells were cultured as previously described [4]. Lentiviral transductions with either empty vector PLKO (control) or shRNA targeting MARK2 transcripts were used to stably modulate MARK2 expression levels (Thermo Fisher, Canada). RT-qPCR was performed as a validation of transduction efficiency.
1.2. Patient samples
In order to filter results of gene-set enrichment analysis, human NSCLC was used. Seventy-seven matched tumor/non-malignant samples from the British Columbia Cancer Research Centre (BCCRC) cohort were considered. Tissues were obtained from both male and female subjects under written, informed consent approved by the University of British Columbia — BC Cancer Agency Research Ethics Board. Fresh-frozen tissues were micro-dissected to ensure > 80% tumor cell content [5]. A 2-fold change threshold between the tumor/non-malignant pairs was established to define differentially expressed genes.
1.3. Microarray experiments
RNA extraction from two biological replicates of each experimental condition was performed using TRIzol reagent (Life Technologies, Grand Island, USA). The quality and concentration of total RNA was assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Foster City, CA). Biotinylated cRNA was prepared from 500 ng of total RNA and hybridized using the GeneChip Human PrimeView Array platform (Affymetrix, Santa Clara, CA, USA) for 16 h at 45 °C. Array slides were washed and stained in the Affymetrix Fluidics Station 400, and scanned using the Affymetrix GeneChip Scanner 3000. Microarray data analysis was performed using default analysis settings in the Affymetrix® Expression Console™ and Partek Genome Suite™ Software.
1.4. Data QC and normalization
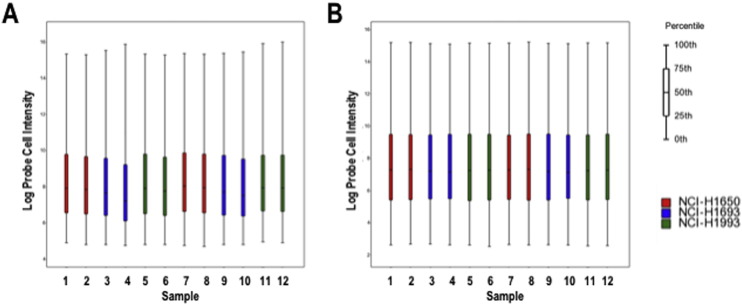
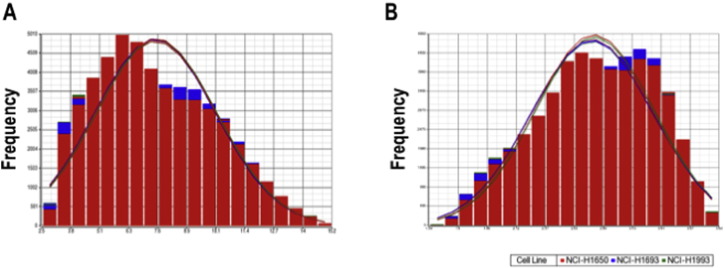
Probe cell intensity (.CEL) files were imported and normalized using the Robust Multiarray Averaging (RMA) normalization method [6]. Normalized probe intensities were summarized in order to display probe-level signal data. Signal intensity before and after log2 RMA signal transformation is displayed in Fig. 1. Distribution of probe intensity values was also assessed, confirming that log2-transformed normalized data is closer to a normal distribution (Fig. 2).
Fig. 1.
Comparison of raw vs. normalized probe intensity. Box-and-whisker plot of A) raw and B) log2 RMA normalized intensity data. Samples are in the following order: 1) NCI-H1650 pLKO control (Replicate 1), 2) NCI-H1650 MARK2 Knockdown (Replicate 1), 3) NCI-H1693 pLKO control (Replicate 1), 4) NCI-H1693 Mark2 Knockdown (Replicate 1), 5) NCI-H1993 pLKO control (Replicate 1), 6) NCI-H1993 Mark2 Knockdown (Replicate 1), 7) NCI-H1650 Mark2 Knockdown (Replicate 2), 8) NCI-H1650 pLKO control (Replicate 2), 9) NCI-H1693 pLKO control (Replicate 2), 10) NCI-H1693 Mark2 Knockdown (Replicate 2), 11) NCI-H1993 pLKO control (Replicate 2), 12) NCI-H1993 Mark2 Knockdown (Replicate 2).
Fig. 2.
Probe intensity distribution: Histogram for intensity distribution on A) raw and B) log2 normalized data is shown (A bell-shaped curve is overlaid for comparison purposes). Box-and-whisker plot of each experimental condition are shown at the bottom of each histogram. Samples are in the following order: 1) NCI-H1650 pLKO control (Replicate 1), 2) NCI-H1650 Mark2 Knockdown (Replicate 1), 3) NCI-H1693 pLKO control (Replicate 1), 4) NCI-H1693 Mark2 Knockdown (Replicate 1), 5) NCI-H1993 pLKO control (Replicate 1), 6) NCI-H1993 Mark2 Knockdown (Replicate 1), 7) NCI-H1650 Mark2 Knockdown (Replicate 2), 8) NCI-H1650 pLKO control (Replicate 2), 9) NCI-H1693 pLKO control (Replicate 2), 10) NCI-H1693 Mark2 Knockdown (Replicate 2), 11) NCI-H1993 pLKO control (Replicate 2), 12) NCI-H1993 Mark2 Knockdown (Replicate 2).
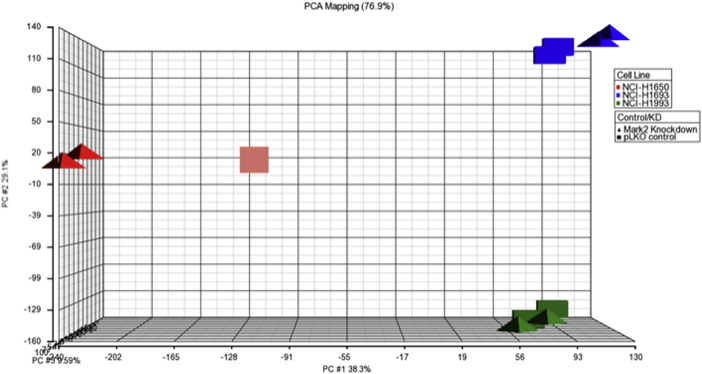
A principal component analysis (PCA) was performed on RMA normalized expression data. First, the analysis reveled that biological replicates showed a high degree of correlation for each cell line. The NCI-H1650 MARK2-KD experiments clustered further apart from PLKO control in the PCA graph, suggesting that the knockdown of MARK2 had the strongest effect in this cell line. In NCI-H1693 and NCI-H1993 MARK2, knockdown cell lines are closely related to their respective control counterparts (Fig. 3).
Fig. 3.
Principal component analysis. A PCA analysis was performed on normalized intensity values on NCI-H1650 (red), NCI-H1693 (blue) and NCI-H1993 (green). The experimental conditions (knock-down — triangles and pLKO controls, squares) are shown.
1.5. Statistical analysis
All statistical analyses were conducted using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). P values for cell viability and colony formation for shRNA knockdown of MARK2 were calculated using two-tailed t-tests. All experiments were performed in triplicate.
Significant analysis of microarrays (SAM v4.0) [7] was conducted using R statistical software (v3.0.1) to determine genes whose expression is significantly changed as a result of MARK2 manipulation. SAM analysis includes a multiple-testing correction, whereby p-values are adjusted to calculate the false-discovery rate (FDR) q-value [8]. Genes with an estimated FDR q-value < 0.05 were used.
1.6. Gene set enrichment analysis
Gene set enrichment analysis (GSEA) algorithm was used to identify function significantly enriched in shRNA MARK2 cell lines compared to their controls [9]. The expression profiles of the ten highest and lowest MARK2 expressing tumor-normal pairs from the BCCRC dataset were assessed to identify genes significantly associated with MARK2 expression.
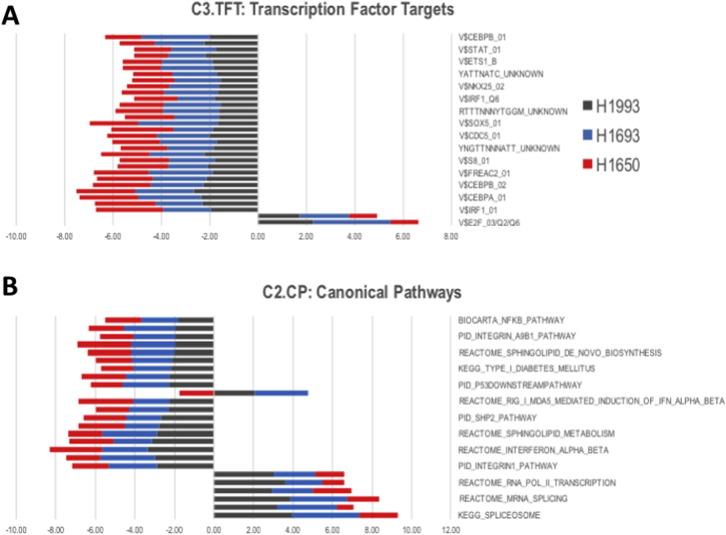
The C3.TFT (Transcription Factor Targets) and C2.CP (Canonical Pathways) collection from the Molecular Signatures Database (MSigDB) were selected to preferentially acquire information from canonical pathway and transcription factor target gene sets in each cell line. In cell line and tumor analysis, fifty-six gene sets were be significantly enriched. This cohort includes genes associated with NF-κB, DNA repair, E2F, and Myc/Max. Additionally, negative association between NF-κB and MARK2 expression has been previously observed (Fig. 4).
Fig. 4.
Gene set enrichment analysis: A gene-set enrichment analysis (GSEA) was performed on differentially expressed genes between shRNA-MARK2 and controls. Two-fold change genes identified on NCI-H1993 (green), NCI-H1693 (blue) and NCI-H1650 (red) were chosen as inputs for A) Transcription factor targets or B) canonical pathways gene sets analysis. Negatively and positively enriched gene sets are shown in bars to the left and right of the zero line. A q-value ≤ 0.05 was used a significance threshold. Names of significantly enriched gene sets are shown on the y-axis.
2. Discussion
The role of MAP/microtubule affinity-regulating kinases have been extensively studied in multiple cellular processes, however their involvement in cancer development and patient outcome remains to be deciphered. Results from Hubaux et al. reveal recurrent overexpression of MARK2 in independent NSCLC cohorts, irrespective of lung cancer histological subtype. Moreover, the manipulation of MARK2 expression in lung cancer cell models revealed its involvement in cell viability and anchorage-independent growth, and provided evidence of its association with DNA damage and cisplatin sensitivity [4].
The present study describes the detailed analytical procedures used to determine the involvement of MARK2 gene expression in the development of lung cancer malignant phenotypes including DNA damage and proliferation. We have presented a thorough characterization of the nature of the raw data obtained from the transcriptome analysis, and have illustrated the quality control and normalization procedures. We also describe in detail the data input used for the gene set enrichment analysis that was conducted to elucidate potential biological pathways affected by lack of MARK2 expression.
In conclusion, we have provided additional information supporting the robustness of the data generated as well as the analytical approach used to analyze the role of MARK2 as a potential lung cancer oncogene.
Funding sources
This work was supported by grants from the Canadian Institutes for Health Research (CIHR) (MOP-97839 and MOP-123273). E.A.M. and C.A. are supported by a BC Cancer Studentship, C.A. by CBCF Studentship, K.L.T. by Vanier Canada Scholarship.
References
- 1.Jemal A. Global cancer statistics. CA Cancer J. Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Matenia D., Mandelkow E.M. The tau of MARK: a polarized view of the cytoskeleton. Trends Biochem. Sci. 2009;34(7):332–342. doi: 10.1016/j.tibs.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Drewes G. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89(2):297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 4.Hubaux R. Microtubule affinity-regulating kinase 2 is associated with DNA damage response and cisplatin resistance in non-small cell lung cancer. Int. J. Cancer. 2015 doi: 10.1002/ijc.29577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson I.M. EYA4 is inactivated biallelically at a high frequency in sporadic lung cancer and is associated with familial lung cancer risk. Oncogene. 2014;33(36):4464–4473. doi: 10.1038/onc.2013.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irizarry R.A. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 7.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storey J.D. The positive false discovery rate: A bayesian interpretation and the q-value. Ann. Stat. 2003;31(6):2013–2035. [Google Scholar]
- 9.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]