Abstract
The study of homologous recombination has its historical roots in meiosis. In this context, recombination occurs as a programmed event that culminates in the formation of crossovers, which are essential for accurate chromosome segregation and create new combinations of parental alleles. Thus, meiotic recombination underlies both the independent assortment of parental chromosomes and genetic linkage. This review highlights the features of meiotic recombination that distinguish it from recombinational repair in somatic cells, and how the molecular processes of meiotic recombination are embedded and interdependent with the chromosome structures that characterize meiotic prophase. A more in-depth review presents our understanding of how crossover and noncrossover pathways of meiotic recombination are differentiated and regulated. The final section of this review summarizes the studies that have defined defective recombination as a leading cause of pregnancy loss and congenital disease in humans.
Meiotic recombination is essential for the accurate segregation and genetic mixing of chromosomes. It differs from recombinational repair in somatic cells in several ways (e.g., nonsister vs. sister chromatid interactions).
MEIOSIS AND THE ROOTS OF RECOMBINATION RESEARCH
The concept of recombination emerged during the early 20th century, following the post-Mendel era of heredity research. Thomas Hunt Morgan’s formal theory of gene linkage and crossing-over (Morgan 1913) was a synthesis of three key concepts: “the chromosome theory of inheritance,” imparted by Wilhelm Roux, Walther Flemming, Theodor Boveri, and Walter Sutton; “gene linkage,” an exception to Mendel’s law of independent assortment, first reported by Carl Correns; and the “chiasmatype theory,” derived from Frans Janssens' cytological observations of meiotic chromosomes. The first proof of the crossover theory came from Harriet Creighton and Barbara McClintock (Creighton and McClintock 1931), who were able to correlate cytological and genetic exchanges in maize.
Experiments aimed at understanding the mechanism of meiotic recombination became dominated by fungal genetics because of the huge advantage afforded by being able to recover all four meiotic products. These elegant studies culminated in four key concepts that formed the foundation of molecular models of recombination: gene conversion, an exception to Mendel’s principle of segregation, signaled a local nonreciprocal transfer of genetic information (Winkler 1930; Lindergren 1953; Mitchell 1955); postmeiotic segregation (PMS) indicated the presence of heteroduplex DNA (Olive 1959; Kitani et al. 1962); polarity gradients of gene conversion lead to the idea that recombination initiated from pseudofixed sites (Lissouba and Rizet 1960; Murray 1960); and the strong correlation between gene conversion/PMS events and crossing-over led to the proposal that these processes were mechanistically linked (Kitani et al. 1962; Perkins 1962; Whitehouse 1963).
MOLECULAR MODELS OF MEIOTIC RECOMBINATION
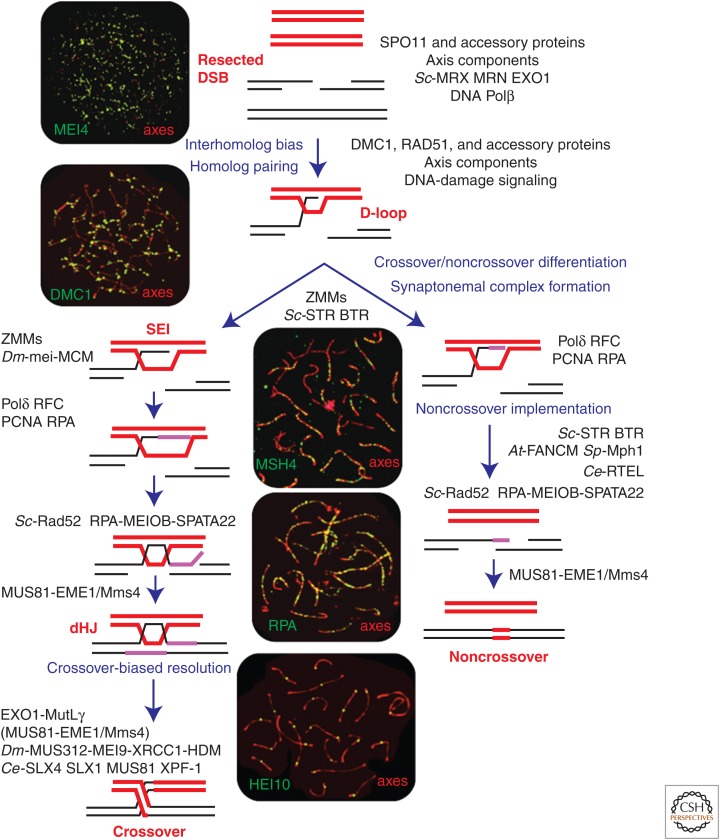
Holliday’s classic model reconciled gene conversion, PMS, and crossing-over into a single mechanism with the key features of hybrid (heteroduplex) DNA formed via strand exchange, mismatch correction of hybrid DNA to yield gene conversion, and a four-way exchange junction that could be resolved to yield either crossover or noncrossover duplex products (Holliday 1964). The meticulous testing and revision of models of meiotic recombination ensued over the next 20 years (Haber 2008), culminating in the formulation of the double-strand break repair (DSBR) model of Szostak et al. (1983), which proposed that meiotic recombination is initiated by DNA double-strand breaks and the ensuing strand exchanges result in the formation of double-Holliday junctions (dHJs) (Fig. 1).
Figure 1.
Model of meiotic recombination. Schematic diagram showing the DNA intermediates of meiotic recombination highlighting the major pathways and key transitions (for more comprehensive models, see Martini et al. 2011; Kaur et al. 2015; Tang et al. 2015). Magenta lines indicate new DNA synthesis. During double-Holliday junction (dHJ) formation along the crossover branch, only one possible mechanism for engagement of the second double-strand break repair (DSB) end is shown, involving end-first displacement of the extended invading strand and annealing (Allers and Lichten 2001b; Lao et al. 2008). Alternative models include annealing of the second DSB end to an expanded D-loop, or a second strand invasion analogous to the first DSB end (Szostak et al. 1983; Martini et al. 2011). Proteins implicated in each step are shown; prefixes indicate proteins specific to a given organism: At, Arabidopsis thaliana, Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe (see main text for details). Accompanying images show surface-spread mouse spermatocyte nuclei at the corresponding prophase stages, immunostained for pertinent recombination factors. In all images, homolog axes are visualized using SYCP3 antibodies. MEI4 is a SPO11-accessory protein required for DSB formation (Kumar et al. 2010). PCNA, Proliferating cell nuclear antigen.
Identification of the key DNA intermediates of the DSBR model was enabled by techniques to synchronize meiosis in budding yeast cultures, and a series of Southern blot assays to monitor events at defined recombination sites. These approaches provided direct confirmation that meiotic recombination initiates with double-strand breaks (DSBs) and gives rise to dHJs (Sun et al. 1989; Cao et al. 1990; Schwacha and Kleckner 1994, 1995). These powerful approaches also revealed several important spatial and temporal features of meiotic recombination. DSBs form after bulk chromosome replication and are rapidly processed to form long single-stranded tails with 3′-termini (Padmore et al. 1991; Sun et al. 1991; Zakharyevich et al. 2010). Distinct from the original DSBR model, there is no evidence that a significant gap is formed at the DSB site. Detection of recombination intermediates by Southern analysis was dependent on the fact that DSBs at the assayed loci were confined to very narrow regions or hotspots, a conserved feature that was subsequently shown to apply genome-wide and has been the subject of intense studies (e.g., Baudat and Nicolas 1997; Gerton et al. 2000; Blitzblau et al. 2007; Buhler et al. 2007; Hwang and Hunter 2011; Pan et al. 2011; Smagulova et al. 2011; de Massy 2013, 2014; Pratto et al. 2014).
Formation of metastable one-ended strand-exchange intermediates, called single-end invasions (SEIs), is coincident with chromosome synapsis, that is, the intimate connection of homologs along their lengths by zipper-like structures called synaptonemal complexes (Fig. 2) (Hunter and Kleckner 2001). Given that chromosome pairing and synapsis requires recombination in most organisms, including budding yeast (but not Drosophila or Caenorhabditis elegans), detected SEIs must be preceded by less stable nascent strand-pairing intermediates (presumably D-loops) that are not readily detected by current approaches. The timing of SEI formation reflects the interdependence between the initiation of synapsis and the initial differentiation of crossover and noncrossover pathways, with SEIs being the earliest detectable crossover-specific joint molecules (see below) (Hunter and Kleckner 2001; Borner et al. 2004; Reynolds et al. 2013; Zhang et al. 2014a). Along the crossover pathway, SEIs give rise to dHJs, which must be resolved exclusively into crossovers, in contrast to the equal mixture of crossovers and noncrossovers originally envisioned by the DSBR model (Allers and Lichten 2001a; Clyne et al. 2003; Zakharyevich et al. 2012).
Figure 2.
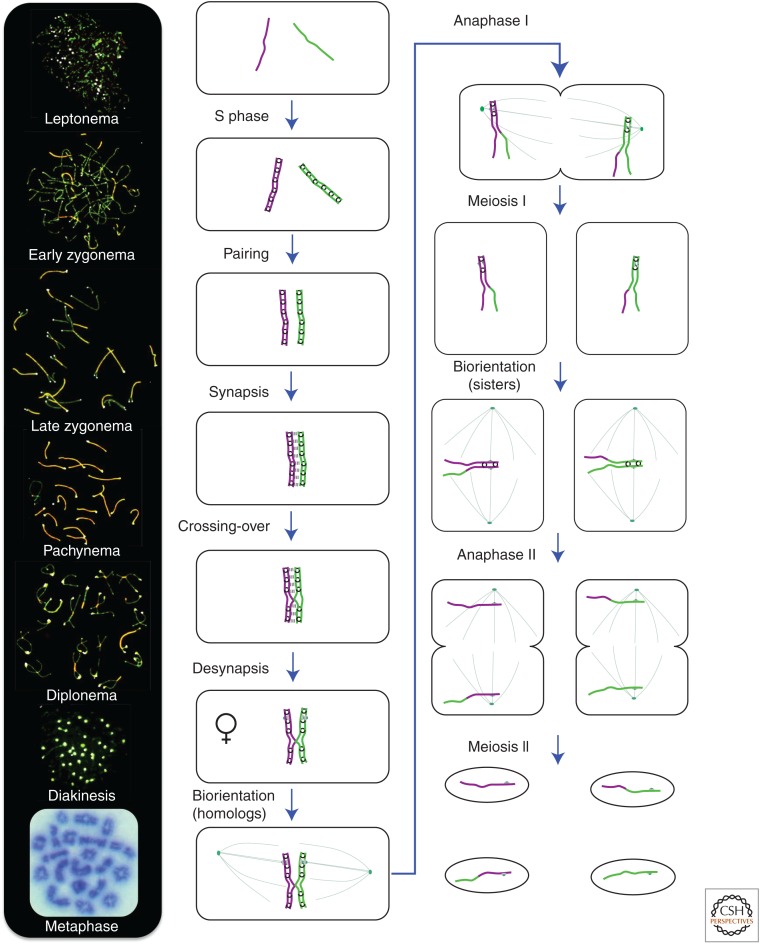
Chromosomal events of meiosis. The main stages of meiosis are shown for a single pair of homologous chromosomes (green and purple lines). Black rings indicate cohesion complexes connecting the sister chromatids. The synaptonemal complex is indicated by the triple-dashed line located between the homologs. Gray disks represent the kinetochores. Spindle microtubules are shown as thin green lines. The Venus symbol indicates the dictyotene stage at which prophase arrests in females. The images on the left-hand side show the corresponding prophase-I stages in surface-spread mouse spermatocyte nuclei immunostained for homolog axes (SYCP3, green) and the central region of the synaptonemal complex (SYCP1, red, but appears orange because of signal overlap). The final image shows a metaphase-I nucleus stained with Giemsa.
These findings are incorporated into contemporary models of meiotic recombination that propose early differentiation of crossover and noncrossover pathways, with a majority of noncrossovers arising from D-loops via synthesis-dependent strand annealing, and crossover-designated events going on to form stable dHJs (Fig. 1) (McMahill et al. 2007; Martini et al. 2011; Tang et al. 2015). Fine-scale analysis of crossover and noncrossover products in yeast, mouse, human, and Arabidopsis support the tenet that crossovers and noncrossovers arise from distinct intermediates (Jeffreys and May 2004; Guillon et al. 2005; Mancera et al. 2008, 2011; Cole et al. 2010, 2014; Martini et al. 2011; Drouaud et al. 2013; Rockmill et al. 2013; Wijnker et al. 2013). However, this feature may not be universal; for example, patterns of heteroduplex in Drosophila are consistent with both crossovers and noncrossovers arising from a common intermediate (Crown et al. 2014).
INTERHOMOLOG INTERACTIONS
In most organisms, recombination is coerced to do work in meiosis, specifically to bring each pair of homologous chromosomes into close end-to-end juxtaposition and thereby facilitate their synapsis (Fig. 2) (e.g., Baudat et al. 2000; Romanienko and Camerini-Otero 2000; Peoples et al. 2002; Tesse et al. 2003; Henderson and Keeney 2004; Zickler and Kleckner 2015). Interhomolog crossing-over occurs within the context of the synapsed chromosomes. Crossovers combine with sister-chromatid cohesion, which was established during premeiotic S phase, to create connections called chiasmata that supersede synaptonemal complexes and are only resolved when cohesion between the chromosome arms is cleaved at anaphase I. Chiasmata are essential for efficient homolog disjunction at the first meiotic division because they allow homolog pairs to stably biorient on the meiosis-I spindle in much the same way that cohesion allows sister chromatids to biorient on the mitotic spindle (Fig. 2) (Petronczki et al. 2003; Hunter 2013).
Specific organizational features of meiotic prophase chromosomes and their coupling to meiotic recombination enable local DNA interactions to effect the juxtaposition of whole chromosomes. A central feature is the organization of sister chromatids into linear arrays of chromatin loops connected by a common cohesin-based axis that is augmented by meiosis-specific proteins (Zickler and Kleckner 1999; Pelttari et al. 2001; Blat et al. 2002; Syrjanen et al. 2014; Zickler and Kleckner 2015). This organization produces semicondensed prophase chromosomes with relatively rigid axes that define the pairing and interaction faces of each homolog pair. Direct physical and functional coupling of recombination complexes to the axes (Blat et al. 2002; Panizza et al. 2011; Borde and de Massy 2013) enables local DNA interactions to juxtapose associated homolog axes and ultimately bring about end-to-end chromosome pairing. Locally, recombinational interactions mediate the formation of visible bridges between pairs of axes, a subset of which nucleate the polymerization of the synaptonemal complex central region to lock in nascent pairing interactions (Albini and Jones 1987; Zickler and Kleckner 1999, 2015). Morphogenesis of homolog axes and synaptonemal complexes define the stages of meiotic prophase (Fig. 2). Axes develop during leptonema coincident with DSB formation and homolog pairing; during zygonema, synaptonemal complexes polymerize between aligned axes; when synapsis is complete, meiocytes enter pachynema and crossing-over occurs; finally, during diplonema, synaptonemal complexes are disassembled and bivalents connected by chiasmata emerge (Hunter 2013).
To efficiently promote pairing, synapsis and formation of chiasmata, recombinational interactions during meiosis are biased to occur between homologs, sharply contrasting somatic recombination, which occurs almost exclusively between sister chromatids (Fig. 1) (Kadyk and Hartwell 1992; Johnson and Jasin 2000; Bzymek et al. 2010; Lao and Hunter 2010; Humphryes and Hochwagen 2014; Brown and Bishop 2015). Although the precise mechanism of this meiotic interhomolog bias remains unknown, coupling of recombination to homolog axes also provides a means for communication between sister chromatids to limit intersister recombination. In budding yeast, where interhomolog bias has been most clearly elucidated, an important aspect of interhomolog bias is modulation of the recombination complex by inhibiting Rad51 (which nonetheless retains an essential supporting role) and switching to Dmc1-mediated DNA pairing and strand exchange (Tsubouchi and Roeder 2006; Niu et al. 2009; Cloud et al. 2012; Hong et al. 2013; Lao et al. 2013; Liu et al. 2014b). Evidence suggests that this feature may be conserved, at least in Arabidopsis (Kurzbauer et al. 2012; Da Ines et al. 2013). Local axis-based checkpoint signaling is also inferred to locally remove a constraint to interhomolog strand exchange that is imposed by cohesin complexes (Kim et al. 2010), and to limit use of the sister-chromatid template (Wan et al. 2004; Niu et al. 2005; Goldfarb and Lichten 2010).
Feedback between homolog pairing and recombination coordinates the two processes such that synapsis is constrained to occur between homologous chromosomes, and progression of recombination beyond the nascent strand-exchange step is prevented until synapsis is ongoing (Hunter and Kleckner 2001). In this way, the reversibility of pairing interactions is maintained until homologous interactions become stabilized by synapsis, thereby limiting chromosome entanglement and facilitating the resolution of interlocked chromosomes as synapsis ensues (Storlazzi et al. 2010).
PROGRAMMED DSB FORMATION
In stark contrast to recombinational repair in somatic cells, meiotic recombination is a programmed event initiated by the deliberate induction of DSBs throughout the genome. Consequently, all chromosomes simultaneously engage in recombination at multiple positions. DSB formation is intimately associated with meiotic chromosome architecture, and is subject to exquisite spatial and temporal regulation (Keeney et al. 2014; Subramanian and Hochwagen 2014; Lam and Keeney 2015; Székvölgyi et al. 2015).
The DSB-forming machinery, centered around the SPO11 transesterase, has been reviewed extensively (e.g., Keeney 2001, 2008; Hunter 2006; Keeney and Neale 2006; Borde and de Massy 2013; de Massy 2013; Yamada and Ohta 2013; Lam and Keeney 2015; Székvölgyi et al. 2015). Local determinants of DSB location include cis-acting DNA sequences, transcription factors, histone modifications, and chromatin accessibility (Hwang and Hunter 2011; Lichten and de Massy 2011; Tischfield and Keeney 2012; Baudat et al. 2013; Yamada and Ohta 2013; Fowler et al. 2014). DSB formation is facilitated by factors that are components of the homolog axes (e.g., Schwacha and Kleckner 1994; Mao-Draayer et al. 1996; Ellermeier and Smith 2005; Goodyer et al. 2008; Kugou et al. 2009; Daniel et al. 2011), and the ensuing recombination complexes are visibly associated with these structures (Zickler and Kleckner 1999). However, the DSB sites themselves map to DNA sequences located in chromatin loops, whereas axis-associated sequences define DSB cold spots (Blat et al. 2002; Ito et al. 2014). These observations inspired the “tethered loop-axis complex” (TLAC) model, in which DSB sites located in loops must interact with the axis to trigger DSB formation (Blat et al. 2002). This dependency is inferred to establish axis association of recombination complexes effectively coupling interhomolog interactions at the DNA and axis levels. Although direct proof remains elusive, the TLAC model is supported by a number of observations. For example, essential DSB factors localize to the axes before DSB activation (Panizza et al. 2011), and bridging proteins, which connect DSB sites located in loops with axis-associated DSB factors, have recently been identified in budding and fission yeasts (Miyoshi et al. 2012; Acquaviva et al. 2013; Sommermeyer et al. 2013).
Global and local coupling to S-phase cell-cycle kinases, together with a meiotic S-phase checkpoint, confine DSB formation to replicated chromatids (Borde et al. 2000; Henderson et al. 2006; Ogino et al. 2006; Sasanuma et al. 2008; Wan et al. 2008; Blitzblau and Hochwagen 2013; Murakami and Keeney 2014). Local feedback regulation, involving kinase signaling by components of the DNA-damage response, also helps to optimize the DSB distribution by promoting even spacing, and locally limiting DSBs to one per four chromatids (Lange et al. 2011; Zhang et al. 2011; Carballo et al. 2013; Cooper et al. 2014; Garcia et al. 2015). As homolog pairing ensues, feedback regulation locally attenuates DSB formation, while delayed pairing appears to locally up-regulate DSBs (Kauppi et al. 2013; Lao et al. 2013; Thacker et al. 2014). Finally, full synapsis and satisfactory progression toward crossing-over appears to shut down the ability to make DSBs (Argunhan et al. 2013; Rosu et al. 2013; Stamper et al. 2013; Thacker et al. 2014).
CROSSOVER CONTROL
Crossover Assurance and Interference
Despite the stochastic nature of crossing-over, which to a first approximation can occur at any site along the chromosomes, crossovers are subject to tight regulation (Jones 1984; Jones and Franklin 2006). Although large numbers of DSBs form per nucleus, the total number of crossovers is very low, typically in the range of one per chromosome to one per chromosome arm. Thus, the popular view that meiotic recombination “scrambles” the genome is misleading. On the contrary, meiotic recombination is relatively conservative and as many as half of all chromatids can emerge from meiosis with parental haplotypes. However, each homolog pair obtains at least one crossover, as a prerequisite for accurate segregation, defining a regulatory feature termed “crossover assurance” (also known as the obligatory crossover). Also, when a single chromosome pair experiences multiple crossovers they tend to be widely and evenly spaced—the classical phenomenon of “crossover interference” that was already noted in the first genetic map constructed by Alfred Sturtevant in 1913 (1913a,b). Most models of interference assume that an inhibitory zone is established around DSB sites that have become committed to crossing-over (Hillers 2004; Berchowitz and Copenhaver 2010; Zhang et al. 2014b). Other DSBs within this zone of inhibition are prevented from becoming crossovers and instead mature as noncrossovers.
Together, crossover assurance and interference dictate the lower and upper limits for crossover numbers and underpin the phenomenon of crossover homeostasis: a low variation in crossover numbers per meiosis despite much larger variations in the numbers of recombinational interactions (Martini et al. 2006; Rosu et al. 2011; Cole et al. 2012; Yokoo et al. 2012). Interference (intercrossover) distances are generally large relative to chromosome length, which drives minimization of crossover numbers. In extreme cases, such as C. elegans, interference is effective over the entire length of the chromosomes such that the combined output of crossover assurance and interference is precisely one crossover per chromosome pair (Hillers and Villeneuve 2003). Not only does minimization of crossover numbers create a unique regulatory challenge for meiotic cells, its raison d’être remains unclear. Excess crossovers, per se, do not appear to interfere with homolog segregation (e.g., Seguela-Arnaud et al. 2015), favoring the idea that limiting exchange has an adaptive benefit, such as preserving favorable haplotypes and/or minimizing deleterious nonallelic recombination.
The mechanisms responsible for crossover assurance and interference remain elusive in part because of the complexity of studying these processes. Crossover assurance comprises temporally and functionally distinct designation/commitment and implementation/execution steps. As such, perturbation of any biochemical process required to implement crossing-over at a designated site (such as dHJ resolution) will reduce the efficiency of crossover assurance without being informative about the crossover designation process per se. Similarly, readouts of crossover interference are not readily amenable to high throughput screening and defining mutants that unambiguously perturb this process has proven both challenging and labor intensive (e.g., Zhang et al. 2014d). Moreover, interpretation of mutant phenotypes is complicated by the existence of a (typically minor) class of crossovers that does not show an interference distribution (Berchowitz and Copenhaver 2010). Thus, mutants that specifically diminish interfering crossovers (termed class I events) can show an apparent loss of interference caused by increasing prevalence of noninterfering class II crossovers (Stahl 2012).
Assuming a model in which interference establishes an inhibitory zone around designated crossover sites, a true interference mutant is expected to show a hypercrossover phenotype (Zhang et al. 2014d). However, hypercrossover mutants do not a priori define genes involved in crossover patterning. For example, hyperrecombination is seen for a number of DNA helicase mutants, but this is because of an interference-independent defect in implementing noncrossovers via synthesis-dependent strand annealing. In these cases, the distribution of class I crossovers remains largely unperturbed, whereas class II events are increased at the expense of noncrossovers (Rockmill et al. 2003; Youds et al. 2010; Crismani et al. 2012; Yokoo et al. 2012; Seguela-Arnaud et al. 2015).
Despite these impediments, important advances in our understanding of crossover control have been made in recent years. Several studies have established that the metric of crossover interference is the physical lengths of prophase chromosomes (as opposed to genetic distance or genomic distance, i.e., bps of DNA) (Martini et al. 2006; Drouaud et al. 2007; Petkov et al. 2007; Zhang et al. 2014b). Thus, crossover rates for the same chromosomes vary coordinately with changes in axis lengths, which reflect differences in chromatin packaging with respect to the size and density of chromatin loops (Lynn et al. 2002; Hillers and Villeneuve 2003; Kleckner et al. 2003; Tease and Hulten 2004; Qiao et al. 2012b; Gruhn et al. 2013; Baier et al. 2014). For example, in humans, variation in the lengths of prophase chromosomes account for long-known differences in recombination rates between males and females. Oocyte prophase chromosomes are about twice as long as their spermatocyte counterparts, have shorter, denser chromatin loops, and experience ∼60% more crossovers (Gruhn et al. 2013). However, interference distances (expressed as µm of synaptonemal complex) are comparable between the sexes (Petkov et al. 2007). Although the basis for such variation in chromatin organization remains unclear, levels of cohesin subunits and other axis components may be important (Novak et al. 2008; Mets and Meyer 2009; Vranis et al. 2010; Murdoch et al. 2013).
A number of studies indicate that axis integrity and continuity are important for regulating crossover rate and interference (Hillers and Villeneuve 2003; Kleckner et al. 2004; Nabeshima et al. 2004; Novak et al. 2008; Storlazzi et al. 2008; Tsai et al. 2008; Joshi et al. 2009; Mets and Meyer 2009; Thacker and Keeney 2009; Zanders and Alani 2009; Qiao et al. 2012b; Libuda et al. 2013; Murdoch et al. 2013; Zhang et al. 2014d). These data suggest that homolog axes may be the conduits for transmission of interference signaling, or at least that axis integrity is important for spreading of interference (e.g., Zhang et al. 2014d). However, it is notable that mice lacking the major axis component, SYCP3, show apparently normal interference despite overt changes in axis length (de Boer et al. 2007).
Crossover homeostasis is, at least in part, a consequence of crossover interference (Martini et al. 2006; Rosu et al. 2011; Cole et al. 2012; Yokoo et al. 2012; Wang et al. 2015). When DSBs are made at a low density relative to the effective interference distance, a given recombination site has a lower probability of being subject to interference from an adjacent site, whereas, for a high density of DSBs, the opposite is true such that similar numbers of crossovers will emerge in both cases (Zickler and Kleckner 2015). These effects are locally manifested as changes in the ratios of crossovers to noncrossovers observed at assayed loci (Martini et al. 2006; Yokoo et al. 2012; Lao et al. 2013). The other essential component of homeostasis is crossover assurance, which requires regulatory inputs at each step of recombination. Preconditions for crossover assurance include forming sufficient numbers of DSBs and efficiently converting them into stable interhomolog interactions (Martini et al. 2006; Rosu et al. 2011; Kauppi et al. 2013; Lao et al. 2013). Faced with conditions that delay stable homolog engagement—such as inadequate DSB numbers, inefficient interhomolog template bias, or delayed/defective progression toward crossing-over—cells attempt to compensate by continuing to form DSBs (Sourirajan and Lichten 2008; Argunhan et al. 2013; Gray et al. 2013; Kauppi et al. 2013; Lao et al. 2013; Rosu et al. 2013; Stamper et al. 2013; Thacker et al. 2014). Successful synapsis and formation of crossover-competent joint molecules are inferred to attenuate both DSB formation and interhomolog bias (Rosu et al. 2011, 2013; Kauppi et al. 2013; Stamper et al. 2013; Thacker et al. 2014).
In the absence of interference from neighboring recombination sites, the crossover outcome is specifically implemented with extremely high efficiency (Martini et al. 2006; Rosu et al. 2011; Lao et al. 2013). However, the processes that ensure that a crossover outcome is triggered for at least one DSB site per chromosome pair (the obligatory crossover) remain unknown. Evidence from a variety of organisms indicates that the patterning processes that designate crossover sites occur before and independently of synapsis (Page and Hawley 2001; Borner et al. 2004; Fung et al. 2004; Higgins et al. 2005; de Boer and Heyting 2006; de Boer et al. 2006; Zhang et al. 2014a). These data rule out the idea that bidirectional polymerization of synaptonemal complex is the major mode of interference signaling (King and Mortimer 1990). However, in C. elegans, subtle perturbation of synaptonemal complex composition elevates the crossover rate and attenuates, but does not abolish, interference (Hayashi et al. 2010; Libuda et al. 2013). Crossing-over is also elevated in the synapsis-defective zep1 mutant in Oryza (Wang et al. 2010). Thus, synaptonemal complex is clearly important for limiting crossing-over in at least some organisms. Notably, in budding yeast, although synaptonemal complex is required for the majority of crossovers, it also acts to suppress centromere-proximal crossing-over (Chen et al. 2008).
Cytological analysis of crossover-specific proteins (that mark class I crossovers) circumvents problems associated with genetic analysis of interference, and permits the study of mutants that are unable to complete meiosis. This approach has been exploited in budding yeast to identify an interference pathway involving topoisomerase II, the axis component Red1, and posttranslational modification of both proteins by SUMO (small ubiquitin-like modifier) (Zhang et al. 2014d). Importantly, this analysis indicates that several mutants that show diminished interference when assayed genetically have normal patterning of crossover markers when assayed cytologically, and are, therefore, unlikely to be involved directly in interference. Involvement of topoisomerase II is compatible with a model in which expansion of axis-constrained chromatin drives crossover designation, which subsequently triggers chromatin contraction that emanates from the crossover site and spreads bidirectionally to inhibit crossing-over at neighboring recombination sites (Kleckner et al. 2004; Zhang et al. 2014b). Direct evidence of the predicted mechanical properties is currently lacking. However, consonant with such effects is the observation that crossover designation in C. elegans induces a local expansion and elongation of the chromosome (Libuda et al. 2013).
Crossover/Noncrossover Differentiation
The crossover control processes described above indicate that the outcome of meiotic DSB repair is tightly regulated. In a number of organisms, the initial differentiation of crossover and noncrossover pathways is temporally and functionally coupled to the onset of synapsis (Bojko 1985; Zickler et al. 1992; Hunter and Kleckner 2001; Borner et al. 2004; Fung et al. 2004; Reynolds et al. 2013; Zhang et al. 2014a). Moreover, in budding yeast, there appears to be a 1:1 correlation between designated crossover sites (marked by crossover-specific markers) and initiation sites for polymerization of synaptonamal complex (Fung et al. 2004; Henderson and Keeney 2004). In other organisms, these sites are also correlated, but synaptonemal complex-initiation sites outnumber crossover sites (Zickler et al. 1992; Zickler and Kleckner 1999; Brown et al. 2005; Gruhn et al. 2013; Zhang et al. 2014a).
At the DNA level, the appearance of metastable crossover-correlated joint molecules, the SEIs, is coincident with formation of synaptonemal complexes (Fig. 1) (Hunter and Kleckner 2001; Borner et al. 2004). In budding yeast, both transitions require the ZMM factors, which include Zip1, Zip2, Zip3, Zip4, Msh4-Msh5 (the MutSγ complex), Mer3, and Spo16 (also defined as synapsis initiation factors [SICs]) (Fung et al. 2004; Hunter 2006; Lynn et al. 2007). The ZMMs act to stabilize nascent joint molecules and coordinately promote polymerization of synaptonemal complexes, ultimately being required for the formation of class I crossovers. Although ZMMs were initially defined as crossover-specific factors in budding yeast, it seems likely that in most organisms a much larger fraction of all recombinational interactions benefit from at least transient stabilization by some or all of the ZMM factors, perhaps as a prerequisite for crossover designation. For example, in mouse, plants, C. elegans, and Sordaria, initial numbers of MutSγ immunostaining foci greatly outnumber final crossover numbers (de Vries et al. 1999; Edelmann et al. 1999b; Kneitz et al. 2000; Higgins et al. 2008b; Yokoo et al. 2012; De Muyt et al. 2014; Zhang et al. 2014c).
The ZMMs define a functionally diverse set of proteins that act on distinct facets of recombination and chromosome synapsis. MutSγ (Msh4–Msh5) and Mer3 interact directly with DNA to stabilize nascent joint molecules. MutSγ is related to the MutS-family of DNA mismatch-repair proteins, and binds specifically to model D-loops and Holliday junctions (HJs) in vitro (Snowden et al. 2004). In vivo analysis supports the inference that MutSγ stabilizes joint molecules by embracing the involved duplexes (Borner et al. 2004; Snowden et al. 2004; Jessop et al. 2006; Oh et al. 2007). MutSγ (and other ZMMs) also appears to protect joint molecules from being dissociated by the anticrossover activity of the Sgs1–Top3–Rmi1 complex (see below) (Jessop et al. 2006; Oh et al. 2007; Kaur et al. 2015; Tang et al. 2015). Mer3 is a DNA helicase that can promote heteroduplex extension during D-loop formation and unwinds a number of DNA structures in vitro (Nakagawa and Kolodner 2002; Mazina et al. 2004). In vivo, Mer3 is required for efficient formation of SEIs, limits chromosome entanglement, and promotes efficient synapsis (Nakagawa and Ogawa 1999; Borner et al. 2004; Chen et al. 2005; Mercier et al. 2005; Tanaka et al. 2006; Sugawara et al. 2009; Wang et al. 2009; Guiraldelli et al. 2013). Localization studies in Sordaria reveal numerous pairs of opposing Mer3 foci associated with coaligned chromosome axes, suggesting that recombination complexes have a defined architecture at this stage, with Mer3 engaging both ends of a DSB. In this scenario, one DSB end engages the homolog, associating with its axis; while the second DSB end remains associated with its axis of origin (Storlazzi et al. 2010).
Whether the other ZMM members directly interact with DNA is unknown. Zip2-related proteins share homology with the XPF family of structure-selective endonucleases, although the active site motif is not conserved (Macaisne et al. 2008). Moreover, in Arabidopsis, the Zip2-relative SHOC1 interacts with an ERCC1-related protein called PTD (parting dancer), implying formation of an XPF–ERCC1-like complex that might bind nascent joint molecules (Macaisne et al. 2011). In budding yeast, Zip2 forms a functional unit together with Zip4, predicted to be an extensive TPR-repeat protein (Perry et al. 2005; Tsubouchi et al. 2006), and the small coiled-coil motif protein, Spo16 (Shinohara et al. 2008). Although the putative Zip2–Zip4–Spo16 complex is important for synapsis in budding yeast, mutation of Zip2 and Zip4 homologs in both plants and mouse causes relatively minor synapsis defects, although synapsis is inferred to initiate from fewer sites than in wild type (Chelysheva et al. 2007; Kuromori et al. 2008; Macaisne et al. 2008; Yang et al. 2008; Shen et al. 2012). Thus, synapsis initiation appears to be less strictly coupled to crossover designation in organisms other than yeast.
Budding yeast Zip3 is a RING-domain E3 ligase inferred to catalyze SUMO conjugation and is required for the normal localization of all other ZMMs (Agarwal and Roeder 2000; Cheng et al. 2006; Shinohara et al. 2008). Zip3 and the proline isomerase Fpr3, ensure that synapsis is rendered dependent on recombination and the Zip2–Zip4–Spo16 complex (Macqueen and Roeder 2009). This layer of regulation may be absent from other organisms as mutation of Zip3-related proteins—ZHP-3, RNF212, and HEI10—do not cause overt synapsis defects (Jantsch et al. 2004; Ward et al. 2007; Bhalla et al. 2008; Chelysheva et al. 2012; Wang et al. 2012a; Reynolds et al. 2013; Qiao et al. 2014). Also, although yeast Zip3 localizes primarily to crossover sites, other family members show dynamic localization patterns along synaptonemal complexes. Abundant focal/linear staining patterns are observed as synapsis ensues, followed by the loss of most staining, with retention/concentration only at designated crossover sites (Agarwal and Roeder 2000; Henderson and Keeney 2004; Jantsch et al. 2004; Bhalla et al. 2008; Chelysheva et al. 2012; Wang et al. 2012a; Yokoo et al. 2012; Reynolds et al. 2013; De Muyt et al. 2014; Qiao et al. 2014).
With the exception of mammalian HEI10, a function that appears to be shared by Zip3-family proteins is the stabilization of recombination factors at designated crossover sites (Agarwal and Roeder 2000; Shinohara et al. 2008; Yokoo et al. 2012; Reynolds et al. 2013; De Muyt et al. 2014). For example, in mouse, only a subset of the abundant RNF212 complexes detected along newly synapsed chromosomes actually localize with recombination sites marked by MutSγ (Reynolds et al. 2013). Sites of stable RNF212–MutSγ colocalization persist and ultimately go on to recruit crossover-specific factors and mature into crossovers, whereas the majority of RNF212 and MutSγ complexes are lost from the chromosomes. Thus, the differentiation of crossover and noncrossover pathways seen at the DNA level (Hunter and Kleckner 2001; Borner et al. 2004) is underpinned by differential stabilization of recombination factors at the protein level.
Whether selective localization of RNF212 (and other Zip3-family proteins) to precrossover sites represents the crossover designation process, per se, or is a downstream manifestation of that process is unclear. In mouse, normal patterning of RNF212 and MutSγ, dissociation of these factors from synapsed chromosomes, and progression of DSB repair require HEI10 and the cyclin-related protein, CNTD1 (see below) (Holloway et al. 2014; Qiao et al. 2014). Mouse HEI10 is not detected along chromosomes at early stages of synapsis, but ultimately concentrates at crossover sites where it supersedes RNF212 (Fig. 1) (Qiao et al. 2014). Although Zip3 and RNF212 are inferred to catalyze SUMO conjugation, human HEI10 has been implicated as a ubiquitin E3-ligase (Toby et al. 2003; Cheng et al. 2006; Reynolds et al. 2013; D Kulkarni and N Hunter, unpubl.).
Together, these observations suggest a model in which association of RNF212 with recombination sites is limited by a process involving HEI10-mediated ubiquitylation (Qiao et al. 2014). At most recombination sites, the absence of RNF212 renders factors such as MutSγ unstable leading to their dissociation or degradation. Consequently, nascent joint molecules are destabilized and dissociated by DNA helicases (see below) to promote a noncrossover outcome. At designated crossover sites a positive-feedback loop, involving SUMOylation, promotes the mutual stabilization of RNF212 and MutSγ, which in turn stabilizes nascent joint molecules to facilitate dHJ formation. Finally, HEI10 accumulates at these sites, where it acts to displace RNF212 and MutSγ to allow the final steps of crossing-over to be implemented (Qiao et al. 2014).
Identification of COSA-1 as a cyclin-related protein that is essential for crossing-over in C. elegans implies that CDK-driven phosphorylation is another facet of crossover regulation via posttranslational protein modification (Yokoo et al. 2012; Holloway et al. 2014). Consistently, in mouse, CDK proteins localize to sites of meiotic recombination (Ashley et al. 2001); CDK4 concentrates at DSB sites as chromosomes synapse and is succeeded by a much smaller number of CDK2 complexes that localize specifically at crossover sites (CDK2 also concentrates at telomeres). C. elegans COSA-1 also accumulates specifically to crossover sites, and is required for the local retention and concentration of ZHP-3 and MSH-5 (Yokoo et al. 2012). The phenotypes of mice lacking the COSA-1 ortholog, CNTD1, are remarkably similar to those of Hei10 mutants (Holloway et al. 2014). Moreover, in somatic cells, human HEI10 interacts with cyclin B1 and can down-regulate cyclin B levels (Toby et al. 2003; Singh et al. 2007). These data suggest that phosphorylation, driven by a CNTD1/COSA-1–CDK complex, and ubiquitylation, targeted by HEI10, are coupled to effect crossover regulation.
Additional complexity to the regulatory circuitry defined by mammalian RNF212–HEI10 is suggested by the existence of multiple RNF212 spliceforms, and an RNF212 ortholog (RNF212B) (Kong et al. 2008; Reynolds et al. 2013). Similarly, the C. elegans genome encodes multiple ZHP-3-related proteins; however, it lacks an obvious HEI10 ortholog (M Zetka, personal communication). Curiously, Sordaria and plants encode only a single HEI10-like protein, with localization characteristics of both RNF212 and HEI10 in mammals (Chelysheva et al. 2012; Wang et al. 2012a; De Muyt et al. 2014). Similarly, budding yeast contains a single Zip3 homolog. Despite this variation and complexity, the relationships between Zip3-family proteins, selective stabilization of MutSγ, and crossing-over appear to be conserved (Agarwal and Roeder 2000; Henderson and Keeney 2004; Jantsch et al. 2004; Bhalla et al. 2008; Chelysheva et al. 2012; Wang et al. 2012a; Yokoo et al. 2012; Reynolds et al. 2013; De Muyt et al. 2014; Qiao et al. 2014).
Pro-Crossover Role of the Synaptonemal Complex
Although not essential for interhomolog DSB repair, synaptonemal complex is required for efficient interhomolog crossing-over in most organisms (Zickler and Kleckner 2015). In mutants lacking components of the synaptonemal complex central region recombination stalls at an intermediate stage and, in mouse, markers such as CDK2, HEI10, RNF212, and MutLγ fail to localize to designated crossover sites (Sym et al. 1993; Borner et al. 2004; de Vries et al. 2005; Reynolds et al. 2013; Qiao et al. 2014). Synaptonemal complex is inferred to act locally to stabilize procrossover factors and at later stages may facilitate the exchange of homolog axes that must accompany exchange at the DNA level (Storlazzi et al. 1996; Borner et al. 2004; de Boer et al. 2007; Shinohara et al. 2008; Qiao et al. 2012a; Voelkel-Meiman et al. 2015). As described above, an inhibitory role for synaptonemal complex that limits crossovers has also been demonstrated (Chen et al. 2008; Hayashi et al. 2010; Wang et al. 2010; Libuda et al. 2013). In Arabidopsis, synapsis has been shown to help ensure the fidelity of meiotic recombination, probably by limiting nonallelic recombination (Higgins et al. 2005).
Recombination-Associated DNA Synthesis
De novo DNA synthesis must accompany all recombinational repair, to allow DSB ends to anneal during noncrossover formation, and for the formation of dHJs during crossover formation (Fig. 1) (Lao et al. 2008). Recombination-associated DNA synthesis is assumed to be distinct from replicative DNA synthesis in that it is: (1) primed by the 3′ terminus of the invading DSB end; (2) uncoupled from lagging strand synthesis; and (3) only limited synthesis is required, sufficient to replace the sequences removed by DSB resection (Haber 2013).
The molecular details of the DNA synthesis associated with meiotic recombination remain poorly characterized. Classic electron microscope (EM) autoradiography studies showed that DNA synthesis is indeed associated with sites of meiotic recombination (Carpenter 1981; Moses et al. 1984). Mapping of newly synthesized DNA in budding yeast confirmed the predicted patterns and timing of DNA synthesis associated with crossover and noncrossover products. Crossover-correlated synthesis occurs on either side of the assayed DSB site, whereas synthesis associated with noncrossovers spans the DSB site. Also, crossover-associated synthesis tracts were longer and occurred later than those associated with noncrossovers (Fig. 1) (Terasawa et al. 2007). These observations are consonant with data from a variety of organisms showing that gene-conversion tracts associated with crossovers are generally much longer than those associated with noncrossovers (Jeffreys and May 2004; Guillon et al. 2005; Cole et al. 2010, 2014; Drouaud et al. 2013; Wijnker et al. 2013).
Mutant phenotypes implicate a number of replication factors in meiotic recombination. In budding yeast, pol3-ct, an allele of the lagging-strand polymerase Polδ, reduces crossing-over and results in shorter gene-conversion tract lengths, indicative of shorter stretches of heteroduplex (Maloisel et al. 2004). In vitro, D-loop extension by Polδ requires the proliferating cell nuclear antigen (PCNA) replicative clamp (Li et al. 2009). Consistently, pol30-201, an allele of budding yeast PCNA confers a phenotype similar to that of the pol3-ct mutation, with shorter heteroduplex tracts and reduced crossing-over (Stone et al. 2008). These phenotypes suggest that the extent of initial DNA synthesis is an important aspect of crossover/noncrossover differentiation that may influence the stability of nascent D-loops, the efficiency of second-end capture to form a dHJ, and/or the structure of the ensuing dHJs and their propensity to be resolved specifically into crossovers (Fig. 1).
Additional mutant phenotypes are consistent with the idea that the mode and processivity of DNA synthesis associated with crossing-over is distinct from that associated with noncrossovers. In Arabidopsis, a hypomorphic mutation in the replication factor C (RFC) clamp-loader complex diminishes class I crossovers (Wang et al. 2012b). Given that one role of RFC is polymerase switching during Okazaki fragment initiation (switching Polα primase for Polδ) (Maga et al. 2000; Mossi et al. 2000), Wang et al. (2012b) suggested that lagging-strand synthesis might occur during crossover formation, where it could function to stabilize the expanding D-loop to promote dHJ formation. However, the primary function of RFC in recombination-associated DNA synthesis could be to load PCNA.
Arabidopsis and Oryza possess multiple paralogs of the replication protein A (RPA) complex subunits and several rpa mutant alleles have been shown to specifically reduce class I crossovers, whereas overall DSB repair remains efficient (Osman et al. 2009; Li et al. 2013). In budding yeast, meiotic DSB repair shows a general dependence on RPA (Soustelle et al. 2002), but RPA phosphorylation modulates crossover patterning (Bartrand et al. 2006). Drosophilia HDM (Hold’em) encodes an OB-fold protein related to RPA70 that is required for a majority of crossover and interacts with the joint-molecule resolving endonuclease complex, MEI9–ERCC1–MUS312 (Joyce et al. 2009). In mammals, the RPA1-related MEIOB protein and associated factor, SPATA22, form an RPA-associated complex that is required for intermediate steps of recombination and normal synapsis (La Salle et al. 2012; Ishishita et al. 2013, 2014; Luo et al. 2013; Souquet et al. 2013). MEIOB–SPATA22 is inferred to act after initial strand exchange, perhaps to promote strand annealing in both SDSA and dHJ pathways, acting analogously to the budding yeast Rad52 protein (Lao et al. 2008; Luo et al. 2013). A 3′-5′ exonuclease activity associated with MEIOB has led to the proposal that MEIOB–SPATA22–RPA also mediates digestion of 3′-flaps that may form on annealing of DSB ends, as a consequence of excess DNA synthesis relative to DSB resection (Fig. 1) (Luo et al. 2013).
The MCM complex has essential functions in the initiation of replication and acts as the replicative helicase during elongation (Bochman and Schwacha 2009). However, roles in a variety of other chromosomal processes have been invoked, including checkpoint signaling and DNA repair (Forsburg 2004; Shukla et al. 2005; Bailis et al. 2008). Unexpectedly, a hypomorph of Drosophila mcm5 specifically reduces crossing-over without perturbing DSB repair (Lake et al. 2007). Three additional MCM-related proteins in Drosophila, REC/MCM8, MEI217, and MEI218, are also required for crossing-over and rec mutants have shorter gene conversion tracts suggesting that a meiotic MCM complex, Mei-MCM, comprising REC–MEI217–MEI218 and perhaps MCM5, enhances the processivity of recombination-associated DNA synthesis (Manheim et al. 2002; Matsubayashi and Yamamoto 2003; Bhagat et al. 2004; Blanton et al. 2005; Kohl et al. 2012). Suppression of the rec/mcm8 crossover defect by mutation of the BLM helicase ortholog, MUS309, led Sekelsky and colleagues to propose that mei-MCM is the functional analog of the MutSγ complex, which is absent from the Drosophila genome (Kohl et al. 2012). Consistent with this idea, absence of MCM8 or MCM9 in species that possess MutSγ confers a general defect in DSB repair, but not the crossover-specific phenotype seen for Drosophila mei-MCM mutants (Lutzmann et al. 2012; Crismani et al. 2013).
Alternative DNA polymerases are involved in a variety of damage-repair processes and mediate damage tolerance by facilitating the bypass of template lesions (Goodman and Woodgate 2013). Several studies have suggested a role for such polymerases in meiotic recombination (Leem et al. 1994; Plug et al. 1997; Garcia-Diaz et al. 2000; Uchiyama et al. 2004; Arbel-Eden et al. 2013; Rattray et al. 2015). Notably, the nucleotide-excision repair factor, DNA polymerase β, localizes to synaptonemal complexes in mouse spermatocytes and is required for normal synapsis and DSB repair (Plug et al. 1997; Kidane et al. 2010). Moreover, the early defects of Polβ-deficient spermatocytes imply an unanticipated function for DNA Polβ in DSB processing (Kidane et al. 2010).
The translesion synthesis (TLS) DNA polymerases allow the replication machinery to bypass template lesions (Goodman and Woodgate 2013). In budding yeast, deficiency for the three TLS factors Rev1, Polζ (zeta), and Polη (eta) reduces the frequencies of both allelic and nonallelic gene conversion (Arbel-Eden et al. 2013). Moreover, direct interaction between Rev1, Polζ, and components of the DSB-forming machinery (Spo11, Mei4, Rec114) is suggested by yeast two-hybrid interactions (Arbel-Eden et al. 2013). Further support for a role of TLS polymerases in meiosis comes from the observation that Polζ also makes a major contribution to the elevated mutation rate that is associated with meiotic recombination (Arbeithuber et al. 2015; Rattray et al. 2015). Recruitment of alternative DNA polymerases to damage sites is mediated by ubiquitylation of PCNA at K164 (Dieckman et al. 2012). In mice expressing a K164R substitution allele, chromosome synapsis and crossover differentiation appear to occur normally, but meiosis arrests during pachytene, which may reflect defective DSB repair (Roa et al. 2008).
RESOLVING, DISSOLVING, AND UNWINDING JOINT MOLECULES TO IMPLEMENT CROSSOVER AND NONCROSSOVER FATES
The last 15 years have witnessed major advances in our understanding of joint-molecule resolution in eukaryotes (Schwartz and Heyer 2011; Bizard and Hickson 2014; Matos and West 2014; Wyatt and West 2014). During meiosis, joint-molecule resolution must achieve efficient implementation of crossing-over to promote accurate homolog disjunction, and the timely and efficient resolution of all remaining joint molecules to allow chromosomes to cleanly separate (De Muyt et al. 2012; Zakharyevich et al. 2012). These two biological imperatives are achieved through precise spatial and temporal regulation of joint-molecule-processing enzymes (Fig. 1).
Differential Timing and Regulation of Crossover and Noncrossover Formation
In budding yeast, crossover and noncrossover pathways of joint-molecule processing show differential dependence on the transcription factor, Ndt80, which defines a late-pachytene transition that commits cells to crossing-over and meiotic divisions (Allers and Lichten 2001a; Winter 2012). ndt80 mutants arrest in pachytene with unresolved joint molecules and very low levels of crossovers, whereas noncrossovers form at wild-type levels (Allers and Lichten 2001a). Thus, the joint molecules that accumulate in ndt80 mutants give rise primarily to crossovers, indicating the existence of an Ndt80-activated, crossover-biased resolution factor. This inference is consonant with the early designation of crossover sites, described above. Although Ndt80 regulates the transcription of hundreds of genes, expression of just one protein, the polo-like kinase Cdc5, is sufficient to induce joint-molecule resolution and crossing-over (Clyne et al. 2003; Sourirajan and Lichten 2008).
MutLγ and EXO1 Define a Crossover-Specific Resolving Factor
In budding yeast, plants, and mammals, the vast majority of crossovers is dependent on two conserved endonuclease activities defined by the mismatch-repair factors, MutLγ (comprising MLH1 and MLH3) and EXO1, and the structure-selective endonuclease, MUS81–EME1/Mms4 (Mus81–Mms4 in budding yeast) (Baker et al. 1996; Hunter and Borts 1997; Wang et al. 1999; Khazanehdari and Borts 2000; Kirkpatrick et al. 2000; Tsubouchi and Ogawa 2000; Lipkin et al. 2002; Wei et al. 2003; Jackson et al. 2006; Berchowitz et al. 2007; Higgins et al. 2008a; Holloway et al. 2008; Jessop and Lichten 2008; Nishant et al. 2008; Oh et al. 2008; Zakharyevich et al. 2010; Hunter 2011; Zakharyevich et al. 2012; Ranjha et al. 2014; Rogacheva et al. 2014). Joint-molecule formation occurs normally in budding yeast exo1 and mlh1/3 mutants, but crossing-over is specifically diminished, indicating that Exo1 and MutLγ define the anticipated crossover-specific resolution factor (Zakharyevich et al. 2010, 2012). EXO1 is a member of the XPG/Rad2 nuclease superfamily and plays a major role in meiotic DSB resection in addition to its role in DNA mismatch repair (Keelagher et al. 2010; Zakharyevich et al. 2010). Importantly, the nuclease activity of EXO1 is not required for its meiotic crossover function, although interaction with MutLγ is important (Zakharyevich et al. 2010). Consistent with a late function in meiotic recombination, MutLγ localizes specifically to future crossover sites in a number of organisms, including humans (Kolas and Cohen 2004), and has become an invaluable marker for the cytological analysis of crossing-over. Like other MutL-family members, MutLγ is an endonuclease and this activity is essential for its procrossover function, consistent with a direct role in dHJ resolution (Nishant et al. 2008; Zakharyevich et al. 2012; Ranjha et al. 2014; Rogacheva et al. 2014). Moreover, MutLγ preferentially binds to HJs in vitro, although targeted incision of joint molecules has not yet been demonstrated (Ranjha et al. 2014).
A General Role for MUS81 Enzymes in Meiotic Joint Molecule Processing
MUS81–EME1/Mms4 is an XPF-family endonuclease capable of cleaving a variety of branched DNA structures in vitro including 3′-flaps, D-loops, and nicked-HJs, favoring substrates with a nick or gap adjacent to the branch point that is ultimately incised (Schwartz and Heyer 2011; Mukherjee et al. 2014). Genetic studies provide compelling evidence that MUS81 enzymes can process meiotic joint molecules in vivo, although the exact substrate(s) remains uncertain (Boddy et al. 2001; Osman et al. 2003; Cromie et al. 2006; Berchowitz et al. 2007; Higgins et al. 2008a; Holloway et al. 2008; Jessop and Lichten 2008; Oh et al. 2008; Hickson and Mankouri 2011; Oke et al. 2014). The cleavage patterns observed with joint molecule substrates in vitro and the predominant role of Mus81–Eme1 in promoting crossovers in fission yeast has led to a sequential D-loop nicking model that specifically produces crossovers (Osman et al. 2003; Gaskell et al. 2007). However, MUS81–EME1/Mms4 is responsible for only a minority of crossovers in most organisms (∼5%–25%), although its activity is generally important for efficient joint molecule resolution (Jessop and Lichten 2008; Oh et al. 2008). Analysis in budding yeast reveals an early function for Mus81–Mms4 in the efficient formation of interhomolog joint molecules, in addition to a late function in joint-molecule resolution (De Los Santos et al. 2003; Oh et al. 2008; Matos et al. 2011). A general, early function for Mus81–Mms4 is further supported by the increased frequency of gene conversions and longer gene-conversion tracts seen in mus81/mms4 mutants (De Los Santos et al. 2003; Oke et al. 2014). These phenotypes are suggested to derive from aberrant processing of 3′-flaps formed when extended DSB ends anneal (Fig. 1). Aberrant processing in the absence of Mus81–Mms4 may result in longer heteroduplex tracts through assimilation of the 3′-flap, or because uncleaved flaps provoke reinvasion of the template chromosome. The late resolution function of Mus81–Mms4 involves its hyperactivation by Cdc5-mediated phosphorylation (Matos et al. 2011; Matos and West 2014).
The STR/BTR Ensembles Are Master Regulators of Meiotic Joint Molecule Metabolism
The third major joint-molecule-processing enzyme in meiosis is the STR “dissolvase” complex in budding yeast, or the analogous BTR complex in metazoans (Bizard and Hickson 2014). These complexes comprise RecQ helicases, Sgs1/BLM, their cognate type-I topoisomerases, Top3/TOPIIIα, and accessory factors Rmi1/RMI1–RMI2. In isolation, Sgs1/BLM can migrate HJs and unwind protein-free D-loops (Karow et al. 2000; van Brabant et al. 2000; Bachrati et al. 2006; Cejka and Kowalczykowski 2010; Fasching et al. 2015), but in combination with the single-strand passage activity of Top3–Rmi1/TOPIIIα–RMI1–RMI2, the STR/BTR complexes perform a unique reaction to dissociate dHJs into component duplexes. This convergent branch migration and decatenation reaction yields exclusively noncrossover products and has been termed dissolution to distinguish it from endonuclease-mediated resolution (Wu and Hickson 2003; Wu et al. 2006; Bussen et al. 2007; Singh et al. 2008; Xu et al. 2008; Cejka et al. 2010; Bocquet et al. 2014). Consistent with this activity, STR/BTR has potent anticrossover activity in vivo and is required for efficient homolog separation at meiosis I (Jessop et al. 2006; Oh et al. 2007; Chu and Hickson 2009; Bzymek et al. 2010; Holloway et al. 2010; Mankouri et al. 2011; Seguela-Arnaud et al. 2015). However, the decatenase activity of Top3–Rmi1/TOPIIIα–RMI1–RMI2 is not limited to dHJ dissolution, but is also required for the dissociation of D-loops formed by Rad51-mediated strand exchange (Fasching et al. 2015). These in vitro data imply that topological complexity impedes simple unwinding of D-loops. Consistent with a general requirement for a decatenase activity in vivo, Top3–Rmi1 is required for all functions previously defined for Sgs1 during meiotic recombination (Kaur et al. 2015; Tang et al. 2015). These include channeling joint molecules into physiological crossover and noncrossover pathways, and suppressing nonallelic recombination by dissociating the products of promiscuous strand exchange. In the absence of STR, intersister joint molecules and aberrant “multi-chromatid” structures comprising three and four interconnected chromosomes become prominent intermediates (Oh et al. 2007). Consequently, joint-molecule resolution and formation of both crossovers and noncrossovers becomes deregulated, acutely dependent on Ndt80 and the structure-selective endonucleases (Mus81–Mms4, Slx1–Slx4 and Yen1, discussed further below), and independent of MutSγ and MutLγ, which define the class I crossover pathway (Jessop et al. 2006; Oh et al. 2007, 2008; Jessop and Lichten 2008; De Muyt et al. 2012; Zakharyevich et al. 2012; Oke et al. 2014; Kaur et al. 2015; Tang et al. 2015). Thus, STR plays both a procrossover role, specifically facilitating formation of class I crossovers, and promotes noncrossover formation via synthesis-dependent strand annealing (Fig. 1). Similar inferences have been made for the C. elegans BLM ortholog, HIM-6 (Schvarzstein et al. 2014). Consistent with this conclusion, in budding yeast, the high levels of crossovers seen when all three structure-selective endonucleases (Mus81–Mms4, Slx1–Slx4 and Yen1) are absent are dependent on both MutLγ and Sgs1 (De Muyt et al. 2012; Zakharyevich et al. 2012). At designated crossover sites, the anticrossover function of STR is thought to be antagonized by MutSγ and other ZMM factors (Jessop et al. 2006; Oh et al. 2007; Chelysheva et al. 2008; Kaur et al. 2015; Seguela-Arnaud et al. 2015; Tang et al. 2015). Thus, the concerted action of stabilizing and destabilizing activities guides dHJ formation along the class-I crossover pathway.
Top3–Rmi1/TOPIIIα–RMI1–RMI2, but apparently not Sgs1/BLM, is also required during joint-molecule resolution to remove recombination-dependent chromosome entanglements (Hartung et al. 2008; Kaur et al. 2015; Tang et al. 2015). This late role of Top3–Rmi1/TOPIIIα–RMI1–RMI2 is absolutely essential for chromosome separation and again consistent with topological complexity in meiotic joint molecules that can only be eliminated with the aid of a decatenase activity.
SLX4-Associated Endonucleases and the GEN1/Yen1 Resolvase
Three other structure-selective endonucleases make variable contributions to meiotic joint-molecule processing depending on the context and the organism. SLX1 comprises an UvrC-intron (URI)-endonuclease domain and carboxy-terminal PHD-type zinc finger characteristic of the URI–YIG family of endonucleases (Dunin-Horkawicz et al. 2006). Nuclease activity depends on interaction with the scaffold protein, SLX4 (also called BTBD12), which mediates transition from an inactive SLX1 homodimer to an active SLX1–SLX4 complex (Gaur et al. 2015). Recombinant SLX1–SLX4 complexes from budding yeast and human can cleave Y-junctions, 5′-flaps, and HJs (Fricke and Brill 2003; Munoz et al. 2009; Svendsen et al. 2009). In metazoans, SLX4 also interacts with MUS81–EME1 and XPF–ERCC1 to assemble and regulate composite nuclease ensembles (budding yeast Slx4 interacts with XPF–ERCC1 ortholog, Rad1–Rad10, but not Mus81–Mms4) (Flott et al. 2007; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009; Castor et al. 2013; Wyatt et al. 2013).
In budding yeast slx1/4 single mutants, slight delays in joint-molecule resolution can be inferred but meiotic recombination is largely unperturbed. Only when SGS1 is also mutated, does Slx1–Slx4 become essential for resolution of a subset of joint molecules (De Muyt et al. 2012; Zakharyevich et al. 2012). In mouse, Slx4/Btbd12 mutation delays meiotic DSB repair, and while final crossover numbers are normal, the fraction of events that are processed via the class-I pathway defined by MutLγ appears to be increased (Holloway et al. 2011). SLX4 orthologs are important for crossing-over in Drosophila and C. elegans (Yildiz et al. 2002; Saito et al. 2009). The crossover function of Drosophila SLX4 ortholog MUS312 involves not SLX1, but the XPF–ERCC1-family nuclease, MEI9–ERCC1, which is responsible for the vast majority of crossovers in this organism (Andersen et al. 2009). In C. elegans, XPF-1 also promotes crossing-over, functioning in parallel with a second pathway defined by MUS81 and SLX1 (Agostinho et al. 2013; Bellendir and Sekelsky 2013; O’Neil et al. 2013; Saito et al. 2013). However, even when both pathways are mutated, ≥50%–70% of normal crossover levels can still form indicating the existence of a third major pathway of joint molecule resolution in C. elegans meiosis. SLX1 also plays an intriguing and unanticipated anticrossover role to suppress exchange in the central regions of C. elegans chromosomes (Saito et al. 2013).
GEN1/Yen1 (Yen1 in budding yeast) was identified as a Rad2/XPG endonuclease family member with HJ-resolving activity in vitro (Furukawa et al. 2003; Ishikawa et al. 2004; Ip et al. 2008). GEN1/Yen1 meets the criteria for a bona fide HJ-resolvase, cutting via concerted symmetrical cleavages analogous to the archetypal prokaryotic RuvC resolvase (Ip et al. 2008; Rass et al. 2010). Although GEN1/Yen1 are not essential for resolving recombination intermediates in mitotically cycling cells, Yen1 partially suppresses the recombination and damage-sensitivity phenotypes of mus81 mutants (Blanco et al. 2010; Ho et al. 2010; Tay and Wu 2010; Agmon et al. 2011). Similarly, Yen1 is not normally important for meiotic recombination, but its function becomes essential specifically when Mus81–Mms4 is absent (Matos et al. 2011; De Muyt et al. 2012; Zakharyevich et al. 2012). Yen1 is subject to inhibitory phosphorylation and only activated very late in meiosis, at the second division (Matos et al. 2011). Thus, GEN1/Yen1 may define the resolvase of last resort that can be called on to resolve rare joint molecules that escape Mus81–Mms4.
The SMC5/6 Complex Facilitates Joint Molecule Formation and Resolution
SMC5 and SMC6 form a large ring-like structure characteristic of the “structural maintenance of chromosome” proteins and combine with up to six non-SMC subunits (Nse1-6 in budding and fission yeasts) to form the SMC5/6 complex (Jeppsson et al. 2014). The SMC5/6 complex is essential for chromosome metabolism, promoting normal replication, cohesion, repair, and segregation. A primary role for SMC5/6 occurs following chromosome replication, where it helps alleviate topological entanglement of sister chromatids, working in concert with topoisomerases (Torres-Rosell et al. 2007; Tapia-Alveal et al. 2010; Kegel et al. 2011). Specifically, SMC5/6 is suggested to embrace nascent intertwinings between sister chromatid to create topologically isolated domains that help to limit supercoiling stress ahead of replication forks (Kegel et al. 2011).
In budding yeast meiosis, Smc5/6 influences early and late steps of recombination. Similar to cells lacking STR function, mutation of the Smc5/6 complex increases the levels of intersister and multichromatid joint molecules and the expense of interhomolog dHJs (Copsey et al. 2013; Lilienthal et al. 2013; Xaver et al. 2013). Analysis in both budding and fission yeasts shows that additional aberrant joint-molecule structures arise when Smc5/6 is defective (Wehrkamp-Richter et al. 2012; Copsey et al. 2013). Notably, in fission yeast, a subset of the joint molecules that accumulates in an nse6Δ mutant contain single-stranded DNA and is suggested to contain hemicatenane structures (Wehrkamp-Richter et al. 2012).
In addition to its early role, Smc5/6 is also essential for resolution of a subset of joint molecules and mutants show a complete block to chromosome segregation (Farmer et al. 2011; Wehrkamp-Richter et al. 2012; Copsey et al. 2013; Lilienthal et al. 2013; Xaver et al. 2013). However, crossovers and noncrossovers are only slightly reduced in budding yeast smc5/6 mutants and remain dependent on the physiological pathways defined by STR and MutLγ (Copsey et al. 2013; Lilienthal et al. 2013; Xaver et al. 2013). These observations are consistent with the idea that Smc5/6 influences alternative resolvase pathways defined by the structure-selective endonucleases, Mus81–Mm4, Slx1–Slx4, and/or Yen1. Although the relationship between Smc5/6 and the structure-selective endonucleases appears to be complex, existing evidence indicates that Smc5/6 acts locally at recombination sites to recruit and regulate the activity of at least one resolvase, Mus81–Mms4 (Wehrkamp-Richter et al. 2012; Copsey et al. 2013; Xaver et al. 2013).
The Nse2/Mms21 component of the budding yeast Smc5/6 complex is a SUMO E3 ligase (Zhao and Blobel 2005). Intriguingly, the early and late functions of the Smc5/6 complex are separated by the ligase-defective mms21-11 allele, which causes aberrant joint molecule formation, but is proficient for resolution (Xaver et al. 2013). Application of the temperature-sensitive smc6-56 allele confirms that the early and late functions of the Smc5/6 complex are separable: aberrant joint molecules caused by early inactivation of Smc5/6 function can still be resolved if Smc5/6 function is restored at the time of resolution (Lilienthal et al. 2013; Xaver et al. 2013).
Collectively, yeast studies point to models in which the Smc5/6 complex embraces DNA to demarcate the borders of ongoing recombination events, where it influences joint molecule formation and resolution involving Mus81–Eme1/Mms4 and perhaps other factors, such as Mph1 and its cofactors Mhf1–Mhf2 (see below), which interact genetically and physically with Smc5/6 (Chen et al. 2009; Chavez et al. 2011; Xue et al. 2015).
Whether the meiotic functions of the Smc5/6 complex defined in yeast are conserved in metazoans is currently unclear. In C. elegans, smc-5/6 mutations are required for efficient meiotic DSB repair, but defects appear to be specific to intersister recombination (Bickel et al. 2010). This phenotype may reflect the limited role of MUS81 in meiosis in C. elegans (Bellendir and Sekelsky 2013). In Arabidopsis, analysis of an mms21 mutant reveals phenotypes consistent with defective joint molecule processing including anaphase bridges, chromosome missegregation, and fragmentation (Liu et al. 2014a). In mammals, SMC5/6 localizes to synaptonemal complex, consistent with a role in meiotic recombination (Gomez et al. 2013; Verver et al. 2014), but mutant analysis is currently lacking.
Implementing Noncrossover Formation
As discussed above, the STR/BTR complexes are essential for the physiological noncrossover pathway, indicating that active disassembly of extended D-loops is required for efficient synthesis-dependent strand annealing (Chelysheva et al. 2008; Kaur et al. 2015; Seguela-Arnaud et al. 2015; Tang et al. 2015). Analysis in Arabidopsis and S. pombe reveals that orthologs of the FANCM (Fanconi anemia of complementation group M) DNA translocase also promote noncrossover formation (Crismani et al. 2012; Knoll et al. 2012; Lorenz et al. 2012). FANCM limits the formation of class-II crossovers involving Mus81–Eme1, but functions in parallel to STR/BTR suggesting that these enzymes act on distinct DNA substrates (Seguela-Arnaud et al. 2015). Analogous to STR/BTR and FANCM, the DEAH-family helicase, RTEL (regulator of telomere length) limits MUS81-dependent crossovers in C. elegans, but is thought to act via a distinct mechanism, by limiting interhomolog strand invasion as opposed to implementing interhomolog noncrossover formation (Youds et al. 2010; Rosu et al. 2011).
CLINICAL SIGNIFICANCE OF MEIOTIC RECOMBINATION
Aneuploidy, Crossing-Over, and Advancing Maternal Age
Human reproduction is prone to errors: ∼15% of couples are affected by infertility and, even for fertile couples, around a quarter of all pregnancies will end in miscarriage. At least a third of miscarriages are caused by fetal aneuploidy (trisomy or monosomy) that derives from defects in meiosis. Aneuploidy is also the leading single cause of congenital birth defects, occurring at a frequency of ∼0.3% (Nagaoka et al. 2012; Herbert et al. 2015). Less than 5% of human aneuploidies originate from errors in the male germline, consistent with the relatively low levels of aneuploidy detected in sperm (Templado et al. 2013). In sharp contrast, an estimated 20% to 70% of eggs are aneuploid (Nagaoka et al. 2012). Although the factors that contribute to the high rate of aneuploidy in the human female germline are clearly complex, two key factors have been identified: suboptimal crossing-over and advancing maternal age.
In humans, the association between altered crossover patterns and ensuing aneuploidy has been known for more than 20 years (Fisher et al. 1995; Hassold et al. 1995; Lamb et al. 1996; Hassold and Hunt 2001; Oliver et al. 2008). Two patterns are observed, a simple failure to crossover and suboptimal placement of crossovers, located either close to telomeres or close to the centromeres. Crossover failure creates an obvious risk for missegregation because unconnected homologs cannot attain a stable bipolar orientation on the meiosis-I spindle (Fig. 2). Distal crossovers are an aneuploidy risk because they are less efficient at promoting biorientation than more proximal exchanges (Lacefield and Murray 2007; Sakuno et al. 2011). In addition, connection of homologs by a single distal crossover is dependent on the short stretches of cohesion between the exchange point and the telomeres. Such “weak” chiasmata may be susceptible to premature resolution (Hassold et al. 1995; Koehler et al. 1996; Lamb et al. 1996; Ross et al. 1996).
Cohesion between sister centromeres is important for the monopolar behavior of sister-kinetochore pairs that enables homolog biorientation on the meiosis-I spindle. In addition, centromeric cohesion must be maintained until the second meiotic division when sister kinetochores now biorient on meiosis-II spindles (Fig. 2) (Watanabe 2012). Centromere-proximal crossovers are associated with the premature separation of sister chromatids at meiosis I, suggesting that centromeric cohesion is compromised by local crossing-over (Koehler et al. 1996; Lamb et al. 1996; Hassold and Hunt 2001; Rockmill et al. 2006). The local disruption of cohesion seen at chiasmata supports this idea (e.g., Kleckner 2006; Garcia-Cruz et al. 2010).
Inherent variability during meiotic prophase in human oocytes also appears to contribute to the highly elevated risk of aneuploidy seen in females (Lenzi et al. 2005). Although oocytes form ∼60% more crossovers than spermatocytes, their distribution appears to be less well regulated, resulting in an estimated ∼10% of oocytes that contain an achiasmate chromosome pair (Cheng et al. 2009). Moreover, crossover rate varies significantly between individuals and has a strong heritable component (Broman et al. 1998; Kong et al. 2002, 2008, 2014; Lenzi et al. 2005; Coop et al. 2008; Chowdhury et al. 2009; Fledel-Alon et al. 2011). Intriguingly, correlations between female crossover rate and fecundity have emerged from population studies. Mothers inheriting a higher crossover rate have slightly more children (Kong et al. 2004; Stefansson et al. 2005; Coop et al. 2008). Moreover, the crossover rate appears to increase with advancing maternal age. Specifically, more crossovers are detected in children born to older mothers, implying that oocytes that give rise to viable offspring in older mothers tend to have more crossovers (Kong et al. 2004; Coop et al. 2008; Campbell et al. 2015; Ottolini et al. 2015). Extra crossovers are proposed to buffer against the effects of advancing maternal age, in particular, the erosion of sister-chromatid cohesion (discussed below). Thus, crossover rate may be under selection in human populations.
Both common and rare human alleles have now been linked to heritable variation in crossover rate (Stefansson et al. 2005; Kong et al. 2008, 2014; Chowdhury et al. 2009; Fledel-Alon et al. 2011; Campbell et al. 2015). These include alleles of genes known to regulate meiotic recombination: the crossover regulators, RNF212, HEI10 (CCNB1IP1), and MSH4 (described above); RAD21L and REC8, which encode meiosis-specific cohesin subunits (Uhlmann 2011); and the DSB regulator, PRDM9 (Baudat et al. 2013).
Selection for oocytes with higher crossover numbers is inferred to be a manifestation of the maternal-age effect—the exponential increase in oocyte-derived aneuploidy that begins after ∼35 years of age (Hassold and Hunt 2001). Advancing maternal age is the major risk factor for human aneuploidy because of the deterioration that occurs during the extremely protracted dictyotene stage that is peculiar to oogenesis (Herbert et al. 2015). Meiosis in females initiates during fetal development, but arrests around birth after crossing-over has been completed and synaptonemal complexes have disassembled (Fig. 2). Resumption of meiosis and completion of the first division occurs only in the mature oocytes that are about to be ovulated. These oocytes will have been in the dictyotene stage for somewhere between ∼11 and 50 years (menarche through menopause). Thus, chiasmata must be maintained for decades in resting oocytes. In contrast, spermatocytes proceed directly into the meiotic divisions.
Although aging clearly impacts oocyte chromosome segregation via multiple pathways (Hassold and Hunt 2001; Selesniemi et al. 2011; Nagaoka et al. 2012), a number of studies indicate that progressive weakening of sister-chromatid cohesion is likely to be a major mechanism (Wolstenholme and Angell 2000; Jeffreys et al. 2003; Jessberger 2010, 2012; Nagaoka et al. 2012). Mouse studies indicate that cohesion is not replenished during dictyotene (Revenkova et al. 2010; Tachibana-Konwalski et al. 2010) and that both cohesion subunits and factors that function to protect centromeric cohesion until meiosis II are depleted in older animals (Hodges et al. 2005; Liu and Keefe 2008; Chiang et al. 2010; Lister et al. 2010; Shomper et al. 2014; Yun et al. 2014). Although definitive proof of analogous “cohesion exhaustion” in aging human oocytes has been harder to obtain, the weight of evidence suggests that this is the case (Angell 1991; Angell et al. 1994; Pellestor et al. 2003; Garcia-Cruz et al. 2010; Fragouli et al. 2011; Tsutsumi et al. 2014; Ottolini et al. 2015).
The effects of cohesin depletion in aging oocytes appear to be several-fold. Chiasmata may be prematurely lost, or become terminalized, both of which will impact the ability of homologs to biorient at meiosis I (Ross et al. 1996; Lacefield and Murray 2007; Sakuno et al. 2011; Watanabe 2012). Diminished centromeric cohesion weakens the normally tight association of sister kinetochores, increasing the frequency and stability of aberrant sister biorientations on the meiosis-I spindle, thereby elevating the risk of premature separation of sister chromatids (Watanabe 2012; Tachibana-Konwalski et al. 2013). Angell inferred that premature separation of sister chromatids is the major route to nondisjunction (Angell 1991; Angell et al. 1994), a proposal that is strongly supported by more recent studies (Pellestor et al. 2003; Fragouli et al. 2011; Handyside et al. 2012; Hou et al. 2013; Ottolini et al. 2015). Unexpectedly, it appears that achiasmate homologs, which were predicted to undergo canonical meiosis-I nondisjunction, instead tend to biorient their sister chromatids on the meiosis-I spindle, thereby evading the spindle-assembly checkpoint (LeMaire-Adkins and Hunt 2000; Kouznetsova et al. 2007; Nagaoka et al. 2011; Ottolini et al. 2015). The ensuing separation of sister chromatids at meiosis I is followed by segregation of nonsister chromatids at meiosis II in a process termed reverse meiosis (Ottolini et al. 2015). Aneuploidy may result from nondisjunction of the nonsister chromatids in meiosis II, whereas successful disjunction effectively rescues the original defect, producing a euploid ovum. Finally, nonexchange chromatids appear to be selected against via an unknown mechanism that preferentially segregates them into the polar body at meiosis II (Ottolini et al. 2015). Thus, suboptimal crossing-over, which is established in fetal oocytes, impacts events that occur decades later, affecting chromosome segregation during both meiosis-I and meiosis-II divisions in unexpected ways.
Meiotic Recombination and Genomic Disorders
Nonallelic meiotic recombination between low-copy dispersed repeats is frequent and generates reciprocal chromosome rearrangements that are generally deleterious. For example, copy-number variants resulting from the reciprocal deletion and duplication products of nonallelic recombination are associated with dozens of recurrent human disorders (Stankiewicz and Lupski 2002; Shaw and Lupski 2004; Sharp et al. 2005; Turner et al. 2008; Abyzov et al. 2015; Martin et al. 2015). These include hereditary neuropathy with liability to pressure palsies and Charcot–Marie–Tooth disease type 1A (Chance et al. 1994), Prader–Willi syndrome/Angelman syndrome and 15q11q13 duplication (Long et al. 1998), velocardiofacial syndrome and dup22(q11.2q11.2) (Edelmann et al. 1999a), Smith–Magenis syndrome and Potocki–Lupski syndrome (Potocki et al. 2000), and Williams–Beuren syndrome and dup7(q11.23) (Somerville et al. 2005). Other rearrangements, such as translocations, may create gene imbalances that are incompatible with fetal development, as seen for most aneuploidies. Thus, selection for higher crossover rates to enhance the fidelity of meiotic chromosome segregation may be countered by selection to preserve genomic stability.
Defective Recombination and Infertility
Meiotic recombination represents a huge mutational target and variants of recombination genes are likely to contribute to infertility. However, infertility defies conventional pedigree-based genetic analysis. A number of variants in recombination genes have been identified in infertile individuals, but causal relationships have proven hard to establish (e.g., Miyamoto et al. 2003, 2008, 2012; Stouffs et al. 2005; Martinez et al. 2007; Hikiba et al. 2008; Xu et al. 2010; Ji et al. 2012; Ghalkhani et al. 2014; Llano et al. 2014; Sazegari et al. 2014). However, a recent survey of azoospermic men revealed that 1% carried mutations in the X-linked TEX11 gene (encoding the Zip4 ortholog) and causality could be confirmed by functional analysis in the mouse (Yang et al. 2015). Thus, rare and spontaneously arising mutations in meiotic recombination genes are likely to be a significant factor for human infertility.
CONCLUDING REMARKS
The centrality to sexual reproduction and heredity places meiotic recombination at the forefront of biological research. Many pressing and outstanding questions remain. For example: why does DSB formation require a dozen or more accessory proteins; how are elastic DNA interactions bolstered to bring homologs into close juxtaposition; how is recombination functionally integrated with chromosome morphogenesis and the progression of meiotic prophase; how are crossover sites selected and how is exchange implemented with high efficiency at these sites; why do crossover numbers tend to be minimized and what conditions might select for altered recombination rates; why is recombination less well regulated in human females; and what contribution does defective recombination make to human infertility? Despite intrinsic challenges to studying meiosis, the advances made in recent years, highlighted here and in the associated collection of reviews (Subramanian and Hochwagen 2014; Brown and Bishop 2015; Herbert et al. 2015; Lam and Keeney 2015; Székvölgyi et al. 2015; Zickler and Kleckner 2015), promise rapid progress toward answering these and other important questions.
ACKNOWLEDGMENTS
Many thanks to Wolf Heyer and Steve Kowalczykowski, and the authors cited in this work. I am also indebted to the members of my laboratory for their hard work and dedication. Research in my laboratory is supported by grants from the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) (GM074223 and GM084955), and by the Howard Hughes Medical Institute (HHMI).
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abyzov A, Li S, Kim DR, Mohiyuddin M, Stutz AM, Parrish NF, Mu XJ, Clark W, Chen K, Hurles M, et al. 2015. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat Commun 6: 7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V. 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Roeder GS. 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. 2011. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res 39: 7009–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinho A, Meier B, Sonneville R, Jagut M, Woglar A, Blow J, Jantsch V, Gartner A. 2013. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini SM, Jones GH. 1987. Synaptonemal complex spreading in Allium cepa and A. fistulosum. I: The initiation and sequence of pairing. Chromosoma 95: 324–338. [Google Scholar]
- Allers T, Lichten M. 2001a. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M. 2001b. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell 8: 225–231. [DOI] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angell RR. 1991. Predivision in human oocytes at meiosis I: A mechanism for trisomy formation in man. Hum Genet 86: 383–387. [DOI] [PubMed] [Google Scholar]
- Angell RR, Xian J, Keith J, Ledger W, Baird DT. 1994. First meiotic division abnormalities in human oocytes: Mechanism of trisomy formation. Cytogenet Cell Genet 65: 194–202. [DOI] [PubMed] [Google Scholar]
- Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. 2015. Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proc Natl Acad Sci 112: 2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel-Eden A, Joseph-Strauss D, Masika H, Printzental O, Rachi E, Simchen G. 2013. Trans-lesion DNA polymerases may be involved in yeast meiosis. G3 (Bethesda) 10.1534/g3.113.005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argunhan B, Farmer S, Leung WK, Terentyev Y, Humphryes N, Tsubouchi T, Toyoizumi H, Tsubouchi H. 2013. Direct and indirect control of the initiation of meiotic recombination by DNA damage checkpoint mechanisms in budding yeast. PLoS ONE 8: e65875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T, Walpita D, de Rooij DG. 2001. Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J Cell Sci 114: 685–693. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. 2006. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res 34: 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier B, Hunt P, Broman KW, Hassold T. 2014. Variation in genome-wide levels of meiotic recombination is established at the onset of prophase in mammalian males. PLoS Genet 10: e1004125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Luche DD, Hunter T, Forsburg SL. 2008. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote S-phase genome stability. Mol Cell Biol 28: 1724–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, et al. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13: 336–342. [DOI] [PubMed] [Google Scholar]
- Bartrand AJ, Iyasu D, Marinco SM, Brush GS. 2006. Evidence of meiotic crossover control in Saccharomyces cerevisiae through Mec1-mediated phosphorylation of replication protein A. Genetics 172: 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Nicolas A. 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. 2013. Meiotic recombination in mammals: Localization and regulation. Nat Rev Genet 14: 794–806. [DOI] [PubMed] [Google Scholar]
- Bellendir SP, Sekelsky J. 2013. An elegans solution for crossover formation. PLoS Genet 9: e1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. 2010. Genetic interference: Don’t stand so close to me. Curr Genom 11: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. 2007. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat R, Manheim EA, Sherizen DE, McKim KS. 2004. Studies on crossover-specific mutants and the distribution of crossing over in Drosophila females. Cytogenet Genome Res 107: 160–171. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet 4: e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel JS, Chen L, Hayward J, Yeap SL, Alkers AE, Chan RC. 2010. Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet 6: e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID. 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 6: a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SC, West SC. 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81–Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair 9: 394–402. [DOI] [PubMed] [Google Scholar]
- Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Hochwagen A. 2013. ATR/Mec1 prevents lethal meiotic recombination initiation on partially replicated chromosomes in budding yeast. eLife 2: e00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. 2007. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol 17: 2003–2012. [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2009. The Mcm complex: Unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73: 652–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Bunker RD, Kowalczykowski SC, Cejka P, Hickson ID, et al. 2014. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIα and RMI1. Nat Struct Mol Biol 21: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR III, Russell P. 2001. Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Bojko M. 1985. Human meiosis IX: Crossing over and chiasma formation in oocytes. Carlsberg Res Commun 50: 43–72. [Google Scholar]
- Borde V, de Massy B. 2013. Programmed induction of DNA double strand breaks during meiosis: Setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev 23: 147–155. [DOI] [PubMed] [Google Scholar]
- Borde V, Goldman AS, Lichten M. 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. [DOI] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, Hunter N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. 1998. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet 63: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Brown MS, Bishop DK. 2015. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol 7: a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PW, Judis L, Chan ER, Schwartz S, Seftel A, Thomas A, Hassold TJ. 2005. Meiotic synapsis proceeds from a limited number of subtelomeric sites in the human male. Am J Hum Genet 77: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M. 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussen W, Raynard S, Busygina V, Singh AK, Sung P. 2007. Holliday junction processing activity of the BLM–Topo IIIα–BLAP75 complex. J Biol Chem 282: 31484–31492. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Furlotte NA, Eriksson N, Hinds D, Auton A. 2015. Escape from crossover interference increases with maternal age. Nat Commun 6: 6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, Borde V, Klein F, Cha RS. 2013. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet 9: e1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AT. 1981. EM autoradiographic evidence that DNA synthesis occurs at recombination nodules during meiosis in Drosophila melanogaster females. Chromosoma 83: 59–80. [DOI] [PubMed] [Google Scholar]
- Castor D, Nair N, Declais AC, Lachaud C, Toth R, Macartney TJ, Lilley DM, Arthur JS, Rouse J. 2013. Cooperative control of Holliday junction resolution and DNA repair by the SLX1 and MUS81–EME1 nucleases. Mol Cell 52: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC. 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J Biol Chem 285: 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. 2010. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1–Top3. Nat Struct Mol Biol 17: 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR. 1994. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3: 223–228. [DOI] [PubMed] [Google Scholar]
- Chavez A, Agrawal V, Johnson FB. 2011. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J Biol Chem 286: 5119–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M. 2007. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Belcram K, Gendrot G, Grelon M. 2008. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet 4: e1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, Vrielynck N, Le Guin S, Novatchkova M, Grelon M. 2012. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 8: e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang W, Timofejeva L, Gerardin Y, Ma H. 2005. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J 43: 321–334. [DOI] [PubMed] [Google Scholar]
- Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. 2008. Global analysis of the meiotic crossover landscape. Dev Cell 15: 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. 2009. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci 106: 21252–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev 20: 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EY, Hunt PA, Naluai-Cecchini TA, Fligner CL, Fujimoto VY, Pasternack TL, Schwartz JM, Steinauer JE, Woodruff TJ, Cherry SM, et al. 2009. Meiotic recombination in human oocytes. PLoS Genet 5: e1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. 2010. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 20: 1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. 2009. Genetic analysis of variation in human meiotic recombination. PLoS Genet 5: e1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. 2009. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer 9: 644–654. [DOI] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 5: 480–485. [DOI] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M. 2010. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell 39: 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14: 424–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Baudat F, Grey C, Keeney S, de Massy B, Jasin M. 2014. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet 46: 1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319: 1395–1398. [DOI] [PubMed] [Google Scholar]
- Cooper TJ, Wardell K, Garcia V, Neale MJ. 2014. Homeostatic regulation of meiotic DSB formation by ATM/ATR. Exp Cell Res 329: 124–131. [DOI] [PubMed] [Google Scholar]
- Copsey A, Jordan PW, Tang S, Blitzblau H, Chan AC, Newcombe S, Newnham L, Li A, Arumugam P, Hochwagen A, et al. 2013. Smc5/6 coordinates formation and resolution o fjoint molecules with chromosome morphology to ensure meotic divisions. PLoS Genet 9: e1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton HB, McClintock B. 1931. A correlation of cytological and genetical crossing-over in Zea Mays. Proc Natl Acad Sci 17: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Crismani W, Portemer V, Froger N, Chelysheva L, Horlow C, Vrielynck N, Mercier R. 2013. MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana. PLoS Genet 9: e1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127: 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown KN, McMahan S, Sekelsky J. 2014. Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet 10: e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, White CI. 2013. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet 9: e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, et al. 2011. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Heyting C. 2006. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115: 220–234. [DOI] [PubMed] [Google Scholar]
- de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. 2006. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci 103: 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Dietrich AJ, Hoog C, Stam P, Heyting C. 2007. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J Cell Sci 120: 731–736. [DOI] [PubMed] [Google Scholar]
- De Los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. 2003. The mus81/mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B. 2013. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu Rev Genet 47: 563–599. [DOI] [PubMed] [Google Scholar]
- de Massy B. 2014. Human genetics. Hidden features of human hotspots. Science 346: 808–809. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. 2014. E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev 28: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P, te Riele H. 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev 13: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. 2005. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19: 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckman LM, Freudenthal BD, Washington MT. 2012. PCNA structure and function: Insights from structures of PCNA complexes and post-translationally modified PCNA. Subcell Biochem 62: 281–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Mercier R, Chelysheva L, Berard A, Falque M, Martin O, Zanni V, Brunel D, Mezard C. 2007. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet 3: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Khademian H, Giraut L, Zanni V, Bellalou S, Henderson IR, Falque M, Mezard C. 2013. Contrasted patterns of crossover and non-crossover at Arabidopsis thaliana meiotic recombination hotspots. PLoS Genet 9: e1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunin-Horkawicz S, Feder M, Bujnicki JM. 2006. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics 7: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. 1999a. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8: 1157–1167. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, Kolodner R, Pollard JW, Kucherlapati R. 1999b. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 21: 123–127. [DOI] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR. 2005. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci 102: 10952–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S, San-Segundo PA, Aragon L. 2011. The Smc5–Smc6 complex is required to remove chromosome junctions in meiosis. PLoS ONE 6: e20948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD. 2015. Top3–Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol Cell 57: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR III, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JM, Harvey JF, Morton NE, Jacobs PA. 1995. Trisomy 18: Studies of the parent and cell division of origin and the effect of aberrant recombination on nondisjunction. Am J Hum Genet 56: 669–675. [PMC free article] [PubMed] [Google Scholar]
- Fledel-Alon A, Leffler EM, Guan Y, Stephens M, Coop G, Przeworski M. 2011. Variation in human recombination rates and its genetic determinants. PLoS ONE 6: e20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J. 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27: 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. 2004. Eukaryotic MCM proteins: Beyond replication initiation. Microbiol Mol Biol Rev 68: 109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res 24: 1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Goodall NN, Sanchez-Garcia JF, Colls P, Wells D. 2011. The cytogenetics of polar bodies: Insights into female meiosis and the diagnosis of aneuploidy. Mol Hum Reprod 17: 286–295. [DOI] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. 2003. Slx1–Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1–Top3. Genes Dev 17: 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JC, Rockmill B, Odell M, Roeder GS. 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kimura S, Ishibashi T, Mori Y, Hashimoto J, Sakaguchi K. 2003. OsSEND-1: A new RAD2 nuclease family member in higher plants. Plant Mol Biol 51: 59–70. [DOI] [PubMed] [Google Scholar]
- Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ. 2015. Tel1-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 520: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cruz R, Brieno MA, Roig I, Grossmann M, Velilla E, Pujol A, Cabero L, Pessarrodona A, Barbero JL, Garcia Caldes M. 2010. Dynamics of cohesin proteins REC8, STAG3, SMC1β and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Hum Reprod 25: 2316–2327. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, de Lera LT, Saniger ML, Ruiz JF, Parraga M, Garcia-Ortiz MJ, Kirchhoff T, del Mazo J, et al. 2000. DNA polymerase lambda (Polλ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol 301: 851–867. [DOI] [PubMed] [Google Scholar]
- Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. 2007. Mus81 cleavage of Holliday junctions: A failsafe for processing meiotic recombination intermediates? EMBO J 26: 1891–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V, Wyatt HD, Komorowska W, Szczepanowski RH, de Sanctis D, Gorecka KM, West SC, Nowotny M. 2015. Structural and mechanistic analysis of the Slx1–Slx4 endonuclease. Cell Rep 10.1016/j.celrep.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. 2000. Inaugural article: Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalkhani E, Sheidai M, Gourabi H, Noormohammadi Z, Bakhtari N, Malekasgar AM. 2014. Study of single nucleotide polymorphism (rs28368082) in SPO11 gene and its association with male infertility. J Assist Reprod Genet 31: 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Lichten M. 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez R, Jordan PW, Viera A, Alsheimer M, Fukuda T, Jessberger R, Llano E, Pendas AM, Handel MA, Suja JA. 2013. Dynamic localization of SMC5/6 complex proteins during mammalian meiosis and mitosis suggests functions in distinct chromosome processes. J Cell Sci 126: 4239–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Woodgate R. 2013. Translesion DNA polymerases. Cold Spring Harb Perspect Biol 5: a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell 14: 263–274. [DOI] [PubMed] [Google Scholar]
- Gray S, Allison RM, Garcia V, Goldman AS, Neale MJ. 2013. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol 3: 130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn JR, Rubio C, Broman KW, Hunt PA, Hassold T. 2013. Cytological studies of human meiosis: Sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS ONE 8: e85075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. 2005. Crossover and noncrossover pathways in mouse meiosis. Mol Cell 20: 563–573. [DOI] [PubMed] [Google Scholar]
- Guiraldelli MF, Eyster C, Wilkerson JL, Dresser ME, Pezza RJ. 2013. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet 9: e1003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. 2008. Evolution of models of homologous recombination. In Recombination and meiosis (ed. Lankenau D-H), pp. 1–64. Springer, Berlin. [Google Scholar]
- Haber JE. 2013. Genome stability: DNA repair and recombination. Garland Science, Abingdon, England. [Google Scholar]
- Handyside AH, Montag M, Magli MC, Repping S, Harper J, Schmutzler A, Vesela K, Gianaroli L, Geraedts J. 2012. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. Eur J Hum Genet 20: 742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. 2008. Topoisomerase 3α and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2001. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Hassold T, Merrill M, Adkins K, Freeman S, Sherman S. 1995. Recombination and maternal age-dependent nondisjunction: Molecular studies of trisomy 16. Am J Hum Genet 57: 867–874. [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Mlynarczyk-Evans S, Villeneuve AM. 2010. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci 101: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. 2006. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Herbert M, Kalleas D, Cooney D, Lamb M, Lister L. 2015. Meiosis and maternal aging: Insights from aneuploid oocytes and trisomy births. Cold Spring Harb Perspect Biol 7: a017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson ID, Mankouri HW. 2011. Processing of homologous recombination repair intermediates by the Sgs1–Top3–Rmi1 and Mus81–Mms4 complexes. Cell Cycle 10: 3078–3085. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. 2005. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FC, Jones GH. 2008a. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54: 152–162. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Vignard J, Mercier R, Pugh AG, Franklin FC, Jones GH. 2008b. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J 55: 28–39. [DOI] [PubMed] [Google Scholar]
- Hikiba J, Hirota K, Kagawa W, Ikawa S, Kinebuchi T, Sakane I, Takizawa Y, Yokoyama S, Mandon-Pepin B, Nicolas A, et al. 2008. Structural and functional analyses of the DMC1–M200V polymorphism found in the human population. Nucleic Acids Res 36: 4181–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers KJ. 2004. Crossover interference. Curr Biol 14: R1036–R1037. [DOI] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM. 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol 13: 1641–1647. [DOI] [PubMed] [Google Scholar]
- Ho CK, Mazon G, Lam AF, Symington LS. 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA. 2005. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet 37: 1351–1355. [DOI] [PubMed] [Google Scholar]
- Holliday R. 1964. A mechanism for gene conversion in fungi. Genet Res 5: 282–304. [DOI] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. 2008. MUS81 generates a subset of MLH1–MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4: e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. 2010. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol 188: 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. 2011. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 7: e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Sun X, Yokoo R, Villeneuve AM, Cohen PE. 2014. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J Cell Biol 205: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP. 2013. The logic and mechanism of homologous recombination partner choice. Mol Cell 51: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, et al. 2013. Genome analyses of single human oocytes. Cell 155: 1492–1506. [DOI] [PubMed] [Google Scholar]
- Humphryes N, Hochwagen A. 2014. A non-sister act: Recombination template choice during meiosis. Exp Cell Res 329: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2006. Meiotic recombination. In Molecular genetics of recombination (ed. Aguilera A, Rothstein R), pp. 381–442. Springer, Heidelberg. [Google Scholar]
- Hunter N. 2011. Double duty for Exo1 during meiotic recombination. Cell Cycle 10: 2607–2609. [DOI] [PubMed] [Google Scholar]
- Hunter N. 2013. Meiosis. Elsevier, Amsterdam. [Google Scholar]
- Hunter N, Borts RH. 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev 11: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. 2001. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Hwang PY, Hunter N. 2011. Mapping meiotic breaks: Spo11 oligonucleotides precisely mark the spots. Genome Biol 12: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361. [DOI] [PubMed] [Google Scholar]
- Ishikawa G, Kanai Y, Takata K, Takeuchi R, Shimanouchi K, Ruike T, Furukawa T, Kimura S, Sakaguchi K. 2004. DmGEN, a novel RAD2 family endo-exonuclease from Drosophila melanogaster. Nucleic Acids Res 32: 6251–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishishita S, Inui T, Matsuda Y, Serikawa T, Kitada K. 2013. Infertility associated with meiotic failure in the tremor rat (tm/tm) is caused by the deletion of spermatogenesis associated 22. Exp Anim 62: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishishita S, Matsuda Y, Kitada K. 2014. Genetic evidence suggests that Spata22 is required for the maintenance of Rad51 foci in mammalian meiosis. Sci Rep 4: 6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kugou K, Fawcett JA, Mura S, Ikeda S, Innan H, Ohta K. 2014. Meiotic recombination cold spots in chromosomal cohesion sites. Genes Cells 19: 359–373. [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FC. 2006. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J 25: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. 2004. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol 24: 7998–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet 36: 151–156. [DOI] [PubMed] [Google Scholar]
- Jeffreys CA, Burrage PS, Bickel SE. 2003. A model system for increased meiotic nondisjunction in older oocytes. Curr Biol 13: 498–503. [DOI] [PubMed] [Google Scholar]
- Jeppsson K, Kanno T, Shirahige K, Sjogren C. 2014. The maintenance of chromosome structure: Positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol 15: 601–614. [DOI] [PubMed] [Google Scholar]
- Jessberger R. 2010. Deterioration without replenishment—The misery of oocyte cohesin. Genes Dev 24: 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R. 2012. Age-related aneuploidy through cohesion exhaustion. EMBO Rep 13: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. 2008. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell 31: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M. 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet 2: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G, Long Y, Zhou Y, Huang C, Gu A, Wang X. 2012. Common variants in mismatch repair genes associated with increased risk of sperm DNA damage and male infertility. BMC Med 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19: 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GH. 1984. The control of chiasma distribution. Symp Soc Exp Biol 38: 293–320. [PubMed] [Google Scholar]
- Jones GH, Franklin FC. 2006. Meiotic crossing-over: Obligation and interference. Cell 126: 246–248. [DOI] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, Borner GV. 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet 5: e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, Tanneti SN, McKim KS. 2009. Drosophila hold’em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics 181: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID. 2000. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci 97: 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, De Muyt A, Lichten M. 2015. Top3–Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell 57: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keelagher RE, Cotton VE, Goldman AS, Borts RH. 2010. Separable roles for Exonuclease I in meiotic DNA double-strand break repair. DNA Repair 10: 126–137. [DOI] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney S. 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn Stab 2: 81–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Neale MJ. 2006. Initiation of meiotic recombination by formation of DNA double-strand breaks: Mechanism and regulation. Biochem Soc Trans 34: 523–525. [DOI] [PubMed] [Google Scholar]
- Keeney S, Lange J, Mohibullah N. 2014. Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu Rev Genet 48: 187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A, Betts-Lindroos H, Kanno T, Jeppsson K, Strom L, Katou Y, Itoh T, Shirahige K, Sjogren C. 2011. Chromosome length influences replication-induced topological stress. Nature 471: 392–396. [DOI] [PubMed] [Google Scholar]
- Khazanehdari KA, Borts RH. 2000. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma 109: 94–102. [DOI] [PubMed] [Google Scholar]
- Kidane D, Jonason AS, Gorton TS, Mihaylov I, Pan J, Keeney S, de Rooij DG, Ashley T, Keh A, Liu Y, et al. 2010. DNA polymerase β is critical for mouse meiotic synapsis. EMBO J 29: 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JS, Mortimer RK. 1990. A polymerization model of chiasma interference and corresponding computer simulation. Genetics 126: 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. 2000. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics 156: 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani Y, Olive LS, El-Ani AS. 1962. Genetics of Sordaria fimicola V. aberrant segregation at the G locus. Am J Bot 49: 697–706. [Google Scholar]
- Kleckner N. 2006. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Storlazzi A, Zickler D. 2003. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet 19: 623–628. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. 2004. A mechanical basis for chromosome function. Proc Natl Acad Sci 101: 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H Jr, Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 14: 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Higgins JD, Seeliger K, Reha SJ, Dangel NJ, Bauknecht M, Schropfer S, Franklin FC, Puchta H. 2012. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24: 1448–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KE, Boulton CL, Collins HE, French RL, Herman KC, Lacefield SM, Madden LD, Schuetz CD, Hawley RS. 1996. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat Genet 14: 406–414. [DOI] [PubMed] [Google Scholar]
- Kohl KP, Jones CD, Sekelsky J. 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Cohen PE. 2004. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet Genome Res 107: 216–231. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Kong A, Barnard J, Gudbjartsson DF, Thorleifsson G, Jonsdottir G, Sigurdardottir S, Richardsson B, Jonsdottir J, Thorgeirsson T, Frigge ML, et al. 2004. Recombination rate and reproductive success in humans. Nat Genet 36: 1203–1206. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, Jonsdottir GM, Gudjonsson SA, Sverrisson S, Thorlacius T, et al. 2008. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319: 1398–1401. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Masson G, Gudbjartsson DF, Villemoes R, Magnusdottir E, Olafsdottir SB, Thorsteinsdottir U, Stefansson K. 2014. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet 46: 11–16. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Lister L, Nordenskjold M, Herbert M, Hoog C. 2007. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat Genet 39: 966–968. [DOI] [PubMed] [Google Scholar]
- Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T, et al. 2009. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell 20: 3064–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Bourbon HM, de Massy B. 2010. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24: 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Azumi Y, Hayakawa S, Kamiya A, Imura Y, Wada T, Shinozaki K. 2008. Homologous chromosome pairing is completed in crossover defective atzip4 mutant. Biochem Biophys Res Commun 370: 98–103. [DOI] [PubMed] [Google Scholar]
- Kurzbauer MT, Uanschou C, Chen D, Schlogelhofer P. 2012. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24: 2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield S, Murray AW. 2007. The spindle checkpoint rescues the meiotic segregation of chromosomes whose crossovers are far from the centromere. Nat Genet 39: 1273–1277. [DOI] [PubMed] [Google Scholar]
- Lake CM, Teeter K, Page SL, Nielsen R, Hawley RS. 2007. A genetic analysis of the Drosophila mcm5 gene defines a domain specifically required for meiotic recombination. Genetics 176: 2151–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lam I, Keeney S. 2015. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7: a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, Gu Y, Shen J, Saker D, May KM, et al. 1996. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet 14: 400–405. [DOI] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. 2011. ATM controls meiotic double-strand-break formation. Nature 479: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Hunter N. 2010. Trying to avoid your sister. PLoS Biol 8: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. 2008. Rad52 promotes post-invasion steps of meiotic double-strand-break repair. Mol Cell 29: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, Dresser ME, Hunter N, Bishop DK. 2013. Meiotic crossover control by concerted action of Rad51–Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet 9: e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Palmer K, O’Brien M, Schimenti JC, Eppig J, Handel MA. 2012. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol Reprod 86: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem SH, Ropp PA, Sugino A. 1994. The yeast Saccharomyces cerevisiae DNA polymerase IV: Possible involvement in double strand break DNA repair. Nucleic Acids Res 22: 3011–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaire-Adkins R, Hunt PA. 2000. Nonrandom segregation of the mouse univalent X chromosome: Evidence of spindle-mediated meiotic drive. Genetics 156: 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE. 2005. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet 76: 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Stith CM, Burgers PM, Heyer WD. 2009. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol Cell 36: 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chang Y, Xin X, Zhu C, Li X, Higgins JD, Wu C. 2013. Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice. Plant Cell 25: 3885–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, de Massy B. 2011. The impressionistic landscape of meiotic recombination. Cell 147: 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienthal I, Kanno T, Sjogren C. 2013. Inhibition of the Smc5/6 complex during meiosis perturbs joint molecule formation and resolution without significantly changing crossover or non-crossover levels. PLoS Genet 9: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindergren CC. 1953. Gene conversion in Saccharomyces. J Genet 51: 625–637. [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31: 385–390. [DOI] [PubMed] [Google Scholar]
- Lissouba P, Rizet G. 1960. Sur l’existence d’une unité génétique polarisée ne subissant que des échanges non réciproques. Compt Rend Acad Sci 250: 3408–3410. [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, et al. 2010. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 20: 1511–1521. [DOI] [PubMed] [Google Scholar]
- Liu L, Keefe DL. 2008. Defective cohesin is associated with age-dependent misaligned chromosomes in oocytes. Reprod Biomed Online 16: 103–112. [DOI] [PubMed] [Google Scholar]
- Liu M, Shi S, Zhang S, Xu P, Lai J, Liu Y, Yuan D, Wang Y, Du J, Yang C. 2014a. SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis. BMC Plant Biol 14: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gaines WA, Callender T, Busygina V, Oke A, Sung P, Fung JC, Hollingsworth NM. 2014b. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet 10: e1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano E, Gomez HL, Garcia-Tunon I, Sanchez-Martin M, Caburet S, Barbero JL, Schimenti JC, Veitia RA, Pendas AM. 2014. STAG3 is a strong candidate gene for male infertility. Hum Mol Genet 23: 3421–3431. [DOI] [PubMed] [Google Scholar]
- Long FL, Duckett DP, Billam LJ, Williams DK, Crolla JA. 1998. Triplication of 15q11-q13 with inv dup(15) in a female with developmental delay. J Med Genet 35: 425–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ. 2013. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun 4: 2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, et al. 2012. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell 47: 523–534. [DOI] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296: 2222–2225. [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV. 2007. ZMM proteins during meiosis: Crossover artists at work. Chromosome Res 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R. 2008. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18: 1432–1437. [DOI] [PubMed] [Google Scholar]
- Macaisne N, Vignard J, Mercier R. 2011. SHOC1 and PTD form an XPF–ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci 124: 2687–2691. [DOI] [PubMed] [Google Scholar]
- Macqueen AJ, Roeder GS. 2009. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr Biol 19: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Stucki M, Spadari S, Hubscher U. 2000. DNA polymerase switching. I: Replication factor C displaces DNA polymerase α prior to PCNA loading. J Mol Biol 295: 791–801. [DOI] [PubMed] [Google Scholar]
- Maloisel L, Bhargava J, Roeder GS. 2004. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics 167: 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Huber W, Steinmetz LM. 2011. Genome-wide survey of post-meiotic segregation during yeast recombination. Genome Biol 12: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manheim EA, Jang JK, Dominic D, McKim KS. 2002. Cytoplasmic localization and evolutionary conservation of MEI-218, a protein required for meiotic crossing-over in Drosophila. Mol Biol Cell 13: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ashton TM, Hickson ID. 2011. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc Natl Acad Sci 108: 4944–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE. 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Kirkpatrick BE, Ledbetter DH. 2015. Copy number variants, aneuploidies, and human disease. Clin Perinatol 42: 227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Bonache S, Carvajal A, Bassas L, Larriba S. 2007. Mutations of SYCP3 are rare in infertile Spanish men with meiotic arrest. Fertil Steril 88: 988–989. [DOI] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. 2011. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet 7: e1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, West SC. 2014. Holliday junction resolution: Regulation in space and time. DNA Repair 19: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC. 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi H, Yamamoto MT. 2003. REC, a new member of the MCM-related protein family, is required for meiotic recombination in Drosophila. Genes Genet Syst 78: 363–371. [DOI] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′–5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK. 2007. Synthesis-dependent strand annealing in meiosis. PLoS Biol 5: e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, Nogue F, Doutriaux MP, Horlow C, Grelon M, et al. 2005. Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr Biol 15: 692–701. [DOI] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MB. 1955. Aberrant recombination of pyridoxine mutants of Neurospora. Proc Natl Acad Sci 41: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Hasuike S, Yogev L, Maduro MR, Ishikawa M, Westphal H, Lamb DJ. 2003. Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet 362: 1714–1719. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Koh E, Sakugawa N, Sato H, Hayashi H, Namiki M, Sengoku K. 2008. Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet 25: 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Tsujimura A, Miyagawa Y, Koh E, Namiki M, Horikawa M, Saijo Y, Sengoku K. 2012. Single-nucleotide polymorphisms in HORMAD1 may be a risk factor for azoospermia caused by meiotic arrest in Japanese patients. Asian J Androl 14: 580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K. 2012. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell 47: 722–733. [DOI] [PubMed] [Google Scholar]
- Morgan TH. 1913. Heredity and sex. Columbia University Press, New York. [Google Scholar]
- Moses MJ, Dresser ME, Poorman PA. 1984. Composition and role of the synaptonemal complex. Symp Soc Exp Biol 38: 245–270. [PubMed] [Google Scholar]
- Mossi R, Keller RC, Ferrari E, Hubscher U. 2000. DNA polymerase switching. II: Replication factor C abrogates primer synthesis by DNA polymerase α at a critical length. J Mol Biol 295: 803–814. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Wright WD, Ehmsen KT, Heyer WD. 2014. The Mus81–Mms4 structure-selective endonuclease requires nicked DNA junctions to undergo conformational changes and bend its DNA substrates for cleavage. Nucleic Acids Res 42: 6511–6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127. [DOI] [PubMed] [Google Scholar]
- Murakami H, Keeney S. 2014. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell 158: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, Owen N, Stevense M, Smith H, Nagaoka S, Hassold T, McKay M, Xu H, Fu J, Revenkova E, et al. 2013. Altered cohesin gene dosage affects mammalian meiotic chromosome structure and behavior. PLoS Genet 9: e1003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray NE. 1960. Complementation and recombination between methionine-2 alleles in Neurospora crassa. Heredity 15: 207–217. [Google Scholar]
- Nabeshima K, Villeneuve AM, Hillers KJ. 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. 2011. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol 21: 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. 2012. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat Rev Genet 13: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kolodner RD. 2002. The MER3 DNA helicase catalyzes the unwinding of Holliday junctions. J Biol Chem 277: 28019–28024. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H. 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J 18: 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Plys AJ, Alani E. 2008. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179: 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell 16: 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Hoog C. 2008. Cohesin Smc1β determines meiotic chromatin axis loop organization. J Cell Biol 180: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, Arai K, Masai H. 2006. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc Natl Acad Sci 103: 8131–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. 2008. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell 31: 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke A, Anderson CM, Yam P, Fung JC. 2014. Controlling meiotic recombinational repair—Specifying the roles of ZMMs, Sgs1 and Mus81/Mms4 in crossover formation. PLoS Genet 10: e1004690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive LS. 1959. Aberrant tetrads in Sordaria fimicola. Proc Natl Acad Sci 45: 727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TR, Feingold E, Yu K, Cheung V, Tinker S, Yadav-Shah M, Masse N, Sherman SL. 2008. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet 4: e1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil NJ, Martin JS, Youds JL, Ward JD, Petalcorin MI, Rose AM, Boulton SJ. 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet 9: e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. 2003. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81–Eme1 in meiosis. Mol Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Osman K, Sanchez-Moran E, Mann SC, Jones GH, Franklin FC. 2009. Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J 28: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini CS, Newnham LJ, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK, Sage K, Summers MC, Thornhill AR, et al. 2015. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet 47: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Andreo B, Arnal F, Humeau C, Demaille J. 2003. Maternal aging and chromosomal abnormalities: New data drawn from in vitro unfertilized human oocytes. Hum Genet 112: 195–203. [DOI] [PubMed] [Google Scholar]
- Pelttari J, Hoja MR, Yuan L, Liu JG, Brundell E, Moens P, Santucci-Darmanin S, Jessberger R, Barbero JL, Heyting C, et al. 2001. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol Cell Biol 21: 5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples TL, Dean E, Gonzalez O, Lambourne L, Burgess SM. 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev 16: 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DD. 1962. The frequency in Neurospora tetrads of multiple exchanges within short intervals. Genet Res 3: 315–327. [Google Scholar]
- Perry J, Kleckner N, Borner GV. 2005. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc Natl Acad Sci 102: 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Broman KW, Szatkiewicz JP, Paigen K. 2007. Crossover interference underlies sex differences in recombination rates. Trends Genet 23: 539–542. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. 2003. Un menage a quatre: The molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- Plug AW, Clairmont CA, Sapi E, Ashley T, Sweasy JB. 1997. Evidence for a role for DNA polymerase β in mammalian meiosis. Proc Natl Acad Sci 94: 1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, et al. 2000. Molecular mechanism for duplication 17p11.2—The homologous recombination reciprocal of the Smith–Magenis microdeletion. Nat Genet 24: 84–87. [DOI] [PubMed] [Google Scholar]
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. 2014. DNA recombination. Recombination initiation maps of individual human genomes. Science 346: 1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Hoog C, Paddy M, Hunter N. 2012a. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Offenberg HH, Anderson LK. 2012b. Altered distribution of MLH1 foci is associated with changes in cohesins and chromosome axis compaction in an asynaptic mutant of tomato. Chromosoma 121: 291–305. [DOI] [PubMed] [Google Scholar]
- Qiao H, Rao HBDP, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong ER, Holloway JK, et al. 2014. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet 46: 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjha L, Anand R, Cejka P. 2014. The Saccharomyces cerevisiae Mlh1–Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J Biol Chem 289: 5674–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Compton SA, Matos J, Singleton MR, Ip SC, Blanco MG, Griffith JD, West SC. 2010. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev 24: 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A, Santoyo G, Shafer B, Strathern JN. 2015. Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLoS Genet 11: e1004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Herrmann K, Adelfalk C, Jessberger R. 2010. Oocyte cohesin expression restricted to predictyate stages provides full fertility and prevents aneuploidy. Curr Biol 20: 1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, et al. 2008. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci 105: 16248–16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Fung JC, Branda SS, Roeder GS. 2003. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr Biol 13: 1954–1962. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Voelkel-Meiman K, Roeder GS. 2006. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics 174: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Lefrancois P, Voelkel-Meiman K, Oke A, Roeder GS, Fung JC. 2013. High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet 9: e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. 2014. Mlh1–Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2–Msh3-stimulated endonuclease. J Biol Chem 289: 5664–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Ross LO, Maxfield R, Dawson D. 1996. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc Natl Acad Sci 93: 4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Libuda DE, Villeneuve AM. 2011. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334: 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM. 2013. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet 9: e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. 2009. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet 5: e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Lui DY, Kim HM, Meyer K, Colaiacovo MP. 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Tanaka K, Hauf S, Watanabe Y. 2011. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev Cell 21: 534–545. [DOI] [PubMed] [Google Scholar]
- Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K, Kawasaki Y, Shibata T, Masai H, Ohta K. 2008. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev 22: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazegari A, Kalantar SM, Pashaiefar H, Mohtaram S, Honarvar N, Feizollahi Z, Ghasemi N. 2014. The T657C polymorphism on the SYCP3 gene is associated with recurrent pregnancy loss. J Assist Reprod Genet 31: 1377–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Pattabiraman D, Libuda DE, Ramadugu A, Tam A, Martinez-Perez E, Roelens B, Zawadzki KA, Yokoo R, Rosu S, et al. 2014. DNA helicase HIM-6/BLM both promotes MutSγ-dependent crossovers and antagonizes MutSγ-independent interhomolog associations during Caenorhabditis elegans meiosis. Genetics 198: 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguela-Arnaud M, Crismani W, Larcheveque C, Mazel J, Froger N, Choinard S, Lemhemdi A, Macaisne N, Van Leene J, Gevaert K, et al. 2015. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci 112: 4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selesniemi K, Lee HJ, Muhlhauser A, Tilly JL. 2011. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc Natl Acad Sci 108: 12319–12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, et al. 2005. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR. 2004. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum Mol Genet 13: R57–R64. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tang D, Wang K, Wang M, Huang J, Luo W, Luo Q, Hong L, Li M, Cheng Z. 2012. ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309. [DOI] [PubMed] [Google Scholar]
- Shomper M, Lappa C, FitzHarris G. 2014. Kinetochore microtubule establishment is defective in oocytes from aged mice. Cell Cycle 13: 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Navadgi VM, Mallikarjuna K, Rao BJ. 2005. Interaction of hRad51 and hRad52 with MCM complex: A cross-talk between recombination and replication proteins. Biochem Biophys Res Commun 329: 1240–1245. [DOI] [PubMed] [Google Scholar]
- Singh MK, Nicolas E, Gherraby W, Dadke D, Lessin S, Golemis EA. 2007. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene 26: 4825–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. 2008. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev 22: 2856–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. 2004. hMSH4–hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, et al. 2005. Severe expressive-language delay related to duplication of the Williams–Beuren locus. N Engl J Med 353: 1694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V. 2013. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54. [DOI] [PubMed] [Google Scholar]
- Souquet B, Abby E, Herve R, Finsterbusch F, Tourpin S, Le Bouffant R, Duquenne C, Messiaen S, Martini E, Bernardino-Sgherri J, et al. 2013. MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet 9: e1003784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A, Lichten M. 2008. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev 22: 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustelle C, Vedel M, Kolodner R, Nicolas A. 2002. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics 161: 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. 2012. Defining and detecting crossover-interference mutants in yeast. PLoS ONE 7: e38476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet 9: e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet 18: 74–82. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et al. 2005. A common inversion under selection in Europeans. Nat Genet 37: 129–137. [DOI] [PubMed] [Google Scholar]
- Stone JE, Ozbirn RG, Petes TD, Jinks-Robertson S. 2008. Role of proliferating cell nuclear antigen interactions in the mismatch repair-dependent processing of mitotic and meiotic recombination intermediates in yeast. Genetics 178: 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Schwacha A, Kleckner N. 1996. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci 93: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Poggeler S, Kleckner N, Zickler D. 2008. Coupling meiotic chromosome axis integrity to recombination. Genes Dev 22: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. 2010. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. 2005. SYCP3 mutations are uncommon in patients with azoospermia. Fertil Steril 84: 1019–1020. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. 1913a. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Zool 14: 43–59. [Google Scholar]
- Sturtevant AH. 1913b. A third group of linked genes in Drosophila ampelophila. Science 37: 990–992. [DOI] [PubMed] [Google Scholar]
- *.Subramanian VV, Hochwagen A. 2014. The meiotic checkpoint network: Step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6: a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara H, Iwabata K, Koshiyama A, Yanai T, Daikuhara Y, Namekawa SH, Hamada FN, Sakaguchi K. 2009. Coprinus cinereus Mer3 is required for synaptonemal complex formation during meiosis. Chromosoma 118: 127–139. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Syrjanen JL, Pellegrini L, Davies OR. 2014. A molecular model for the role of SYCP3 in meiotic chromosome organisation. eLife 3: 10.7554/eLife.02963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Székvölgyi L, Ohta K, Nicolas A. 2015. Initiation of meiotic homologous recombination: Flexibility, impact of histone modifications, and chromatin remodeling. Cold Spring Harb Perspect Biol 7: a016527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. 2010. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev 24: 2505–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana-Konwalski K, Godwin J, Borsos M, Rattani A, Adams DJ, Nasmyth K. 2013. Spindle assembly checkpoint of oocytes depends on a kinetochore structure determined by cohesin in meiosis I. Curr Biol 23: 2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Miyamoto N, Shouguchi-Miyata J, Ikeda JE. 2006. HFM1, the human homologue of yeast Mer3, encodes a putative DNA helicase expressed specifically in germ-line cells. DNA Seq 17: 242–246. [DOI] [PubMed] [Google Scholar]
- Tang S, Wu MK, Zhang R, Hunter N. 2015. Pervasive and essential roles of the Top3–Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol Cell 57: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Alveal C, Outwin EA, Trempolec N, Dziadkowiec D, Murray JM, O’Connell MJ. 2010. SMC complexes and topoisomerase II work together so that sister chromatids can work apart. Cell Cycle 9: 2065–2070. [DOI] [PubMed] [Google Scholar]
- Tay YD, Wu L. 2010. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J Biol Chem 285: 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tease C, Hulten MA. 2004. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res 107: 208–215. [DOI] [PubMed] [Google Scholar]
- Templado C, Uroz L, Estop A. 2013. New insights on the origin and relevance of aneuploidy in human spermatozoa. Mol Hum Reprod 19: 634–643. [DOI] [PubMed] [Google Scholar]
- Terasawa M, Ogawa H, Tsukamoto Y, Shinohara M, Shirahige K, Kleckner N, Ogawa T. 2007. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proc Natl Acad Sci 104: 5965–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci 100: 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Keeney S. 2009. PCH’ing together an understanding of crossover control. PLoS Genet 5: e1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield SE, Keeney S. 2012. Scale matters: The spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 11: 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby GG, Gherraby W, Coleman TR, Golemis EA. 2003. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol Cell Biol 23: 2109–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragon L. 2007. Anaphase onset before complete DNA replication with intact checkpoint responses. Science 315: 1411–1415. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. 2008. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev 22: 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. 2006. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev 20: 1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS. 2006. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell 10: 809–819. [DOI] [PubMed] [Google Scholar]
- Tsutsumi M, Fujiwara R, Nishizawa H, Ito M, Kogo H, Inagaki H, Ohye T, Kato T, Fujii T, Kurahashi H. 2014. Age-related decrease of meiotic cohesins in human oocytes. PLoS ONE 9: e96710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. 2008. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet 40: 90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, Kimura S, Yamamoto T, Ishibashi T, Sakaguchi K. 2004. Plant DNA polymerase λ, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur J Biochem 271: 2799–2807. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. 2011. Cohesin subunit Rad21L, the new kid on the block has new ideas. EMBO Rep 12: 183–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39: 14617–14625. [DOI] [PubMed] [Google Scholar]
- Verver DE, Langedijk NS, Jordan PW, Repping S, Hamer G. 2014. The SMC5/6 complex is involved in crucial processes during human spermatogenesis. Biol Reprod 91: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Johnston C, Thappeta Y, Subramanian VV, Hochwagen A, MacQueen AJ. 2015. Separable crossover-promoting and crossover-constraining of Zip1 activity during budding yeast meiosis. PLoS Genet 11: e1005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranis NM, Van der Heijden GW, Malki S, Bortvin A. 2010. Synaptonemal complex length variation in wild-type male mice. Genes 1: 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell 15: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Niu H, Futcher B, Zhang C, Shokat KM, Boulton SJ, Hollingsworth NM. 2008. Cdc28–Clb5 (CDK-S) and Cdc7–Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev 22: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N. 1999. Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci 96: 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, et al. 2009. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122: 2055–2063. [DOI] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z. 2010. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z. 2012a. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng Z, Huang J, Shi Q, Hong Y, Copenhaver GP, Gong Z, Ma H. 2012b. The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet 8: e1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zickler D, Kleckner N, Zhang L. 2015. Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14: 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, Griffin LB, Langlais KK, Backus VL, Schimenti KJ, O’Brien MJ, et al. 2007. Mutation in mouse Hei10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet 3: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. 2012. Geometry and force behind kinetochore orientation: Lessons from meiosis. Nat Rev Mol Cell Biol 13: 370–382. [DOI] [PubMed] [Google Scholar]
- Wehrkamp-Richter S, Hyppa RW, Prudden J, Smith GR, Boddy MN. 2012. Meiotic DNA joint molecule resolution depends on Nse5–Nse6 of the Smc5–Smc6 holocomplex. Nucleic Acids Res 40: 9633–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H Jr, Kneitz B, et al. 2003. Inactivation of exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev 17: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse HL. 1963. A theory of crossing-over by means of hybrid deoxyribonucleic acid. Nature 199: 1034–1040. [DOI] [PubMed] [Google Scholar]
- Wijnker E, Velikkakam James G, Ding J, Becker F, Klasen JR, Rawat V, Rowan BA, de Jong DF, de Snoo CB, Zapata L, et al. 2013. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H. 1930. Die konversion der gene. Gustav Fischer, Jena, Germany. [Google Scholar]
- Winter E. 2012. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 76: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme J, Angell RR. 2000. Maternal age and trisomy—A unifying mechanism of formation. Chromosoma 109: 435–438. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. 2006. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci 103: 4068–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wyatt HDM, West SC. 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 6: a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, Sarbajna S, Matos J, West SC. 2013. Coordinated actions of SLX1–SLX4 and MUS81–EME1 for Holliday junction resolution in human cells. Mol Cell 52: 234–247. [DOI] [PubMed] [Google Scholar]
- Xaver M, Huang L, Chen D, Klein F. 2013. Smc5/6-mms21 prevents and eliminates inappropriate recombination intermediates in meiosis. PLoS Genet 9: e1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. 2008. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev 22: 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Lu T, Zhou H, Bai L, Xiang Y. 2010. The role of MSH5 C85T and MLH3 C2531T polymorphisms in the risk of male infertility with azoospermia or severe oligozoospermia. Clin Chim Acta 411: 49–52. [DOI] [PubMed] [Google Scholar]
- Xue X, Choi K, Bonner JN, Szakal B, Chen YH, Papusha A, Saro D, Niu H, Ira G, Branzei D, et al. 2015. Selective modulation of the functions of a conserved DNA motor by a histone fold complex. Genes Dev 29: 1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ohta K. 2013. Initiation of meiotic recombination in chromatin structure. J Biochem 154: 107–114. [DOI] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, et al. 2008. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 22: 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Silber S, Leu NA, Oates RD, Marszalek JD, Skaletsky H, Brown LG, Rozen S, Page DC, Wang PJ. 2015. TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol Med 10.15252/emmm.201404967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O, Majumder S, Kramer B, Sekelsky JJ. 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ. 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y, Lane SI, Jones KT. 2014. Premature dyad separation in meiosis II is the major segregation error with maternal age in mouse oocytes. Development 141: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol Cell 40: 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, Alani E. 2009. The pch2Δ mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet 5: e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kim KP, Kleckner NE, Storlazzi A. 2011. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci 108: 20036–20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE. 2014a. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc Natl Acad Sci 111: E5059–E5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liang Z, Hutchinson J, Kleckner N. 2014b. Crossover patterning by the beam-film model: Analysis and implications. PLoS Genet 10: e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tang D, Luo Q, Chen X, Wang H, Li Y, Cheng Z. 2014c. Crossover formation during rice meiosis relies on interaction of OsMSH4 and OsMSH5. Genetics 198: 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. 2014d. Topoisomerase II mediates meiotic crossover interference. Nature 511: 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1999. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- *.Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Moreau PJ, Huynh AD, Slezec AM. 1992. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics 132: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]