SUMMARY
In multicellular organisms, cell death is a critical and active process that maintains tissue homeostasis and eliminates potentially harmful cells. There are three major types of morphologically distinct cell death: apoptosis (type I cell death), autophagic cell death (type II), and necrosis (type III). All three can be executed through distinct, and sometimes overlapping, signaling pathways that are engaged in response to specific stimuli. Apoptosis is triggered when cell-surface death receptors such as Fas are bound by their ligands (the extrinsic pathway) or when Bcl2-family proapoptotic proteins cause the permeabilization of the mitochondrial outer membrane (the intrinsic pathway). Both pathways converge on the activation of the caspase protease family, which is ultimately responsible for the dismantling of the cell. Autophagy defines a catabolic process in which parts of the cytosol and specific organelles are engulfed by a double-membrane structure, known as the autophagosome, and eventually degraded. Autophagy is mostly a survival mechanism; nevertheless, there are a few examples of autophagic cell death in which components of the autophagic signaling pathway actively promote cell death. Necrotic cell death is characterized by the rapid loss of plasma membrane integrity. This form of cell death can result from active signaling pathways, the best characterized of which is dependent on the activity of the protein kinase RIP3.
Apoptosis, autophagy, and necrosis are executed through distinct but interconnected signaling pathways. For example, autophagy is often potentiated by caspase activation, whereas RIP-dependent necrosis is antagonized.
1. INTRODUCTION
Although cell death can happen as a result of overwhelming damage, most cell deaths in animals occur in an active manner, as a consequence of specific signaling events. In general, there are three types of cell death, defined in large part by the appearance of the dying cell: apoptosis (also known as type I cell death), autophagic cell death (type II), and necrosis (type III) (Galluzzi et al. 2007).
Apoptosis is characterized by cell shrinkage, membrane blebbing, and condensation of the chromatin (pyknosis) (Kerr et al. 1972). It can be further defined as cell death accompanied by the activation of caspase proteases (Galluzzi et al. 2012). Two major signaling pathways trigger apoptotic cell death: the mitochondrial (the intrinsic) pathway and the death receptor (the extrinsic) pathway. The latter involves a classical ligand–cell-surface-receptor interaction. For example, cytotoxic lymphocytes can kill infected or transformed cells by expressing ligands for death receptors (DRs), a subset of the tumor necrosis factor (TNF) receptor (TNFR) family. These ligands induce apoptotic cell death of the targeted cells provided they express such DRs. DR-induced cell death in general is critical for immune system function and homeostasis. In contrast, the mitochondrial apoptotic pathway is usually initiated in a cell-autonomous manner. Most cellular stresses, such as DNA damage (induced by genotoxic agents or defects in DNA repair) or endoplasmic reticulum (ER) stress (induced by the accumulation of unfolded proteins), actively engage apoptosis when cells are damaged beyond repair.
Conversely, the lack of a signal, such as those activated by growth factors (e.g., cytokines and neurotrophic factors), can lead to cell death. This mechanism is critical for the development of the nervous system in vertebrates and it is estimated that half of the neurons generated die during this process (Buss et al. 2006). This cell death is due, in part, to the failure of some neuronal precursors to properly migrate or innervate their targets and the consequent lack of neurotrophic factor stimulation. Similarly, during an immune response, cytokine deprivation (together with the DR pathway) is responsible for the acute contraction of the lymphocyte population after clearance of the pathogen. Another example of “loss-of-signal”-induced cell death is the particular form of apoptosis called anoikis, which occurs when epithelial or endothelial cells detach from the extracellular matrix (ECM). In this scenario, unligated ECM receptors of the integrin family cease to induce prosurvival signaling pathways, eventually leading to apoptosis. This mechanism prevents cells shedding from their original location from colonizing elsewhere (a characteristic of metastatic cancer cells). Finally, apoptosis can be induced by oncogenes (e.g., Myc) as a safeguard mechanism against cancer development. This process is controlled in part by a p53-dependent apoptotic pathway, which is activated in response to aberrant mitogenic signals resulting from oncogene overexpression or mutation. As a consequence, evasion of apoptotic cell death is often a requisite to sustain oncogene transformation (see Sever and Brugge 2014).
Autophagic cell death is characterized by the appearance of large intracellular vesicles and engagement of the autophagy machinery. Note that although autophagy (i.e., the membrane engulfment and catabolic degradation of parts of the cytoplasm) is a well-defined process, its function as an active cell death mechanism remains highly controversial. Autophagy is mainly a survival process engaged in response to a metabolic crisis (e.g., low ATP levels and nutrient and amino acid deprivation) or to remove damaged organelles (e.g., mitochondria with low membrane potential) and protein aggregates. As a stress response, autophagy accompanies rather than promotes cell death in most scenarios and merely represents a failed survival attempt (Shen et al. 2012). Nevertheless, there are specific examples in which the autophagy machinery is absolutely required for cell death. During Drosophila metamorphosis, obsolete larval tissues such as the midgut and salivary glands regress through massive autophagic cell death, a process triggered by the steroid hormone ecdysone. In this particular case, a deficiency in genes of the autophagic signaling pathway alters the cell death program (Berry and Baehrecke 2007; Denton et al. 2009). Autophagic cell death has also been reported in response to deregulated H-Ras activity and could therefore represent a safeguard mechanism against oncogenic transformation (Elgendy et al. 2011).
Necrosis is characterized by cell swelling and plasma membrane rupture, and a loss of organellar structure without chromatin condensation. Although necrosis can occur as a consequence of irreparable cell damage, at least one pathway of active necrosis exists. This form of cell death, sometimes called necroptosis, is engaged by several signaling pathways that all converge on the activation of receptor-interacting protein kinase 3 (RIP3). RIP3 is activated upon recruitment to macromolecular complexes downstream from various cell-surface receptors: DRs, Toll-like receptors (TLRs), and the T-cell receptor (TCR). Additionally, DNA damage can directly induce the formation of a RIP3-activation platform, independently of cell-surface receptor ligation. Finally, RIP3-dependent necrosis is also triggered by the cytosolic DNA sensor, DNA-dependent activator of interferon (DAI) regulatory factors, following virus infection and the presence, in the cytosol, of double-stranded viral DNA.
Here we are concerned with signaling leading to cell death in vertebrates, and focus on processes that are at least partially understood at the molecular level. We do not discuss the physiology and pathology of cell death in detail or processes involved in the clearance of dying cells. Readers will find a more complete overview of these topics and other aspects of cell death elsewhere (Green 2011b).
2. TYPE I CELL DEATH: APOPTOSIS
Apoptotic cell death is predominantly initiated either by the DR or mitochondrial pathway, although additional pathways exist. DRs—for example, Fas (also known as CD95), Trail receptor (TRAIL-R), or TNFR1—induce apoptosis by directly recruiting a caspase-activation platform upon binding to their respective ligand. The mitochondrial pathway of apoptosis, on the other hand, is triggered upon loss of integrity of the mitochondrial outer membrane, which allows the release of proapoptotic factors (e.g., cytochrome c) from the mitochondria into the cytosol. This process is controlled by the Bcl2 protein family. Once in the cytosol, cytochrome c induces the assembly of a caspase-activation complex: the apoptosome. Both pathways culminate in the activation of caspase proteases and the cleavage of intracellular proteins, ultimately leading to the dismantling of the cell. Below, we explore these processes in detail, starting with the end point: caspase activation.
2.1. Caspase Activation, Function, and Regulation
Apoptosis involves the activation of caspases, which orchestrate all of the morphological changes that characterize this form of cell death. Caspases are cysteine proteases with specificity for aspartic acid residues in their substrates. Although at least 17 different caspases exist in mammals, our focus is on only a subset of these for which the activation is at least partially understood and roles in cell death have been established.
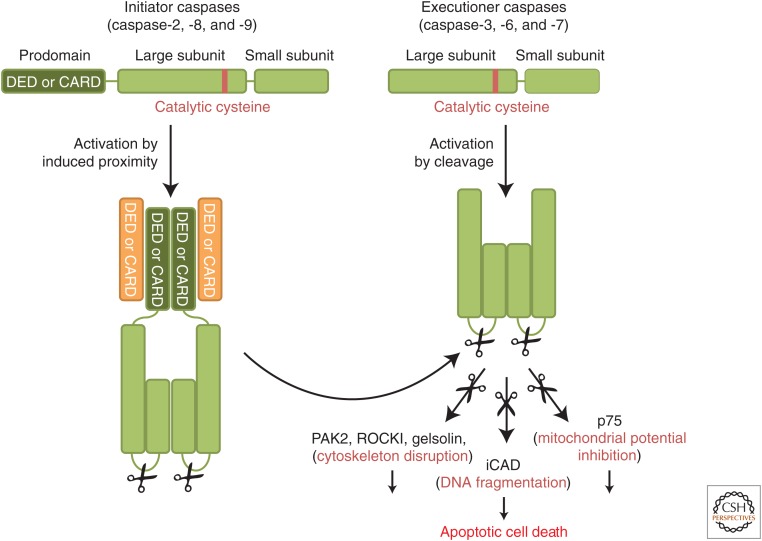
The executioner caspases (caspase-3, caspase-6, and caspase-7) effect the destruction and are produced as inactive dimers that lack protein-interaction domains (Fig. 1). Activation is due to proteolytic cleavage between what will be the large and small subunits of the mature enzyme (Salvesen and Riedl 2008). Upon cleavage, the new ends fold into the dimer interface and promote conformational changes to create two active sites in the mature protease. Cleavage of caspase-6 is mediated by caspase-3 and caspase-7 (Slee et al. 1999), whereas activation of the latter two caspases is generally the function of “initiator” caspases. It is these initiator caspases, and their activation, that define the apoptotic signaling pathways (see below).
Figure 1.
The caspase protein family. Initiator caspases (caspase-2, caspase-8, and caspase-9) are the apical caspases of the apoptotic-signaling cascade. Initiator caspases are produced as inactive zymogens composed of a prodomain (containing a CARD or a death effector domain [DED]) and a large and small subunit. They are recruited through their prodomains into large activation platforms and activated by dimerization. In contrast, executioner caspases (caspase-3, caspase-6, and caspase-7) are activated by cleavage of the zymogen between the large and small subunits and are therefore dependent on initiator caspases for their activation. Catalytically active caspases are composed of a heterotetramer of two small and two large subunits.
Following activation, the executioner caspases, particularly caspase-3 and caspase-7, can process at least 1000 proteins (Crawford and Wells 2011). The cleavage of these caspase substrates can result in either a gain or a loss of function of these proteins and eventually leads to the cellular changes associated with apoptosis. Notably, caspase proteolysis inactivates components of essential physiological processes. For example, caspase cleavage of the p75 subunit of complex I of the electron transport chain disrupts the mitochondrial transmembrane potential, electron transport, and ATP production during apoptosis (Ricci et al. 2004). Conversely, caspase cleavage can also activate specific pathways. This is the case for caspase-activated nuclease (CAD), a DNAse that cuts chromatin between nucleosomes when its inhibitor, iCAD, is cleaved by caspase-3 (Enari et al. 1998; Sakahira et al. 1998). Additionally, caspases can hijack signaling pathways through constitutive activation of some of their components. For example, the characteristic morphology of apoptotic cells is caused by caspase-mediated activation of several actin cytoskeleton modulators: gelsolin, p21-activated kinase 2 (PAK2), and Rho-associated kinase I (ROCKI). Gelsolin, a calcium-regulated actin-severing protein, becomes constitutively active following caspase processing (Kothakota et al. 1997). PAK2 and ROCK I are serine–threonine kinase effectors of the Rho GTPase family (Rac1, Cdc42, and Rho) that regulate actin polymerization and actin–myosin contractility. Upon caspase cleavage, their kinase activity becomes independent of the GTPases, thus inducing an aberrant reorganization of the actin cytoskeleton and the characteristic membrane blebbing observed in apoptotic cells (Rudel and Bokoch 1997; Coleman et al. 2001; Sebbagh et al. 2001) (for more details on cytoskeleton-modulating proteins, see Devreotes and Horwitz 2014).
Unlike the executioner caspases, initiator caspases (Fig. 1) exist as inactive monomers in cells and are not activated by cleavage. Instead, adaptor molecules that assemble into caspase-activation platforms recruit these initiator caspases, forcing monomers into close proximity and causing conformational changes that result in the formation of active sites. In some (but not all) cases, subsequent autocleavage of the caspase is necessary to stabilize the mature, active enzyme (Pop et al. 2007; Oberst et al. 2010). Note that although cleavage of an executioner caspase is indicative of its activation, that of initiator caspases is not necessarily an indication of activation (McStay et al. 2008).
The interactions between caspase-activation platforms and caspases involve “death fold” domains of the proteins (Kersse et al. 2011). Such death fold elements are present in adaptor proteins and caspases: the caspase-recruitment domain (CARD), and the death effector domain (DED). Other death folds, the death domain (DD) and the pyrin (PYR) domain, are involved in the assembly of some caspase-activation platforms but not present on caspases (Kersse et al. 2011). Death fold domains typically mediate protein–protein interaction through homotypic interaction. They do not share sequence similarity but have similar structures composed of six amphipathic α helices.
2.2. Caspase-9 Activation: The Mitochondrial Pathway of Apoptosis
The mitochondrial pathway of apoptosis, also called the intrinsic pathway, is the most common mechanism of apoptosis in vertebrates. It is activated in response to a variety of cellular stresses, including DNA damage, growth factor deprivation, ER stress, and developmental cues. In this pathway, the executioner caspases are cleaved and activated by caspase-9, which is itself activated by a caspase-activation platform called the apoptosome (Fig. 2) (Bratton et al. 2001; Bratton and Salvesen 2010).
Figure 2.
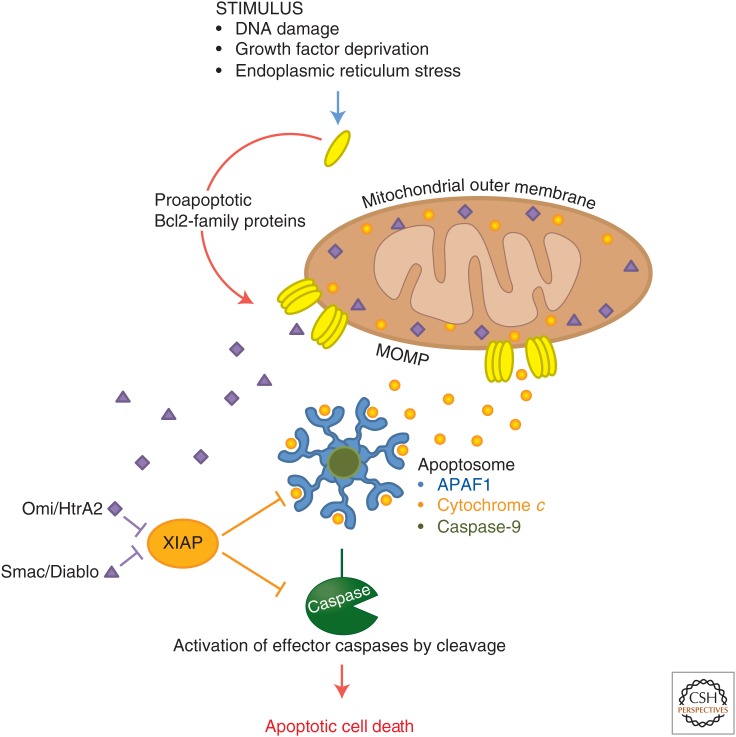
The mitochondrial apoptotic pathway. In response to various cellular stresses, proapoptotic members of the Bcl2 family induce mitochondrial outer membrane permeabilization (MOMP), allowing the release into the cytosol of proapoptotic factors that are normally sequestered in the intermembrane space of the mitochondria (including cytochrome c, Smac, and Omi). In the cytosol, cytochrome c binds to APAF1 and triggers its oligomerization. Caspase-9 is then recruited and activated by this platform, known as the apoptosome. Catalytically active caspase-9 cleaves and activates the executioner caspases-3 and -7. Upon release in the cytosol, Smac and Omi bind to and inhibit the caspase inhibitor X-linked inhibitor of apoptosis (XIAP). In doing so, they relieve XIAP inhibition of caspase-9, caspase-3, and caspase-7, and potentiate overall caspase activation by the apoptosome.
Apoptotic protease-activating factor 1 (APAF1) constitutes the scaffold around which the apoptosome is assembled. During intrinsic apoptosis, cytochrome c is released from mitochondria into the cytosol (see below) and binds to Apaf1 (Zou et al. 1997). This interaction triggers hydrolysis of the Apaf1 cofactor dATP to dADP (Kim et al. 2005). The subsequent exchange of dADP with exogenous dATP allows the oligomerization of seven APAF1–dATP–cytochrome-c units into an active apoptosome. At the center of the apoptosome, exposed CARDs on APAF1 bind to the CARD of caspase-9, thus bringing the inactive caspase-9 monomers into close proximity for activation and autoprocessing (Yu et al. 2005; Yuan et al. 2010). Owing to its higher affinity for the apoptosome, full-length caspase-9 displaces the processed form, creating a continuous cycle of caspase-9 recruitment, activation, processing, and release (Malladi et al. 2009). Because caspase-9 only sustains catalytic activity in this bound state (Rodriguez and Lazebnik 1999; Stennicke et al. 1999; Bratton et al. 2001), the apoptosome functions as a molecular timer in which its lifetime is directly proportional to the amount of unprocessed caspase-9 present (Malladi et al. 2009).
In healthy cells, cytochrome c is found only in the mitochondrial intermembrane space. For it to interact with APAF1, mitochondrial outer membrane permeabilization (MOMP) triggered by apoptotic stimuli must occur (Tait and Green 2010). MOMP induces the release of all soluble proteins of the mitochondrial intermembrane space into the cytosol. In addition to cytochrome c, two other proapoptotic proteins are released during that process: Smac (also known as Diablo) and Omi (also known as HtrA2). Smac and Omi potentiate the apoptosome activity by antagonizing the caspase inhibitor X-linked inhibitor of apoptosis (XIAP) (Fig. 2). In the absence of Smac and Omi, XIAP binds to, and inhibits the catalytic activity of, the initiator caspase-9 and the executioner caspases-3 and -7. XIAP further dampens intrinsic apoptosis induction through direct ubiquitylation and proteasomal degradation of active caspases (Eckelman et al. 2006).
MOMP is a tightly regulated event controlled by members of the Bcl2 family. These share one or more Bcl2 homology (BH) regions defined by sequence, structure, and function (Fig. 3A). There are three broad classes of Bcl2 proteins: the proapoptotic effector proteins (Bax and Bak), which are necessary and sufficient for MOMP; the antiapoptotic Bcl2 proteins (e.g., Bcl2, Bcl-xL, and Mcl1), which block MOMP; and the BH3-only proteins (e.g., Bid, Bim, Bad, and Noxa), which activate the proapoptotic effectors and/or neutralize the antiapoptotic Bcl2 proteins. Bcl2 proteins also control several other cellular processes, including mitochondrial fusion, autophagy, and calcium efflux from the ER (Chipuk et al. 2010).
Figure 3.
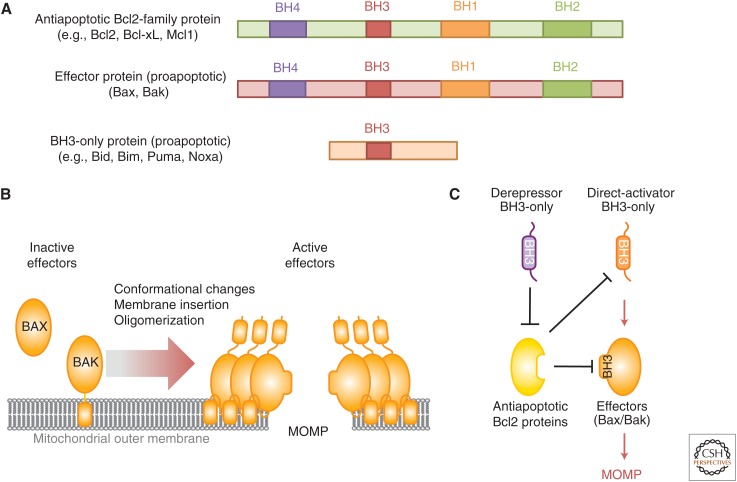
Regulation of mitochondrial outer membrane integrity by the Bcl2 protein family. (A) Members of the Bcl2 protein family are characterized by the presence of one or more Bcl2 homology (BH) region. The antiapoptotic Bcl2 proteins (e.g., Bcl2, Bcl-xL, and Mcl1) and the proapoptotic effectors (e.g., Bax and Bak) share four BH regions and a similar globular structure. BH3-only proteins (e.g., Bid, Bim, Bad, and Noxa) are characterized by a single BH region (BH3). (B) The proapoptotic effectors reside in cells in inactive forms tethered to the outer mitochondrial membrane (Bak) or soluble in the cytosol (Bax). Upon activation, Bax and Bak oligomerize and further insert into the membrane, causing mitochondrial outer membrane permeabilization (MOMP) and thus apoptosis. (C) Bax/Bak activation is triggered following the transient binding of a subset of direct activator BH3-only proteins (e.g., Bid and Bim). Antiapoptotic Bcl2 proteins inhibit MOMP by sequestering the direct activator proteins and/or the effectors. Another group of BH3-only proteins, called sensitizers or derepressors (e.g., Bad and Noxa) promote MOMP by antagonizing antiapoptotic Bcl2 proteins, thereby releasing both direct activator BH3-only proteins and Bax/Bak.
Bax and Bak are directly responsible for the loss of mitochondrial outer membrane integrity (Fig. 3B). Upon activation, they form large oligomers that insert into the mitochondrial outer membrane, disrupting it (Eskes et al. 2000; Korsmeyer et al. 2000; Dewson et al. 2008, 2009). The precise nature of the disruption remains unclear, but it allows the near simultaneous release of all intermembrane space proteins (Goldstein et al. 2000; Munoz-Pinedo et al. 2006).
Bax and Bak act redundantly in MOMP and at least one of them is required to permeabilize mitochondria. In living cells these proteins are generally inactive, but become activated in response to upstream events. At least two of the BH3-only proteins (Bim and active Bid) activate Bax and Bak through transient interaction (Fig. 3C), although other conditions (such as heat, changes in pH, and changes in the lipid milieu) may activate the effectors independently of BH3-only proteins (Wei et al. 2000; Kuwana et al. 2002, 2005; Letai et al. 2002).
MOMP is antagonized by the antiapoptotic Bcl2 proteins, which bind to and inhibit both Bax/Bak and the BH3-only proteins by interacting with their BH3 domains (Llambi et al. 2011) (Fig. 3C). Antiapoptotic Bcl2 proteins are also regulated at both the transcriptional and posttranslational levels. In particular, Mcl1 degradation by the ubiquitin-proteasome system participates in apoptosis induction following several cellular stresses. Upon DNA damage, the BH3-domain-containing protein Mcl1 ubiquitin ligase E3 (MULE) directly binds to Mcl1 and catalyzes its ubiquitylation and degradation (Warr et al. 2005). During antitubulin chemotherapeutic-induced mitotic arrest, Mcl1 is phosphorylated by stress-activated and mitotic kinases such as the MAP kinase (MAPK) Jun amino-terminal kinase (JNK), casein kinase II (CKII), and p38 MAPK. These phosphorylation events unveil a degron on Mcl1 that recruits the SCF-Fbw7 ubiquitin ligase complex, thus targeting Mcl1 for ubiquitylation and degradation (Wertz et al. 2011). Similarly, Mcl1 is degraded following growth factor withdrawal (e.g., IL3) and the subsequent loss of phosphoinositide 3-kinase (PI3K)-Akt signaling. This process relieves glycogen synthase kinase 3 (GSK3) from Akt inhibition. GSK3 then phosphorylates Mcl1, allowing its ubiquitylation by the E3 ligase β-transductin-repeat-containing protein (β-TrCP) and its subsequent degradation (Maurer et al. 2006; Ding et al. 2007). Conversely, cancer cells can increase Mcl1 stability and overall resistance to cellular stress by expressing USP9X, a deubiquitylase that removes polyubiquitin chains from Mcl1 (Schwickart et al. 2010).
Proteins of the Bcl2 family integrate pro- and antiapoptotic signals in healthy and stressed cells and therefore constitute one of the main signaling nodes in the life or death decision. Within this family, BH3-only proteins constitute the main upstream sensors of the mitochondrial apoptotic pathway. A wide variety of signaling pathways converge on the BH3-only family of proteins and regulate their expression level and activity both transcriptionally and posttranscriptionally (see Table 1). For example, Bid is activated upon cleavage by caspase-8 following DR ligation (Li et al. 1998, Luo et al. 1998). In doing so, Bid coordinates the cross-regulation between the extrinsic and intrinsic apoptotic pathways. Additionally, proteolytic activation of Bid can be achieved by granzyme B during the cytotoxic lymphocyte killing process (Heibein et al. 2000; Sutton et al. 2000). The apoptotic response to genotoxic stress is performed, in part, by Puma and Noxa (Jeffers et al. 2003; Villunger et al. 2003). Both are direct transcriptional targets of the tumor suppressor p53 (Oda et al. 2000; Nakano and Vousden 2001; Yu et al. 2003), although other stimuli regulate their expression levels (see Table 1). Similarly, Bim is transcriptionally up-regulated by the forkhead transcription factor FOXO3A upon cytokine deprivation (Dijkers et al. 2000; Gilley et al. 2003; Urbich et al. 2005). Bim is also a major apoptotic factor of the ER stress pathway engaged in response to accumulation of unfolded proteins (Puthalakath et al. 2007). Finally, Bad activity is negatively regulated through phosphorylation by several kinases, such as Akt (Datta et al. 1997; del Peso et al. 1997), which induces its sequestration by 14-3-3 proteins. Upon growth factor deprivation and loss of Akt signaling, Bad is released and antagonizes antiapoptotic Bcl2 proteins (Zha et al. 1996; Datta et al. 1997).
Table 1.
Signaling to BH3-only proteins
| BH3-only protein | Stimulus/input | Type of regulation | Signaling pathway | Output | References |
|---|---|---|---|---|---|
| Bad | Growth factors and cytokines | Phosphorylation | Akt | Cell survival | Datta et al. 1997, 2002; del Peso et al. 1997 |
| Phosphorylation | p70 S6K | Cell survival | Harada et al. 2001 | ||
| Phosphorylation | PKA | Cell survival | Harada et al. 1999 | ||
| Phosphorylation | PIM kinases | Cell survival | Fox et al. 2003; Yan et al. 2003 | ||
| Phosphorylation | PAK | Cell survival | Schürmann et al. 2000; Cotteret et al. 2003 | ||
| Sequestration | 14-3-3 | Cell survival | Zha et al. 1996; Datta et al. 2000 | ||
| TNF | Phosphorylation | IKK | Cell survival | Yan et al. 2013 | |
| Neuronal activity deprivation | Phosphorylation | Cdc2 | Apoptosis | Konishi et al. 2002 | |
| Growth factor or cytokine deprivation | Phosphorylation | JNK | Apoptosis | Donovan et al. 2002 | |
| Dephosphorylation | PP1, PP2A, and PP2C | Apoptosis | Ayllón et al. 2000; Chiang et al. 2001; Klumpp et al. 2003 | ||
| Calcium | Dephosphorylation | Calcineurin | Apoptosis | Wang et al. 1999 | |
| Bid | Cytotoxic T cell | Cleavage | Granzyme B | Apoptosis | Heibein et al. 2000; Sutton et al. 2000 |
| Fas/TNF/TRAIL | Cleavage | Caspase-8 | Apoptosis | Li et al. 1998; Luo et al. 1998 | |
| Heat shock or ER stress | Cleavage | Caspase-2 | Apoptosis | Bonzon et al. 2006; Upton et al. 2008 | |
| Ischemia or cisplatin | Cleavage | Calpain | Apoptosis | Chen et al. 2001; Mandic et al. 2002 | |
| Lysosome permeabilization | Cleavage | Cathepsin | Apoptosis | Stoka et al. 2001; Reiners et al. 2002 | |
| Bim | Growth factors or cytokines | Phosphorylation | ERK1/2 and RSK1/2 | Cell survival | Ley et al. 2003; Hubner et al. 2008; Dehan et al. 2009 |
| Ubiquitylation | βTrCP | Cell survival | Akiyama et al. 2003; Ley et al. 2003; Dehan et al. 2009 | ||
| mRNA stability | Hsc70 | Cell survival | Matsui et al. 2007 | ||
| ? | mRNA stability | miR-17-92 | Cell survival | Xiao et al. 2008 | |
| ER stress | Transcription | CHOP-C/EBPa | Apoptosis | Puthalakath et al. 2007 | |
| Dephosphorylation | PP2A | Apoptosis | Puthalakath et al. 2007 | ||
| Glucocorticoid | Transcription | GR | Apoptosis | Erlacher et al. 2005; Ploner et al. 2008 | |
| Growth factor deprivation or DNA damage | Phosphorylation | JNK | Apoptosis | Lei and Davis 2003; Putcha et al. 2003 | |
| Growth factor or cytokine deprivation | Transcription | FOXO3a | Apoptosis | Dijkers et al. 2000; Gilley et al. 2003 | |
| Noxa | DNA damage | Transcription | p53 | Apoptosis | Oda et al. 2000; Villunger et al. 2003; Naik et al. 2007 |
| DNA damage | Transcription | p63 | Apoptosis | Kerr et al. 2012 | |
| DNA damage | Deubiquitylation | UCH-L1 | Apoptosis | Brinkmann et al. 2013 | |
| Glucose deprivation | Transcription | ? | Apoptosis | Alves et al. 2006 | |
| Hypoxia | Transcription | HIF1α | Apoptosis | Kim et al. 2004 | |
| Proteasome inhibition | Transcription/protein stabilization | Myc, others | Apoptosis | Fernandez et al. 2005; Qin et al. 2005; Nikiforov et al. 2007 | |
| Puma | Cytokine deprivation | Transcription | FOXO3a | Apoptosis | Jeffers et al. 2003; Villunger et al. 2003; You et al. 2006; Ekoff et al. 2007 |
| DNA damage | Transcription | p53 | Apoptosis | Nakano and Vousden 2001; Yu et al. 2001; Jeffers et al. 2003; Villunger et al. 2003 | |
| DNA damage or ER stress | Transcription | p63 | Apoptosis | Pyati et al. 2011; Kerr et al. 2012 | |
| Glucocorticoid | Transcription | GR | Apoptosis | Villunger et al. 2003; Erlacher et al. 2005 | |
| BIK | DNA damage | Transcription | Smad | Proapoptotic | Spender 2009 |
| TGFβ | Transcription | E2F1 | Proapoptotic | Real 2006 | |
| Ubiquitylation? | Proapoptotic | Nikrad 2005; Zhu 2005 | |||
| Proteasome inhibition | Cleavage | RHBDD1 | Antiapoptotic | Wang et al. 2008a | |
| Phosphorylation | Casein kinase | Proapoptotic | Verma 2001; Li 2003 | ||
| Transcription | Smad | Proapoptotic | Ramjaun 2007 | ||
| BMF | TGFβ | Sequestration | DLC2 | Antiapoptotic | Puthalakath 2001 |
| Transcription | HIFα | Proapoptotic | Bruick 2000; Guo 2001 | ||
| BNIP3 | Hypoxia | Transcription | E2F1 | Proapoptotic | Yurkova 2008 |
| Transcription inhibition | NF-κB | Antiapoptotic | Shaw 2006, 2008 | ||
| Transcription | Jun | Proapoptotic | Ma 2007 | ||
| HRK | Potassium deprivation | Transcription | E2F1 | Proapoptotic | Hershko 2004 |
| Transcription inhibition | DREAM | Antiapoptotic | Sanz 2001 | ||
| Cytokines | Transcription | HIFα | Proapoptotic | Sowter 2001 | |
| NIX | Hypoxia |
MOMP often condemns a cell to death even if caspase activation is blocked or disrupted (Tait and Green 2010). This is probably the consequence of a catastrophic loss of mitochondrial function that results in bioenergetic failure (Lartigue et al. 2009). Under these conditions, some cells can recover from MOMP, however, and survive (Tait et al. 2010). Therefore, the regulation of caspase activation downstream from MOMP may be important in some settings. For example following cytochrome c binding to APAF1, the failure to replace hydrolyzed dADP by exogenous dATP triggers nonfunctional aggregation and irreversible inactivation of APAF1 (Kim et al. 2005). Nucleotide exchange, and therefore functional apoptosome assembly, is facilitated by a complex composed of heat shock protein 70 (Hsp70), cellular apoptosis susceptibility (CAS) protein, and putative HLA-DR-associated protein I (PHAPI) (Kim et al. 2008). Accordingly, cellular levels of these proteins modulate caspase-activation efficiency during intrinsic apoptosis. tRNA levels also regulate apoptosome activity by binding to cytochrome c, blocking its interaction with APAF1 (Mei et al. 2010). Finally, intrinsic apoptosis can be perturbed downstream from MOMP through phosphorylation and inhibition of caspase-9 (see below) (Allan and Clarke 2009).
2.3. Caspase-8 Activation: The Death Receptor Pathway
Caspase-8 is activated predominantly by the DR pathway of vertebrate apoptosis. DRs are a subset of the TNFR superfamily and include TNFR1, Fas, and TRAIL-R1/2 (Dickens et al. 2012). They contain a DD in their intracellular regions. Through a series of homotypic interactions, these initiate the assembly of large macromolecular complexes that recruit and activate caspase-8 for apoptotic signaling, as well as other signaling molecules that control processes such as inflammation and cell adhesion (Newton and Dixit 2012).
When Fas or a TRAIL-R is bound by its ligand, clustering of the receptors recruits a DD-containing adaptor molecule, FADD, through DD-DD interactions (Fig. 4A). This exposes another death fold domain in FADD, its DED. The complex represents a caspase-activation platform called a death-inducing signaling complex (DISC), as the FADD DEDs bind to DEDs in the prodomain of caspase-8. This brings caspase-8 monomers into close proximity, triggering their protease activity. Caspase-8 then undergoes autocatalytic cleavage both between the large and small subunits and between the large subunit and the prodomain. The first cleavage stabilizes the active dimer (and is required for homodimer activity in this pathway); the second cleavage releases it from the DISC (Dickens et al. 2012).
Figure 4.
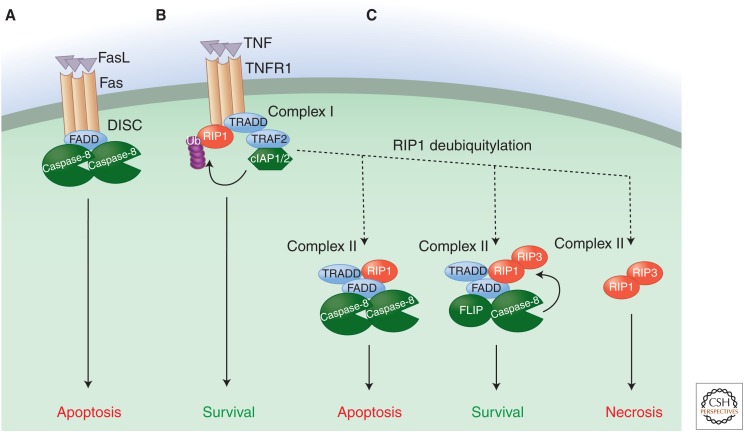
Death receptor signaling pathway. The death receptor signaling pathway is triggered by ligation of a death receptor (TNFR1, Fas, or TRAIL-R1/2) and potentially leads to three different outcomes: survival, apoptosis, or necrosis. (A) Upon ligation (by FasL and TRAIL, respectively), Fas/CD95 and TRAIL-Rs assemble a caspase-activation platform called the DISC. This platform recruits and activates caspase-8 via the adaptor protein FADD and thus engages the extrinsic apoptotic pathway. (B) Upon ligation to TNF, TNFR1 recruits the adaptor protein TRADD and the kinase RIP1, which in turn mobilizes additional partners, such as the ubiquitin ligases TRAF2 and cIAP1/2. These catalyze the nondegradative ubiquitylation of RIP1 as well as other components of the protein complex, resulting in the stabilization of a prosurvival and proinflammatory signaling platform called complex I. (C) The removal of ubiquitin chains of RIP1 by inhibition of cIAP1/2 or through the action of deubiquitylases destabilizes complex I, thus allowing the release of TRADD and RIP1. TRADD and RIP1 then promote the formation of a series of cytosolic complexes (complex II) that can initiate either apoptotic or necrotic cell death. In the proapoptotic configuration of the complex, TRADD and RIP1 recruit FADD and caspase-8, resulting in caspase activation and apoptosis. When FADD or caspase-8 are absent, or when caspase activity is blocked, RIP1 binds to and activates RIP3, thus triggering necrotic cell death. Expression of FLICE-like inhibitory protein (FLIP) inhibits the activity of both complexes. FLIP forms a heterodimer with caspase-8 that cannot induce apoptosis but still displays catalytic activity, and this activity antagonizes the activation of RIP3. Therefore, complexes containing FLIP–caspase-8 heterodimers simultaneously block apoptosis and RIP-dependent necrosis.
In some cells (called type I cells), active caspase-8 then promotes apoptosis by cleaving and activating caspase-3 and caspase-7. However, in many cell types (type II cells), the active executioner caspases are inhibited by XIAP, and thus apoptosis is blocked (Jost et al. 2009). In these cases, another caspase-8 substrate comes into play, the BH3-only protein Bid (see above). Caspase-8-mediated cleavage activates Bid, which in turn activates Bax and Bak to promote MOMP. The IAP antagonists Smac and Omi are then released, and these neutralize XIAP to allow apoptosis to proceed.
The signaling pathways engaged downstream from TNFR1 are more complex and potentially lead to three different outcomes: survival, apoptosis or necrosis (Fig. 4B). Upon TNFR1 ligation, a different DD-containing adaptor, TNFR-associated death domain protein (TRADD), is first recruited. TRADD does not directly bind or activate caspase-8 but instead acts as a membrane-bond scaffold for the recruitment of additional signaling molecules, including a kinase, receptor-interacting protein 1 (RIP1), and the ubiquitin ligases TRAF2 and cIAP1/2. Components of this complex, including RIP1, are modified by a complex series of nondegradative ubiquitylation events performed by both the cIAPs and the linear ubiquitin assembly complex (LUBAC). The assembly and ubiquitylation of this signaling platform (referred to as complex I) culminates in the activation of the NF-κB signaling pathway and an inflammatory response rather than cell death (Newton and Dixit 2012).
TNFR1 ligation leads to cell death when ubiquitylation of RIP1 is compromised either by inhibition of ubiquitin ligases contained in complex I (e.g., cIAP1/2) or through direct de-ubiquitylation of RIP1 by enzymes such as CYLD (and probably other signaling events). RIP1 and TRADD are then released from TNFR1 to form a series of dynamic cytosolic signaling platforms (collectively referred to as complex II) that have the potential to cause both apoptotic and necrotic cell death (Fig. 4C). Cytosolic TRADD recruits FADD (via DD-DD interaction), which in turn can bind to and activate caspase-8 similarly to the DISC (Fig. 4C, left panel) (Christofferson and Yuan 2010; Declercq et al. 2009). In this configuration, complex II formation leads to apoptotic cell death. In many cases, however, apoptosis does not proceed upon TNFR1 ligation, at least in part because the activation of NF-κB induces the expression of the caspase-8-like protein FLICE-like inhibitory protein (FLIP) (Micheau et al. 2001). FLIP has a domain structure similar to that of caspase-8, but it lacks a catalytic cysteine. It preferentially binds to FADD (via DED-DED interactions) and a monomer of caspase-8. The caspase-8–FLIP heterodimer is catalytically active (Micheau et al. 2002; Pop et al. 2011), but does not promote apoptosis. Why this is remains obscure, but may relate to more rapid degradation of the complex (Geserick et al. 2009). Consequently, the assembly of complex II under conditions of active NF-κB signaling, and therefore FLIP expression, leads to a survival outcome (Fig. 4C, middle panel). Finally, complex II formation can engage a necrotic form of cell death (see section below). As we will see below, the activity of the caspase-8–FLIP heterodimer is also important for preventing this necrotic cell death signaling engaged by ligated TNFR1.
2.4. Caspase -1 and Caspase-5/-11 Activation: The Inflammasome Pathway
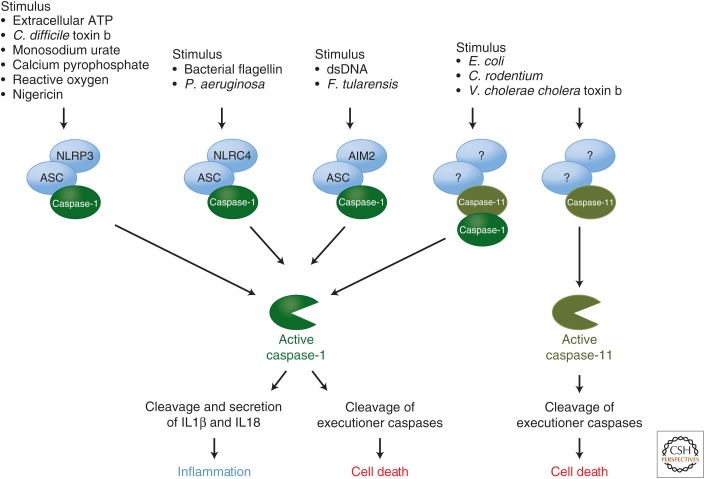
Caspase-1 and human caspase-5 (caspase-11 in rodents) have large prodomains containing CARDs. These are engaged by caspase-activation platforms called inflammasomes. Generally these platforms form in response to infectious agents (e.g., viruses, bacteria, and fungi) and inert substances that induce inflammation (e.g., uric acid crystals, calcium phosphate crystals, alum, asbestos) (Franchi et al. 2012).
The platforms that engage caspase-1 are established by activated NOD-like receptors (NLRs), which bear either a CARD or, more often, a PYR domain (Fig. 5). In most cases, the NLR engages the adaptor molecule ASC via PYR–PYR interactions. ASC also contains a CARD that binds to and activates caspase-1 (Martinon et al. 2002; Faustin et al. 2007; Franchi et al. 2009).
Figure 5.
The inflammasomes. The inflammasomes are caspase-activation platforms that assemble in response to infectious agents (e.g., viruses, bacteria, and fungi) and inert substances that induce inflammation (e.g., uric acid crystals, calcium phosphate crystals, alum, asbestos). They recruit and activate the inflammatory caspase-1 and -11. The caspase-1 activation platforms are supported by the NOD-like receptors (NLRs; e.g., NLRP3 and NLRC4) and AIM. Upon activation by inflammatory agents, these proteins recruit the adaptor molecule ASC and caspase-1. The subsequent activation of caspase-1 induces cleavage and secretion of two inflammatory cytokines, interleukin (IL) 1β and IL18.
Another caspase-1 inflammasome involves the sensor AIM2, which binds to cytosolic DNA (e.g., from viruses) and also engages ASC (Fig. 5) (Hornung et al. 2009). Caspase-1 cleaves and promotes the secretion of two inflammatory cytokines, interleukin (IL) 1β and IL18. It can also cleave and activate Bid, as well as caspase-3 and caspase-7 to promote apoptosis. This form of caspase-1-mediated cell death is sometimes called pyroptosis (Brennan and Cookson 2000; Cookson and Brennan 2001; Fink and Cookson 2006; Bergsbaken and Cookson 2007).
Little is known about the activation platform for caspase -5/-11, except that it does not involve ASC or the NLRs that function in caspase-1 activation (Kayagaki et al. 2011). Nevertheless, caspase-11 activation can result in apoptosis (Kayagaki et al. 2011). Caspase-11 can probably also bind to and activate caspase-1 (Green 2011a). In addition, caspase-11, but not caspase-1, is involved in the lethal effects of bacterial lipolysaccharides in mice in vivo (endotoxemia) (Kayagaki et al. 2011), which may also involve one of the executioner caspases, caspase-7 (Lamkanfi et al. 2009).
2.5. Caspase-2 Activation: The PIDDosome Pathway
The functions of caspase-2 in mammalian apoptosis remain somewhat obscure (Krumschnabel et al. 2009). Caspase-2 is activated in response to heat shock, microtubule disruption and DNA damage (Bouchier-Hayes et al. 2009), and it has been implicated in apoptosis in oocytes (Nutt et al. 2005) and degenerating neurons (Troy et al. 1997, 2000, 2001).
The activation platform for caspase-2 includes the adaptor molecule RAIDD (Duan and Dixit 1997; Hofmann et al. 1997; Chou et al. 1998), which bears a CARD that binds to the CARD in the prodomain of the caspase. Like caspase-8, caspase-2 is activated by induced proximity, following which intrachain autocleavage stabilizes the mature enzyme (Baliga et al. 2004).
In addition to a CARD, RAIDD also has a DD. This binds to the DD-containing protein PIDD (Lin et al. 2000; Telliez et al. 2000) to form what has been called the PIDDosome (Tinel and Tschopp 2004). PIDD contains two intein regions that promote its autocleavage. The first cleavage produces PIDD-C, a molecule that functions in NF-κB activation (Janssens et al. 2005; Tinel et al. 2007). The second generates PIDD-CC, which binds to RAIDD (Tinel and Tschopp 2004; Tinel et al. 2007). However, caspase-2 activation occurs in the absence of PIDD (Manzl et al. 2009; Ribe et al. 2012), and it is not clear when or if PIDD is required (Manzl et al. 2012). The interaction of caspase-2 with RAIDD is regulated by phosphorylation of the caspase (see below).
2.6. Regulation of Caspase Activation by Kinases
Much of the regulation of apoptosis inevitably occurs upstream of caspase-activation platforms. But other signaling events can modify caspases such that, even when a suitable activation platform forms, their activation is inhibited. XIAP and FLIP are two examples. Below we consider some others.
Regulation of caspase-2 activation is particularly well described. When NADPH is plentiful in the cell (e.g., owing to the activity of the pentose phosphate pathway) calcium/calmodulin-dependent protein kinase II (CaMKII) phosphorylates its CARD (Nutt et al. 2005). This allows the binding of 14-3-3ζ, which prevents association with the PIDDosome (Nutt et al. 2009). If NADPH levels decrease, protein phosphatase 1 (PP1) dephosphorylates caspase-2, permitting its activation by the PIDDosome (Nutt et al. 2009). SIRT1-mediated acetylation of 14-3-3ζ inhibits its binding to phospho-caspase-2, reinforcing this signal (Andersen et al. 2011). Intriguingly, the Drosophila initiator caspase Dronc (see Box 1) is also regulated by NADPH and phosphorylation by CaMKII (Yang et al. 2010; Rhind and Russel 2012).
BOX 1. CASPASE ACTIVATION IN INVERTEBRATE ORGANISMS.
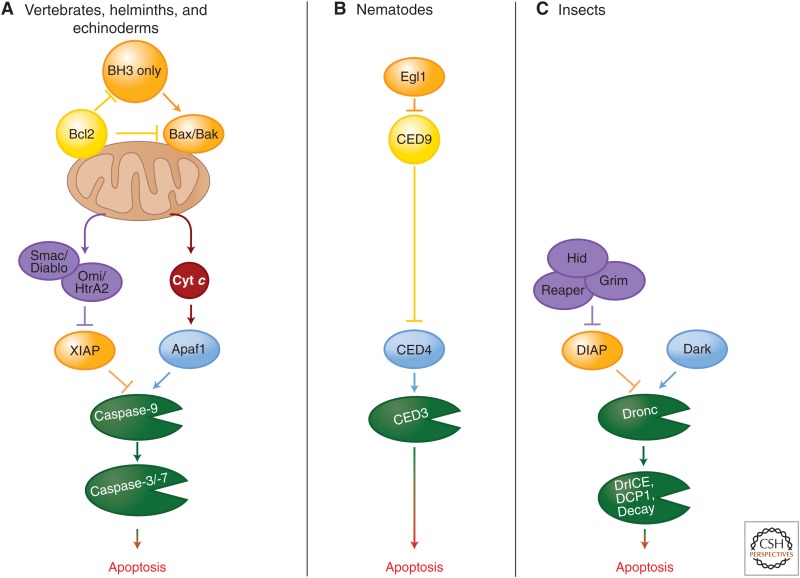
Homologs of mammalian components of the mitochondrial pathway (e.g., caspases, APAF1 and Bcl2 proteins) are found throughout the animal kingdom, including invertebrates. However, in neither of the two invertebrates studied extensively—Drosophila and Caenorhabditis elegans—has a role for the mitochondrial pathway involving MOMP and cytochrome-c-mediated activation of the apoptosome been unambiguously shown. Recent studies indicate that mitochondrial pathway may exist in helminths (Lee et al. 2011; Bender et al. 2012), and cytochrome c (Cyt c) triggers caspase activation in cytosolic extracts of a helminth and several echinoderms (Bender et al. 2012). Therefore, it is possible that the insect and nematode pathways outlined below are derived from an ancestral mitochondrial pathway resembling that of mammals (see the figure below, part A).
In C. elegans, the single caspase involved in apoptosis, CED3, contains a CARD and is activated by a platform composed of the APAF1 ortholog CED4 (Yuan et al. 1993; Chinnaiyan et al. 1997a). Monomeric CED4 is held inactive in living cells by the Bcl2 ortholog CED9 (Chinnaiyan et al. 1997b; Seshagiri and Miller 1997; Spector et al. 1997; Wu et al. 1997a,b). (Recall that this is not the case in mammals, in which Bcl2 proteins do not sequester APAF1, but instead restrict access to cytochrome c [Newmeyer et al. 2000].) In cells that are triggered to undergo apoptosis, the BH3-only protein Egl1 binds to CED9, releasing CED4, which then assembles in the caspase-activation platform for CED3 (Conradt and Horvitz 1998; Yan et al. 2004). The control of expression of Egl1 is the major way in which apoptosis in this organism is regulated (see the figure above, part B).
In Drosophila, there are several initiator and executioner caspases. The initiator caspase Dronc contains a CARD and is activated on a platform composed of the APAF1 ortholog, Dark (Kanuka et al. 1999; Rodriguez et al. 1999; Zhou et al. 1999). The Dark apoptosome appears to be constitutively active (Yu et al. 2006) but the activity of Dronc is inhibited by an IAP, DIAP1 (Rodriguez et al. 2002). The expression of one of several DIAP1 inhibitors, Reaper, Hid, Grim, or Sickle (Goyal et al. 2000; Lisi et al. 2000; Yoo et al. 2002), leads to displacement of DIAP1, allowing Dronc to cleave and thereby activate the executioner caspases DCP1, drICE, and Decay. Therefore, the expression of the DIAP1 inhibitors appears to directly control apoptosis in flies (see the figure above, part C). The roles of MOMP and cytochrome c release in Drosophila apoptosis are controversial. Although MOMP occurs, it appears to be predominantly caspase dependent and therefore is probably an effect rather than a cause of caspase activation.
The cell-cycle regulator cyclin-dependent kinase 1 (CDK1) bound to cyclin B1 also phosphorylates caspase-2 (Andersen et al. 2009). In this case, the phosphorylation is in the region between what will be the large and small subunits of the active caspases, and appears to directly inhibit the generation of the mature, stable enzyme.
Caspase-9 is phosphorylated on a threonine residue in the region between the prodomain and the large subunit by several kinases such as the ERK2 MAPK and CDK1 (Allan and Clarke 2009). Inhibition of caspase-9 activity by ERK2 occurs downstream from Ras in response to prosurvival stimuli such as growth factor stimulation (Allan et al. 2003). CDK1–cyclin-B1 controls caspase-9 phosophorylation and activity during mitosis. This process is thought to dampen the threshold for intrinsic apoptosis signals during the cell cycle, especially upon prolonged mitotic arrest (Allan and Clarke 2007). The mechanism by which this phosphorylation event inhibits caspase-9 activity remains obscure because it does not affect binding to APAF1.
3. TYPE II CELL DEATH AND AUTOPHAGY
3.1. The Autophagy Pathway
Macroautophagy, referred to here simply as autophagy, is a cellular process that is highly conserved in eukaryotes.1 It is a catabolic process engaged under metabolic stress (such as nutrient starvation and bioenergetics failure), to ensure availability of critical metabolic intermediates. It is also important for the removal of damaged organelles (including mitochondria), protein aggregates, and infecting organisms (Levine and Kroemer 2008; Kroemer et al. 2010).
Autophagy, in general, is a survival process. Its presence during so-called autophagic (type II) cell death usually represents a failed attempt to overcome lethal stress, and disruption of this process promotes rather than inhibits cell death in many cases (but see below). This form of cell death is therefore often referred to as caspase-independent cell death accompanied by autophagy (Kroemer and Levine 2008; Shen et al. 2012).
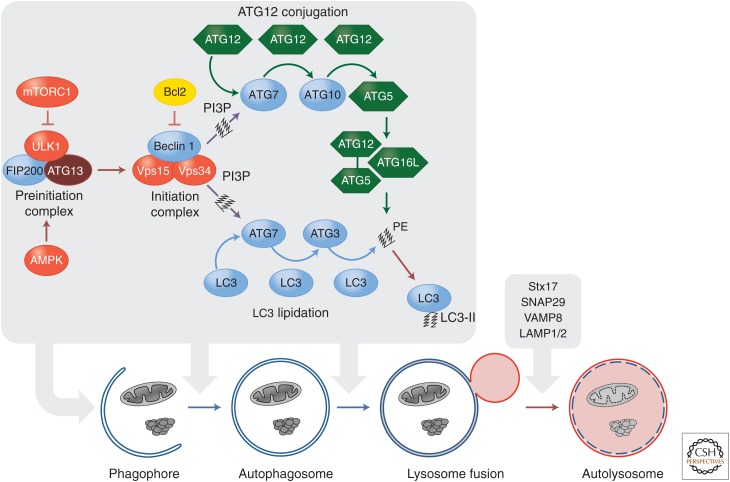
The autophagy pathway culminates in the formation of a double membrane structure, the autophagosome. This envelops intracellular material and ultimately fuses with lysosomes allowing degradation of the enveloped material (Fig. 6). Autophagosome formation and maturation is regulated by the sequential function of multiple autophagy-related (ATG) proteins. The process starts with the activation of a preinitiation complex composed of the kinase Unc-51-like kinase 1 (ULK1), FIP200, and ATG13. The activity of this kinase complex is directly regulated by two major metabolic checkpoints: mammalian target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK) (see Hardie 2012; Laplante and Sabatini 2012). In healthy cells, mTORC1 inhibits the preinitiation complex through phosphorylation of ULK1 and ATG13. Upon metabolic stress (e.g., amino acid deprivation), mTORC1-mediated inhibition of ULK1 and ATG13 is abolished (Ganley et al. 2009; Hosokawa et al. 2009; Jung et al. 2009). In contrast, AMPK activates the preinitiation complex (Egan et al. 2011; Kim et al. 2011). When ATP synthesis is unable to meet the demands of ATP consumption in a cell, AMP and ADP accumulate and activate AMPK. Active AMPK promotes autophagy indirectly through suppression of mTORC1 activity and directly by phosphorylating and activating ULK1. Therefore, conditions under which mTORC1 is inhibited and/or AMPK is activated engage the activity of the preinitiation complex.
Figure 6.
The autophagic signaling pathway. Under metabolic stress, AMPK activation and/or mTORC1 inhibition lead to the activation of the preinitiation complex (ULK1, FIP200, and ATG13). The latter activates the initiation complex (beclin 1, VPS15, and VPS34) that generates PI3P and recruits ATG7 to the phagophore. ATG7 functions similarly to an E1-ubiquitin ligase and initiates two conjugation pathways necessary for membrane elongation and closure of the autophagosome. In the ATG5-ATG12 conjugation pathway, ATG12 is sequentially transferred to ATG7, ATG10, and ATG12. The ATG5-12 conjugate recruits ATG16L and forms a complex necessary to stabilize the phagophore and to complete the second conjugation pathway. In the LC3-PE pathway, LC3 is cleaved by ATG4 and sequentially conjugated to ATG7 and ATG3. The ATG5-ATG12-ATG16L complex carries out the final step by transferring LC3 to PE to form an LC3-PE conjugate (also called LC3-II). LC3-II associates with the autophagosomal membrane and is crucial for the targeting of autophagosomes to lysosomes, as well as for the selective autophagy of organelles and protein aggregates.
The preinitiation complex recruits and activates an initiation complex composed of beclin 1, a class III PI3K (Vps34), and the protein kinase Vps15, which results in the generation of the lipid phosphatidylinositol 3-phosphate (PI3P) (He and Levine 2010). The activity of the initiation complex is negatively regulated by several independent signaling pathways. Akt directly phosphorylates beclin 1, thus allowing it to bind to 14-3-3 (Wang et al. 2012). Beclin-1–14-3-3 complexes are sequestered by vimentin, a component of intermediate filaments. This Akt-mediated cytoskeletal sequestration dampens the lipid kinase activity of the initiation complex. Therefore, growth factors and the PI3K-AKT pathway can inhibit autophagy both directly through beclin 1 and indirectly through the mTOR pathway. Beclin 1 is a scaffold protein that recruits several additional partners (in addition to Vps34 and Vps15) that regulate the lipid kinases activity of the preinitiation complex (for a more comprehensive review of beclin-1-interacting proteins, see Kroemer et al. 2010). For example, beclin-1 binds to autophagy/beclin-1 regulator 1 (AMBRA1), a protein that tethers the beclin-1–Vps34 complex to microtubules through the dynein motor complex (Di Bartolomeo et al. 2010; Fimia et al. 2012). Upon autophagy induction, AMBRA1 is phosphorylated by ULK1 and released from the cytoskeleton to allow autophagosome formation.
Interestingly, members of the Bcl2 family can regulate the initiation complex. Beclin 1 contains a bona fide BH3 domain, which allows its sequestration by antiapoptotic proteins such as Bcl2 and Bcl-xL (Pattingre et al. 2005; Maiuri et al. 2007b; Oberstein et al. 2007). This dampens the activity of the initiation complex. Under stress conditions (e.g., starvation or hypoxia), the BH3-only proteins Bad and Bnip3 release beclin 1 to permit its participation in the autophagic process (Maiuri et al. 2007a,b; Bellot et al. 2009). Alternatively, dissociation of beclin 1 from Bcl2 and Bcl-xL can be achieved through phosphorylation of the beclin–1 BH3 domain by the death-associated protein kinase (DAPK) (Zalckvar et al. 2009a,b). Finally, kinases of the JNK family disrupt this complex by directly phosphorylating Bcl2, thus decreasing its affinity for beclin 1 (Wei et al. 2008). This process is essential for regulation of exercise-induced autophagy and glucose metabolism in muscles (He et al. 2012).
The elongation and ultimate closure of the autophagosome is regulated by two distinct but complementary ubiquitin-like protein conjugation systems: the ATG5-12 and LC3-PE conjugation pathways (Fig. 6). Both pathways are initiated by a single E1-ligase-like activating enzyme, ATG7, recruited to the phagophore through PI3P generated by the initiation complex. First, a small protein, ATG12, is covalently attached to the active site cysteine residue of ATG7 by a thioester linkage. ATG12 is then transferred via thioester exchange to an E2-like conjugating enzyme, ATG10, which further transfers ATG12 to ATG5. Finally, the ATG5-ATG12 conjugate noncovalently recruits ATG16L to form a large multimeric complex necessary to stabilize the forming phagophore and to complete the second conjugation pathway (Hanada et al. 2007). The LC3-PE conjugation pathway is initiated following cleavage of LC3 by the cysteine protease ATG4. Cleaved LC3 binds to ATG7 and is subsequently transferred to the E2 enzyme ATG3. The ATG5-ATG12-ATG16L complex then acts as an E3 ligase, conjugating LC3 to phosphatidylethanolamine (PE) to form an LC3-PE conjugate (also referred to as LC3-II) (Hanada et al. 2007).
This lipidated, membrane-associated form, LC3-II, is crucial for the targeting of autophagosomes to lysosomes. Once completed, the autophagosome fuses with lysosomes to form an autolysosome, thus degrading the engulfed material. This fusion event is achieved when membranes from both organelles are brought into close proximity through interaction between the autophagosomal protein syntaxin 17 (Stx17), SNAP29, and VAMP8 on the lysosome (Itakura et al. 2012). Additionally, two lysosomal proteins, LAMP1 and LAMP2, are required for the fusion process, and in their absence the autophagosome fails to fuse to lysosomes (Tanaka et al. 2000; Eskelinen et al. 2002, 2004).
During starvation (e.g., amino acid or serum deprivation), large portions of the bulk cytosol are randomly engulfed and degraded by autophagy. Additionally, autophagy can be a selective process and remove specific subcellular structures (e.g., protein aggregates and damaged or excess organelles) in the absence of metabolic stress. During selective autophagy, substrate specificity is conferred by adaptor proteins that tether the targeted structure to the nascent phagophore. Aggregates formed by unfolded proteins are ubiquitylated and linked to the phagophore by proteins such as p62 and neighbor of breast cancer 1 (NBR1) that bind both LC3 and ubiquitin (Shaid et al. 2013). Similarly, autophagic removal of mitochondria (a process called mitophagy) is controlled by mitochondrion-anchored LC3-binding proteins such as NIP-like protein X (NIX, also called Bnip3L) and FUN14-domain-containing 1 (FUNDC1). NIX, a Bcl2-family-related protein, promotes removal of the entire mitochondrial pool during terminal erythrocyte differentiation (Schweers et al. 2007; Sandoval et al. 2008), and FUNDC1 participates in hypoxia-induced mitochondrial clearance (Liu et al. 2012). Additionally, damaged mitochondria that display low membrane potential are also removed by mitophagy following ubiquitylation of mitochondrial proteins by the ubiquitin ligase parkin (Narendra and Youle 2011), although the exact mechanism by which the phagophore is recruited in that scenario is not clear.
It is relatively easy to see how autophagy is engaged under conditions of cellular stress and can ameliorate the stress to protect the cells. Two questions persist: what kills the cells in type II cell death, and can autophagy itself ever be a mechanism that promotes cell death? Currently, we do not have a satisfactory answer to the first question but we are closer to answering the second.
3.2. Can Autophagy Kill Cells?
Autophagy often accompanies type II cell death without participating in it. There are cases, however, in which it appears to actively promote cell death. This is best exemplified during Drosophila metamorphosis, in which autophagy-dependent cell death drives the involution of the salivary gland (Berry and Baehrecke 2007) and of the midgut (Denton et al. 2009). This physiological form of cell death is triggered by the steroid hormone ecdysone and occurs (in the salivary gland) following growth arrest and inhibition of the PI3K signaling pathway. In both cases, mutation of any of several essential ATG genes reduces these developmental cell deaths. Reciprocally, forced expression of the ULK1 ortholog ATG1 is sufficient to drive this form of cell death.
Autophagy-dependent cell death also occurs in mammalian cells transformed with a constitutively active H-Ras (Elgendy et al. 2011). In this setting, Ras-dependent cell death is inhibited if ATG5, ATG7, or beclin 1 are silenced. Intriguingly, this form of autophagic cell death appears to be regulated by Bcl2 proteins. In particular, displacement of beclin 1 from Mcl1 is thought to be triggered by the proapoptotic BH3-only protein Noxa. Again, how autophagy components promote the death of the cell remains to be elucidated.
Untransformed breast epithelial cells that lose attachment to a substrate can engulf each other, resulting in the death of the engulfed cell by entosis (Overholtzer et al. 2007). The engulfed cell undergoes apoptosis, which can be inhibited by expression of Bcl2, but this does not rescue the cell. However, if the engulfing cell lacks components of the autophagy pathway, the engulfed cell can survive (Krajcovic et al. 2011). Recent studies have shown that components of the autophagy pathway can be recruited to phagosomes that have engulfed organisms or cells, facilitating the degradation of the phagocytosed cargo (Sanjuan et al. 2007; Martinez et al. 2011). The preinitiation complex does not appear to be required. In the case of entosis for example, FIP200 is dispensable (Krajcovic et al. 2011). Thus, in these cases, autophagy may act as a “murder weapon” allowing one cell to efficiently kill another.
4. TYPE III CELL DEATH, NECROSIS
4.1. Types of Necrotic Cell Death
Cellular necrosis or necrotic cell death encompasses a wide variety of cell death processes with one common denominator: the loss of plasma membrane integrity followed by cytoplasmic leakage (Yuan and Kroemer 2010). Necrosis can occur simply as a consequence of such extensive damage that cell integrity is disrupted—for example, at high temperature, following freeze-thaw, or upon mechanical stress. In such cases, cell death is passive and does not require the activation of any particular signaling pathway. Note that a necrotic morphology (i.e., rupture of the plasma membrane) can also be observed at late stages of an apoptotic or autophagic cell death program, when dead cells fail to be cleared from the system by phagocytosis. This process is referred to as secondary necrosis and is independent of any other signaling event than those initially engaged (apoptotic or autophagic). However, necrotic cell death is not always an accidental or passive process and can also be the result of a directed signaling cascade.
The best-characterized form of programmed necrosis is RIP-kinase-dependent necrosis (also referred to as “necroptosis”) (Degterev et al. 2005). This requires the kinase activity of RIP3 and can result in a rapid cell death with the features of necrosis (Cho et al. 2009; He et al. 2009; Zhang et al. 2009). Necroptosis can be engaged downstream from TNFR1 (Schulze-Osthoff et al. 1994; Vercammen et al. 1998; Holler et al. 2000; Matsumura et al. 2000). Following TNFR1 ligation, RIP3 is activated through recruitment to complex II by RIP1 (Fig. 4C). The interaction between RIP1 and RIP3 is mediated by their respective RIP homotypic interaction motifs (RHIMs). In this configuration, complex II induces necrosis rather than apoptosis and is referred to as the RIPoptosome (Feoktistova et al. 2011a; Tenev et al. 2011). The assembly and activation of the necrotic signaling platform are controlled by a series of posttranslational modifications of components of this complex. RIPoptosome formation is negatively regulated by ubiquitylation of RIP1. Consequently, treatments with cIAP1/2 inhibitors or expression of the deubiquitylase CYLD promote TNFR1-induced necrosis (Wang et al. 2008b; O’Donnell et al. 2011; Moulin et al. 2012). Additionally, RIP3 activation depends on the kinase activity of RIP1 (Cho et al. 2009; He et al. 2009; Zhang et al. 2009), which can be inhibited by the drug necrostatin (Degterev et al. 2008). Accordingly, RIP-dependent necrosis is blocked by necrostatin (Degterev et al. 2005). In addition, necrosis induction by the RIPoptosome is negatively regulated by the catalytic activity of the caspase-8–FLIP heterodimers (see below).
RIP-dependent necrosis can be triggered by TLR3 and TLR4 (for more details on TLR signaling pathways, see Lim and Staudt 2013). TLR3 and TLR4 promote the assembly of the RIPoptosome by recruiting the RHIM-containing protein TIR-domain-containing adaptor-inducing interferon-β (TRIF) (Imtiyaz et al. 2006; Feoktistova et al. 2011b). TRIF recruits RIP1 and RIP3 to induce RIP-dependent necrosis (Feoktistova et al. 2011a). TRIF might, however, recruit RIP3 independently of RIP1 through direct RHIM–RHIM interaction. RIP-dependent necrosis can also be engaged during viral infection (Cho et al. 2009; Upton et al. 2010) via the RHIM-containing cytosolic DNA sensor, DNA-dependent activator of interferon regulatory factors (DAI). DAI is activated by double-stranded viral DNA and recruits RIP3 through its RHIM domain (Upton et al. 2012). Finally, DNA damage signaling can also cause RIPoptosome formation independently of any membrane receptor signaling (Tenev et al. 2011).
The mechanism by which RIP3 causes cell death remains obscure. Several mechanisms have been proposed, including the generation of reactive oxygen species (either mitochondrial or via NADPH oxidase) and elevation of metabolic processes to deplete ATP and/or NADH, but the evidence for them is not compelling (Hitomi et al. 2008). Recent studies have implicated the pseudokinase mixed lineage kinase-like (MLKL) (Sun et al. 2012; Zhao et al. 2012) as a critical player in the execution of RIP-dependent necrosis. Phosphorylation by RIP3 triggers MLKL oligomerization and translocation to the plasma membrane (Cai et al. 2014; Chen et al. 2014). The exact mechanism by which MLKL impairs the plasma membrane integrity is not known but is thought to involve a disruption of ionic homeostasis.
4.2. Control of RIPK-Dependent Necrosis by Caspases
Stimuli that activate RIP1 and the RIPoptosome (e.g., ligation of TNFR1, ligation of TLR3/4, and possibly DNA damage) engage caspase-8 and FLIP. The catalytic activity of caspase-8–FLIP heterodimers antagonizes activation of RIP3 and the ensuing necrosis (Pop et al. 2011). As a result, cells that are treated with TNF or TLR3/4 ligands do not generally undergo cell death. However, if caspase-8–FLIP activity is blocked or disrupted, this can engage necrosis in a manner that is dependent on both RIP1 and RIP3 (Cho et al. 2009; He et al. 2009; Zhang et al. 2009; Oberst et al. 2010). RIP1, RIP3, and CYLD are substrates of caspase-8 (Lin et al. 1999; Feng et al. 2007; Lu et al. 2011; O’Donnell et al. 2011). However, in the absence of FLIP, activation of caspase-8 is not sufficient to prevent RIPK-dependent necrosis, even if apoptosis downstream from caspase-8 activation is blocked (Oberst et al. 2011).
4.3. Other Necrosis Signaling Pathways
Several other forms of necrosis signaling can occur in cells, although, like RIP-dependent necrosis, none of these is fully characterized. Ischemia/reperfusion injury (I/R), for example, causes cell death when a tissue is transiently deprived of blood supply and is then restored. High levels of calcium influx and generation of reactive oxygen species (ROS) have been associated with this form of cell death (Halestrap and Pasdois 2009). These trigger cell death, in part, by causing a mitochondrial permeability transition (mPT), in which the mitochondrial inner membrane becomes permeable to small solutes (Ricchelli et al. 2011). This results in a rapid dissipation of the transmembrane potential, followed by matrix swelling and often rupture of the mitochondrial outer membrane. Cyclophilin D (CypD), a peptidyl-proline trans-isomerase, plays a major role in mPT. Mice lacking CypD display defective mPTs (Baines et al. 2005; Basso et al. 2005; Nakagawa et al. 2005; Schinzel et al. 2005) and display diminished tissue damage under conditions of I/R (Baines et al. 2005; Nakagawa et al. 2005; Schinzel et al. 2005). Importantly, mice lacking CypD have normal apoptosis (Baines et al. 2005; Nakagawa et al. 2005; Schinzel et al. 2005) and RIP-dependent necrosis (Ch’en et al. 2011).
Another form of active necrosis involves NADPH oxidase1 (Nox1) and can be triggered by TNF (Kim et al. 2007). Nox1 generates a ROS burst that may be important. There may be a relationship between Nox1 and RIP-dependent necrosis (Vanden Berghe et al. 2007), but further evidence supporting this connection is needed.
In neurons, engagement of glutamate receptors can trigger a form of necrotic cell death called excitotoxicity (Wang and Qin 2010). This appears to involve a calcium influx that triggers an ROS burst and also activates calpain (Higuchi et al. 2005; Vosler et al. 2008). Excitotoxicity is associated with I/R and may occur in other pathological conditions (Szydlowska and Tymianski 2010).
5. BEYOND TYPE III—OTHER FORMS OF CELL DEATH
There are additional forms of cell death that appear to require the participation of the cell in its demise. These include mitotic catastrophe (Castedo et al. 2004), ferroptosis (Dixon et al. 2012), and others. Although outside the scope of our discussion, a useful distinction might be made between cellular suicide and cellular sabotage. Suicide mechanisms (including apoptosis, necroptosis, and perhaps autophagic cell death, as outlined above) represent signaling pathways that appear to have evolved to execute the cell. In contrast, sabotage can be thought of as the consequence of disruption of normal cellular processes that have not evolved for the purpose of cell death. Much as a train must be moving if removal of a railroad tie (a sabot) is to damage it, these forms of cell death occur when cellular machinery confronts a disruption that results in cellular destruction. For example, if chromosomes are cross-linked, mitosis may sufficiently damage them that the cell dies, even if mechanisms of active cell suicide cannot be engaged (mitotic catastrophe, although it is not clear that mitotic catastrophe proceeds in this way [Castedo et al. 2004]).
6. CONCLUSION
Active or programmed cell death is essential to maintain homeostasis in multicellular organisms as well as for the selective elimination of potentially harmful or infected cells. Accordingly, deregulation of the signaling pathways that trigger cell death can lead to the development of catastrophic diseases such as cancer and autoimmunity (too little cell death) as well as degenerative diseases (too much cell death). Therefore, the existence of tightly controlled and efficient means to induce cell death can be interpreted as the logical consequence of the evolution of multicellular organisms. However, the necessity for so many different death-signaling pathways might appear counterintuitive. Taken together, cell death induction could be viewed as a simple signaling process with multiple inputs/stimuli and one outcome: the death of the cell. However, the way by which a cell dies has important consequences for neighboring cells and sometimes the entire organism. For example, apoptotic and necrotic cells display divergent inflammatory properties and trigger different immune responses. Additionally, particular death programs include the release of proliferative signals that trigger compensatory proliferation in surrounding tissues. These signals might differ from one type of cell death to the next. Finally, death-signaling pathways are clearly interconnected. For example, autophagic cell death is often potentiated by caspase activation, whereas RIP-dependent necrosis is antagonized by a caspase-dependent activity. Cross talk between these pathways potentially provides numerous backup mechanisms for cell death programs and could explain why inhibition of a single program often has minor consequences for the organism. A better understanding of the impact of each type of cell death on surrounding tissues and of the interplay between these cell death programs might help to answer some of these questions.
Two other processes, microautophagy and chaperone-mediated autophagy, also exist, but are not further discussed here.
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung U-I, Fukuda A, Hikita A, Seto H, Okada T, et al. 2003. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J 22: 6653–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan LA, Clarke PR. 2007. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell 26: 301–310. [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR. 2009. Apoptosis and autophagy: Regulation of caspase 9 by phosphorylation. FEBS J 276: 6063–6073. [DOI] [PubMed] [Google Scholar]
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. 2003. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5: 647–654. [DOI] [PubMed] [Google Scholar]
- Alves NL, Derks IA, Berk E, Spijker R, van Lier RA, Eldering E. 2006. The Noxa/Mcl-1 axis regulates susceptibility to apoptosis under glucose limitation in dividing T cells. Immunity 24: 703–716. [DOI] [PubMed] [Google Scholar]
- Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, Nutt LK, Thompson JW, Moseley MA, Kornbluth S. 2009. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase 2. EMBO J 28: 3216–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, Thompson JW, Lindblom KR, Johnson ES, Yang CS, Lilley LR, Freel CD, Moseley MA, Kornbluth S. 2011. A biotin switch-based proteomics approach identifies 14–3-3ζ as a target of Sirt1 in the metabolic regulation of caspase 2. Mol Cell 43: 834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayllón V, Martínez-A C, García A, Cayla X, Rebollo A. 2000. Protein phosphatase 1α is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J 19: 2237–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. 2005. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434: 658–662. [DOI] [PubMed] [Google Scholar]
- Baliga BC, Read SH, Kumar S. 2004. The biochemical mechanism of caspase 2 activation. Cell Death Differ 11: 1234–1241. [DOI] [PubMed] [Google Scholar]
- Basso E, Fante L, Fowlkes J, Petronilli V, Forte MA, Bernardi P. 2005. Properties of the permeability transition pore in mitochondria devoid of Cyclophilin D. J Biol Chem 280: 18558–18561. [DOI] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. 2009. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29: 2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CE, Fitzgerald P, Tait SWG, Llambi F, McStay GP, Tupper DO, Pellettieri J, Sánchez Alvarado A, Salvesen GS, Green DR. 2012. Mitochondrial pathway of apoptosis is ancestral in metazoans. Proc Natl Acad Sci 109: 4904–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Cookson BT. 2007. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog 3: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzon C, Bouchier-Hayes L, Pagliari LJ, Green DR, Newmeyer DD. 2006. Caspase-2-induced apoptosis requires bid cleavage: A physiological role for bid in heat shock-induced death. Mol Biol Cell 17: 2150–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SWG, Dillon CP, Flanagan JM, Beere HM, Green DR. 2009. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell 35: 830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. 2010. Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci 123: 3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM. 2001. Recruitment, activation and retention of caspases 9 and 3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J 20: 998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MA, Cookson BT. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol 38: 31–40. [DOI] [PubMed] [Google Scholar]
- Brinkmann K, Zigrino P, Witt A, Schell M, Ackermann L, Broxtermann P, Schull S, Andree M, Coutelle O, Yazdanpanah B, et al. 2013. Ubiquitin C-terminal hydrolase-L1 potentiates cancer chemosensitivity by stabilizing NOXA. Cell Rep 3: 881–891. [DOI] [PubMed] [Google Scholar]
- Bruick RK. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci 97: 9082–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss RR, Sun W, Oppenheim RW. 2006. Adaptive roles of programmed cell death during nervous system development. Annu Rev Neurosci 29: 1–35. [DOI] [PubMed] [Google Scholar]
- Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J, Ward Y, Wu LG, Liu ZG. 2014. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 16: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. 2004. Cell death by mitotic catastrophe: A molecular definition. Oncogene 23: 2825–2837. [DOI] [PubMed] [Google Scholar]
- Chen M, He H, Zhan S, Krajewski S, Reed JC, Gottlieb RA. 2001. Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia/reperfusion. J Biol Chem 276: 30724–30728. [DOI] [PubMed] [Google Scholar]
- Ch’en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. 2011. Mechanisms of necroptosis in T cells. J Exp Med 208: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li W, Ren J, Huang D, He WT, Song Y, Yang C, Zheng X, Chen P, Han J. 2014. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res 24: 105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CW, Harris G, Ellig C, Masters SC, Subramanian R, Shenolikar S, Wadzinski BE, Yang E. 2001. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin- 3-dependent lymphoid cells by a mechanism requiring 14–3-3 dissociation. Blood 97: 1289–1297. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan AM, Chaudhary D, O’Rourke K, Koonin EV, Dixit VM. 1997a. Role of CED-4 in the activation of CED-3. Nature 388: 728–729. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan AM, O’Rourke K, Lane BR, Dixit VM. 1997b. Interaction of CED-4 with CED-3 and CED-9: A molecular framework for cell death. Science 275: 1122–1126. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. 2010. The BCL-2 family reunion. Mol Cell 37: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. 2009. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JJ, Matsuo H, Duan H, Wagner G. 1998. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell 94: 171–180. [DOI] [PubMed] [Google Scholar]
- Christofferson DE, Yuan J. 2010. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol 22: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. 2001. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nature Cell Biol 3: 339–345. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93: 519–529. [DOI] [PubMed] [Google Scholar]
- Cookson BT, Brennan MA. 2001. Proinflammatory programmed cell death. Trends Microbiol 9: 113–114. [DOI] [PubMed] [Google Scholar]
- Cotteret S, Jaffer ZM, Beeser A, Chernoff J. 2003. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol 23: 5526–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. 2011. Caspase substrates and cellular remodeling. Annu Rev Biochem 80: 1055–1087. [DOI] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241. [DOI] [PubMed] [Google Scholar]
- Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 2000. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell 6: 41–51. [PubMed] [Google Scholar]
- Datta SR, Ranger AM, Lin MZ, Sturgill JF, Ma Y-C, Cowan CW, Dikkes P, Korsmeyer SJ, Greenberg ME. 2002. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev Cell 3: 631–643. [DOI] [PubMed] [Google Scholar]
- Declercq W, Vanden Berghe T, Vandenabeele P. 2009. RIP kinases at the crossroads of cell death and survival. Cell 138: 229–232. [DOI] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. 2005. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 1: 112–119. [DOI] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. 2008. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DCS, Taunton J, Pagano M. 2009. βTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell 33: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Peso L, González-García M, Page C, Herrera R, Nuñez G. 1997. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278: 687–689. [DOI] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. 2009. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 19: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Devreotes P, Horwitz AR. 2014. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Sim HW, Puthalakath H, Adams JM, Colman PM, Kluck RM. 2008. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:Groove interactions. Mol Cell 30: 369–380. [DOI] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM. 2009. Bak activation for apoptosis involves oligomerization of dimers via their α6 helices. Mol Cell 36: 696–703. [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. 2010. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 191: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens LS, Powley IR, Hughes MA, MacFarlane M. 2012. The “complexities” of life and death: Death receptor signalling platforms. Exp Cell Res 318: 1269–1277. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. 2000. Expression of the proapoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol 10: 1201–1204. [DOI] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, Lee DF, Liu JC, Zhong Q, Wang X, et al. 2007. Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 27: 4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. 2012. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan N, Becker EBE, Konishi Y, Bonni A. 2002. JNK phosphorylation and activation of BAD couples the stress-activated signaling pathway to the cell death machinery. J Biol Chem 277: 40944–40949. [DOI] [PubMed] [Google Scholar]
- Duan H, Dixit VM. 1997. RAIDD is a new “death” adaptor molecule. Nature 385: 86–89. [DOI] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. 2006. Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Rep 7: 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekoff M, Kaufmann T, Engstrom M, Motoyama N, Villunger A, Jonsson JI, Strasser A, Nilsson G. 2007. The BH3-only protein Puma plays an essential role in cytokine deprivation induced apoptosis of mast cells. Blood 110: 3209–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy M, Sheridan C, Brumatti G, Martin SJ. 2011. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell 42: 23–35. [DOI] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. 1998. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391: 43–50. [DOI] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, Adams JM, Strasser A, Villunger A. 2005. BH3-only proteins Puma and Bim are rate-limiting for γ-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 106: 4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. 2002. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell 13: 3355–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, et al. 2004. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 15: 3132–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes R, Desagher S, Antonsson B, Martinou JC. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol 20: 929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC. 2007. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell 25: 713–724. [DOI] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M. 2007. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal 19: 2056–2067. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M. 2011a. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 43: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Häcker G, Leverkus M. 2011b. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 43: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Y, Verhaegen M, Miller TP, Rush JL, Steiner P, Opipari AW Jr, Lowe SW, Soengas MS. 2005. Differential regulation of noxa in normal melanocytes and melanoma cells by proteasome inhibition: Therapeutic implications. Cancer Res 65: 6294–6304. [DOI] [PubMed] [Google Scholar]
- Fimia GM, Corazzari M, Antonioli M, Piacentini M. 2012. Ambra1 at the crossroad between autophagy and cell death. Oncogene 32: 3311–3318. [DOI] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol 8: 1812–1825. [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. 2003. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev 17: 1841–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. 2009. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol 10: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Munoz-Planillo R, Nunez G. 2012. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 13: 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Maiuri MC, Vitale I, Zischka H, Castedo M, Zitvogel L, Kroemer G. 2007. Cell death modalities: Classification and pathophysiological implications. Cell Death Differ 14: 1237–1243. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. 2012. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 19: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. 2009. ULK1. ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B, Gollnick H, Silke J, Leverkus M. 2009. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol 187: 1037–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J. 2003. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 162: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. 2000. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol 2: 156–162. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 19: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. 2011a. Immunology: A heavyweight knocked out. Nature 479: 48–50. [DOI] [PubMed] [Google Scholar]
- Green DR. 2011b. Means to an end. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, Yu KT, Jaye M, Ivashchenko Y. 2001. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ 8: 367–376. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, Pasdois P. 2009. The role of the mitochondrial permeability transition pore in heart disease. Biochim Biophys Acta 1787: 1402–1415. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302. [DOI] [PubMed] [Google Scholar]
- Harada H, Becknell B, Wilm M, Mann M, Huang LJ, Taylor SS, Scott JD, Korsmeyer SJ. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell 3: 413–422. [DOI] [PubMed] [Google Scholar]
- Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. 2001. p70S6 kinase signals cell survival as well as growth, inactivating the proapoptotic molecule BAD. Proc Natl Acad Sci 98: 9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hardie DG. 2012. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb Perspect Biol 4: a006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. 2010. The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137: 1100–1111. [DOI] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, et al. 2012. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibein JA, Goping IS, Barry M, Pinkoski MJ, Shore GC, Green DR, Bleackley RC. 2000. Granzyme B-mediated cytochrome c release is regulated by the Bcl-2 family members bid and Bax. J Exp Med 192: 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D. 2004. Up-regulation of Bcl-2 homology 3 BH3-only proteins by E2F1 mediates apoptosis. J Biol Chem 279: 8627–8634. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Tomioka M, Takano J, Shirotani K, Iwata N, Masumoto H, Maki M, Itohara S, Saido TC. 2005. Distinct mechanistic roles of calpain and caspase activation in neurodegeneration as revealed in mice overexpressing their specific inhibitors. Biol Chem 280: 15229–15237. [DOI] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. 2008. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Tschopp J. 1997. The CARD domain: A new apoptotic signalling motif. Trends Biochem Sci 22: 155–156. [DOI] [PubMed] [Google Scholar]
- Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. 2000. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 1: 489–495. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Barrett T, Flavell RA, Davis RJ. 2008. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell 30: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiyaz HZ, Rosenberg S, Zhang Y, Rahman ZSM, Hou Y-J, Manser T, Zhang J. 2006. The Fas-associated death domain protein (FADD) is required in apoptosis and TLR-induced proliferative responses in B cells. J Immunol 176: 6852–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151: 1256–1269. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. 2005. PIDD mediates NF-κB activation in response to DNA damage. Cell 123: 1079–1092. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328. [DOI] [PubMed] [Google Scholar]
- Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DC, Bouillet P, Thomas HE, Borner C, Silke J, et al. 2009. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 460: 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. 1999. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell 4: 757–769. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. 2011. Non-canonical inflammasome activation targets caspase 11. Nature 479: 117–121. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. 1972. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26: 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JB, Hutt KJ, Michalak EM, Cook M, Vandenberg CJ, Liew SH, Bouillet P, Mills A, Scott CL, Findlay JK, et al. 2012. DNA damage-induced primordial follicle oocyte apoptosis and loss of fertility require TAp63-mediated induction of Puma and Noxa. Mol Cell 48: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. 2011. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci 36: 541–552. [DOI] [PubMed] [Google Scholar]
- Kim J-Y, Ahn H-J, Ryu J-H, Suk K, Park J-H. 2004. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1α. J Exp Med 199: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HE, Du F, Fang M, Wang X. 2005. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci 102: 17545–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Morgan MJ, Choksi S, Liu ZG. 2007. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol Cell 26: 675–687. [DOI] [PubMed] [Google Scholar]
- Kim HE, Jiang X, Du F, Wang X. 2008. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell 30: 239–247. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Selke D, Krieglstein J. 2003. Protein phosphatase type 2C dephosphorylates BAD. Neurochem Int 42: 555–560. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Lehtinen M, Donovan N, Bonni A. 2002. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell 9: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. 2000. Proapoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ 7: 1166–1173. [DOI] [PubMed] [Google Scholar]
- Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry TJ, Kirschner MW, Koths K,, Kwiatkowski DJ et al. 1997. Caspase-3–generated fragment of gelsolin: Effector of morphological change in apoptosis. Science 278: 294–298. [DOI] [PubMed] [Google Scholar]
- Krajcovic M, Johnson NB, Sun Q, Normand G, Hoover N, Yao E, Richardson AL, King RW, Cibas ES, Schnitt SJ, et al. 2011. A non-genetic route to aneuploidy in human cancers. Nat Cell Biol 13: 324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Levine B. 2008. Autophagic cell death: The story of a misnomer. Nat Rev Mol Cell Biol 9: 1004–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. 2010. Autophagy and the integrated stress response. Mol Cell 40: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumschnabel G, Manzl C, Villunger A. 2009. Caspase 2: Killer, savior and safeguard—emerging versatile roles for an ill-defined caspase. Oncogene 28: 3093–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111: 331–342. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. 2005. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 17: 525–535. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Moreira LO, Makena P, Spierings DC, Boyd K, Murray PJ, Green DR, Kanneganti TD. 2009. Caspase-7 deficiency protects from endotoxin-induced lymphocyte apoptosis and improves survival. Blood 113: 2742–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Laplante M, Sabatini DM. 2012. mTOR signaling. Cold Spring Harb Perspect Biol 4: a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue L, Kushnareva Y, Seong Y, Lin H, Faustin B, Newmeyer DD. 2009. Caspase-independent mitochondrial cell death results from loss of respiration, not cytotoxic protein release. Mol Biol Cell 20: 4871–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EF, Clarke OB, Evangelista M, Feng Z, Speed TP, Tchoubrieva EB, Strasser A, Kalinna BH, Colman PM, Fairlie WD. 2011. Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes. Proc Natl Acad Sci 108: 6999–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Davis RJ. 2003. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci 100: 2432–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. 2002. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2: 183–192. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. 2003. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 278: 18811–18816. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94: 491–501. [DOI] [PubMed] [Google Scholar]
- Li YM, Wen Y, Zhou BP, Kuo H-P, Ding Q, Hung M-C. 2003. Enhancement of Bik antitumor effect by Bik mutants. Cancer Res 63: 7630–7633. [PubMed] [Google Scholar]
- *.Lim K-H, Staudt LM. 2013. Toll-like receptor signaling. Cold Spring Harb Perspect Biol 5: a011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Devin A, Rodriguez Y, Liu ZG. 1999. Cleavage of the death domain kinase RIP by caspase 8 prompts TNF-induced apoptosis. Genes Dev 13: 2514–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet 26: 122–127. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154: 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. 2012. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 14: 177–185. [DOI] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SWG, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. 2011. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 44: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JV, Weist BM, van Raam BJ, Marro BS, Nguyen LV, Srinivas P, Bell BD, Luhrs KA, Lane TE, Salvesen GS, et al. 2011. Complementary roles of Fas-associated death domain (FADD) and receptor interacting protein kinase-3 (RIPK3) in T-cell homeostasis and antiviral immunity. Proc Natl Acad Sci 108: 15312–15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94: 481–490. [DOI] [PubMed] [Google Scholar]
- Ma C, Ying C, Yuan Z, Song B, Li D, Liu Y, Lai B, Li W, Chen R,, Ching Y-P et al. 2007. dp5/HRK is a c-Jun target gene and required for apoptosis induced by potassium deprivation in cerebellar granule neurons. J Biol Chem 282: 30901–30909. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. 2007a. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-XL. Autophagy 3: 374–376. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. 2007b. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J 26: 2527–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. 2009. The Apaf-1•procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J 28: 1916–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandic A, Viktorsson K, Strandberg L, Heiden T, Hansson J, Linder S, Shoshan MC. 2002. Calpain-mediated Bid cleavage and calpain-independent Bak modulation: Two separate pathways in cisplatin-induced apoptosis. Mol Cell Biol 22: 3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, Logette E, Tschopp J, Villunger A. 2009. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol 185: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, Newbold A, Johnstone R, Villunger A. 2012. PIDDosome-independent tumor suppression by Caspase 2. Cell Death Differ 19: 1722–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. 2011. Microtubule-associated protein 1 light chain 3α (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci 108: 17396–17401. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinon F, Burns K, Tschopp J. 2002. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell 10: 417–426. [DOI] [PubMed] [Google Scholar]
- Matsui H, Asou H, Inaba T. 2007. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell 25: 99–112. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Shimizu Y, Ohsawa Y, Kawahara A, Uchiyama Y, Nagata S. 2000. Necrotic death pathway in Fas receptor signaling. J Cell Biol 151: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. 2006. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell 21: 749–760. [DOI] [PubMed] [Google Scholar]
- McStay GP, Salvesen GS, Green DR. 2008. Overlapping cleavage motif selectivity of caspases: Implications for analysis of apoptotic pathways. Cell Death Differ 15: 322–331. [DOI] [PubMed] [Google Scholar]
- Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. 2010. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell 37: 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. 2001. NF-κB signals induce the expression of c-FLIP. Mol Cell Biol 21: 5299–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grutter MG. 2002. The long form of FLIP is an activator of caspase 8 at the Fas death-inducing signaling complex. J Biol Chem 277: 45162–45171. [DOI] [PubMed] [Google Scholar]
- Moulin M, Anderton H, Voss AK, Thomas T, Wong WW, Bankovacki A, Feltham R, Chau D, Cook WD,, Silke J et al. 2012. IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J 31: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Pinedo C, Guio-Carrion A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. 2006. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc Natl Acad Sci 103: 11573–11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik E, Michalak EM, Villunger A, Adams JM, Strasser A. 2007. Ultraviolet radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J Cell Biol 176: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. 2005. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature 434: 652–658. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694. [DOI] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. 2011. Targeting mitochondrial dysfunction: Role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal 14: 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer DD, Bossy-Wetzel E, Kluck RM, Wolf BB, Beere HM, Green DR. 2000. Bcl-xL does not inhibit the function of Apaf-1. Cell Death Differ 7: 402–407. [DOI] [PubMed] [Google Scholar]
- *.Newton K, Dixit VM. 2012. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4: a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Riblett M, Tang W-H, Gratchouck V, Zhuang D, Fernandez Y, Verhaegen M, Varambally S, Chinnaiyan AM, Jakubowiak AJ, et al. 2007. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci 104: 19488–19493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikrad M, Johnson T, Puthalalath H, Coultas L, Adams J, Kraft AS. 2005. The proteasome inhibitor bortezomib sensitizes cells to killing by death receptor ligand TRAIL via BH3–only proteins Bik and Bim. Mol Cancer Ther 4: 443–449. [DOI] [PubMed] [Google Scholar]
- Nutt LK, Margolis SS, Jensen M, Herman CE, Dunphy WG, Rathmell JC, Kornbluth S. 2005. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase 2. Cell 123: 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt LK, Buchakjian MR, Gan E, Darbandi R, Yoon SY, Wu JQ, Miyamoto YJ, Gibbons JA, Andersen JL, Freel CD, et al. 2009. Metabolic control of oocyte apoptosis mediated by 14-3-3ζ-regulated dephosphorylation of caspase 2. Dev Cell 16: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Pop C, Tremblay AG, Blais V, Denault J-B, Salvesen GS, Green DR. 2010. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J Biol Chem 285: 16632–16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. 2011. Catalytic activity of the caspase-8-FLIPL complex inhibits RIPK3-dependent necrosis. Nature 471: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, Shi Y. 2007. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem 282: 13123–13132. [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058. [DOI] [PubMed] [Google Scholar]
- O’Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Green DR, Ting AT. 2011. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol 13: 1437–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. 2007. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131: 966–979. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939. [DOI] [PubMed] [Google Scholar]
- Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, Kofler R. 2008. The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia 22: 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Fitzgerald P, Green DR, Salvesen GS. 2007. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 46: 4398–4407. [DOI] [PubMed] [Google Scholar]
- Pop C, Oberst A, Drag M, Van Raam BJ, Riedl SJ, Green DR, Salvesen GS. 2011. FLIPL induces caspase-8 activity in the absence of interdomain caspase-8 cleavage and alters substrate specificity. Biochem J 433: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B, Alix S, Youle RJ, LaMarche A, Maroney AC, et al. 2003. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron 38: 899–914. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, et al. 2007. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Pyati UJ, Gjini E, Carbonneau S, Lee JS, Guo F, Jette CA, Kelsell DP, Look AT. 2011. p63 mediates an apoptotic response to pharmacological and disease-related ER stress in the developing epidermis. Dev Cell 21: 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JZ, Ziffra J, Stennett L, Bodner B, Bonish BK, Chaturvedi V, Bennett F, Pollock PM, Trent JM, Hendrix MJ, et al. 2005. Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res 65: 6282–6293. [DOI] [PubMed] [Google Scholar]
- Ramjaun AR, Tomlinson S, Eddaoudi A, Downward J. 2007. Upregulation of two BH3–only proteins, Bmf and Bim, during TGF β-induced apoptosis. Oncogene 26: 970–981. [DOI] [PubMed] [Google Scholar]
- Real PJ, Sanz C, Gutierrez O, Pipaon C, Zubiaga AM, Fernandez-Luna JL. 2006. Transcriptional activation of the proapoptotic bik gene by E2F proteins in cancer cells. FEBS Lett 580: 5905–5909. [DOI] [PubMed] [Google Scholar]
- Reiners JJ, Caruso JA, Mathieu P, Chelladurai B, Yin X-M, Kessel D. 2002. Release of cytochrome c and activation of procaspase 9 following lysosomal photodamage involves Bid cleavage. Cell Death Differ 9: 934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rhind N, Russel P. 2012. Signaling pathways that regulate cell division. Cold Spring Harb Perspect Biol 4: a005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribe EM, Jean YY, Goldstein RL, Manzl C, Stefanis L, Villunger A, Troy CM. 2012. Neuronal caspase-2 activity and function requires RAIDD, but not PIDD. Biochem J 444: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchelli F, Sileikyte J, Bernardi P. 2011. Shedding light on the mitochondrial permeability transition. Biochim Biophys Acta 1807: 482–490. [DOI] [PubMed] [Google Scholar]
- Ricci JE, Munoz-Pinedo C, Fitzgerald P, Bailly-Maitre B, Perkins GA, Yadava N, Scheffler IE, Ellisman MH, Green DR. 2004. Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell 117: 773–786. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. 1999. Caspase 9 and APAF-1 form an active holoenzyme. Genes Dev 13: 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. 1999. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol 1: 272–279. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Chen P, Oliver H, Abrams JM. 2002. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J 21: 2189–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Bokoch GM. 1997. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 276: 1571–1574. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. 1998. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391: 96–99. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Riedl SJ. 2008. Caspase mechanisms. Adv Exp Med Biol 615: 13–23. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454: 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450: 1253–1257. [DOI] [PubMed] [Google Scholar]
- Sanz C, Mellstrom B, Link WA, Naranjo JR, Fernandez-Luna JL. 2001. Interleukin 3-dependent activation of DREAM is involved in transcriptional silencing of the apoptotic Hrk gene in hematopoietic progenitor cells. EMBO J 20: 2286–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel AC, Takeuchi O, Huang Z, Fisher JK, Zhou Z, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ. 2005. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc Natl Acad Sci 102: 12005–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Osthoff K, Krammer PH, Droge W. 1994. Divergent signalling via APO-1/Fas and the TNF receptor, two homologous molecules involved in physiological cell death. EMBO J 13: 4587–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann A, Mooney AF, Sanders LC, Sells MA, Wang HG, Reed JC, Bokoch GM. 2000. p21-Activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol 20: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, et al. 2007. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci 104: 19500–19505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O’Rourke K, Bazan F, Eastham-Anderson J, et al. 2010. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463: 103–107. [DOI] [PubMed] [Google Scholar]
- Sebbagh M, Renvoizé C, Hamelin J, Riché N, Bertoglio J, Bréard J. 2001. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nature Cell Biol 3: 346–352. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Miller LK. 1997. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr Biol 7: 455–460. [DOI] [PubMed] [Google Scholar]
- *.Sever R, Brugge JS. 2014. Signaling in cancer. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaid S, Brandts CH, Serve H, Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ 20: 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Zhang T, Rzeszutek M, Yurkova N, Baetz D, Davie JR, Kirshenbaum LA. 2006. Transcriptional silencing of the death gene BNIP3 by cooperative action of NF-κB and histone deacetylase 1 in ventricular myocytes. Circ Res 99: 1347–1354. [DOI] [PubMed] [Google Scholar]
- Shaw J, Yurkova N, Zhang T, Gang H, Aguilar F, Weidman D, Scramstad C, Weisman H, Kirshenbaum LA. 2008. Antagonism of E2F-1 regulated Bnip3 transcription by NF-κB is essential for basal cell survival. Proc Natl Acad Sci 105: 20734–20739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Kepp O, Kroemer G. 2012. The end of autophagic cell death? Autophagy 8: 1–3. [DOI] [PubMed] [Google Scholar]
- Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, et al. 1999. Ordering the cytochrome-c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 144: 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. 2001. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res 61: 6669–6673. [PubMed] [Google Scholar]
- Spector MS, Desnoyers S, Hoeppner DJ, Hengartner MO. 1997. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 385: 653–656. [DOI] [PubMed] [Google Scholar]
- Spender LC, O'Brien DI, Simpson D, Dutt D, Gregory CD, Allday MJ, Clark LJ, Inman GJ. 2009. TGF-β induces apoptosis in human B cells by transcriptional regulation of BIK and Bcl-xL. Cell Death Differ 16: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. 1999. Caspase 9 can be activated without proteolytic processing. J Biol Chem 274: 8359–8362. [DOI] [PubMed] [Google Scholar]
- Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, Ellerby LM, Bredesen D, Freeze H, Abrahamson M, et al. 2001. Lysosomal protease pathways to apoptosis. Cleavage of bid, not procaspases, is the most likely route. J Biol Chem 276: 3149–3157. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, et al. 2012. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148: 213–227. [DOI] [PubMed] [Google Scholar]
- Sutton VR, Davis JE, Cancilla M, Johnstone RW, Ruefli AA, Sedelies K, Browne KA, Trapani JA. 2000. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J Exp Med 192: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowska K, Tymianski M. 2010. Calcium, ischemia and excitotoxicity. Cell Calcium 47: 122–129. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. 2010. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11: 621–632. [DOI] [PubMed] [Google Scholar]
- Tait SWG, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Muñoz-Pinedo C, Green DR. 2010. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell 18: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406: 902–906. [DOI] [PubMed] [Google Scholar]
- Telliez JB, Bean KM, Lin LL. 2000. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta 1478: 280–288. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, et al. 2011. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 43: 432–448. [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. 2004. The PIDDosome, a protein complex implicated in activation of caspase 2 in response to genotoxic stress. Science 304: 843–846. [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J. 2007. Autoproteolysis of PIDD marks the bifurcation between prodeath caspase 2 and prosurvival NF-κB pathway. EMBO J 26: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Stefanis L, Greene LA, Shelanski ML. 1997. Nedd2 is required for apoptosis after trophic factor withdrawal, but not superoxide dismutase (SOD1) downregulation, in sympathetic neurons and PC12 cells. J Neurosci 17: 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Rabacchi SA, Friedman WJ, Frappier TF, Brown K, Shelanski ML. 2000. Caspase-2 mediates neuronal cell death induced by β-amyloid. J Neurosci 20: 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Rabacchi SA, Hohl JB, Angelastro JM, Greene LA, Shelanski ML. 2001. Death in the balance: Alternative participation of the caspase-2 and -9 pathways in neuronal death induced by nerve growth factor deprivation. J Neurosci 21: 5007–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JP, Austgen K, Nishino M, Coakley KM, Hagen A, Han D, Papa FR, Oakes SA. 2008. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol Cell Biol 28: 3943–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. 2010. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7: 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. 2012. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11: 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Knau A, Fichtlscherer S, Walter DH, Brühl T, Potente M, Hofmann WK, de Vos S, Zeiher AM, Dimmeler S. 2005. FOXO-dependent expression of the proapoptotic protein Bim: Pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J 19: 974–976. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T, Declercq W, Vandenabeele P. 2007. NADPH oxidases: New players in TNF-induced necrotic cell death. Mol Cell 26: 769–771. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W, Grooten J, Fiers W, Vandenabeele P. 1998. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 187: 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Zhao LJ, Chinnadurai G. 2001. Phosphorylation of the pro-apoptotic protein BIK: Mapping of phosphorylation sites and effect on apoptosis. J Biol Chem 276: 4671–4676. [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- Vosler PS, Brennan CS, Chen J. 2008. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol Neurobiol 38: 78–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin ZH. 2010. Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis 15: 1382–1402. [DOI] [PubMed] [Google Scholar]
- Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke TF, Reed JC. 1999. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 284: 339–343. [DOI] [PubMed] [Google Scholar]
- Wang Y, Guan X, Fok KL, Li S, Zhang X, Miao S, Zong S, Koide SS, Chan HC, Wang L. 2008a. A novel member of the Rhomboid family, RHBDD1, regulates BIK-mediated apoptosis. Cell Mol Life Sci 65: 3822–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. 2008b. TNF-α induces two distinct caspase-8 activation pathways. Cell 133: 693–703. [DOI] [PubMed] [Google Scholar]
- Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. 2012. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338: 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Acoca S, Liu Z, Germain M, Watson M, Blanchette M, Wing SS, Shore GC. 2005. BH3-ligand regulates access of MCL-1 to its E3 ligase. FEBS Lett 579: 5603–5608. [DOI] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev 14: 2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. 2008. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 30: 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. 2011. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471: 110–114. [DOI] [PubMed] [Google Scholar]
- Wu D, Wallen HD, Inohara N, Nunez G. 1997a. Interaction and regulation of the Caenorhabditis elegans death protease CED-3 by CED-4 and CED-9. J Biol Chem 272: 21449–21454. [DOI] [PubMed] [Google Scholar]
- Wu D, Wallen HD, Nunez G. 1997b. Interaction and regulation of subcellular localization of CED-4 by CED-9. Science 275: 1126–1129. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, Lilly M. 2003. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem 278: 45358–45367. [DOI] [PubMed] [Google Scholar]
- Yan N, Gu L, Kokel D, Chai J, Li W, Han A, Chen L, Xue D, Shi Y. 2004. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell 15: 999–1006. [DOI] [PubMed] [Google Scholar]
- Yan J, Xiang J, Lin Y, Ma J, Zhang J, Zhang H, Sun J, Danial NN, Liu J, Lin A. 2013. Inactivation of BAD by IKK inhibits TNFα-induced apoptosis independently of NF-κB activation. Cell 152: 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Thomenius MJ, Gan EC, Tang W, Freel CD, Merritt TJ, Nutt LK, Kornbluth S. 2010. Metabolic regulation of Drosophila apoptosis through inhibitory phosphorylation of Dronc. EMBO J 29: 3196–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 4: 416–424. [DOI] [PubMed] [Google Scholar]
- You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, Villunger A, Mak TW. 2006. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med 203: 1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 7: 673–682. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. 2003. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci 100: 1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. 2005. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure 13: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey CW. 2006. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol 355: 577–589. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kroemer G. 2010. Alternative cell death mechanisms in development and beyond. Genes Dev 24: 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 β-converting enzyme. Cell 75: 641–652. [DOI] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. 2010. Structure of an apoptosome-procaspase-9 CARD complex. Structure 18: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkova N, Shaw J, Blackie K, Weidman D, Jayas R, Flynn B, Kirshenbaum LA. 2008. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ Res 102: 472–479. [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Eisenstein M, Kimchi A. 2009a. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 5: 720–722. [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. 2009b. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 10: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell 87: 619–628. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. 2009. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325: 332–336. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu Z-G. 2012. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci 109: 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Song Z, Tittel J, Steller H. 1999. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol Cell 4: 745–755. [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhang L, Dong F, Guo W, Wu S, Teraishi F, Davis JJ, Chiao PJ, Fang B. 2005. Bik/NBK accumulation correlates with apoptosis-induction by bortezomib PS-341, Velcade and other proteasome inhibitors. Oncogene 24: 4993–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome-c-dependent activation of caspase 3. Cell 90: 405–413. [DOI] [PubMed] [Google Scholar]